Abstract
We previously found that TYK2 tyrosine kinase signaling through its downstream effector phospho-STAT1 (p-STAT1) acts to upregulate BCL2, which in turn mediates aberrant survival of T-cell acute lymphoblastic leukemia (T-ALL) cells. Here we show that pharmacologic inhibition of heat shock protein 90 (HSP90) with a small-molecule inhibitor, NVP-AUY922 (AUY922), leads to rapid degradation of TYK2 and apoptosis in T-ALL cells. STAT1 protein levels were not affected by AUY922 treatment, but p-STAT1 (Tyr 701) levels rapidly became undetectable, consistent with a block in signaling downstream of TYK2. BCL2 expression was downregulated after AUY922 treatment, and although this effect was necessary for AUY922-induced apoptosis, it was not sufficient because many T-ALL cell lines were resistant to ABT-199, a specific inhibitor of BCL2. Unlike ABT-199, AUY922 also upregulated the proapoptotic proteins BIM and BAD, whose increased expression was required for AUY922-induced apoptosis. Thus, the potent cytotoxicity of AUY922 involves the synergistic combination of BCL2 downregulation coupled with upregulation of the proapoptotic proteins BIM and BAD. This two-pronged assault on the mitochondrial apoptotic machinery identifies HSP90 inhibitors as promising drugs for targeting the TYK2-mediated prosurvival signaling axis in T-ALL cells.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is caused by the malignant transformation of thymocyte progenitors. Its prognosis has improved substantially with the introduction of intensified chemotherapy, with cure rates exceeding 75% in children and about 50% in adults.1,2 Nonetheless, the clinical outcome in T-ALL patients with primary resistant or relapsed disease remains poor,1,3,4 indicating an urgent need for new therapeutic approaches based on more effective and less toxic antileukemic drugs.5
We recently reported a novel oncogenic pathway in T-ALL that involves aberrant activation of tyrosine kinase 2 (TYK2) and its downstream effector, STAT1, which ultimately promotes T-ALL cell survival through upregulation of the prosurvival protein BCL2.6 This finding was the first to implicate TYK2, a member of the Janus-activated kinase (JAK) tyrosine kinase family, in T-ALL pathogenesis. Indeed, our gene knockdown experiments showed TYK2 dependency in 14 (88%) of 16 T-ALL cell lines and 5 (63%) of 8 patient-derived T-ALL xenografts, while pharmacologic inhibition of TYK2 with a small-molecule pan-JAK inhibitor, JAK inhibitor I, induced apoptosis in multiple T-ALL cell lines.6 We concluded from these findings that in many T-ALL cases, the leukemic cells depend upon the TYK2-STAT1-BCL2 pathway to maintain cell survival, suggesting that inhibition of TYK2 would be beneficial in patients with T-ALL. Unfortunately, effective inhibitors of TYK2 are not available for clinical use, leading us to seek alternative approaches to target TYK2 in T-ALL cells.
Because TYK2 is a client protein of heat shock protein 90 (HSP90),7,8 we considered that pharmacologic inhibition of HSP90 would be a reasonable strategy to disrupt TYK2 protein stability. As an ATP-dependent molecular chaperone, HSP90 participates in stabilizing and activating its client proteins, many of which are essential for cell signaling and adaptive response to stress.9,10 Since cancer cells exploit this chaperone mechanism to support activated oncoproteins with important functions in the development and promotion of malignancy, targeting HSP90 has emerged as a promising approach to cancer therapy.11,12 Small-molecule HSP90 inhibitors now under clinical evaluation occupy the ATP-binding pocket of HSP90, where they block ATP binding and stop the chaperone cycle, leading to ubiquitin proteasome–mediated degradation of its client proteins.11 Early reports on the therapeutic efficacy of HSP90 inhibitors against widely different cancers have been encouraging.13,14 Such drugs have shown both in vitro and in vivo activity in myeloproliferative malignancies 15 and in a subset of B-cell acute lymphoblastic leukemias with rearrangements of the cytokine receptor-like factor 2 gene (CRLF2).16 The antitumor effects of the HSP90 inhibitors in these studies were mediated through degradation of JAK2, a member of the JAK tyrosine kinase family, and treatment with an HSP90 inhibitor overcame genetic resistance to JAK enzymatic inhibitors.15,16 Together, these observations provide a compelling rationale for testing an HSP90 inhibitor as a means to target the activity of TYK2 in T-ALL.
Here we report striking activity against T-ALL by a small-molecule HSP90 inhibitor, NVP-AUY922 (AUY922),17 which has recently been in clinical trials against breast cancer, non-small cell lung cancer, gastric cancer, advanced solid tumors and multiple myeloma. We show that AUY922 induces apoptosis in human T-ALL cell lines at concentrations associated with TYK2 degradation and downregulation of its downstream pathways, including dephosphorylation of STAT1 and downregulation of BCL2 expression. Our results also demonstrate that the proapoptotic BH3-only proteins BIM and BAD are upregulated by AUY922 treatment, simultaneously with the reduction of BCL2 levels. These results provide a mechanism for the induction of apoptosis by AUY922 in leukemic T-ALL cells, justifying further evaluation of this novel targeting strategy.
MATERIALS AND METHODS
Reagents
NVP-AUY922 (AUY922) was provided by Novartis (Basel, Switzerland). ABT-199 was purchased from Chemie Tek (Indianapolis, IN, USA).
Cell culture
All human T-ALL cell lines (KOPT-K1, DND41, JURKAT, SUP-T13, HPB-ALL, MOLT-4, MOLT-16, RPMI-8402, TALL-1, and LOUCY) were obtained from ATCC (Manassas, VA, USA) or DSMZ (Braunschweig, Germany). The creation of Ba/F3 cells transformed by TYK2-E957D was described previously.6 JURKAT and KOPT-K1 cells overexpressing BCL2 were generated with the MSCV-IRES-GFP retroviral expression system. JURKAT and KOPT-K1 cells overexpressing BCLXL or MCL1 cDNA were generated with the pHAGE-CMV-IRES-ZsGreen lentiviral expression system. For additional information, see Supplementary Materials and Methods. These cells were maintained in RPMI-1640 medium (GIBCO, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA) and 1% penicillin/streptomycin (Invitrogen, Waltham, MA, USA).
shRNA knockdown experiments
All shRNA constructs cloned into the lentiviral vector pLKO.1-puro were obtained from the RNAi Consortium (Broad Institute, Cambridge, MA, USA). Target sequences for each shRNA are listed in Supplementary Table 2. For additional information, see Supplementary Materials and Methods.
Cell viability and growth analysis
Cell Titer Glo assay (Promega, Fitchburg, WI, USA) was used to assess relative cell viability and cell growth upon treatment. Cells were plated at a density of 5000 - 10000 cells per well in a 96-well plate and incubated with DMSO or increasing concentrations of drug. The relative cell viability was measured after different treatment intervals and reported as a percentage of the DMSO control. The concentration of drug required for 50% inhibition of cell viability (IC50) was determined by substituting values in the following equation: IC50=10 ^ (LOG[A/B]*(50-C)/(D-C) + LOG[B]), where A= higher concentration near 50%; B= lower concentration near 50%; C= inhibition rate at B; D= inhibition rate at A. Cell growth after treatment with a drug is reported as the fold change from day 0.
