Summary
The origin recognition complex (ORC) proteins, ORC1-6, are the first proteins that bind DNA replication origins to mark the competency for the initiation of DNA synthesis. These proteins have complex mechanisms of assembly into the ORC complex, and unexpected localizations in the mitotic chromosomes, cytoplasm, and nuclear structures. The mammalian zygote is a potentially important model that may contribute to our understanding of the mechanisms and features influencing origin establishment, and in identifying other functions of the ORC proteins. Along with expected localizations to the chromatin during G1, we found unexpected distribution in the cytoplasm that appeared to accumulate ORC proteins, which suggest potential roles for ORC subunits in mitosis and chromatin segregation. ORC1, 2, 3 and 5 all localize to the area between the separating maternal chromosomes shortly after fertilization. ORC4 forms a cage around the set of chromosomes that will be extruded during polar body formation before it binds to the chromatin shortly before zygotic DNA replication. These data suggest that the ORC proteins may also play roles in preparing the cell for DNA replication in addition to their direct role in establishing functional replication origins.
Introduction
The replication of the mammalian zygote's DNA is one of the most important events in reproduction because that single copy of the combined parental genome will be replicated an estimated 5 trillion times during the lifetime of the individual (DePamphilis et al. 2006). This initial replication event has two important unique features. The first is that each parental genome is replicated independently in different pronuclei before the maternal and paternal chromosomes fuse into a single nucleus just before zygotic mitosis (Sirlin and Edwards 1959). This occurs more or less synchronously (Aoki and Schultz 1999, Howlett and Bolton 1985, Yamauchi et al. 2009), but it is possible to experimentally disrupt this synchrony and still obtain viable embryos (Kishigami et al. 2004, Yamauchi et al. 2009). The second unique feature of zygotic DNA replication is that each parental genome is prepared for replication through very different states of chromatin configuration. After fertilization, maternal DNA exits from metaphase II (M-II), while paternal DNA undergoes a more dramatic transformation by exiting G0 and exchanging its protamines for histones (Balhorn et al. 1984, Marushige and Marushige 1975, McLay and Clarke 2003, Yanagimachi 1994). This asymmetric development suggests the zygotic cytoplasm is permissive to the licensing of unique cell-cycle chromatin states. These two unique features of zygotic DNA replication require special scrutiny when applying what is known about mammalian DNA synthesis to the zygote.
We recently embarked on an investigation of one aspect of this process during early development, the licensing of DNA origins by origin recognition complex (ORC) proteins. These studies identified several unique features that may point to new functions, and thereby to potentially new control mechanisms for DNA replication. Here we summarize these findings and speculate on future directions for studying the roles of ORC proteins in both fertilization and in somatic cell replication.
DNA Replication Licensing
Eukaryotic origins of replication are established by a conserved ensemble of pre-replication complex (pre-RC) proteins. The process of pre-RC formation is initiated by the origin recognition complex binding to potential origins (Coster et al. 2014, Ticau et al. 2015). Nearly 103-105 of these replication origins assemble during the M/G1 transition (Kara et al. 2015). The assembly of ORC subunits which initiates pre-RC formation is a highly orchestrated event that can vary between different cell types. In general, three of the subunits, ORC2, ORC3 and ORC5, form a complex in the cytoplasm, which are then transported into the nucleus where they combine with ORC4 and ORC6 (Ghosh et al. 2011). This origin recognition complex may bind to the DNA replication origin either before (Takeda et al. 2005) or possibly after (Ghosh et al. 2011) ORC1 binds to the complex. ORC1 is transiently expressed, and is later digested via the ubiquitin proteasome pathway during S-phase to prevent additional licensing of origins (Li and DePamphilis 2002, Tatsumi et al. 2003). The formation of the functional ORC on the replication origin initiates the binding CDC6 and CDT1, and concludes with the loading of two heterohexameric minichromosome maintenance (MCM2-7) helicase complexes (Coster et al. 2014, Ticau et al. 2015). This fully formed pre-RC at the origin represents a licensed origin, marking it as competent for replication. The genome contains many more potential origins than are needed for each round of replication. Different cell types use different combinations of origins to ensure complete replication so that not all origins are activated during any one given replication event. For each round of replication only those origins that are licensed are replicated. Once S-phase begins, no new licensing is permitted by the cell (Takeda et al. 2005). This prevents the cell from re-replicating segments of the DNA by initiating replication at points along the chromosome that have already been copied. In this way, licensing provides the cell with an accounting system to ensure that the entire genome is replicated but that none of it is replicated more than once (Blow and Laskey 1986).
