Abstract
Computed tomography (CT) imaging is an essential tool in various clinical diagnoses and radiotherapy treatment planning. Since CT image intensities are directly related to positron emission tomography (PET) attenuation coefficients, they are indispensable for attenuation correction (AC) of the PET images. However, due to the relatively high dose of radiation exposure in CT scan, it is advised to limit the acquisition of CT images. In addition, in the new PET and magnetic resonance (MR) imaging scanner, only MR images are available, which are unfortunately not directly applicable to AC. These issues greatly motivate the development of methods for reliable estimate of CT image from its corresponding MR image of the same subject. In this paper, we propose a learning-based method to tackle this challenging problem. Specifically, we first partition a given MR image into a set of patches. Then, for each patch, we use the structured random forest to directly predict a CT patch as a structured output, where a new ensemble model is also used to ensure the robust prediction. Image features are innovatively crafted to achieve multi-level sensitivity, with spatial information integrated through only rigid-body alignment to help avoiding the error-prone inter-subject deformable registration. Moreover, we use an auto-context model to iteratively refine the prediction. Finally, we combine all of the predicted CT patches to obtain the final prediction for the given MR image. We demonstrate the efficacy of our method on two datasets: human brain and prostate images. Experimental results show that our method can accurately predict CT images in various scenarios, even for the images undergoing large shape variation, and also outperforms two state-of-the-art methods.
Index Terms: CT Prediction, PET Attenuation Correction, Dose Planning, Random Forest, Auto-context
I. Introduction
Computed tomography (CT) imaging is widely used in various medical practices, e.g., for detecting infarction, tumors, calcifications, etc. Besides, CT images are also indispensable for attenuation correction (AC) of positron emission tomography (PET) images [1] in the PET/CT system. AC is a process of deriving attenuation map from the CT image to correct the corresponding PET image, based on the radiation-attenuation properties of tissues revealed by the attenuation map.
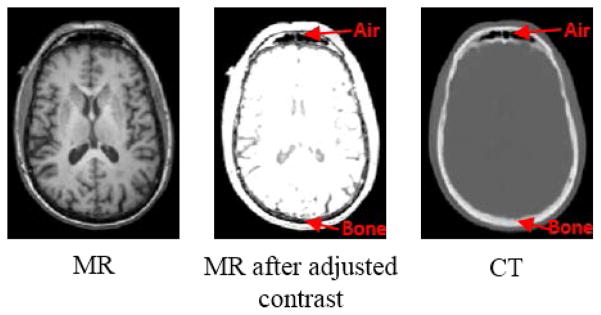
However, the use of CT scan is advised to be limited, considering the risk of radiation exposure. For example, it has been shown that as many as 0.4% of cancers in the US are due to CT scanning performed in the past, and this number may increase to as high as 1.5 to 2% in the future [2]. Besides, as an emerging imaging tool, the recent PET and magnetic resonance imaging (MRI) system replaces CT by MRI. However, it is intrinsically hard to predict attenuation coefficients from MR images, since the MRI signals of individual voxels are related to proton density, not the electron density information that is required for AC. For example, using most standard MRI sequences, both air and compact bone generate very low signals, whereas their attenuation coefficients are highly different, as shown in Fig. 1.
Fig. 1.
A pair of corresponding MR and CT images from the same human brain. Both air and bone have very low responses in MR image, but they are highly differentiable in CT image.
Hence, there is a crucial need for predicting a CT image from an MR image. The existing works can be roughly classified into the following four categories:
Tissue segmentation based methods. The basic idea is to first segment an MR image into different tissue classes, and then assign each class with a known attenuation property. However, this type of methods may fail due to the existence of some ambiguous tissue classes, such as air and bone. In particular, Zaidi et al. [3] proposed a fuzzy logic based method to segment MRI into five tissue classes, and manually fixed failures of automatic segmentation. Hsu et al. [4] performed segmentation using multiple MRI modalities, as well as considering the fat and water image volumes with Dixon MRI sequence. They found that a single MRI volume is insufficient to separate all tissue classes. Similarly, Berker et al [5] combined UTE/Dixon MRI sequence to segment tissues for AC.
Atlas-based methods. These methods estimate the attenuation map of a given subject by warping the attenuation map of an atlas to this subject [6, 7], using the deformation field estimated by registration of the MR images between the atlas and the subject. However, the performance of these methods is highly dependent on the registration accuracy.
Learning-based methods. The MR-CT relationship can be learned from a training set and then applied to a target MR image for CT image prediction. Since it is not easy to learn such relationship from a single modality, Johansson et al. [8] proposed to build a Gaussian mixture regression model for learning from multiple modalities, i.e., two UTE images and one T2-weighted MR image. The idea of using Gaussian mixture model was also adopted by Roy et al. [9] to synthesize CT image from two UTE MR images. The same technique was also applied to estimate CT image from one MR image [10]. However, the spatial information was disregarded and then the quality of estimated CT image was modest, thus often used as the intermediate means to facilitate the subsequent registration task.
Integration of atlas-based and pattern recognition methods. After warping atlases to the target image, a local regression model (characterized by both spatial locations and image patch intensity) can be built and applied to the target image. For example, Hofmann et al. [7] used a Gaussian process regression model. Although this type of techniques can produce promising results, their performance still highly depends on the accuracy of the deformable registration between the atlas and target MR images.
Another closely related research field is image synthesis, which has a similar goal of synthesizing one image modality from other modalities, although with different applications. Most of such studies fall under the following two categories:
Learning-based methods. Jog et al. used random forest to reconstruct the high-resolution T2-weighted MR image from both low-resolution T2-weighted and high-resolution T1-weighted MR images [11]. A similar technique was also used to estimate Fluid Attenuated Inversion Recovery (FLAIR) sequence from T1, T2, and PD-weighted MR sequences [12]. The random forests used in these works are rather general and simple, which neither used spatial information nor had specific enhancements for CT image prediction. In [13], Li et al. used the convolutional neural networks to estimate PET image from MR image. However, neural network approach suffers from long training time, ranging from days to weeks, and its performance highly depends on the successful tuning of many parameters.