Apoptosis and cell-cycle analysis
The TUNEL assay and propidium iodide (PI) staining were performed with the APOBrdU™ TUNEL assay kit (Invitrogen) according to the manufacturer's recommendation. Additional information can be found in Supplementary Materials and Methods. Annexin V and PI double staining was also used for detecting apoptosis. 2 × 105 cells of each treated sample were washed with PBS, incubated in staining buffer containing fluorescein isothiocynate (FITC)-conjugated anti-Annexin V antibody and PI (MBL International, Nagoya, Japan), and then analyzed by BD FACSCalibur (BD Biosciences, San Jose, CA, USA).
Western blot and immunoprecipitation
Whole-cell lysates were prepared in RIPA buffer (Cell Signaling Tech, Danvers, MA, USA) with FOCUS™ ProteaseArrest™ (G-Biosciences, St. Louis, MO, USA) and Phosphatase Inhibitor Cocktail Set II (EMD Millipore, Billerica, MA, USA). ). Immunoblotting was performed with each of the specific antibodies to TYK2, STAT1, phospho-STAT1 (Y701), BCL2, BCLXL, MCL1, HSP70, PARP, BIM, BAX, BAK, BID, PUMA, BIK, α-tubulin (Cell Signaling Tech), HSP90 (Abcam, Cambridge, UK, #ab1429) and BAD (Santa Cruz, Dallas, TX, USA, #sc-8044). For immunoprecipitation experiments, the cells were lysed in Pierce IP lysis buffer (Thermo Scientific) with Halt™ protease inhibitor cocktail (Thermo Scientific). Rabbit BCL2 antibody (Cell Signaling Tech #4223) coupled to Dynabeads Protein A (Life technologies, Waltham, MA, USA) was incubated with 100 to 180 μl lysate on a rotator for 10 min at room temperature. Beads were washed three times on a magnet, and then proteins were eluted and immunoblotted with each of specific antibodies.
RNA extraction, cDNA synthesis and gene expression analysis
Total RNA was extracted by TRIzol (Invitrogen) followed by a column purification using the RNeasy Mini kit (QIAGEN, Venlo, Netherlands). Purified RNA was reverse-transcribed with the SuperScript™ III First-Strand Synthesis system (Invitrogen) according to the manufacturer's protocol. Quantitative real-time PCR analysis was performed with the Applied Biosystems 7500 or ViiA 7 Real-Time PCR system (Applied Biosystems, Waltham, MA, USA), using specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems) according to the manufacturer's protocol. The primer sequences are listed in Supplementary Table 3.
Statistical analysis
Statistical significance in assays with identical cell lines was assessed with Student's t test (two-tailed). GraphPad Prism 6.02 (GraphPad Software, La Jolla, CA, USA) was used for all statistical analyses.
RESULTS
The small-molecule HSP90 inhibitor NVP-AUY922 inhibits the growth of multiple TALL cell lines
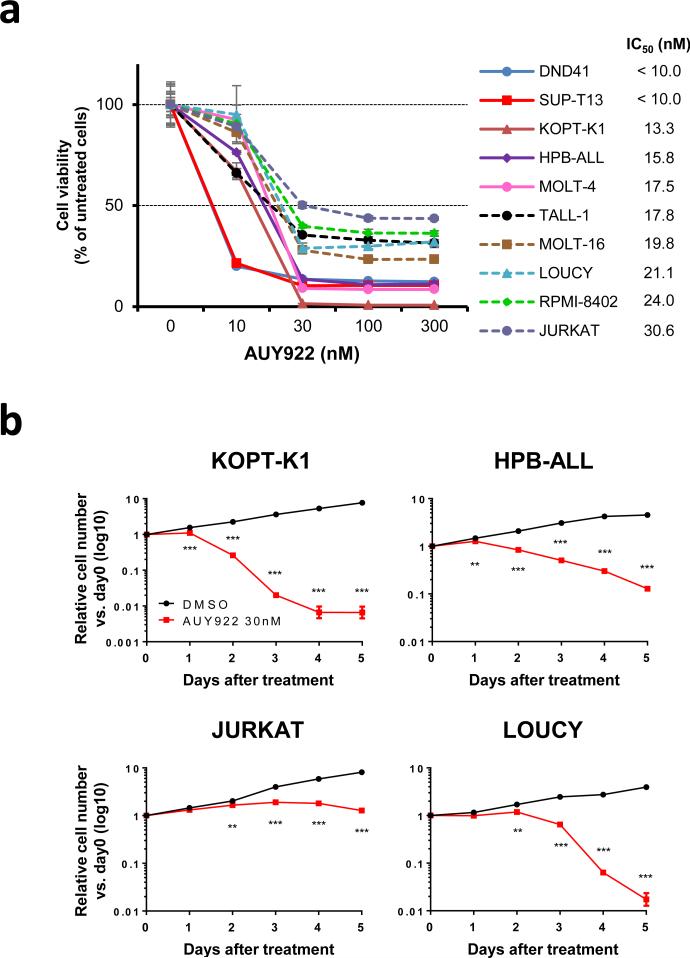
To assess the antitumor potency of HSP90 inhibition against T-ALL cells, we first tested the inhibitory effect of AUY922 on the growth of multiple T-ALL cell lines. This agent induced remarkable cytotoxicity in each of the 10 human T-ALL cell lines tested, which harbor a range of different molecular aberrations typical of T-ALL (Supplementary Table 1) and express substantial levels of HSP90 and TYK2 (Supplementary Figure 1). In each of the T-ALL cell lines, the 50% inhibitory concentration (IC50) of AUY922 was less than or equal to 30.6 nM after 72 h of exposure (Figure 1a and Supplementary Table 1). The relative cell viability after 96 h exposure to the drug was decreased in most cell lines when compared to that after 72 h of treatment, although the decrease of viability appeared to plateau at 30 nM in JURKAT cells (Supplementary Figure 2 and Supplementary Table 1). To confirm these results, we studied the cell growth kinetics of 4 representative T-ALL cell lines (KOPT-K1, HPB-ALL, JURKAT and LOUCY) treated with 30 nM of AUY922. The cell numbers in each cell line were significantly lower in the AUY922-treated samples than in the controls after 1 or 2 days of treatment, indicating that the drug induces cytotoxicity rather than just delaying cell proliferation (Figure 1b). These results demonstrate that AUY922 potently inhibits the growth of T-ALL cell lines, regardless of the genetic abnormalities that contribute to leukemic transformation.
Figure 1. AUY922 inhibits the growth of T-ALL cell lines.