Despite progressive advancement in the field of replication, the mechanisms that demarcate origins in metazoans remain unclear. Mammalian ORC proteins bind to all DNA without sequence specificity (Takeda and Dutta 2005), so the origin complex, itself, does not identify which DNA sequences will be used as origins. In eukarya, origins have been postulated to include CpG islands (Cadoret et al. 2008), A-T rich sites, and more recently histone post-translational modifications to name a few features that might be involved in recruiting pre-RC formation (reviewed by (Rivera et al. 2014). The tissue-specific spatiotemporal initiation of DNA replication at the licensed origins in some cases has bolstered the possibility of understanding origin specificity in terms of cell-cycle inheritance and gene expression. However, the molecular rules that govern why different origins are used for DNA replication in different cell types, and what sequences can be origins are not understood (Cleary et al. 2010, Pope et al. 2014).
The mammalian zygote is a potentially important model that may contribute to our understanding of the mechanisms and features influencing origin establishment. The meiotic maternal chromosomes appear to be already partially licensed (see below (Ortega et al. 2012)), while the G0 paternal chromosomes are almost certainly licensed de novo, or at least they can be experimentally manipulated to be so. We demonstrated that the mouse spermatazoon can have most major components removed with a high-salt and DTT wash, which leaves only the most basic scaffolding and DNA-loop structures, termed the sperm halo. When the sperm halo is microinjected injected into an embryo it can form a pronucleus (PN) that is capable of replicating (Mohar et al. 2002, Shaman et al. 2007, Yamauchi et al. 2009). ORC proteins, if present, would not remain bound to DNA under these conditions, indicating that the zygote is capable of supporting de novo licensing. Because sperm halos can reorganize into replication competent pronuclei, it appears that ORC2 can be recruited de novo by paternal DNA (Ortega et al. 2012) and that sperm halos can be reorganized into replication competent pronuclei. When taken together, these features of sperm halos suggest ORC proteins may recognize critical salt resistant structures in sperm when establishing origins. The process of pronuclear formation by sperm haloes may provide an experimental system in which to study ORC functions in more detail.
ORC behavior in the mammalian zygote
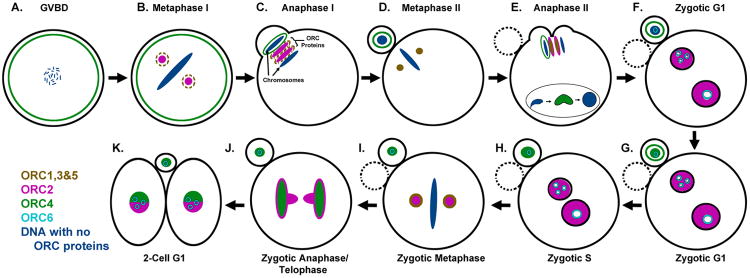
Previous research has examined the roles of ORC proteins in C. elegans and Xenopus oocytes or egg extracts, and these studies have helped establish clear roles for these licensing factors in origin establishment (Carpenter et al. 1996, Rowles et al. 1996, Sonneville et al. 2012). We have recently completed an immunocytochemical (ICC) survey of all six ORC proteins in the mouse zygote, and the results are diagrammed in Fig. 1. Along with expected localizations to the chromatin during G1, we found unexpected areas in the cytoplasm that appeared to accumulate ORC proteins that suggest potential roles for ORC subunits in mitosis and chromatin segregation. During mouse M-II, ORC2 is located at the spindle poles of the oocyte (Ortega et al. 2012) in a similar pattern to γ-tubulin (Meng et al. 2004) (Fig. 1D). This spindle-like formation and behavior observed by ORC2 is mirrored by ORC1, ORC3, and ORC5 (Nguyen et al. 2015). ORC2 also localizes at the spindles in metaphase I (M-I) during oocyte meiosis, but it is not yet known whether ORC1, ORC3 and ORC5 are similarly present at the M-I spindles (Fig. 1B). Shortly after the sperm we injected into the egg, the M-II chromosomes begin to separate, and ORC1-3, and ORC5, can be found between the separating chromosomes in two distinct patches (Fig. 1E). Simultaneously, ORC4 asymmetrically surrounds the extruding chromosomes that will become the second polar body. Once again, this pattern was repeated by ORC4 during the first meiotic division during anaphase I (Fig. 1C). The localization of ORC2 to the spindles and to the space between the separating chromosomes in both meiotic divisions without an intervening DNA replication step has implications for possible functions outside of origin licensing (discussed below).