-
Examplar-based methods. A dominant line of research in this category is the sparse representation based methods. First, the target image patch is sparsely represented by a set of atlas patches of the same modality. Then, the resulting sparse coefficients are used to integrate the corresponding atlas patches of another modality to estimate the desired image patch in that modality for the target subject. Roy et al. used this technique to solve multiple problems, i.e., predicting magnetization prepared rapid gradient echo sequence from spoiled gradient recalled sequence and vice versa [14, 15], as well as predicting FLAIR image from T1- and T2-weighted MRI [16]. Ye et al. [17] estimated T2- and diffusion-weighted images from T1-weighted MRI, while Iglesias et al. [18] predicted T1-weighted MRI from PD-weighted MRI. But one main drawback of these methods is that the prediction is often very computationally expensive due to huge optimization needed in the testing phase.
In this paper, we will employ an enhanced random forest to specifically tackle the problem of estimating CT image from MRI data, with the following contributions:
Spatial information is effectively incorporated to enhance the CT prediction quality through only rigid registration of atlas and subject, instead of deformable registration.
Image features are innovatively crafted with multi-level sensitivity to balance the need for achieving some degrees of invariance to image deformation, yet still being sensitive to small structural changes. Furthermore, features are extracted at multiple resolutions to incorporate information from wider neighborhood and also combine both global and local information, thus making the algorithm more robust to different image resolutions as well.
Structured random forest is used to predict a CT image patch as a structured output, thus preserving the neighboring structures in the predicted CT image. This cannot be achieved by the traditional methods, which often predict individual CT values independently.
A novel ensemble model is proposed to combine the results from multiple decision trees in the random forest for improving both robustness and accuracy of CT prediction.
An auto-context model is applied to incorporate the context information in the predicted CT image for iterative refinement.
Below, we first describe our method in Section II, and then evaluate it extensively in Section III. We finally give discussions and conclusions in Sections IV and V, respectively.
II. Method
A. System Overview
Suppose we have a set of pairs of MR and CT training images. For each pair, the CT image is used as the regression target of the MR image. We further assume that the training data have been preprocessed by removing noise and uninformative regions (e.g., device tools), and have been aligned (see below for details). In the training stage, we first extract multi-scale features from each MR image and use them together with the corresponding CT image to train an initial structured random forest. We then use the resulting forest to predict the CT image for each MR image in the training set, leading to an initial set of predictions. Together with the features from MR images, we can further extract context features from the predicted CT images to train a new structured random forest and perform prediction again. By repeating this process until convergence, we can finally obtain a sequence of trained forests. In the testing stage, we extract features from the new (target) MR image and feed them into the trained forests for CT image prediction.
B. Intra-subject and Inter-subject Alignment
The intra-subject registration is to align each pair of CT and MR images of the same subject. For the brain images, linear registration is sufficient due to small deformation, and thus can be achieved by FLIRT [19] with 12 degrees of freedom. As there is often a large deformation on soft tissues for the prostate images, we perform intra-subject deformable registration with B-Splines (Elastix [20]), using mutual information as a similarity measure. Finally, we perform rigid-body inter-subject registration to roughly bring all subjects onto a common space.
C. Multi-scale Feature Extraction
1) Feature Types
We use the following four types of features to effectively characterize each MR image patch:
Spatial coordinates (x, y, z): They are the coordinates of the center of an image patch in the common space, and are used as features input to the structured random forest, to provide the spatial information.
Pairwise voxel differences: This is a voxel-level feature, which is generated by randomly choosing a pair of voxels at different locations within an image patch and then computing their intensity difference.
Haar-like features: This type of features operates on the sub-region level in an image patch. It was originally proposed by Viola and Jones [21] for object detection, and has been applied to many applications due to its efficiency. Here, we use a variant of the Haar-like features calculated based on the difference between mean values of two sub-regions within an image patch. The size and position of each sub-region can be randomly chosen [22].
Discrete Cosine Transform (DCT) coefficients: DCT can characterize an image in the frequency space, and was successfully applied in image compression [23]. DCT coefficients were also proved to be efficient invariant features for image retrieval and recognition [24, 25], which can provide some degrees of invariance to image deformation in the spatial domain. Here, we can use DCT coefficients of image patch as features, which essentially operate on the whole-patch level.
Note that the last three types of features are carefully chosen to characterize information of an image patch at different levels, i.e., from voxel level, sub-region level, to whole-patch level. Their sensitivities to image changes vary at different levels, i.e., from fine, local, to global, respectively. By integrating them into structured random forest for automatic feature selection, they help balance the need of not only having some degrees of invariance to image deformation but also being sensitive to small structural changes for preserving image details.
2) Multi-scale Approach
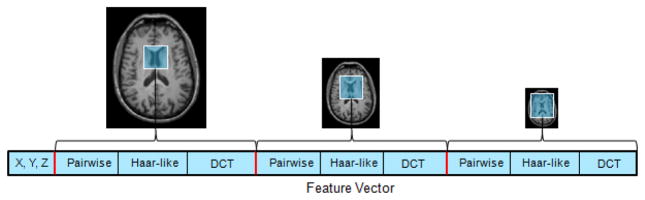
To cover a wider neighborhood and combine both global and local characteristics of the image region, we also extract features at multiple resolutions. Specifically, each original MR image is down-sampled to obtain its corresponding images at coarser scales. Then, we fix the size of the image patch to extract features at corresponding positions across all scales. Finally, we group all the extracted features, together with the coordinates of the center of the image patch, to form the final feature vector for each location in the MR image (see Fig. 2).
Fig. 2.
Multi-scale feature extraction. At each location, the feature vector is comprised of the spatial coordinates of the location and also the features extracted from an image patch centered at that location with fixed size across all scales.
D. Structured Random Forest
1) Random Forest
Random forest is a learning-based method for classification and regression problems, and has been widely adopted in medical imaging applications [26]. Random forest comprises of multiple decision trees. At each internal node of a tree, a feature is chosen to split the incoming training samples to maximize the information gain. Specifically, let u ∈ U ⊂ ℝq be an input feature vector, and ν ∈ V ⊂ ℝ be its corresponding target value for regression. For a given internal node j and a set of samples Sj ⊂ U × V, the information gain achieved by choosing the k-th feature to split the samples in the regression problem is computed by:
| (1) |
| (2) |
| (3) |
where L and R denote the left and right child nodes, , uk is the k-th feature of feature vector u, is the splitting, threshold chosen to maximize the information gain for the k-th feature uk, and |·| is the cardinality of the set. H(S) denotes the variance of all target values in our regression problem.
