Abstract
Purpose of review
This review highlights recent advances in our understanding of the interactions between genetic polymorphisms in genes that metabolize choline and the dietary requirements of choline and how these interactions relate to human health and disease.
Recent findings
The importance of choline as an essential nutrient has been well established but our appreciation of the interaction between our underlying genetic architecture and dietary choline requirements is only beginning. It has been shown in both human and animal studies that choline deficiencies contribute to diseases such as non-alcoholic fatty liver disease and various neurodegenerative diseases. An adequate supply of dietary choline is important for optimum development, highlighted by the increased maternal requirements during fetal development and in breast-fed infants. We discuss recent studies investigating variants in PEMT and MTHFR1 that are associated with a variety of birth defects. In addition to genetic interactions, we discuss several recent studies that uncover changes in fetal global methylation patterns in response to maternal dietary choline intake that result in changes in gene expression in the offspring. In contrast to the developmental role of adequate choline, there is now an appreciation of the role choline has in cardiovascular disease through the gut microbiota-mediated metabolite trimethylamine N-oxide. This pathway highlights some of our understanding of how the microbiome affects nutrient processing and bioavailability. Finally, in order to better characterize the genetic architecture regulating choline requirements, we discuss recent results focused on identifying polymorphisms that regulate choline and its derivative products.
Summary
Here we discuss recent studies that have advanced our understanding of how specific alleles in key choline metabolism genes are related to dietary choline requirements and human disease.
Keywords: choline, polymorphisms, gene by diet interactions
Introduction
Many diseases have both genetic and environmental risk factors and there is expanding interest in identifying interactions between dietary factors and genetic polymorphisms that alter either nutrient requirements or susceptibility to disease. This renewed interest in diet has been partially prompted by improved –omic technologies used to identify metabolites and genetic variants. The interest in environmental effects, such as diet, on disease has been in part driven by the surprisingly low heritability uncovered by current genome-wide association studies (GWAS). There is now an appreciation that the identification of how food (and other environmental factors) interacts with genetic polymorphisms may be key to improving health. Clearly, we are moving beyond a “one-size fits all” mentality and ultimately a better understanding of gene by diet interactions may help to refine our current dietary recommendations.
Of particular interest is the nutrient choline which is found in nuts, dairy, and meat products, and was first classified as an essential nutrient based on its role in the prevention of non-alcoholic fatty liver disease (NAFLD) [1]. An individual’s response to choline deficient diets is varied and perhaps an indication of complex genetic regulation of choline requirements [2]. Additionally, the requirements for choline may vary across an individual’s lifetime as maternal choline deficiency is associated with certain birth defects [3] which need to be balanced with the recent evidence that choline-derived metabolites increase CVD risk [4–6].
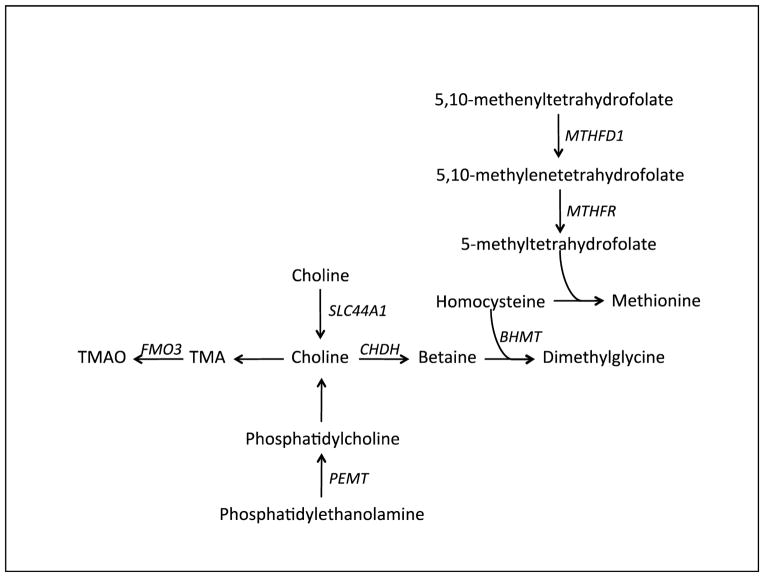
The liver is a primary site of choline metabolism where several metabolic pathways regulate maintenance of cell membranes. For cardiovascular risk, phosphatidylcholine synthesis in the liver is also important for the assembly of very low-density lipoprotein (VLDL) particles that are responsible for transporting fat and cholesterol from the liver to peripheral organs. Thus, one effect of choline deficiency is non-alcohol fatty liver disease (NAFLD) and this may be a direct result of reduced levels of hepatic phosphatidylcholine leading to hepatic accumulation of fat and cholesterol. Genetic polymorphisms in genes regulating choline metabolism have been extensively studied as they have been hypothesized to modify requirements for choline. These are outlined in Figure 1 and described in detail below.