Figure 1. Diagram of ORC Protein Localization during Oocyte Maturation, Fertilization and the First Zygotic Cell Cycle.
All six of the ORC proteins have been localized during these stages of development. Dashed lines in B and C for ORCs 1, 3 & 5 represent suspected, but not proven localizations. See text for details.
After fertilization, ORC2 is not initially detectable on sperm (Ortega et al. 2012). In the mouse, as the sperm decondenses it donates centrosome proteins (Goto et al. 2010) and RNA (Krawetz et al. 2011) to the zygote during protamine-histone exchange. Concurrently, ORC4 can be transiently visualized during decondensation prior to pronuclear formation (Figure 1 B) (Nguyen et al. 2015). However, after pronuclear formation only ORC2 and ORC6 are detectable by ICC with ORC2 binding throughout the PN while ORC6 localizes around the nucleoli. ORC2 becomes salt-resistant in both pronuclei and thus tightly bound to the DNA during G1. This is similar to chromatin affinity assays performed in somatic G1 cells. As the embryo then progresses to G1, the decreased CDK activity at this stage in development permits dephosphorylation of ORC2 (Lee et al. 2014), and facilitates chromatin binding during early pronuclear formation. At the onset of S-phase, cyclin-A activity promotes phosphorylation of ORC2 (Lee et al. 2012) which reduces some of the ORC2 in the pronuclei but much of it remains bound to the DNA (Figure 1H) (Lee et al. 2012). As the pronuclei synapse and condense to enter zygotic metaphase, ORC1-3, and ORC5 appear to be evicted to the cytoplasm (Hemerly et al. 2009, Ortega et al. 2012, Prasanth et al. 2004). While some ORC1 is targeted for ubiquitation in somatic studies (Li and DePamphilis 2002), recent studies have also demonstrated ORC1 may maintain a presence on the metaphase chromatin that could act to recall the other ORC proteins for pre-RC formation (Kara et al. 2015). Thus, the remaining ORC proteins that resist eviction are associated with zygotic metaphase chromosomes and may facilitate the reestablishment of topologically associated origin domains.
The displacement patterning of the ORC subunits during cellular division may be relevant with respect to the inheritance of origin domains. In zygotic telophase, ORC2 surrounds the separating chromosomes as a pocket with spindle-like streamers directing toward the cleavage furrow, while ORC4 more directly associates with the anaphase chromosomes (Fig. 1J). All ORC proteins can thereafter be detected in the nuclei of the two-cell embryo. Taken together, these observations suggest that ORC proteins asymmetrically load onto the parental pronuclei of the zygote as each is conditioned for replication during pronuclear formation. However, as all of these conclusions are based on ICC, it is important to keep in mind that low levels of ORC proteins below the detectable limits of ICC may remain associated with the DNA.
ORC2 in Zygotic DNA Synthesis
In addition to the unique ORC behavior in zygotes, MCM7 also demonstrates unpredicted localization behavior. MCM7 is one of the minichromosome maintenance proteins (Coster et al. 2014, Ticau et al. 2015), which are the set of helicases that are the last component of the licensing complex to be loaded onto the origins. MCM7 is present on the M-II chromatin which implies partial licensing (Ortega et al. 2012). It is possible that transient ORC association in the maturing oocyte prior to M-II might explain the observed helicase deposit (Nguyen et al. 2015). If true, ORC might recruit MCM to the origins in late meiosis-I, then leave the chromatin before M-II. The presence of MCM7 without clear ORC association on this meiotic metaphase DNA is particularly interesting because it has also been shown that MCM complexes are capable of recruiting a second complex without further aid from ORC (Ticau et al. 2015). Thus, further licensing of particular sites in the maternal pronucleus (MPN) may be completed in an ORC independent manner, while other sites may go on to recruit ORC through the normal pathway in G1. Whatever the early state of ORC deposition is, licensing seems to be incomplete, as there is a clear build-up and association of ORC2 in G1 after the PN are formed that correlates with an increase in salt-resistant MCM signal. This suggests that licensing in the female pronucleus may not be completed until late in zygotic G1.