In the training stage, the splitting process is performed recursively until the information gain is not significant, or the number of training samples falling into one node is less than a pre-defined threshold.
2) Structured Random Forest
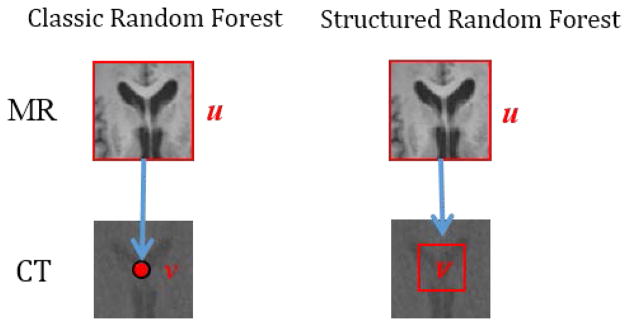
In the classic random forest, its output is just a target value for our case of regression, i.e., predicting voxel-wise CT value by features computed from an MR image patch. Inspired by the work of Dollár and Zitnick [27] on deriving the structured output for 2D edge detection, we propose here to construct the structured random forest for patch-wise CT prediction, i.e., predicting target values in the whole CT patch from its corresponding MR image patch. The difference between structured and classic random forests is illustrated in Fig. 3. There are two notable advantages by directly predicting all target values in the whole CT patch, compared to the voxel-wise CT value prediction. First, the neighborhood information can be preserved in each predicted CT patch, which cannot be achieved by the voxel-wise CT value prediction. Second, more target values can be conveyed in the resulting outputs, thus reducing the number of decision trees in the random forest and improving the efficiency of the entire prediction.
Fig. 3.
Illustration of classic random forest and structured random forest. The top row shows an MR image patch, used to predict a corresponding CT value ν or a CT patch ν (shown in the bottom row). In the classic random forest, the input feature vector derived from MR image patch is used to predict a target value ν for a voxel (red point) in the CT image, while, in the structured random forest, the same input feature vector u is used to predict all values ν in a target CT patch (marked by the red rectangle).
Despite the above advantages, a major problem with patch-wise prediction is how to efficiently characterize the similarity of the structured outputs for defining the information gain. One naïve solution is to define the similarity of CT patches based on the similarity of voxel-wise intensities in the CT patches. However, this approach is computationally expensive and also too sensitive to individual voxel changes to capture the high-level image patch structure. For example, a small CT patch of size a×a×a can decrease the speed in cubic exponent, i.e., a3 times, for each attempt to evaluate the information gain at one node alone. It will thus cause significant delay in the training process due to multiple splitting evaluations at each node and also the exponential growth of the number of nodes. A more effective solution is to find a mapping that can effectively capture the information from each image patch, and then compare just the mapping coefficients. Let the regression target set be V ⊂ ℝg, where each element ν represents the target values of all voxels in the target CT patch. A desired mapping should be able to map ν ∈ V ⊂ ℝg to a new coefficient space ℂ ⊂ ℝd:
| (4) |
such that d < g, and also we can measure the dissimilarity of two CT patches by computing the Euclidian distance of their corresponding coefficients in ℂ. In this paper, we characterize the image patches by principal component analysis (PCA), i.e., using the first d eigenvectors of all CT patches in V as a mapping function and the mapped coefficients of ν in ℂ represent the first d PCA coefficients. This mapping is efficient to compute, and also effective in deriving the most significant information in each CT patch. Suppose that w = Π(ν) denotes the mapped coefficients of ν, where w ∈ ℂ, then H(S) from Eqs. (2) and (3) can be computed as:
| (5) |
| (6) |
Although we only use the first d PCA coefficients to compute information gain, we do not discard the rest coefficients. Instead, we store and use all the coefficients for inverse mapping into image patches in the reconstruction phase, thus preserving the quality of output image patches. Note that the proposed structured random forest leads to a predicted CT patch at each voxel. We fuse all prediction results (i.e., by averaging) to get the final predicted CT image.
3) Ensemble Model
For simplicity, we use the notion of voxel-wise prediction in the classic random forest to explain our proposed ensemble model. The similar concept and steps can be directly applied to patch-wise prediction in our structured random forest.
Each leaf node of the trees in random forest stores multiple training samples that can be used to derive the final prediction result. In the testing stage, feature vector of each MR image patch is passed through each tree, and finally reaches one leaf node per decision tree. To derive the final prediction for the input MR image patch, we have to combine all information stored in those arrived leaf nodes. A common approach is to average all the training samples in those arrived leaf nodes across all decision trees. Specifically, let be the CT value of the l-th training sample in the arrived leaf node of decision tree t ∈ T, where l ∈ {1,…,Lt } and Lt is the number of training samples in the current arrived leaf node of decision tree t. The final prediction result is given by:
| (7) |
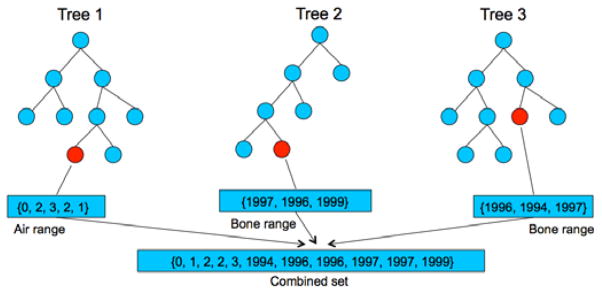
However, this ensemble method is not robust, especially when the variation of the results from different decision trees is large. An example is given in Fig. 4, where the forest has three decision trees. For the input MR image patch, the arrived leaf nodes in the two decision trees (tree 2 and tree 3) contain training samples with their corresponding CT values in the intensity range of bone, while the arrived leaf node in the third tree (tree 1) contains CT values in the intensity range of air. This is a typical case in our application, where, for bone and air, they are ambiguous in the MR image, but highly differentiated in the CT image. As predictions of most decision trees (i.e., 2 trees) are bone, the correct output value should be similar to bone. However, if using the classic ensemble model in Eq. 7, the final prediction is μ =1089.7, which is neither bone nor air, and will be misclassified into the intensity range of white matter.
Fig. 4.
An example of the output results from 3 different decision trees in the random forest.