Figure 1. Genes involved in Choline Metabolism: We discuss genetic variants in several genes known to affect choline metabolism.
Known genes regulating choline metabolism and dietary choline requirements. PEMT catalyzes the synthesis of phosphatidylcholine from phosphatidylethanolamine, a precursor of endogenously derived choline. The transporter SLC44A1 can transport choline into the mitochondria for further processing. In the gut, choline is the precursor of trimethylamine which is absorbed and transported to the liver for oxidation by FMO3 (flavinmonooxygenase 3) to TMAO. Choline is the precursor of betaine, which serves as a source of methyl groups for homocysteine remethylation to dimethylglycine and methionine. Alternatively, homocysteine can be remethylated with methyl groups from 5-methyltetrahydrofolate, which is derived from 5,10-methylenetetrahydrofolate. MTHFD1 (methylenetetrahydrofolate dehydrogenase 1), an enzyme that catalyzes the production of 5,10-methylenetetrahydrofolate from 5,10-methenyltetrahydrofolate.
Genetic polymorphism, dietary choline, and pregnancy outcomes
Much of the interest in choline as an essential nutrient is due to previous reports of low choline being associated with certain birth defects [7–9]. In addition to choline intake, there has been substantial interest in either the mother’s genotype or fetal genotype influencing fetal choline requirements or metabolism. The premise is that identifying mothers with altered choline need or metabolism may reduce negative outcomes.
Certain polymorphisms may be particularly deleterious in the presence of dietary choline deficiencies [10, 11]. For example, polymorphisms in the gene methylenetetrahydrofolate dehydrogenase 1 (MTHFD1), specifically the A allele of MTHFD1 G1958A, may affect homocysteine methylation when folate availability is low [12, 13]. Thus polymorphisms of MTHFD1 have been associated with various congenital abnormalities such as neural tube defects [14], Down’s Syndrome, cleft lip, cleft palate [15], and congenital heart defects. There is some heterogeneity in these data as studies have inconsistently reported the association of specific effect alleles (or underlying polymorphisms), so additional work is necessary to clarify the risk variants and their effect size. More recently, meta-analyses have identified associations between MTHFD1 polymorphisms, such as the MTHFD1 G1958A variant, as being associated with increased risk for birth defects [16], [17], [18]. Additionally, two missense mutations, rs7946 and rs897453, in PEMT, an enzyme required for de novo production of choline, have been associated with altered homocysteine levels and neural tube defects, respectively [19, 20].
However, data directly linking neural tube defects (NTDs) and maternal choline levels and specific polymorphisms are inconsistent. Recently, Mills et al conducted a study to investigate the link between NTDs and maternal choline levels and associated SNPs. They measured choline and betaine levels and looked at SNPs in PEMT in women with a current NTD-affected pregnancy, women with a previous NTD-affected pregnancy currently pregnant and unaffected, and unaffected control women. They reported that the levels of choline and betaine did not differ in women who had a current NTD-affected pregnancy. Cases were significantly more likely to have the G allele of PEMT rs7946 (V175M, +5465 G.A) [15]. It is quite possible that these polymorphisms affect fetal or maternal choline requirements but it was not possible to measure umbilical cord metabolite levels in this particular study. Another group recently compared maternal plasma levels of choline, betaine, dimethylglycine, and trimethylamine N-oxide (TMAO) in pregnant women at 16 weeks of gestation, at delivery, and in umbilical cord plasma [21]. Mean maternal plasma free choline, dimethylglycine, and TMAO concentrations increased during pregnancy by 49%, 17%, and 13%, respectively, while betaine concentrations decreased by 21%. Curiously, cord plasma concentrations of free choline and betaine were 3.2 and 2.0 times higher than maternal plasma values at delivery, indicating perhaps an altered requirement for these metabolites by the fetus. The authors did not find an association between maternal diet, as assessed by a food frequency questionnaire, and cord plasma concentrations of choline nor did they find any associations between fetal genotype of 10 candidate SNPs and cord plasma concentrations of choline. Improving our understanding of how maternal dietary requirements and genetics interact to affect fetal health has important public health implications.