(a) Human T-ALL cell lines were cultured with graded concentrations of AUY922 for 72 h. Cell viability values are mean ± s.d. percentages of the untreated control value in triplicate experiments. The IC50 value of each cell line in this assay is indicated on the right. (b) KOPT-K1, HPB-ALL, JURKAT, and LOUCY cells were cultured with 30 nM of AUY922 or DMSO, and their growth was measured. Values are mean ± s.d fold changes relative to day 0 in triplicate experiments. **, P < 0.01; ***, P < 0.001 by two-sample, two-tailed t test.
AUY922 treatment induces apoptosis in T-ALL cell lines
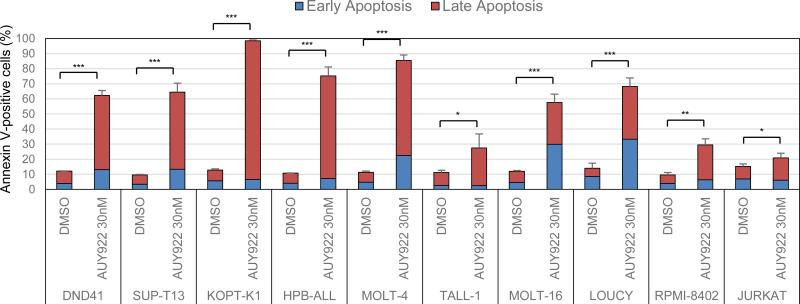
To gain insight into the cytotoxic mechanism triggered by AUY922, we next assessed the effect of the inhibitor on apoptosis by Annexin V and PI double staining. In each cell line tested, 30 nM of AUY922 significantly increased the percentage of Annexin V-positive cells after 72 h of treatment by comparison with results for DMSO-treated cells (Figure 2; Supplementary Figure 3). We also observed significantly increased levels of apoptotic cells at 48 h after treatment with 30 nM of AUY922 by flow cytometric analysis after TUNEL and PI double staining to detect the fragmented DNA of apoptotic cells and changes in cell-cycle progression (Supplementary Figures 4a and b). The results show an excess of apoptotic cells in the G2 and S phases of the cell cycle, as well as in G1 phase (Supplementary Figure 4a), indicating that AUY922 treatment kills T-ALL cells by inducing apoptosis in a non-cell-cycle-specific manner.
Figure 2. AUY922 treatment induces apoptosis in T-ALL cell lines.
T-ALL cell lines were treated with DMSO or 30 nM of AUY922 for 72 h, and assessed for apoptosis by flow cytometric analysis following Annexin V-FITC and PI double staining. Early and late apoptosis represent the fraction of Annexin V-positive / PI-negative cells and Annxin V-positive / PI-positive cells, respectively. Values are mean ± s.d. percentages of Annexin V-positive cells in triplicate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-sample, two-tailed t test. The representative dot plots images in this assay are shown in Supplementary Figure 1.
HSP90 inhibition leads to TYK2 degradation and subsequent downregulation of its downstream pathway in T-ALL cells
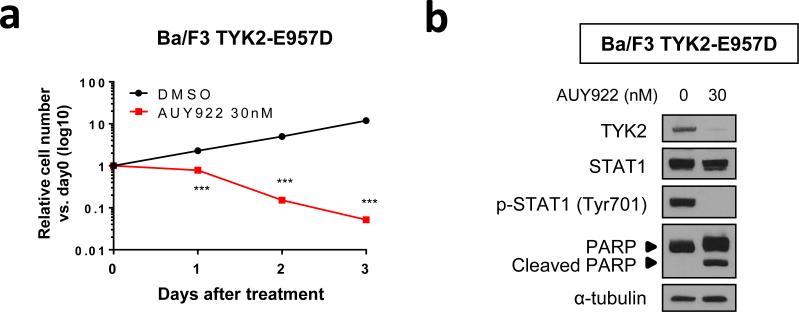
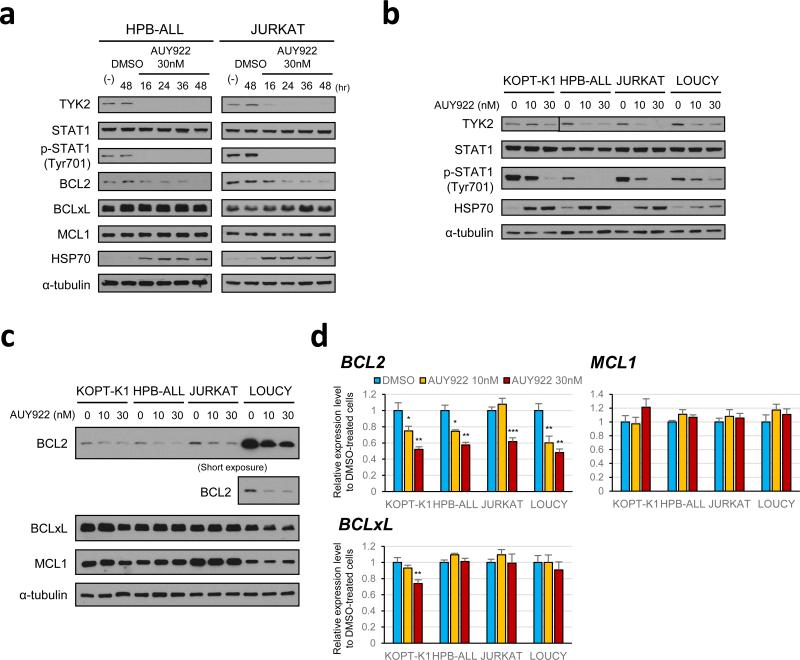
As an HSP90 client protein, TYK2 offers an attractive target for AUY922 treatment.7,8 Indeed, Ba/F3 cells transformed by constitutively activated TYK2 (TYK2-E957D) 6 were highly sensitive to AUY922 treatment (Figure 3a), which was associated with AUY922-mediated TYK2 degradation (Figure 3b). Next, to assess the effect of HSP90 inhibition on TYK2-STAT1-BCL2 prosurvival signaling in T-ALL cells, we examined the status of this pathway in T-ALL cell lines treated with AUY922. Time-course analyses with HPB-ALL and JURKAT cells demonstrated a dramatic reduction in TYK2 levels and the elimination of STAT1 Tyr-701 phosphorylation after 16 h of treatment with 30 nM of AUY922, while levels of the STAT1 protein remained unaffected in both cell lines (Figure 4a). Progressively lower levels of BCL2 were observed starting at 16 h after treatment with AUY922 in both cell lines, with the lowest levels seen at 48 h, whereas the levels of BCLxL and MCL1 proteins were unaffected in this context (Figure 4a). AUY922 promoted HSP70 upregulation, a known response to HSP90 inhibition mediated by heat shock factor 1 (Figure 4a).18,19 Rapid reduction of TYK2 and elimination of STAT1 Tyr-701 phosphorylation were also observed in multiple T-ALL cell lines treated with AUY922 for 16 h (Figure 4b), with decreased levels of BCL2 protein confirmed by western blot analysis in each of these cells after 48 h of treatment (Figure 4c). BCLxL and MCL1 protein levels were unaffected, except that BCLxL levels were slightly reduced in KOPT-K1 cells (Figure 4c), a T-ALL cell line with a marked sensitivity to AUY922 (Figures 1 and 2; Supplementary Figures 3 and 4). Quantitative RT-PCR showed that treatment with AUY922 also significantly decreased BCL2 mRNA levels (Figure 4d), indicating that the AUY922-induced decrease of BCL2 is at least partly due to downregulation of its upstream signaling that promotes BCL2 transcription. BCLxL and MCL1 mRNA levels were unaffected by AUY922, except for decreased expression of BCLxL in KOPT-K1 cells treated with AUY922 (Figure 4d). These results indicate that pharmacologic inhibition of HSP90 leads to TYK2 degradation and subsequent downregulation of its downstream signaling pathway in T-ALL cells.