To address the above problem, we propose a new ensemble model. Specifically, instead of using the mean, we use the median of prediction results, thus guaranteeing the final prediction to be either air or bone for the above example in Fig. 4. But, if there are more training samples included in the current leaf node of decision tree 1, by taking the median of all prediction results from all decision trees, we could get the final result in the intensity range of air, which is incorrect since most decision trees have the prediction results in the intensity range of bone.
To this end, we use 1) median of medians and 2) median of means to avoid potential bias. For the former model, we take the median for results in each decision tree, and then take the median again across the obtained medians of all decision trees. For the latter, we compute the mean for each tree, and then take the median across the obtained means of all decision trees. Both models ensure the final result to be in the range of the outputs of all decision trees, and also favor the contributions from results of most of trees. The difference between the two models is that the former provides the output value only from those observed in the decision trees, while the latter can infer a new value within the output range of all decision trees.
For the above example, the former model leads to a prediction value of 1996, which is an existing value belonging to the bone, while the latter gets 1995.7, which is a new value but still belonging to the bone.
E. Auto-context Model
It has been known that the context, i.e., the surrounding information with respect to an object of interest, plays a vital role in interpreting image content [28]. Similarly, the prediction of an image element could be enhanced by the information from its surrounding neighbors. Among the methods leveraging context information, auto-context model (ACM) [29] has been shown highly effective. ACM uses the prediction result from the learned model to characterize the context information, and then uses such information as context features, together with the appearance features extracted from the input MR image, to recursively train a series of prediction models. Here we integrate ACM into our method, as summarized in Framework 1, where Ω is the total number of training MR and CT image pairs, Xω represents an MR image while Yω is its corresponding CT image, and is the predicted CT map corresponding to the input MR image Xω at iteration ϕ.
Framework 1.
Training procedure with integrated auto-context model
For each training MR image Xω, initialize its predicted CT map
with zeros on all the voxels. For ϕ = 1,…Φ:
|
In the initial stage, as there is no context information derived yet, the CT maps are initialized to zeros. It has been proven that ACM monotonically decreases the training error in each stage [29]. Thus, the prediction result is expected to converge after a number of iterations in Framework 1.
In the testing stage, to predict the CT image, the new given MR image follows the same sequence of models as learned during the training stage, to generate the final prediction.
III. Experiments
A. Datasets
We tested our algorithm on two datasets, as described below:
The brain dataset was acquired from 16 subjects with both MRI and CT scans in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (see www.adni-info.org for details). CT images were acquired on a Siemens Somatom scanner, with voxel size 0.59×0.59×3 mm3; The MR images were acquired using a Siemens Triotim scanner, with voxel size 1.2×1.2×1 mm3, TE 2.95 ms, TR 2300 ms, and flip angle 9°.
The prostate dataset is our in-house data, which has 22 subjects, each with the corresponding MR and CT scans. CT images (voxel size 1.17×1.17×1 mm3) were acquired on a Philips scanner, while MR images (voxel size 1×1×1 mm3, TE 123 ms, TR 2000 ms, and flip angle 150°) were acquired using a Siemens Avanto scanner. The preprocessing and alignment process of the data is presented in Section II.B, in which the Dice Similarity Coefficients (DSC) for prostate, bladder and rectum after intra-subject deformable registration are 0.91, 0.95, and 0.92, respectively.
We performed leave-one-out cross validation on each dataset. In all experiments throughout this paper, if not mentioned specifically, the following parameters were used:
Input patch size in MR image: 15×15×15.
Output patch size in CT image: 3×3×3.
Number of PCA coefficients used in the structured random forest: 10.
Number of DCT coefficients extracted in each input MR image patch = Number of Haar-like features = Number of pairwise voxel difference features = 200.
Feature length: (3 spatial coordinates) + (200 pairwise voxel difference features + 200 Haar-like features + 200 DCT coefficients) x (3 image scales) = 1803.
Number of trees: 20; Maximum tree depth: 19; Minimum number of training samples at each leaf node: 5.
B. Quantitative Measurements
To quantitatively characterize the prediction accuracy, we used two popular metrics: 1) mean absolute error (MAE) and 2) peak signal-to-noise ratio (PSNR):
| (8) |
where E is the ground truth CT image, Ê is the corresponding estimated CT image, Q is the maximal intensity value of E and Ê, and C is the number of voxels in the image. In general, a good prediction result has lower MAE and higher PSNR.
C. Comparison between Classic Random Forest and the Proposed Random Forest
We first compare our method with the classic random forest. In Section II.D, we introduced two enhancements over the classic random forest, i.e., structured random forest and new ensemble models. For the new ensemble models, the median of medians and median of means are similar and thus expected to give similar results. Hence, we chose to use the median of means for the experiment due to its computational efficiency. By alternatively integrating these enhancements with the traditional approach, we have four variants of random forest: classic random forest with mean ensemble model (C-M), structured random forest with mean ensemble model (S-M), classic random forest with median of means ensemble model (C-MM), and structured random forest with median of means ensemble model (S-MM).
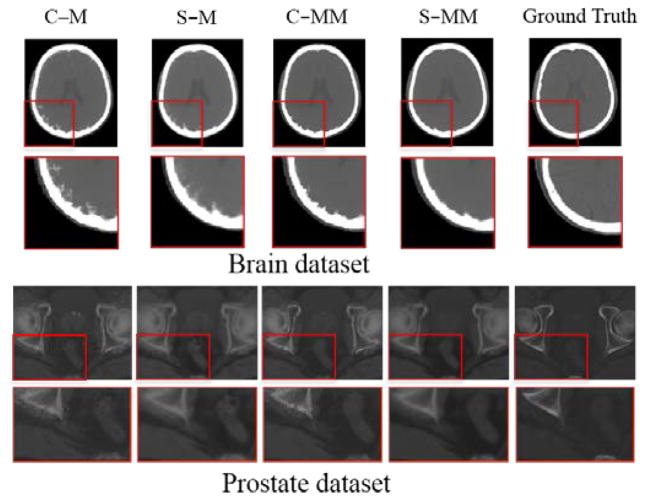
Fig. 5 provides detailed visual comparison for all four variants. The results are rather consistent across the two datasets. We can see that the structured random forest can better preserve the continuity, coalition and smoothness in the prediction results, since it uses local neighborhood constraint in the image patch, as discussed in Section II.D.2). This is observed from the comparison between C-M and S-M, and also between C-MM and S-MM. Besides, the effects of different ensemble models can be clearly seen by differences between the results from C-M and C-MM, and also between results from S-M and S-MM. The median of means model always provides cleaner and less noisy results compared to the traditional mean model, indicating that the latter is more prone to generating the odd result which does not exist in the outputs of the trained trees, as discussed in Section II.D.3).