Epigenetic effects
An area of intense interest is the effect of maternal diet on offspring due to altered epigenetic marks that control gene expression [22]. Choline is particularly interesting as it is a methyl donor and thus differences in choline status could affect global DNA methylation. Studies in rodents have found that the gestational availability of choline has a large impact on DNA methylation and researchers are focusing on understanding how these specific changes in methylation may affect disease risk. A study using a toxic milk (tx-j) mouse model of Wilson’s Disease on a C3H/HeJ background found that maternal choline supplementation altered transcript levels of methionine and lipid metabolism genes. In this study, maternal choline supplementation increased global DNA methylation measured in fetal livers, indicating the importance of dietary choline during fetal development [23]. Additional studies have argued that the availability of methyl donors is tightly regulated, and that high levels of methylation are not universally protective as increased DNA methylation may be linked to behavioral disorders [24–26].
Understanding the effects of choline and 1-carbon metabolites on the human epigenome is an area of active investigation. Interestingly, although not surprisingly, the effects of choline deficiency have been identified as population specific, likely due to differences in gene allele frequency in certain populations as well as dietary differences among populations. In a recent study focused on women in rural Gambia, researchers showed that seasonal variations in methyl-donor nutrient intake in mothers as early as conception influenced maternal plasma concentrations of several key methyl-donor pathway substrates, including choline, betaine, methionine, and homocysteine, which in turn were shown to influence the methylation status of fetal alleles [27].
Genetic polymorphism, dietary choline, and metabolic disease
Initial interest in choline metabolites and CVD arose via the association between homocysteine levels and CVD, but this association is now considered inconclusive [28]. To better understand these data, recent research has focused on specific polymorphisms that alter homocysteine levels such as those in MTHFR, the enzyme that converts 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, which is a substrate required for conversion of homocysteine to methionine. The SNP rs1801133 has been linked to elevated plasma homocysteine levels and increased risk of CAD [29]; however, the mechanisms by which elevated homocysteine levels promote CAD are still not well understood and other studies have failed to replicate this result [30]. Studies in different ethnicities have identified a similar association between A1298C and CVD in two independent Indian populations [31, 32]. A meta-analysis of 6912 individuals confirmed that the C677T polymorphism is also associated with premature CAD [33]. It is not clear from these studies if different choline intake (dietary substrate) is the source of these disparate results. It is important to note that some of the heterogeneity in the literature may in fact be due to dramatic differences in folate levels supplied in the diet. In 1997 folate supplementation was mandated and thus it is possible that certain results are discordant due to this difference in substrate availability for these enzymatic reactions.
More importantly, a recent randomized clinical trial to evaluate the efficacy of folic acid therapy in the prevention of strokes grouped participants based on MTHFR C677T genotype. This study found that folic acid therapy significantly reduced stroke risk among participants with the CC or CT genotypes, but not among those with the TT genotype [34] and these results may extend to renal disease [35]. The association of MTHFR is complicated as SNPs, including rs13306561, rs1801133, and rs17367504, in the gene have been associated with increases in diastolic blood pressure, but these associations have not replicated across studies [36, 37]. There is a possibility that some of the effects of these polymorphisms mediate expression of MTHFR from the recent identification of a polymorphism at the locus that affects its expression, also known as a cis-eQTL. This variant has also been associated with CVD related traits [38].