Figure 3. Ba/F3 cells transformed by constitutively activated TYK2 are highly sensitive to AUY922.
(a) Ba/F3 cells transformed by TYK2-E957D were cultured with 30 nM of AUY922 or DMSO, and their growth was measured. Values are mean ± s.d. fold changes relative to day 0 in triplicate experiments. ***, P < 0.001 by two-sample, two-tailed t test. (b) Western blot analysis using Ba/F3 TYK2-E957D cells treated with 30 nM of AUY922 or DMSO (AUY922 0 nM) for 24 h.
Figure 4. HSP90 inhibition by AUY922 leads to TYK2 degradation and subsequent downregulation of its downstream pathway in T-ALL cells.
(a) Time-course analysis to assess the effect of AUY922 on the TYK2-STAT1-BCL2 pro-survival pathway in HPB-ALL and JURKAT cells. The cells were treated with DMSO or 30 nM of AUY922, and the proteins were harvested at the indicated times. Western blot analysis was conducted with each specific antibody. (b) Western blot analysis to assess the effect of AUY922 on the TYK2-STAT1 signaling in multiple T-ALL cell lines. KOPT-K1, HPB-ALL, JURKAT and LOUCY cells were treated with the indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 16 h, and subjected to immunoblot analysis with antibodies specific for TYK2, STAT1, phospho-STAT1 (Tyr701), HSP70 and α-tubulin. (c) Western blot analysis to assess the effect of AUY922 on the expression of antiapoptotic BCL2 family proteins in multiple T-ALL cell lines. KOPT-K1, HPB-ALL, JURKAT and LOUCY cells were treated with indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 48 h, and subjected to immunoblot analysis with antibodies specific for BCL2, BCLxL, MCL1 and α-tubulin. (d) BCL2, BCLxL and MCL1 mRNA expressions after AUY922 treatment in T-ALL cell lines. KOPT-K1, HPB-ALL, JURKAT and LOUCY cells were treated with the indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 16 h. BCL2, BCLXL and MCL1 mRNA levels were measured by quantitative RT-PCR and normalized by GAPDH expression. Expression levels relative to DMSO-treated cells are shown as mean ± s.d. of triplicate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-sample, two-tailed t test.
Loss of BCL2 expression is necessary for AUY922-induced apoptosis in T-ALL cells
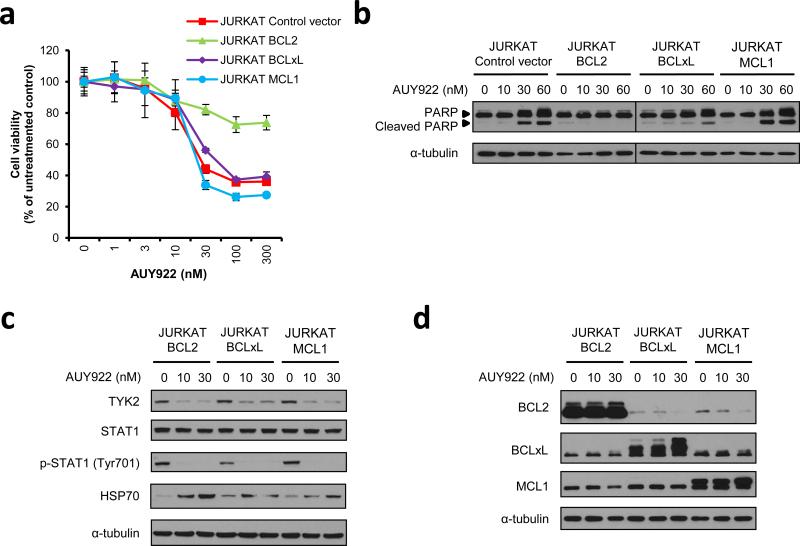
Because AUY922 treatment efficiently reduces BCL2 expression in T-ALL cells (Figure 4), with subsequent induction of apoptosis in each of the cell lines tested (Figure 2; Supplementary Figures 3 and 4), we hypothesized that reduced BCL2 levels may be necessary for AUY922-mediated apoptosis in T-ALL cells. Thus, we prepared JURKAT TALL cell lines that overexpress each of three different antiapoptotic BCL2 family proteins: BCL2, BCLxL, or MCL1. Overexpression of each antiapoptotic protein rescued daunomycin-induced decrease of cell viability (Supplementary Figure 5), indicating that each overexpressed protein is functional in JURKAT cells. By contrast, only BCL2 overexpression was able to partially rescue an AUY922-induced decrease of cell viability, while overexpression of BCLxL or MCL1 had no effect (Figure 5a). BCL2 overexpression protected the cells from AUY922-induced apoptosis, as indicated by the absence of cleaved PARP (Figure 5b) and the absence of an increase in TUNEL positivity (Supplementary Figures 6a and b) in the BCL2-overexpressing JURKAT cells. AUY922 rapidly reduced TYK2 protein levels and eliminated STAT1 Tyr701 phosphorylation in each of the JURKAT cell lines overexpressing an antiapoptotic BCL2 family protein (Figure 5c), as observed in parental cells (Figure 4b). Expression of the BCL2 protein was decreased after AUY922 treatment in BCLxL- or MCL1-overexpressing cells, but was maintained in cells that overexpress BCL2 (Figure 5d). BCLxL and MCL1 levels were unaffected or upregulated by AUY922 treatment in this context (Figure 5d). We also showed that overexpression of BCL2, but not BCLxL or MCL1, partially rescued the KOPT-K1 T-ALL cell line from AUY922-induced apoptosis, as best shown by its ability to block PARP cleavage (Supplementary Figures 7a - e). These results demonstrate that exogenous expression of BCL2 can rescue T-ALL cells from apoptosis induced by AUY922, suggesting a central role for decreased levels of BCL2 in the apoptotic response induced by this drug.
Figure 5. BCL2 overexpression can rescue AUY922-induced apoptosis in JURKAT T-ALL cell line.