Fig. 5.
Qualitative comparison of the prediction results from different variants of random forest for two datasets: 1) brain (upper panel) and 2) prostate (lower panel). In each panel, from left to right, the top row shows the prediction results using classic random forest with the mean ensemble model (C-M), structured random forest with the mean ensemble model (S-M), classic random forest with the median of means ensemble model (C-MM), structured random forest with the median of means ensemble model (S-MM), and the CT ground truth; The bottom row shows the close-ups of the regions indicated by red rectangles in the top row.
Tables 1–2 further demonstrate the above observations quantitatively. As the results from different variants differ mainly in the soft tissue regions (especially in the regions close to the bone, as seen in Fig. 5), we only provide results within such region. Tables 1–2 clearly show the consistent improvement of the structured random forest over the classic random forest (S-M vs. C-M, and S-MM vs. C-MM), and the improvement of the median of means ensemble model compared to the traditional mean model (C-MM vs. C-M, and S-MM vs. S-M). To further show the statistical significance of the improvement, we performed the null hypothesis tests using obtained results. The resulting p-values in both datasets are well below 0.05.
Table 1.
Quantitative comparison of the prediction results from 4 different variants of random forest on the soft tissue regions of the brain dataset, in terms of MAE and PSNR.
| MAE | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| C-M | 36.3±9.8 | 37.0 | 20.0 | 49.2 |
| S-M | 33.5±9.0 | 34.7 | 18.8 | 45.3 |
| C-MM | 22.5±6.5 | 21.6 | 12.9 | 30.9 |
| S-MM | 21.4±6.3 | 20.6 | 12.0 | 30.0 |
| PSNR | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| C-M | 22.0 ± 2.1 | 21.7 | 19.9 | 26.0 |
| S-M | 22.7 ± 2.1 | 22.6 | 20.5 | 26.7 |
| C-MM | 23.6 ± 2.3 | 23.6 | 21.0 | 27.7 |
| S-MM | 24.4 ± 2.2 | 24.9 | 21.6 | 28.4 |
Table 2.
Quantitative comparison of the prediction results from different variants of random forest on the soft tissue regions of the prostate dataset, in terms of MAE and PSNR.
| MAE | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| C-M | 45.9±6.7 | 45.4 | 37.4 | 53.9 |
| S-M | 38.3±5.6 | 37.6 | 31.6 | 45.1 |
| C-MM | 35.4±3.6 | 35.4 | 30.9 | 40.1 |
| S-MM | 31.8±4.5 | 31.7 | 25.3 | 37.8 |
| PSNR | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| C-M | 28.2±0.5 | 28.3 | 27.3 | 28.8 |
| S-M | 28.7±0.6 | 28.8 | 27.9 | 29.4 |
| C-MM | 28.8±0.5 | 28.9 | 28.2 | 29.3 |
| S-MM | 29.1±0.6 | 29.3 | 28.3 | 29.7 |
D. Contribution and Convergence of Auto-context Model
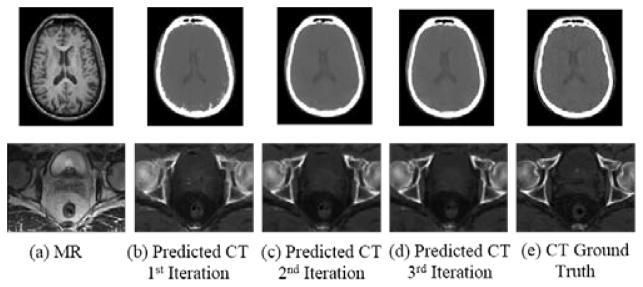
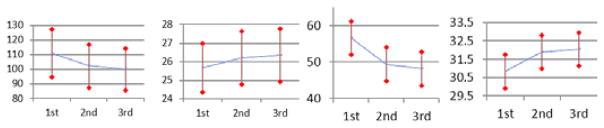
We now show the contribution of ACM and its convergence. As shown in Fig. 6, the prediction quality improves notably with the use of context information by comparing Fig. 6b and Fig. 6c. The subsequent iterations (Fig. 6d) keep improving the quality with the refined context information, though it may be less noticeable. This can also be seen from the quantitative results given in Fig. 7, where both MAE and PSNR are improved gradually and consistently with iterations, in both datasets. The ACM still produces an improved prediction in the final iteration, though not as significantly as it does in the first two iterations. The statistical tests show that the p-values after each iteration are well below 0.05. We stopped at the third iteration, since the improvement is minor after this iteration (PSNR<0.1).
Fig. 6.
Contribution of auto-context model. Top: results for the brain dataset. Bottom: results for the prostate dataset.
Fig. 7.
Mean and standard deviation of MAE and PSNR measurements at different iterations of auto-context models in two datasets (left two: brain, right two: prostate). Both MAE and PSNR are measured in the entire image domain.
E. Comparison with Atlas-based and Sparse Representation based Methods
In this section, we compare our method with two popular approaches: an atlas-based method and a sparse representation based method. While there are many variants of atlas-based [6, 7, 16, 30] and sparse representation based methods [14–16, 18], we only chose one standard implementation for each approach for demonstration purpose:
Atlas-based method: An atlas has an MR image and its corresponding CT image. To predict the CT image for a target MR image, the MR image of each atlas is first aligned [31] onto the target MR image, and the resulting deformation field is used to warp the CT image of each atlas. Averaging the resulting warped CT images leads to the final prediction.
Sparse representation based method: After warping the atlases to the target image space, a local sparse representation is then performed. Specifically, a local patch in the target MR image is represented as a sparse linear combination of neighboring patches from the warped atlas MR images, similar to [32]. The resulting sparse coefficients are then applied to the warped atlas CT images to obtain the final prediction.