In both the MPN and PPN, MCM7 is tightly bound to the DNA in G1 and becomes soluble during S-phase (Figure 1H) (Ortega et al. 2012). Taken together, these observations suggest zygotic licensing is a dynamic event in which ORC and MCM demonstrate some previously unexpected behaviors in the zygote. The asymmetric licensing of the maternal and paternal DNA implies a model that permits some licensing to occur early while other origins are established later. This may support similar behavior observed in C. elegans where ORC2 oscillates in and out of the nucleus to load multiple MCMs rather than remaining statically bound to a single origin (Sonneville et al. 2012). Our data suggest that while some licensing occurs early zygotic development, it is completed in late G1. The two phases of zygotic DNA licensing may be related to the changes in DNA topology, particularly in the paternal genome, that occur just after sperm injection.
Implications of ORC subunit localizations outside of the chromatin
The dynamic observations of ORC behavior in the zygote support a long argued hypothesis that ORC proteins have non-replicative functions in the cell-cycle (Chesnokov et al. 2003, Huang et al. 2005, Prasanth et al. 2002, Scholefield et al. 2011). For example, ORC2 has been observed at centrosomes in HeLa cells and C. cerevisae, These findings, along with the increased occurrence of metaphase arrest in ORC2 knockdown models suggest a role for ORC2 in cellular division (Machida et al. 2005, Prasanth et al. 2004). Furthermore, while not observed in the zygote, ORC1 has also been shown to localize to the centrosomes in other systems and the perturbation of ORC1 is enough to inhibit centrosome reduplication and effects spindle assemble checkpoints (Gibson et al. 2006, Hemerly et al. 2009). Here, it should be noted that unlike somatic cells, oocytes do not have centrosomes. Instead, anaphase II (A-II) segregation is directed by self-organizing microtubule organizing centers (MTOCs) (Schuh and Ellenberg 2007). The imminent question then is how and why does ORC localize to MTOCs with or without the presence of centrosomes? One could speculate the cytoplasmic presence of ORC1-3 and ORC5 proteins might play a role in spindle formation, which may explain the described effects of ORC2 knockdown. Another potential explanation for the behavior observed in the mammalian zygote is that ORC spindle midzone association may act as a depository for the reestablishment of origins during subsequent PN formation. Yet, ultimately this aspect of ORC2 localization has no clear explanation (Fig. 1J).
Some of the strongest evidence for non-replicative ORC functions can be observed in ORC6 in D. melanogaster. ORC6 is the least conserved ORC subunit and it demonstrates quite unique behavior in the fly. In D. melanogaster studies, ORC6 associates with Pnut in forming a septin-collar in dividing embryos and it has been argued that ORC6 may therefore be directly involved in cytokinesis (Chesnokov et al. 2003). In HeLa cells, ORC6 localizes to the area between the separating chromosomes (Prasanth et al. 2002), just as ORC1-3 and ORC5 were found to do during A-II of the mouse zygote (Nguyen et al. 2015). Disruption of ORC6 in human cells results in cytokinetic defects further supporting the argument for a role of ORC6 in this process (Prasanth et al. 2002).
Though we are not yet able to define the non-licensing functions for ORC, the behavior of its subunits argues for the existence of one or more. ORC2, and likely ORC1, ORC3, and ORC5, localizes to both the spindle poles and then the space between the separating chromosomes in M-I and A-I respectively, and then re-organize to these same places during the second division of meiosis without an intervening S-phase. Thus, the presence of ORC2 at these two sites is probably not related to DNA synthesis initiation. One possibility is that sequestering the ORC proteins away from the DNA prevents the inappropriate initiation of DNA synthesis during M-I but there are likely to be other functions as yet undiscovered.