There is evidence that genetic variations in the PEMT gene also affect choline metabolism. Polymorphisms such as the variant tagged by the C allele of rs12325817 in PEMT were found to be associated with development of organ dysfunction in individuals fed a choline deficient diet [2]. Furthermore, previous results have shown an interesting interaction of PEMT and estrogen signaling. For this reason, of particular interest are two potential functional variants: an amino acid substitution, Val58Leu (rs897453), in a transmembrane domain of PEMT and rs750546 which is found in the ESR1 binding site of PEMT [39]. The specific effect of these polymorphisms on CVD risk remains unknown but the importance of PEMT in CVD was shown by recent studies in genetically modified mice [40]. For example, Pemt−/− mice have reduced circulating homocysteine levels and resistance to atherosclerotic lesions, signifying the importance of PEMT in CVD [41]. Pemt−/− mice also exhibit low plasma VLDL levels and develop fatty livers in response to a high fat diet, which was partially ameliorated when the diets were supplemented with choline as hepatic cholesterol levels decreased and liver function improved. These data suggest that genetic variants of PEMT alter dietary choline requirements and that choline requirements could vary by an individual’s genotype.
The gut microbiome regulates choline levels and contributes to complex diseases in humans
For several years, researchers have studied the role of the gut microbiome in complex diseases such as cancer, obesity, diabetes, CVD, and NAFLD [42–45]. Direct links have been made between these various diseases and the regulation of choline levels, especially in situations of dietary choline deficiency [46]. TMAO, a derivative of choline, was identified as a predictive biomarker linking the microbiome and CVD risk [47, 48]. TMAO is an oxidation product of trimethylamine (TMA) that is found in both humans and mice. TMAO is synthesized as a conversion product from TMA, which is derived from the breakdown of dietary choline-containing compounds by the gut microbiota.
There are widespread differences in the microbiota composition among humans that could affect the metabolism and bioavailability of dietary choline. For example, Romano et al identified several isolates, from a collection of 79 sequenced human intestinal samples, capable of producing TMA from choline in vitro. They identified nine bacterial strains capable of producing TMA from choline in vitro and tested these using gnotobiotic mice to show that TMAO accumulates in the serum of animals colonized with TMA-producing species. Remarkably, low levels of colonization by TMA-producing bacteria significantly reduced choline levels available to the host which was more pronounced as the abundance of TMA-producing bacteria increased in the gnotobiotic mice [49]. These important studies indicate that the composition of the microbiome affects plasma TMAO and choline concentrations. These results are particularly intriguing given studies that have shown that egg consumption leads to increased TMAO concentrations in the blood [47]. This result was confirmed in a small clinical study that demonstrated that 11–15% of dietary total choline is converted into TMAO after consumption of a meal that contained eggs [6].
Normally, TMA is oxidized to TMAO primarily through the action of flavin mono-oxygenase 3 (FMO3). Studies in mice have shown a direct link between plasma levels of TMAO, Fmo3 expression, and atherosclerosis [5]. More recent studies have identified Fmo3 as a key metabolic gene that affects lipid and glucose metabolism. Studies in LDLr−/− mice treated with antisense Fmo3 oligonucleotides exhibited decreases in hepatic lipids and in levels of plasma lipids, ketone bodies, glucose, and insulin, whereas opposite effects were observed with transgenic mice overexpressing Fmo3 [50]. Knockdown of Fmo3 in cholesterol-fed mice alters biliary lipid secretion, blunts intestinal cholesterol absorption, and limits the production of hepatic oxysterols and cholesteryl esters. Furthermore, FMO3 knockdown stimulates basal and liver X receptor (LXR)-stimulated macrophage RCT, thereby improving cholesterol balance [51].
Novel regulatory variants and choline status
Identification of genetic variants affecting choline requirements could be used to refine our dietary recommendations in pregnancy and in cardiovascular patients. A targeted genetic study of 200 SNPs in ten candidate genes selected for their biochemical importance in choline metabolism identified novel associations with liver and muscle function in subjects fed a low-choline diet [52]. The authors used a combination of clinical studies in which patients were fed a choline deficient diet while under medical supervision and were deemed to have organ dysfunction when they developed increased creatine phosphokinase or aspartate aminotransferase levels. Genotypes were then compared between patients with organ dysfunction and patients that were resistant to choline restriction. Novel SNPs in SLC44A1, CHKA, and CHKB were identified as associated with increased choline requirements, and there was an indication that some of the SNPs in SLC44A1 may differentiate between patients that develop muscle dysfunction or hepatic dysfunction. The authors then tested the allele frequency of these SNPs in Mexican Americans, European Americans, Asian Americans and Africans (n=100 per population) and several of the SNPs associated with choline requirements in the clinical study were dramatically different between these small population cohorts. For example rs7873937, a SNP in SLC44A1, was present in all populations except Asian Americans. They concluded that these differences in allele frequency could alter the choline requirement in a population-specific manner [52]. These data are supported by a case report of a patient with postural orthostatic tachycardia syndrome. This patient had decreased levels of the choline transporter SLC44A1 and reduced plasma choline levels, suggesting an intriguing link between SLC44A1 and plasma choline [53]. These data suggest that genetic variants of choline metabolizing enzymes can alter dietary choline requirements and that choline requirements could vary by an individual’s genotype.