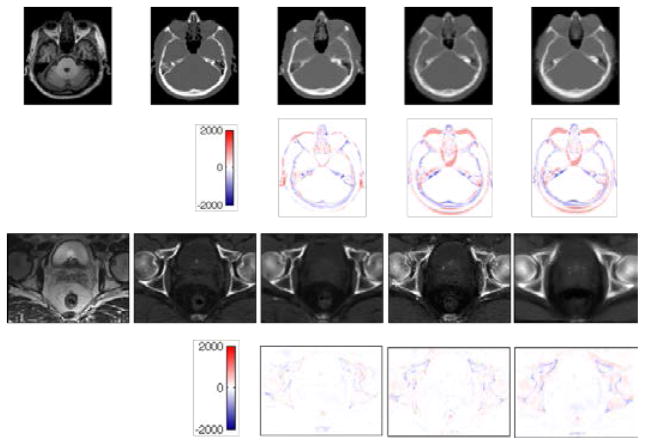
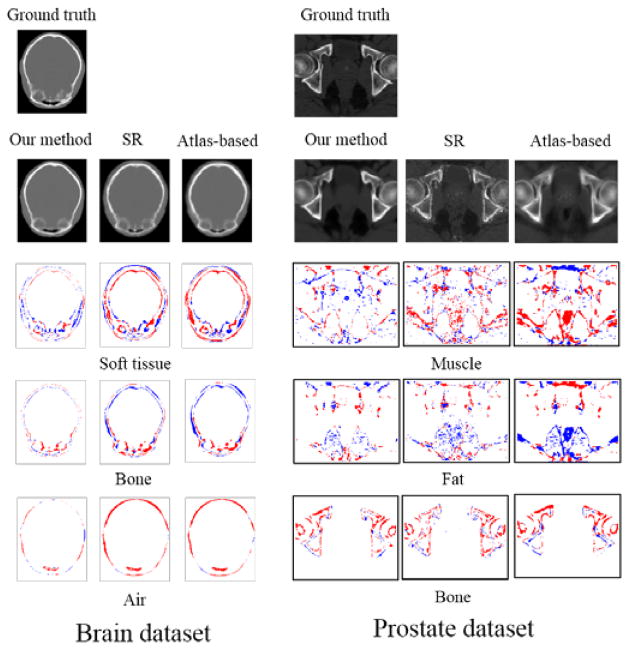
The training time for our method took ~ 4 hours on 2.67GHz Intel Xeon 6-core processor, while the testing time took ~ 20 minutes. The typical prediction results are given in Fig. 8. It can be seen that the CT images generated by our method are quite similar to ground truth in both datasets. Furthermore, our method outperforms other two methods, with higher similarity in shape and lower difference values. The atlas-based method often leads to blurred prediction due to simple averaging of the warped atlases. This can be clearly seen on the prostate dataset, which has large shape variation across different subjects. Although the sparse representation based method works better than the atlas-based method, its prediction is still noisy.
Fig. 8.
Examples of the prediction results from two datasets: 1) brain (upper panel) and 2) prostate (lower panel). In each panel, from left to right, the top row shows the MR image, ground-truth CT image, and the predicted CT images given by our method, the sparse representation (SR) based method, and the atlas-based method, respectively; The bottom row shows the residual images by subtracting the ground-truth CT image from the prediction results given by our method, the sparse representation (SR) based method, and the atlas-based method, respectively.
Quantitative results in Tables 3–4 show that our method outperforms other two methods in terms of MAE and PSNR. For the brain dataset, our method gives an average PSNR of 26.3, significantly better than 21.1 and 20.9 given by the sparse representation based method and the atlas-based method, respectively. A similar result can also be seen on the prostate dataset, with the average PSNR of 32.1, 30.4 and 29.1 for the three methods, respectively. The hypothesis tests between our method and the other two methods led to p-values well below 0.05, showing the statistical significance in the improvement.
Table 3.
Quantitative comparison of our method with the atlas-based method and the sparse representation (SR) based method on the brain dataset, in terms of MAE and PSNR.
| MAE | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| Atlas | 169.5±35.7 | 169.9 | 130.1 | 222.3 |
| SR | 166.3±37.6 | 164.3 | 126.5 | 222.0 |
| Proposed | 99.9±14.2 | 97.6 | 83.8 | 118.8 |
| PSNR | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| Atlas | 20.9±1.6 | 20.5 | 19.3 | 22.6 |
| SR | 21.1±1.7 | 21.1 | 19.4 | 23.0 |
| Proposed | 26.3±1.4 | 26.3 | 24.5 | 28.2 |
Table 4.
Quantitative comparison of our method with the atlas-based method and the sparse representation (SR) based method on the prostate dataset, in terms of MAE and PSNR.
| MAE | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| Atlas | 64.6±6.6 | 65.9 | 56.8 | 71.5 |
| SR | 54.3±10.0 | 54.0 | 44.3 | 61.7 |
| Proposed | 48.1±4.6 | 48.3 | 42.2 | 53.6 |
| PSNR | Mean±s.d. | Med. | 10th-tile | 90th-tile |
|---|---|---|---|---|
| Atlas | 29.1±2.0 | 29.8 | 25.8 | 30.7 |
| SR | 30.4±2.6 | 31.3 | 26.7 | 32.9 |
| Proposed | 32.1±0.9 | 31.8 | 31.1 | 33.4 |
To further evaluate the accuracy of the predicted CT range, we also perform tissue-wise voxel classification test based on known CT ranges for different tissue types, i.e., bone, air, fat, muscle, and soft tissues in head region. Results are shown in Fig. 9 and Table 5. We can see that most misclassifications are around the tissue boundary, and the proposed method clearly outperforms other two methods, especially in brain data, with the mean accuracy of 0.91, compared to 0.84 and 0.82 by the sparse representation and atlas-based methods, respectively.
Fig. 9.
Tissue-wise voxel misclassification maps for two datasets. Red denotes false negative, while blue denotes false positive.
Table 5.
Mean accuracy of tissue-wise voxel classification.
| Proposed | SR | Atlas | |
|---|---|---|---|
| Brain | 0.91 | 0.84 | 0.82 |
| Prostate | 0.79 | 0.74 | 0.60 |
IV. DISCUSSIONS
We have presented a learning-based approach to predicting the CT image from a single MR image. To the best of our knowledge, this is the first work that uses a structured random forest based framework for this task. Several mechanisms are also incorporated to boost the prediction performance. We tested the proposed system on the two challenging datasets and compared its performance with two state-of-the art approaches.