(a) JURKAT cells overexpressing BCL2, BCLXL or MCL1, or infected with a control vector, were treated with graded concentrations of AUY922 for 72 h. Cell viability values are shown as mean ± s.d. percentages of the untreated control value in triplicate experiments. (b) Western blot analysis to examine PARP cleavage in the specific JURKAT cells treated with AUY922. The cells were treated with the indicated concentrations of AUY922 or DMSO control (AUY922 0 nM) for 48 h, and subjected to immunoblot analysis with antibodies specific for PARP and α-tubulin. (c) Western blot analysis of specific JURKAT cells treated with the indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 16 h. (d) Western blot analysis of specific JURKAT cells treated with the indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 48 h.
AUY922 treatment not only downregulates BCL2, but also upregulates BIM and BAD
Recent reports on the efficacy of the BCL2-specific inhibitor ABT-199 have shown that this drug is not active in the killing of typical T-ALL cells, whereas T-ALL cells with an early T-cell progenitor (ETP) phenotype show remarkable sensitivity.20-22 Consistent with these findings, our results also show a lack of sensitivity of most T-ALL cell lines to ABT-199 (Supplementary Figure 8a), whereas the LOUCY cell line, which is distinguished by its ETP phenotype,23 was highly sensitive (Supplementary Figure 8b). Thus, decreased expression of BCL2 alone does not appear sufficient to kill most T-ALL cell lines, implying that mechanisms besides downregulation of BCL2 contribute to the induction of apoptosis by AUY922.
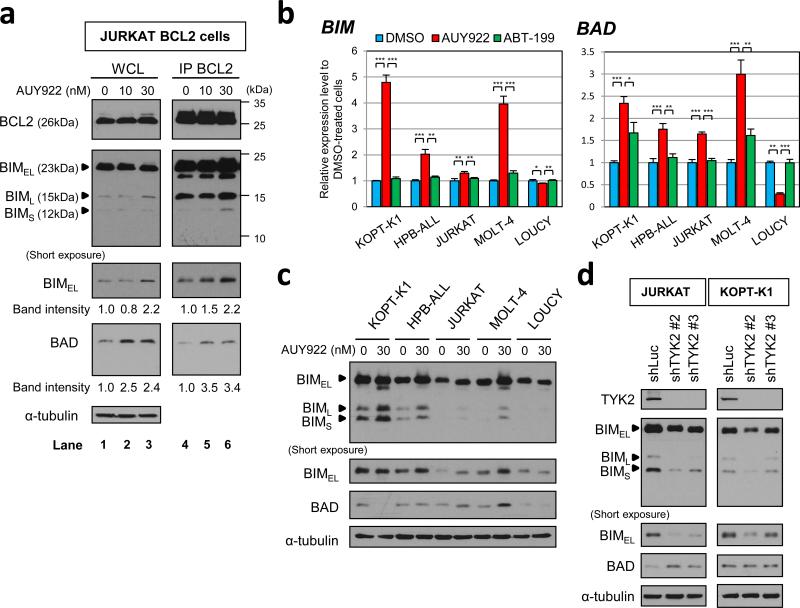
BCL2 and other antiapoptotic BCL2 family proteins can block apoptosis by binding to and sequestering proapoptotic proteins in this family that cooperatively induce mitochondrial outer membrane permeabilization.24,25 To investigate the relevance of this mechanism for AUY922-induced apoptosis in T-ALL cells, we studied the effects of the drug on levels of proapoptotic proteins and their interactions with overexpressed BCL2 in the JURKAT cell line. Treatment with 30 nM of AUY922 for 48 h resulted in increased BIM and BAD levels in the BCL2-overexpressing JURKAT cells, without an effect in the BCL2 level (Figure 6a, lanes 1-3). BAX expression was not detected in these cells, even after AUY922 treatment (Supplementary Figure 9a), as others have shown for JURKAT cells;26 while BAK expression was unchanged (Supplementary Figure 9b). Levels of the BH3-only proteins BID, PUMA and BIK were also unaffected or even downregulated by AUY922 treatment (Supplementary Figures 9c - e). Moreover, an immunoprecipitation assay with BCL2 antibody (IP BCL2) revealed that AUY922 treatment leads to increased binding of both BIM and BAD to BCL2 (Figure 6a, lanes 4-6), demonstrating that AUY922 triggers upregulation of BIM and BAD, and that these proapoptotic proteins are then sequestered in the cells by overexpressed BCL2.
Figure 6. AUY922 upregulates the expression of BIM and BAD proteins, which are sequestered by overexpressed BCL2 in JURKAT cells.
(a) BCL2-overexpressing JURKAT cells were treated with the indicated concentrations of AUY922 or DMSO (AUY922 0 nM) for 48 h. Anti-BCL2 immunoprecipitate from 320 μg of each total cell lysate (IP BCL2) was immunoblotted for BCL2, BIM and BAD (lanes 4-6). Whole-cell lysate (20 μg; WCL) was also loaded and immunoblotted for BCL2, BIM, BAD, and α-tubulin (lanes 1-3). Three isoforms of BIM (BIMEL, BIML and BIMS) are shown. Band intensities of BIMEL and BAD on the short-exposure film were measured by ImageJ, and relative intensities are shown. (b) BIM and BAD mRNA expression in T-ALL cell lines treated with 30 nM of AUY922, 5 μM of ABT-199 or DMSO for 48 h. LOUCY cells were treated with 1 nM of ABT-199 instead of 5 μM of ABT-199, based on the high sensitivity of the cells to ABT-199. BIM and BAD mRNA expression was measured by quantitative RTPCR and normalized by GAPDH expression. Expression levels relative to DMSO-treated cells are shown as mean ± s.d. of triplicate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-sample, two-tailed t test. (c) Western blot analysis to assess the effect of AUY922 on BIM and BAD protein expression levels in T-ALL cell lines. KOPT-K1, HPBALL, JURKAT, MOLT-4 and LOUCY cells were treated with 30 nM of AUY922 or DMSO (AUY922 0 nM) for 48 h, and subjected to immunoblot analysis with antibodies specific for BIM, BAD and α-tubulin. (d) JURKAT and KOPT-K1 cells were lentivirally transduced with each of TYK2-targeting shRNAs (shTYK2 #2 and #3) or a control shRNA targeting luciferase (shLuc). Whole-cell extracts were analyzed by immunoblotting with antibodies specific for TYK2, BIM, BAD and α-tubulin.