The identification of SNPs in key genes regulating choline metabolism as associated with human diseases across multiple general populations and among small isolated populations indicates the importance of this pathway in regulating multiple complex processes contributing to human health. Dietary choline intake varies geographically and different populations exhibit different allele frequencies in key choline metabolism genes. For this reason, a recently published study sought to identify whether key SNPs that increase dependence on dietary choline would be under selective pressure specifically in a geographic location with a prevalent choline-poor diet [10]. They assessed CHDH rs12676, MTHFD1 rs2236225, and PEMT rs12325817 for negative selective pressure in The Gambia, West Africa, known for its choline-deficient diet, versus two other populations with choline-rich diets, including a genetically similar population in Maasai in East Africa and a genetically dissimilar population the United States. The authors used two different genetic models to demonstrate minor allele frequencies of polymorphisms associated with increased choline requirements between the Gambian and the other populations. Although difficult to definitively test in this particular study, the authors hypothesize that the dramatic difference in allele frequency is actually driven by historical dietary differences between these populations. These data do present the intriguing question of whether our genetically programmed needs for specific nutrients may in fact be partially determined by ethnicity and/or selective pressures due to historical diets of our ancestors.
In addition to genes that may affect the metabolism of choline, recent studies have been focused on determining the genetics underlying the association of TMAO to CVD since this metabolite is a by-product of microbial metabolism of choline. Based on the highly variable plasma TMAO levels observed in human studies, it is reasonable to hypothesize that TMAO levels are affected by the intrinsic genetic factors of the host. However, with the exception of FMO3, deficiency of which leads to elevated TMA and decreased TMAO levels [5], the genes that control plasma TMAO levels are not known. A comparative GWAS approach using a large mouse genetic reference population and ~2000 subjects was used to identify candidate genes [4]. These studies identified a genome-wide significant locus on chromosome 3 associated with plasma TMAO concentrations in mice on a chow diet (p= 2.37x10−6). This locus also exhibited highly significant evidence (p=1.07x10−20) for cis-regulation of solute carrier family 30 member 7 (Slc30a7) expression in the liver. The co-localization of loci for plasma TMAO and Slc30a7 mRNA levels suggest that this zinc transporter represents a positional candidate gene responsible for the association signal at this locus. By comparison, no locus exceeding the genome-wide significance threshold was observed in the human GWAS, including the orthologous region on human chromosome 1p21.2 harboring SLC30A7 or at the FMO3 locus, which indicates the complex genetic regulation of TMAO concentrations. Supporting these findings, a GWAS in the Framingham Heart Study (FHS) also failed to detect significant associations for plasma TMAO concentrations, although several suggestively associated loci were also identified [54]. Further GWAS investigations that account for differences in choline or l-carnitine intake may better identify genes regulating plasma levels of TMAO.
Conclusion
We are just beginning to understand the complex interactions between diet and genetic polymorphisms. Here we discuss choline as it has been shown to be a clinically important nutrient that has been associated with diseases of deficiency, in the case of birth defects, while excessively elevated choline levels are associated with CVD risk. Polymorphisms in several candidate genes demonstrate that genetic perturbations can affect the metabolism of choline and presumptively an individual’s dietary requirement for choline. Studies with rural African populations have demonstrated altered methylation status and indicated unique selective pressures on genes regulating choline metabolism. Studies in mice and humans with CVD indicate that gut microbial metabolism, and perhaps genetic polymorphisms of the host, influence the conversion of dietary choline into the pro-atherogenic metabolite TMAO. Clearly additional work to identify the underlying genetic variants and how they relate to dietary choline requirements across a lifetime are necessary before altering our current recommendations.
Key Points.