The efficacy of using structured random forest is demonstrated through Fig. 5 and Tables 1–2. The results show that the structured random forest can generally improve the prediction performance, qualitatively and quantitatively. In particular, it can better preserve the continuity, coalition and smoothness in the predicted results, since more local neighborhood information is leveraged and preserved in the estimated CT image.
Fig. 5 and Tables 1–2 also validate the efficacy of the proposed ensemble model using the median of means approach. The new model notably reduces noisy predictions compared to the traditional mean model. This is because the mean model simply averages the outputs of the trees, which is more likely to render the final result different from the outputs of the trained trees, especially when the outputs of different trees vary dramatically.
Context information does help improve the prediction accuracy remarkably, as shown in Fig. 6 and Fig. 7, where the prediction is gradually improved at each iteration of the auto-context model. One last remark of the proposed method is on the efficacy of the constructed features. The spatial information is efficiently incorporated and learned to improve the prediction performance. Note that such information is only obtained from rigid registration other than deformable registration (which is often required by most of the existing methods). The choice of features with multi-level sensitivity makes the prediction results more robust and accurate. As shown in Fig. 8, our system can predict the CT images that are highly similar to the ground-truth values and can effectively capture very small changes in the image.
Our method works remarkably better than the sparse representation based method and the atlas-based methods, as shown in Fig. 8 and Tables 3–4. For the brain dataset, our method consistently outperforms both methods, while, for the prostate dataset, we achieve the best overall performance, although the sparse representation based method works better occasionally for some subjects. This might be due to the large deformations of the prostate data across subjects, which cause difficulty in generalizing the pattern rules. Despite this, our method still achieves better overall performance by using only the rough spatial information derived from rigid-body alignment, compared to the deformable registration used in the presented sparse representation based method. The efficacy of our method is further consolidated by the tissue-wise voxel classification results presented in Fig. 9 and Table 5. Our method clearly provides better classification of tissue types of voxels compared to other two methods.
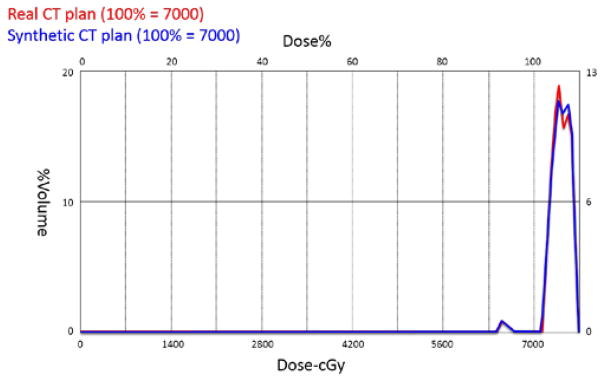
To evaluate the practical value of our method, we have also conducted a preliminary study by using our synthesized CT images for dose planning in the prostate radiation therapy. Specifically, we chose several typical patients from 22 patients. We respectively used their real CT images and the synthetic CT images from our method for dose calculation in the radiotherapy planning. Since physicians care more about regions around the prostate, our method is applied to predict Hounsfield units within those regions. All dose calculations are based on the sub-regions. Fig. 10 shows the differential dose-volume histograms of one patient obtained by real CT image (red) and our synthetic CT image (blue). We can see that two histograms overlap quite well, indicating that the prostate has similar dosimetric coverage when the same treatment beams are calculated on real CT image or synthetic CT image. Quantitatively, the mean doses on the prostate obtained by real and synthetic CT images are 7389.92 cGy and 7393.85 cGy, respectively. The prescription of radiotherapy plan of this case is 7000 cGy on the prostate and allows 10% dose variation. Accordingly, the mean dose obtained with our synthetic CT is clinically acceptable. In our future work, we will evaluate on more patients in prostate radiation therapy, as well as extend our evaluation to PET attenuation correction.
Fig. 10.
Differential Dose-Volume Histograms (DVHs) of the prostate with real CT image (red) and our synthetic CT image (blue) for a typical patient.
V. Conclusion
In this paper, we propose a novel approach to predicting a CT image from a single MR image. The proposed method is based on random forest with multiple improvements, to effectively capture the relationship between the CT and MR images. Specifically, image information is better characterized by introducing spatial information to the crafted feature. The context and neighborhood information are also well preserved by using the structured random forest and the auto-context model, with the final result further improved by the new ensemble model. The proposed method is capable of reliably predicting the CT image from the MR image for different organs of human body. Specifically, we tested our method on two challenging and highly different datasets, with the results better than two state-of-the-art methods.
For future work, we will further investigate the influence of different parameters in our method, and extend it to work on more datasets. The clinical application of the proposed method such as for AC in PET/MRI scanner is also of high interest for further investigation.
Acknowledgments
Part of the data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). This work was partially supported by NIH grants (EB006733, EB008374, EB009634, MH100217, AG041721, AG042599, CA140413).
Contributor Information
Tri Huynh, Email: hquoctri@gmail.com, IDEA lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC, USA.
Yaozong Gao, Email: yzgao@cs.unc.edu, Department of Computer Science, and also with the IDEA lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC, USA.
Jiayin Kang, IDEA lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC, USA.
Li Wang, IDEA lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC, USA.
Pei Zhang, IDEA lab, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, NC, USA.
Jun Lian, Email: jun_lian@med.unc.edu, Department of Radiation Oncology, University of North Carolina at Chapel Hill, NC, USA.
Dinggang Shen, Email: dgshen@med.unc.edu, Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, NC 27599, USA, and also with the Department of Brain and Cognitive Engineering, Korea University, Seoul 136-071, Korea.