ORC4 in polar body formation
The most surprising finding in zygotic ORC behavior is that observed by ORC4. ORC4 forms an ovoid structure surrounding the extruding A-II chromosomes during both meiotic divisions (Fig. 1C and E) (Nguyen et al. 2015). During zygotic anaphase, ORC4 behaves in a way that is more consistent with its role as a DNA replication licensing protein by more clearly associating with both chromosome sets (Fig. 1J). These observations indicate a possible role for ORC4 in the unique, asymmetric cytokinesis that occurs in both female meiotic divisions. As noted above, the ORC proteins have already been shown to be involved in some aspects of cellular division, but ORC4's role in the oocyte's two asymmetric divisions appears to be unique.
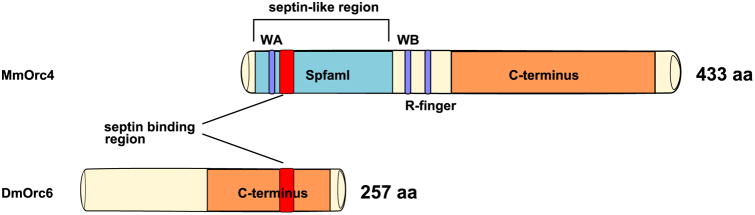
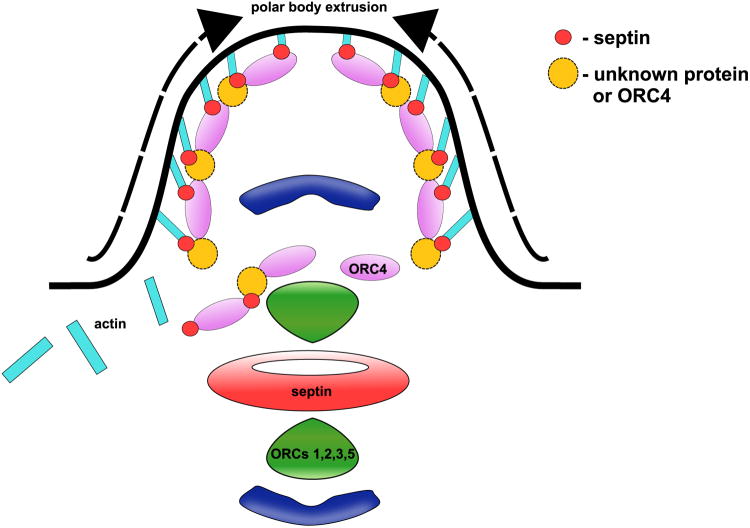
How might ORC4 interact with the cytoskeleton to assist in this? Two interesting studies demonstrated that ORC6 interacts directly with Pnut, an actin organizing protein of the septin family, during cytokinesis in Drosophila embryonic cells (Chesnokov et al. 2003, Huijbregts et al. 2009). During meiosis in mammals, septins bend and organize F-actin into a cap for polar body extrusion (PBE) (Mavrakis et al. 2014). A blast for known motifs (GenomeNet) in the mouse ORC4 reveals a septin-like region in the N-terminal, but not in mouse ORC6. This septin-like region contains a region with close homology to the septin binding region identified Drosophila ORC6 that was shown to bind to Pnut (Fig. 2). One possibility, therefore, is that ORC4 interacts with the actin cap through septin family proteins to encompass one copy of the meiotic chromosome set during each of the PBE events in both meiotic divisions. A second possibility is that ORC4 interacts directly through its septin-like domain with actin.
Figure 2. Comparison of Mouse ORC4 and Drosophila ORC6.
A septin-binding region was identified in DmORC6, and a similar region was found in MmORC4. MmORC4 also contained a much larger septin-like region.
Conclusions
It is clear that in the mouse zygote, ORC proteins are involved in DNA licensing as in other cell types. However, they also appear to have functions in the cytoplasm that may be related to controlling chromatin segregation and cellular division. Furthermore, at least one of the subunits, ORC4, may be involved in the unique, asymmetric divisions the oocyte undergoes during meiosis.
Figure 3. A Model for the Role of MmORC4 in Polar Body Extrusion.