Insufficient intake of dietary choline in pregnant women is associated with developmental disorders.
Certain populations are more susceptible to choline-related diseases due to the low availability of dietary choline and the presence of polymorphism that alter dietary choline requirements.
Choline is a methyl donor and choline deficiency has been shown to alter global methylation patterns.
Understanding how common polymorphisms influence dietary choline requirements may allow for a more personalized approach to dietary recommendations.
Acknowledgments
Financial support and sponsorship; This work was supported in part by NIH grant, R01ES021801, R01ES021801-S3, R01ES025786, P01ES022845, and R01HL128572, and US EPA grant RD83544101.
Footnotes
Conflicts of interest: None
References
- 1.De Witt Stetten J, Juan Salcedo J. The Effect of Chain Length of the Dietary Fatty Acid upon the Fatty Liver of Choline Deficiency. The Journal of Nutrition. 1945:167–170. [Google Scholar]
- 2.da Costa KA, Kozyreva OG, Song J, et al. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 2006;20:1336–1344. doi: 10.1096/fj.06-5734com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shaw GM, Carmichael SL, Yang W, et al. Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. American journal of epidemiology. 2004;160:102–109. doi: 10.1093/aje/kwh187. [DOI] [PubMed] [Google Scholar]
- 4.Hartiala J, Bennett BJ, Tang WH, et al. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and L-carnitine. Arterioscler Thromb Vasc Biol. 2014;34:1307–1313. doi: 10.1161/ATVBAHA.114.303252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett BJ, de Aguiar Vallim TQ, Wang Z, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller CA, Corbin KD, da Costa KA, et al. Effect of egg ingestion on trimethylamine-N-oxide production in humans: a randomized, controlled, dose-response study. Am J Clin Nutr. 2014;100:778–786. doi: 10.3945/ajcn.114.087692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaw GM, Finnell RH, Blom HJ, et al. Choline and risk of neural tube defects in a folate-fortified population. Epidemiology. 2009;20:714–719. doi: 10.1097/EDE.0b013e3181ac9fe7. [DOI] [PubMed] [Google Scholar]
- 8.Zeisel SH. Choline: critical role during fetal development and dietary requirements in adults. Annu Rev Nutr. 2006;26:229–250. doi: 10.1146/annurev.nutr.26.061505.111156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw GM, Vollset SE, Carmichael SL, et al. Nested case-control study of one-carbon metabolites in mid-pregnancy and risks of cleft lip with and without cleft palate. Pediatr Res. 2009;66:501–506. doi: 10.1203/PDR.0b013e3181b9b544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silver MJ, Corbin KD, Hellenthal G, et al. Evidence for negative selection of gene variants that increase dependence on dietary choline in a Gambian cohort. FASEB J. 2015;29:3426–3435. doi: 10.1096/fj.15-271056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pogribny IP, Kutanzi K, Melnyk S, et al. Strain-dependent dysregulation of one-carbon metabolism in male mice is associated with choline- and folate-deficient diet-induced liver injury. FASEB J. 2013;27:2233–2243. doi: 10.1096/fj.12-227116. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov A, Nash-Barboza S, Hinkis S, Caudill MA. Genetic variants in phosphatidylethanolamine N-methyltransferase and methylenetetrahydrofolate dehydrogenase influence biomarkers of choline metabolism when folate intake is restricted. J Am Diet Assoc. 2009;109:313–318. doi: 10.1016/j.jada.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohlmeier M, da Costa KA, Fischer LM, Zeisel SH. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc Natl Acad Sci U S A. 2005;102:16025–16030. doi: 10.1073/pnas.0504285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brody LC, Conley M, Cox C, et al. A polymorphism, R653Q, in the trifunctional enzyme methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase/formyltetrahydrofolate synthetase is a maternal genetic risk factor for neural tube defects: report of the Birth Defects Research Group. Am J Hum Genet. 2002;71:1207–1215. doi: 10.1086/344213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills JL, Molloy AM, Parle-McDermott A, et al. Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82:636–643. doi: 10.1002/bdra.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balduino Victorino D, de Godoy MF, Goloni-Bertollo EM, Pavarino EC. Genetic polymorphisms involved in folate metabolism and maternal risk for down syndrome: a meta-analysis. Dis Markers. 2014;2014:517504. doi: 10.1155/2014/517504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng J, Han L, Zhuang B. Association between MTHFD1 polymorphisms and neural tube defect susceptibility. J Neurol Sci. 2015;348:188–194. doi: 10.1016/j.jns.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Zheng J, Lu X, Liu H, et al. MTHFD1 polymorphism as maternal risk for neural tube defects: a meta-analysis. Neurol Sci. 2015;36:607–616. doi: 10.1007/s10072-014-2035-7. [DOI] [PubMed] [Google Scholar]