References
- 1.Kinahan PE, et al. Attenuation correction for a combined 3D PET/CT scanner. Medical Physics. 1998;25(10):2046–2053. doi: 10.1118/1.598392. [DOI] [PubMed] [Google Scholar]
- 2.Brenner DJ, Hall EJ. Computed Tomography — An Increasing Source of Radiation Exposure. New England Journal of Medicine. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 3.Zaidi H, Montandon ML, Slosman DO. Magnetic resonance imaging-guided attenuation and scatter corrections in three-dimensional brain positron emission tomography. Medical Physics. 2003;30(5):937–948. doi: 10.1118/1.1569270. [DOI] [PubMed] [Google Scholar]
- 4.Hsu S-H, et al. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Physics in medicine and biology. 2013;58(23) doi: 10.1088/0031-9155/58/23/8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berker Y, et al. MRI-Based Attenuation Correction for Hybrid PET/MRI Systems: A 4-Class Tissue Segmentation Technique Using a Combined Ultrashort-Echo-Time/Dixon MRI Sequence. Journal of Nuclear Medicine. 2012;53(5):796–804. doi: 10.2967/jnumed.111.092577. [DOI] [PubMed] [Google Scholar]
- 6.Kops ER, Herzog H. Alternative methods for attenuation correction for PET images in MR-PET scanners. Nuclear Science Symposium Conference Record, 2007. NSS ‘07. IEEE; 2007. [Google Scholar]
- 7.Hofmann M, et al. MRI-Based Attenuation Correction for PET/MRI: A Novel Approach Combining Pattern Recognition and Atlas Registration. Journal of Nuclear Medicine. 2008;49(11):1875–1883. doi: 10.2967/jnumed.107.049353. [DOI] [PubMed] [Google Scholar]
- 8.Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Medical Physics. 2011;38(5):2708–2714. doi: 10.1118/1.3578928. [DOI] [PubMed] [Google Scholar]
- 9.Roy S, et al. PET Attenuation Correction using Synthetic CT from Ultrashort Echo-time MRI. Journal of nuclear medicine: official publication, Society of Nuclear Medicine. 2014;55(12):2071–2077. doi: 10.2967/jnumed.114.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy S, et al. MR to CT Registration of Brains using Image Synthesis. Proceedings of SPIE. 2014:9034. doi: 10.1117/12.2043954. spie.org/Publications/Proceedings/Paper/10.1117/12.2043954. [DOI] [PMC free article] [PubMed]
- 11.Jog A, Carass A, Prince JL. Improving magnetic resonance resolution with supervised learning. Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on; 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jog A, et al. Random forest FLAIR reconstruction from T1, T2, and PD-weighted MRI. Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on; 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, et al. Deep Learning Based Imaging Data Completion for Improved Brain Disease Diagnosis. In: Golland P, et al., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2014. Springer International Publishing; 2014. pp. 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy S, Carass A, Prince J. A Compressed Sensing Approach for MR Tissue Contrast Synthesis. Information Processing in Medical Imaging. 2011;22:371–383. doi: 10.1007/978-3-642-22092-0_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy S, Carass A, Prince JL. Magnetic Resonance Image Example-Based Contrast Synthesis. Medical Imaging, IEEE Transactions on. 2013;32(12):2348–2363. doi: 10.1109/TMI.2013.2282126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy S, et al. MR contrast synthesis for lesion segmentation. Biomedical Imaging: From Nano to Macro, 2010 IEEE International Symposium on; 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye D, et al. Modality Propagation: Coherent Synthesis of Subject-Specific Scans with Data-Driven Regularization. In: Mori K, et al., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Springer; Berlin Heidelberg: 2013. pp. 606–613. [DOI] [PubMed] [Google Scholar]
- 18.Iglesias J, et al. Is Synthesizing MRI Contrast Useful for Inter-modality Analysis? In: Mori K, et al., editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013. Springer; Berlin Heidelberg: 2013. pp. 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 20.Klein S, et al. Elastix: A Toolbox for Intensity-Based Medical Image Registration. Medical Imaging, IEEE Transactions on. 2010;29(1):196–205. doi: 10.1109/TMI.2009.2035616. [DOI] [PubMed] [Google Scholar]
- 21.Viola P, Jones M. Rapid object detection using a boosted cascade of simple features. Computer Vision and Pattern Recognition, 2001. CVPR 2001. Proceedings of the 2001 IEEE Computer Society Conference on; 2001. [Google Scholar]
- 22.Gao Y, et al. Learning Distance Transform for Boundary Detection and Deformable Segmentation in CT Prostate Images. In: Wu G, Zhang D, Zhou L, editors. Machine Learning in Medical Imaging. Springer International Publishing; 2014. pp. 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skodras A, Christopoulos C, Ebrahimi T. The JPEG 2000 still image compression standard. Signal Processing Magazine, IEEE. 2001;18(5):36–58. [Google Scholar]
- 24.Thepade DHBKaTKSaSD. Image Retrieval using Color-Texture Features from DCT on VQ Codevectors obtained by Kekre’s Fast Codebook Generation. Graphics, Vision and Image Processing GVIP. 2009;9(5):1–8. [Google Scholar]
- 25.Dong-Gyu S, Hae-Kwang K, Dae-Il O. Translation, scale, and rotation invariant texture descriptor for texture-based image retrieval. Image Processing, 2000. Proceedings. 2000 International Conference on; 2000. [Google Scholar]
- 26.Criminisi A, Shotton J. Decision Forests for Computer Vision and Medical Image Analysis. Springer Publishing Company, Incorporated; 2013. p. 387. [Google Scholar]
- 27.Dollár P, Zitnick CL. Proceedings of the 2013 IEEE International Conference on Computer Vision. IEEE Computer Society; 2013. Structured Forests for Fast Edge Detection; pp. 1841–1848. [Google Scholar]
- 28.Belongie S, Malik J, Puzicha J. Shape matching and object recognition using shape contexts. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2002;24(4):509–522. doi: 10.1109/TPAMI.2005.220. [DOI] [PubMed] [Google Scholar]
- 29.Tu Z, Bai X. Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans Pattern Anal Mach Intell. 2010;32(10):1744–57. doi: 10.1109/TPAMI.2009.186. [DOI] [PubMed] [Google Scholar]
- 30.Burgos N, et al. Attenuation Correction Synthesis for Hybrid PET-MR Scanners: Application to Brain Studies. Medical Imaging, IEEE Transactions on. 2014;33(12):2332–2341. doi: 10.1109/TMI.2014.2340135. [DOI] [PubMed] [Google Scholar]
- 31.Vercauteren T, et al. Diffeomorphic demons: Efficient non-parametric image registration. NeuroImage. 2009;45(1) Supplement 1:S61–S72. doi: 10.1016/j.neuroimage.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 32.Wright J, et al. Robust Face Recognition via Sparse Representation. Pattern Analysis and Machine Intelligence, IEEE Transactions on. 2009;31(2):210–227. doi: 10.1109/TPAMI.2008.79. [DOI] [PubMed] [Google Scholar]