Based on previous work on DmORC6 and our analysis of the MmORC4 sequence (Fig. 2) we suggest that MmORC4 interacts with the actin cap in the polar body through its septin-like domain.
Acknowledgments
This work was supported by NIH Grant HD060722 to W.S.W.
References
- Aoki E, Schultz RM. DNA replication in the 1-cell mouse embryo: stimulatory effect of histone acetylation. Zygote. 1999;7:165–172. doi: 10.1017/s0967199499000532. [DOI] [PubMed] [Google Scholar]
- Balhorn R, Weston S, Thomas C, Wyrobek AJ. DNA packaging in mouse spermatids. Synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150:298–308. doi: 10.1016/0014-4827(84)90572-x. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci U S A. 2008;105:15837–15842. doi: 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chesnokov IN, Chesnokova ON, Botchan M. A cytokinetic function of Drosophila ORC6 protein resides in a domain distinct from its replication activity. Proc Natl Acad Sci U S A. 2003;100:9150–9155. doi: 10.1073/pnas.1633580100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Tome S, Lopez Castel A, Panigrahi GB, Foiry L, Hagerman KA, Sroka H, Chitayat D, Gourdon G, Pearson CE. Tissue- and age-specific DNA replication patterns at the CTG/CAG-expanded human myotonic dystrophy type 1 locus. Nat Struct Mol Biol. 2010;17:1079–1087. doi: 10.1038/nsmb.1876. [DOI] [PubMed] [Google Scholar]
- Coster G, Frigola J, Beuron F, Morris EP, Diffley JF. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Vassilev AP, Zhang J, Zhao Y, DePamphilis ML. Assembly of the human origin recognition complex occurs through independent nuclear localization of its components. J Biol Chem. 2011;286:23831–23841. doi: 10.1074/jbc.M110.215988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Bell SP, Aparicio OM. Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae. Genes Cells. 2006;11:557–573. doi: 10.1111/j.1365-2443.2006.00967.x. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, O'Brien DA, Eddy EM. Speriolin is a novel human and mouse sperm centrosome protein. Hum Reprod. 2010;25:1884–1894. doi: 10.1093/humrep/deq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett SK, Bolton VN. Sequence and regulation of morphological and molecular events during the first cell cycle of mouse embryogenesis. J Embryol Exp Morphol. 1985;87:175–206. [PubMed] [Google Scholar]
- Huang Z, Zang K, Reichardt LF. The origin recognition core complex regulates dendrite and spine development in postmitotic neurons. J Cell Biol. 2005;170:527–535. doi: 10.1083/jcb.200505075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijbregts RP, Svitin A, Stinnett MW, Renfrow MB, Chesnokov I. Drosophila Orc6 facilitates GTPase activity and filament formation of the septin complex. Mol Biol Cell. 2009;20:270–281. doi: 10.1091/mbc.E08-07-0754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara N, Hossain M, Prasanth SG, Stillman B. Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells. J Biol Chem. 2015;290:12355–12369. doi: 10.1074/jbc.M114.625012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S, Wakayama S, Nguyen VT, Wakayama T. Similar time restriction for intracytoplasmic sperm injection and round spermatid injection into activated oocytes for efficient offspring production. Biol Reprod. 2004;70:1863–1869. doi: 10.1095/biolreprod.103.025171. [DOI] [PubMed] [Google Scholar]
- Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP. A survey of small RNAs in human sperm. Hum Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Bang SW, Yoon SW, Lee SH, Yoon JB, Hwang DS. Phosphorylation of ORC2 protein dissociates origin recognition complex from chromatin and replication origins. J Biol Chem. 2012;287:11891–11898. doi: 10.1074/jbc.M111.338467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Bae JS, Yoon S, Hwang DS. Dephosphorylation of Orc2 by protein phosphatase 1 promotes the binding of the origin recognition complex to chromatin. Biochem Biophys Res Commun. 2014;448:385–389. doi: 10.1016/j.bbrc.2014.04.109. [DOI] [PubMed] [Google Scholar]