We next determined whether BIM and BAD upregulation after AUY922 treatment occurs in the other human T-ALL cell lines that are sensitive to this drug. Based on the IC50 values of T-ALL cell lines after 72 h of exposure to AUY922 or ABT-199 (Figure 1a; Supplementary Figures 8a), we treated the cells with 30 nM of AUY922, 5 μM of ABT-199, or DMSO for 48 h, and then compared BIM and BAD mRNA levels. LOUCY cells were treated with 1 nM of ABT-199 instead of 5 μM, because they are highly sensitive to this drug (Supplementary Figure 8b). Quantitative RT-PCR analysis indicated that treatment with 30 nM of AUY922 resulted in significant upregulation of BIM and BAD expression in each of the T-ALL cell lines included in this study, except for LOUCY cells (Figure 6b; Supplementary Figure 10). BIM protein levels were also increased by AUY922 treatment in multiple T-ALL cell lines excluding LOUCY cells, whereas increased levels of BAD protein were observed in JURKAT and MOLT-4 cells at 48 h of exposure to the drug (Figure 6c). Importantly, in all of the specific JURKAT cell lines and other T-ALL cell lines tested, treatment with ABT-199 had little or no effect on BIM and BAD expression levels as compared with AUY922 treatment (Figure 6b; Supplementary Figure 10), suggesting that AUY922-mediated BIM and BAD upregulation is not associated with a decrease of BCL2 activity. We also showed that silencing of TYK2 by lentiviral shRNA knockdown resulted in increased levels of BAD expression in JURKAT cells, but not in KOPT-K1 cells (Figure 6d). The BIM levels were not upregulated by TYK2 knockdown in either of the cell lines (Figure 6d). Thus, AUY922 treatment not only leads to decreased BCL2 levels, but also to increased levels of either BIM or BAD, or both proteins in T-ALL cell lines.
Increased BIM expression is crucial for AUY922-induced apoptosis in KOPT-K1 cells
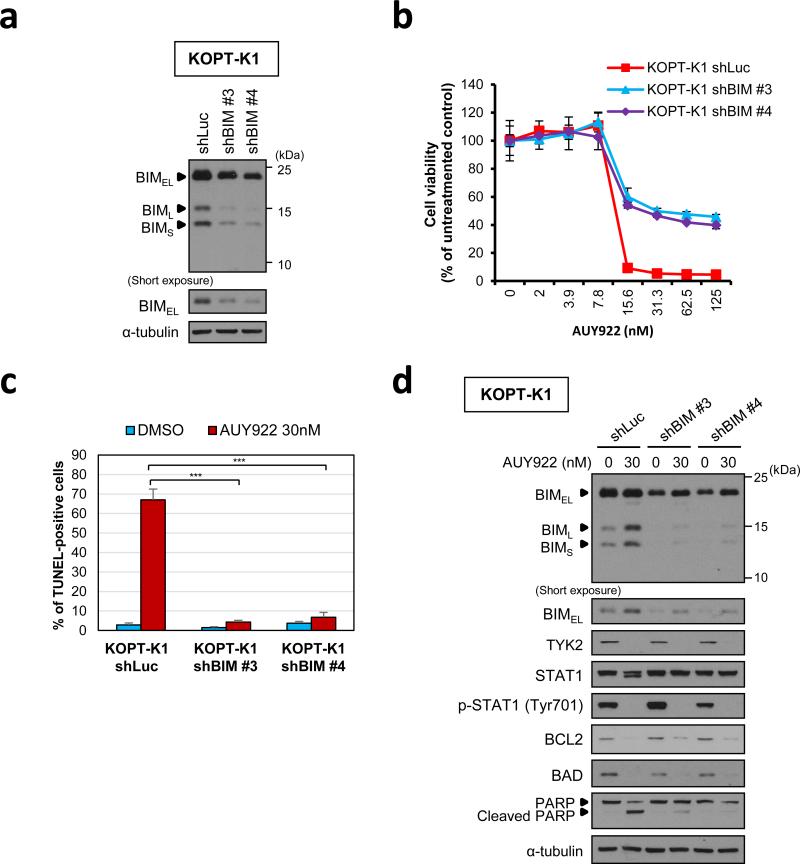
Finally, to assess the importance of elevated proapoptotic BH3-only proteins in AUY922-mediated T-ALL cell death, we transduced KOPT-K1 cells, in which AUY922 had only upregulated BIM expression (Figure 6c), with two independent shRNAs targeting BIM and a control luciferase shRNA, and then treated the cells with AUY922 or DMSO starting 4 days after infection. Cell viability assay after 48 h exposure to the drug demonstrated that incomplete BIM knockdown (Figure 7a) partially rescued the decrease of cell viability induced by AUY922 (Figure 7b). Moreover, flow cytometric analysis following TUNEL/PI double staining indicated that BIM knockdown led to a dramatic decrease in TUNEL positivity among cells treated with 30 nM of AUY922 (Figures 7c; Supplementary Figure 11). BIM expression levels in the cells transfected with BIM-targeting shRNAs were slightly upregulated by AUY922 treatment, but were still lower than in the cells transfected with a control shRNA (Figure 7d). AUY922 treatment did not upregulate BAD levels in all of the shRNA-transfected KOPT-K1 cells (Figure 7d), as observed in parental cells (Figure 6c). We also showed that BIM knockdown was not able to rescue AUY922-induced decrease of viability in JURKAT cells (Supplementary Figures 12a and b), in which BAD protein levels were upregulated after AUY922 treatment unlike in KOPT-K1 cells (Supplementary Figures 12c). These results demonstrate a critical role for increased BIM expression in AUY922-induced apoptosis in KOPT-K1 T-ALL cells, and indicate that upregulation of the BH3-only proteins after AUY922 treatment represents a second activity of the drug that synergistically contributes to cell death when coupled with a decrease of BCL2 expression.
Figure 7. BIM knockdown can rescue AUY922-induced apoptosis in KOPT-K1 T-ALL cell line.
(a) BIM was silenced by lentiviral shRNA knockdown in KOPT-K1 cells and protein expression was assessed by western blotting. Two independent shRNAs (shBIM #3 and #4) were compared with a control shRNA targeting luciferase (shLuc). (b) KOPT-K1 cells transduced with each of BIM-targeting shRNAs (shBIM #3 and #4) or a control shLuc were treated with graded concentrations of AUY922 for 48 h. Cell viability values are shown as mean ± s.d. percentages of the untreated control value in triplicate experiments. (c) KOPT-K1 cells transduced with each of BIM-targeting shRNAs (shBIM #3 and #4) or a control shLuc were treated with DMSO or 30 nM of AUY922 for 36 h. These cells were fixed and assessed for apoptosis and cell-cycle distribution by flow cytometric analysis following TUNEL/PI double labeling. Values represent mean ± s.d. percentages of TUNEL-positive cells in triplicate experiments. ***, P < 0.001 by two-sample, two-tailed t test. The representative dot plots images in this assay are shown in Supplementary Figure 8. (d) KOPT-K1 cells transduced with each of BIM-targeting shRNAs (shBIM #3 and #4) or a control shLuc were treated with 30 nM of AUY922 or DMSO (AUY922 0 nM) for 36 h. Whole-cell extracts were analyzed by immunoblotting with each specific antibody.
DISCUSSION
Here we report the activity of a small-molecule HSP90 inhibitor, NVP-AUY922 (AUY922), against T-ALL cells. This agent efficiently impaired the TYK2-STAT1-BCL2 prosurvival pathway through TYK2 degradation, and then enhanced expression of the potent proapoptotic BH3-only proteins BIM and BAD, leading to apoptosis in T-ALL cells. These findings, in the context of an earlier study in which we demonstrated dependency on the TYK2-STAT1-BCL2 prosurvival signaling pathway in most T-ALL cell lines and more than half of patient-derived T-ALL xenografts,6 suggest that targeting HSP90 would be a beneficial therapeutic strategy for many patients with T-ALL, especially those with high-risk disease.