- 19.Yan J, Winter LB, Burns-Whitmore B, et al. Plasma choline metabolites associate with metabolic stress among young overweight men in a genotype-specific manner. Nutr Diabetes. 2012;2:e49. doi: 10.1038/nutd.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mills JL, Fan R, Brody LC, et al. Maternal choline concentrations during pregnancy and choline-related genetic variants as risk factors for neural tube defects. Am J Clin Nutr. 2014;100:1069–1074. doi: 10.3945/ajcn.113.079319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Visentin CE, Masih S, Plumptre L, et al. Maternal Choline Status, but Not Fetal Genotype, Influences Cord Plasma Choline Metabolite Concentrations. J Nutr. 2015;145:1491–1497. doi: 10.3945/jn.115.211136. This report compares assesses metabolite concentration at different stages of gestation and in umbical cord blood. [DOI] [PubMed] [Google Scholar]
- 22.Carolan-Olah M, Duarte-Gardea M, Lechuga J. A critical review: early life nutrition and prenatal programming for adult disease. J Clin Nurs. 2015 doi: 10.1111/jocn.12951. [DOI] [PubMed] [Google Scholar]
- 23.Medici V, Shibata NM, Kharbanda KK, et al. Maternal choline modifies fetal liver copper, gene expression, DNA methylation, and neonatal growth in the tx-j mouse model of Wilson disease. Epigenetics. 2014;9:286–296. doi: 10.4161/epi.27110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neggers YH. Increasing prevalence, changes in diagnostic criteria, and nutritional risk factors for autism spectrum disorders. ISRN Nutr. 2014;2014:514026. doi: 10.1155/2014/514026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barua S, Chadman KK, Kuizon S, et al. Increasing maternal or post-weaning folic acid alters gene expression and moderately changes behavior in the offspring. PLoS One. 2014;9:e101674. doi: 10.1371/journal.pone.0101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shorter KR, Felder MR, Vrana PB. Consequences of dietary methyl donor supplements: Is more always better? Prog Biophys Mol Biol. 2015;118:14–20. doi: 10.1016/j.pbiomolbio.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez-Salas P, Moore SE, Baker MS, et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun. 2014;5:3746. doi: 10.1038/ncomms4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke R, Halsey J, Lewington S, et al. Effects of lowering homocysteine levels with B vitamins on cardiovascular disease, cancer, and cause-specific mortality: Meta-analysis of 8 randomized trials involving 37 485 individuals. Arch Intern Med. 2010;170:1622–1631. doi: 10.1001/archinternmed.2010.348. [DOI] [PubMed] [Google Scholar]
- 29.Biselli PM, Guerzoni AR, de Godoy MF, et al. Genetic polymorphisms involved in folate metabolism and concentrations of methylmalonic acid and folate on plasma homocysteine and risk of coronary artery disease. J Thromb Thrombolysis. 2010;29:32–40. doi: 10.1007/s11239-009-0321-7. [DOI] [PubMed] [Google Scholar]
- 30.Ghazouani L, Abboud N, Mtiraoui N, et al. Homocysteine and methylenetetrahydrofolate reductase C677T and A1298C polymorphisms in Tunisian patients with severe coronary artery disease. J Thromb Thrombolysis. 2009;27:191–197. doi: 10.1007/s11239-008-0194-1. [DOI] [PubMed] [Google Scholar]
- 31.Ramkaran P, Phulukdaree A, Khan S, et al. Methylenetetrahydrofolate reductase C677T polymorphism is associated with increased risk of coronary artery disease in young South African Indians. Gene. 2015 doi: 10.1016/j.gene.2015.06.044. [DOI] [PubMed] [Google Scholar]
- 32.Matam K, Khan IA, Hasan Q, Rao P. Coronary artery disease and the frequencies of MTHFR and PON1 gene polymorphism studies in a varied population of Hyderabad, Telangana region in south India. Journal of King Saud University. 2015;27:143–150. [Google Scholar]