- Li CJ, DePamphilis ML. Mammalian Orc1 protein is selectively released from chromatin and ubiquitinated during the S-to-M transition in the cell division cycle. Mol Cell Biol. 2002;22:105–116. doi: 10.1128/MCB.22.1.105-116.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Teer JK, Dutta A. Acute reduction of an origin recognition complex (ORC) subunit in human cells reveals a requirement of ORC for Cdk2 activation. J Biol Chem. 2005;280:27624–27630. doi: 10.1074/jbc.M502615200. [DOI] [PubMed] [Google Scholar]
- Marushige Y, Marushige K. Enzymatic unpacking of bull sperm chromatin. Biochim Biophys Acta. 1975;403:180–191. doi: 10.1016/0005-2744(75)90020-0. [DOI] [PubMed] [Google Scholar]
- Mavrakis M, Azou-Gros Y, Tsai FC, Alvarado J, Bertin A, Iv F, Kress A, Brasselet S, Koenderink GH, Lecuit T. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat Cell Biol. 2014;16:322–334. doi: 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XQ, Fan HY, Zhong ZS, Zhang G, Li YL, Chen DY, Sun QY. Localization of gamma-tubulin in mouse eggs during meiotic maturation, fertilization, and early embryonic development. J Reprod Dev. 2004;50:97–105. doi: 10.1262/jrd.50.97. [DOI] [PubMed] [Google Scholar]
- Mohar I, Szczygiel MA, Yanagimachi R, Ward WS. Sperm Nuclear Halos Can Transform Into Normal Chromosomes After Injection Into Oocytes. Mol Reprod Dev. 2002;62:416–420. doi: 10.1002/mrd.10147. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Ortega MA, Ko M, Marh J, Ward WS. ORC4 Surrounds Extruded Chromatin in Female Meiosis. J Cell Biochem. 2015;116:778–786. doi: 10.1002/jcb.25033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega MA, Marh J, Alarcon VB, Ward WS. Unique Pattern of ORC2 and MCM7 Localization During DNA Replication Licensing in the Mouse Zygote. Biol Reprod. 2012;8762:61–69. doi: 10.1095/biolreprod.112.101774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, Thurman RE, Cheng Y, Gulsoy G, Dennis JH, Snyder MP, Stamatoyannopoulos JA, Taylor J, Hardison RC, Kahveci T, Ren B, Gilbert DM. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297:1026–1031. doi: 10.1126/science.1072802. [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 2004;23:2651–2663. doi: 10.1038/sj.emboj.7600255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera C, Gurard-Levin ZA, Almouzni G, Loyola A. Histone lysine methylation and chromatin replication. Biochim Biophys Acta. 2014;1839:1433–1439. doi: 10.1016/j.bbagrm.2014.03.009. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Scholefield G, Veening JW, Murray H. DnaA and ORC: more than DNA replication initiators. Trends Cell Biol. 2011;21:188–194. doi: 10.1016/j.tcb.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Shaman JA, Yamauchi Y, Ward WS. The Sperm Nuclear Matrix is Required for Paternal DNA Replication. J Cell Biochem. 2007;102:680–688. doi: 10.1002/jcb.21321. [DOI] [PubMed] [Google Scholar]
- Sirlin JL, Edwards RG. Timing of DNA synthesis in ovarian oocyte nuclei and pronuclei of the mouse. Exp Cell Res. 1959;18:190–194. doi: 10.1016/0014-4827(59)90308-8. [DOI] [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Dutta A. DNA replication and progression through S phase. Oncogene. 2005;24:2827–2843. doi: 10.1038/sj.onc.1208616. [DOI] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi Y, Ohta S, Kimura H, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem. 2003;278:41528–41534. doi: 10.1074/jbc.M307534200. [DOI] [PubMed] [Google Scholar]
- Ticau S, Friedman LJ, Ivica NA, Gelles J, Bell SP. Single-Molecule Studies of Origin Licensing Reveal Mechanisms Ensuring Bidirectional Helicase Loading. Cell. 2015;161:513–525. doi: 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Ward MA, Ward WS. Asynchronous DNA Replication and Origin Licensing in the Mouse One Cell Embryo. J Cell Biochem. 2009;107:214–223. doi: 10.1002/jcb.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagimachi R. Fertility of mammalian spermatozoa: its development and relativity. Zygote. 1994;2:371–372. doi: 10.1017/s0967199400002240. [DOI] [PubMed] [Google Scholar]