The HSP90 chaperone machinery is central to the conformational maturation and overall stability of numerous client proteins, including oncogenic factors involved in cancer cell growth and survival, such as HER2, EGFR, ALK and JAK2.11,12,15,16 Our finding that pharmacologic inhibition of HSP90 by AUY922 leads to rapid degradation of TYK2 in T-ALL cell lines (Figure 4) underscores the dependency of this kinase on HSP90 for its stability. Indeed, Taipale et al. have shown that the TYK2 kinase strongly binds to HSP90 and that exogenously expressed TYK2 protein in the HEK293T cell line strikingly decreased within 16 h after treatment with the HSP90 inhibitor ganetespib.7
Importantly, AUY922 treatment efficiently reduced STAT1 phosphorylation and BCL2 expression levels in T-ALL cell lines (Figure 4), while forced expression of BCL2 rescued AUY922-induced apoptosis (Figure 5), consistent with the critical role of BCL2 in T-ALL oncogenesis.6,27 We conclude that the AUY922-induced downregulation of BCL2 is due, at least in part, to a decrease in BCL2 mRNA, as indicated by quantitative RT-PCR analysis (Figure 4d). These results agree with our previous work indicating that shRNA knockdown of the TYK2 or STAT1 gene results in a significant decrease of BCL2 mRNA expression in T-ALL cell lines.6 Exceptions are the “TYK2-independent” LOUCY and TALL-1 cell lines, in which the BCL2 expression levels and the cell viability were unaffected by TYK2 shRNA knockdown, unlike other “TYK2-dependent” T-ALL cell lines.6 Importantly, the LOUCY cell line has a transcriptional signature similar to ETP-ALL cells, which are associated with a poor prognosis,28 and has been shown to be highly sensitive to BCL2 inhibition by ABT-199 (Supplementary Figure 8b).20-22 We think it noteworthy that AUY922 treatment efficiently reduced BCL2 expression in LOUCY cells (Figure 4c) and induced apoptosis without upregulation of BIM and BAD in this cell line (Figures 2, 6b and 6c; Supplementary Figure 3), presumably by affecting another pathway that is independent of TYK2/STAT1 and mediated by proteins that are also HSP90 clients.
We also focused on the BH3-only proteins BIM and BAD as important coactivators of AUY922-induced apoptosis in T-ALL cells. BIM acts as a proapoptotic activator protein, while BIM and BCL2 have each been shown to be important determinants of commitment to apoptosis in lymphocytes.29,30 Furthermore, Reynolds et al. have demonstrated that mimicking BIM function with a stapled peptide mimetic of the BIM BH3 domain has therapeutic activity against human T-ALL cells.31 BAD forms a heterodimer with BCL2 and BCLxL, sequestering them to allow BAX/BAK-triggered apoptosis. Our study indicates that AUY922 treatment increases BIM and BAD expression levels in multiple T-ALL cell lines (Figure 6), accounting for the efficient induction of apoptosis, which was not observed after inhibition of BCL2 alone. These upregulated BH3-only proteins were captured and apoptosis was rescued by exogenously overexpressed BCL2 (Figures 5b and 6a; Supplementary Figures 6a and b), demonstrating that AUY922-induced upregulation of BIM and BAD contributes to T-ALL cell death in the context of decreased BCL2 expression. BIM upregulation induced by treatment with an HSP90 inhibitor has been reported in melanoma cells as well, in which concomitant downregulation of MCL1 also occurs after HSP90 inhibition,32,33 suggesting that induction of proapoptotic BH3-only proteins and concomitant downregulation of prosurvival BCL2 family proteins could be generally involved in the antitumor activity of HSP90 inhibitors.
Interestingly, our results in the JURKAT T-ALL cell line demonstrate that the apoptosis induced by AUY922 can be rescued by overexpression of BCL2, but not BCLxL or MCL1 (Figure 5), even though each of these prosurvival proteins is able to rescue daunomycin-induced decrease of viability to a similar degree (Supplementary Figure 5). We interpret these data to indicate an essential role for BCL2 in maintaining cell survival in the face of AUY922 treatment, an effect not provided even by very high levels of BCLxL or MCL1. However, most T-ALL cell lines are not sensitive to BCL2 inhibition by ABT-199 (Supplementary Figure 8a) 20-21, indicating that loss of BCL2 expression alone is not sufficient to induce apoptosis in these cells, and that AUY922-induced apoptosis requires simultaneous upregulation of BIM and sometimes BAD. It is noteworthy that the basal expression levels of BIM protein in T-ALL cell lines highly sensitive to AUY922, such as DND41, KOPT-K1 and HPB-ALL, are relatively high compared to those in other cell lines (Supplementary Figure 1). Although the BIM protein can be directly inactivated by BCLxL and MCL1, as well as BCL2, and the BAD protein also interacts with BCLxL and BCL2 24, overexpression of BCLxL or MCL1 did not protect T-ALL cells from AUY922-induced apoptosis in our experiments (Figure 5; Supplementary Figure 6). Thus, the loss of BCL2 expression appears necessary but not sufficient by itself for AUY922-induced apoptosis in T-ALL cells.
In a phase I dose-escalation study of AUY922,34 101 patients with advanced solid tumors received 1-h i.v. infusions of AUY922 once a week in a 28-day cycle. At the maximal dose of 70 mg/m2, AUY922 had acceptable tolerability. Importantly, the trough concentrations of AUY922 achieved in patients treated at dose levels of 40 mg/m2 or more were higher than 20 ng/ml (42.9 nM), an efficacious plasma concentration in human cancer xenograft models.17,35 Our in vitro study demonstrates that treatment with 30 nM of AUY922 efficiently decreases TYK2 levels, increases BIM and BAD expression, and ultimately induces significant growth arrest and apoptosis in multiple T-ALL cell lines, indicating that the tolerable AUY922 concentrations achieved in the phase I clinical trial would be adequate for the treatment of T-ALL. Thus, our study identifies HSP90 inhibition as a potential therapeutic strategy for T-ALL and provides a rationale for the evaluation of HSP90 inhibitors in patients with T-ALL.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr. Anthony Letai for helpful discussions and critical review. We also thank John R. Gilbert for critical review of the manuscript and editorial suggestions, and Drs. Julia Etchin and Cherry Ng-Dombrowsky for helpful advice and assistance. This research was supported by fellowships and grants from the Leukemia & Lymphoma Society (CDP5014-14; K.A.), the Lauri Strauss Leukemia Foundation (2013 Discovery research grant; K.A.) and NPO Corporation the Gold Ribbon Network of the Japan (K.A.), Bridge grant from Alex's Lemonade Stand Foundation, and grants from the National Cancer Institute (5R01CA176746 and 5P01CA109901) (A.T.L.). M.R.M. is funded by the Claudia Adams Barr Innovative Basic Science Research Program and the Kay Kendall Leukaemia Trust of the UK. T.S. is supported by a grant from the National Research Foundation, Prime Minister's Office, Singapore under its NRF Fellowship Programme (Award No. NRF-NRFF2013-02). We also thank Novartis for the compound NVP-AUY922.