- 33.Hou X, Chen X, Shi J. Genetic polymorphism of MTHFR C677T and premature coronary artery disease susceptibility: A meta-analysis. Gene. 2015;565:39–44. doi: 10.1016/j.gene.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 34.Huo Y, Li J, Qin X, et al. Efficacy of folic acid therapy in primary prevention of stroke among adults with hypertension in China: the CSPPT randomized clinical trial. JAMA. 2015;313:1325–1335. doi: 10.1001/jama.2015.2274. [DOI] [PubMed] [Google Scholar]
- 35.Trovato FM, Catalano D, Ragusa A, et al. Relationship of MTHFR gene polymorphisms with renal and cardiac disease. World J Nephrol. 2015;4:127–137. doi: 10.5527/wjn.v4.i1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ganesh SK, Tragante V, Guo W, et al. Loci influencing blood pressure identified using a cardiovascular gene-centric array. Hum Mol Genet. 2013;22:1663–1678. doi: 10.1093/hmg/dds555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xi B, Shen Y, Zhao X, et al. Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children. J Hum Hypertens. 2014;28:32–36. doi: 10.1038/jhh.2013.50. [DOI] [PubMed] [Google Scholar]
- 38.Benton MC, Lea RA, Macartney-Coxson D, et al. Mapping eQTLs in the Norfolk Island genetic isolate identifies candidate genes for CVD risk traits. Am J Hum Genet. 2013;93:1087–1099. doi: 10.1016/j.ajhg.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoneyama S, Guo Y, Lanktree MB, et al. Gene-centric meta-analyses for central adiposity traits in up to 57 412 individuals of European descent confirm known loci and reveal several novel associations. Hum Mol Genet. 2014;23:2498–2510. doi: 10.1093/hmg/ddt626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Al Rajabi A, Castro GS, da Silva RP, et al. Choline supplementation protects against liver damage by normalizing cholesterol metabolism in Pemt/Ldlr knockout mice fed a high-fat diet. J Nutr. 2014;144:252–257. doi: 10.3945/jn.113.185389. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, Su B, Jacobs RL, et al. Lack of phosphatidylethanolamine N-methyltransferase alters plasma VLDL phospholipids and attenuates atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2009;29:1349–1355. doi: 10.1161/ATVBAHA.109.188672. [DOI] [PubMed] [Google Scholar]
- 42.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–12516. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- 44.Ussher JR, Lopaschuk GD, Arduini A. Gut microbiota metabolism of L-carnitine and cardiovascular risk. Atherosclerosis. 2013;231:456–461. doi: 10.1016/j.atherosclerosis.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Amirian ES, Petrosino JF, Ajami NJ, et al. Potential role of gastrointestinal microbiota composition in prostate cancer risk. Infect Agent Cancer. 2013;8:42. doi: 10.1186/1750-9378-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spencer MD, Hamp TJ, Reid RW, et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest. 2014;124:4204–4211. doi: 10.1172/JCI72331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Tang WH, Buffa JA, et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur Heart J. 2014;35:904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6:e02481. doi: 10.1128/mBio.02481-14. This paper uses studies in gnotobiotic mice to identify gut microbes affecting choline bioavailability and TMAO levels. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shih DM, Wang Z, Lee R, et al. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res. 2015;56:22–37. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warrier M, Shih DM, Burrows AC, et al. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. 2015 doi: 10.1016/j.celrep.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52**.da Costa KA, Corbin KD, Niculescu MD, et al. Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups. FASEB J. 2014;28:2970–2978. doi: 10.1096/fj.14-249557. This paper identifys variants that affect choline metabolism in populations consuming low choline diets. This study suggests that historical nutrient intake may apply selective pressures that influence allele frequency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schenkel LC, Singh RK, Michel V, et al. Mechanism of choline deficiency and membrane alteration in postural orthostatic tachycardia syndrome primary skin fibroblasts. FASEB J. 2015;29:1663–1675. doi: 10.1096/fj.14-258566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhee EP, Ho JE, Chen MH, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]