Footnotes
CONFLICT OF INTEREST
Dr. Radimerski is an employee of Novartis. The other authors declare no conflict of interest.
Supplementary information is available at Leukemia's website.
REFERENCES
- 1.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, et al. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J Clin Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 2.Marks DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, et al. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993). Blood. 2009;114:5136–5145. doi: 10.1182/blood-2009-08-231217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oudot C, Auclerc MF, Levy V, Porcher R, Piguet C, Perel Y, et al. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J Clin Oncol. 2008;26:1496–1503. doi: 10.1200/JCO.2007.12.2820. [DOI] [PubMed] [Google Scholar]
- 4.Schrappe M, Hunger SP, Pui CH, Saha V, Gaynon PS, Baruchel A, et al. Outcomes after induction failure in childhood acute lymphoblastic leukemia. N Engl J Med. 2012;366:1371–1381. doi: 10.1056/NEJMoa1110169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 6.Sanda T, Tyner JW, Gutierrez A, Ngo VN, Glover J, Chang BH, et al. TYK2-STAT1-BCL2 Pathway Dependence in T-Cell Acute Lymphoblastic Leukemia. Cancer Discov. 2013;3:564–577. doi: 10.1158/2159-8290.CD-12-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldas-Lopes E, Cerchietti L, Ahn JH, Clement CC, Robles AI, Rodina A, et al. Hsp90 inhibitor PU-H71, a multimodal inhibitor of malignancy, induces complete responses in triple-negative breast cancer models. Proc Natl Acad Sci U S A. 2009;106:8368–8373. doi: 10.1073/pnas.0903392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wandinger SK, Richter K, Buchner J. The Hsp90 chaperone machinery. J Biol Chem. 2008;283:18473–7. doi: 10.1074/jbc.R800007200. [DOI] [PubMed] [Google Scholar]
- 10.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 11.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 13.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim Biophys Acta. 2012;1823:742–755. doi: 10.1016/j.bbamcr.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marubayashi S, Koppikar P, Taldone T, Abdel-Wahab O, West N, Bhagwat N, et al. HSP90 is a therapeutic target in JAK2-dependent myeloproliferative neoplasms in mice and humans. J Clin Invest. 2010;120:3578–3593. doi: 10.1172/JCI42442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigert O, Lane AA, Bird L, Kopp N, Chapuy B, van Bodegom D, et al. Genetic resistance to JAK2 enzymatic inhibitors is overcome by HSP90 inhibition. J Exp Med. 2012;209:259–273. doi: 10.1084/jem.20111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, et al. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 18.Whitesell L, Bagatell R, Falsey R. The stress response: implications for the clinical development of hsp90 inhibitors. Curr Cancer Drug Targets. 2003;3:349–358. doi: 10.2174/1568009033481787. [DOI] [PubMed] [Google Scholar]
- 19.Mesa RA, Loegering D, Powell HL, Flatten K, Arlander SJ, Dai NT, et al. Heat shock protein 90 inhibition sensitizes acute myelogenous leukemia cells to cytarabine. Blood. 2005;106:318–327. doi: 10.1182/blood-2004-09-3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson NM, Harrold I, Mansour MR, Sanda T, McKeown M, Nagykary N, et al. BCL2-specific inhibitor ABT-199 synergizes strongly with cytarabine against the early immature LOUCY cell line but not more-differentiated T-ALL cell lines. Leukemia. 2014;28:1145–1148. doi: 10.1038/leu.2013.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni Chonghaile T, Roderick JE, Glenfield C, Ryan J, Sallan SE, Silverman LB, et al. Maturation Stage of T-cell Acute Lymphoblastic Leukemia Determines BCL-2 versus BCLXL Dependence and Sensitivity to ABT-199. Cancer Discov. 2014;4:1074–1087. doi: 10.1158/2159-8290.CD-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peirs S, Matthijssens F, Goossens S, Van de Walle I, Ruggero K, de Bock CE, et al. ABT-199 mediated inhibition of BCL-2 as a novel therapeutic strategy in T-cell acute lymphoblastic leukemia. Blood. 2014;124:3738–3747. doi: 10.1182/blood-2014-05-574566. [DOI] [PubMed] [Google Scholar]
- 23.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208:2571–2579. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer cell. 2006;9:351–365. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 25.Chao DT, Linette GP, Boise LH, White LS, Thompson CB, Korsmeyer SJ. Bcl-XL and Bcl-2 repress a common pathway of cell death. J Exp Med. 1995;182:821–828. doi: 10.1084/jem.182.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 27.Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. 2014;46:364–370. doi: 10.1038/ng.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, et al. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 30.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds C, Roderick JE, Labelle JL, Bird G, Mathieu R, Bodaar K, et al. Repression of BIM mediates survival signaling by MYC and AKT in high-risk T-cell acute lymphoblastic leukemia. Leukemia. 2014;28:1819–1827. doi: 10.1038/leu.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paraiso KH, Haarberg HE, Wood E, Rebecca VW, Chen YA, Xiang Y, et al. The HSP90 inhibitor XL888 overcomes BRAF inhibitor resistance mediated through diverse mechanisms. Clin Cancer Res. 2012;18:2502–2514. doi: 10.1158/1078-0432.CCR-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haarberg HE, Paraiso KH, Wood E, Rebecca VW, Sondak VK, Koomen JM, et al. Inhibition of Wee1, AKT, and CDK4 underlies the efficacy of the HSP90 inhibitor XL888 in an in vivo model of NRAS-mutant melanoma. Mol Cancer Ther. 2013;12:901–912. doi: 10.1158/1535-7163.MCT-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sessa C, Shapiro GI, Bhalla KN, Britten C, Jacks KS, Mita M, et al. First-in-human phase I dose-escalation study of the HSP90 inhibitor AUY922 in patients with advanced solid tumors. Clin Cancer Res. 2013;19:3671–3680. doi: 10.1158/1078-0432.CCR-12-3404. [DOI] [PubMed] [Google Scholar]
- 35.Jensen MR, Schoepfer J, Radimerski T, Massey A, Guy CT, Brueggen J, et al. NVPAUY922: a small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 2008;10:R33. doi: 10.1186/bcr1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 40.Shepherd C, Banerjee L, Cheung CW, Mansour MR, Jenkinson S, Gale RE, et al. PI3K/mTOR inhibition upregulates NOTCH-MYC signalling leading to an impaired cytotoxic response. Leukemia. 2013;27:650–660. doi: 10.1038/leu.2012.285. [DOI] [PubMed] [Google Scholar]
- 41.Mansour MR, Sanda T, Lawton LN, Li X, Kreslavsky T, Novina CD, et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J Exp Med. 2013;210:1545–1557. doi: 10.1084/jem.20122516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.