Abstract
The amount of genomic information about leukemia cells currently far exceeds our overall understanding of the precise genetic events that ultimately drive disease development and progression. Effective implementation of personalized medicine will require tools to distinguish actionable genetic alterations within the complex genetic landscape of leukemia. In this study, we performed kinase inhibitor screens to predict functional gene targets in primary specimens from patients with acute myeloid leukemia (AML) and chronic myelomonocytic leukemia (CMML). Deep sequencing of the same patient specimens identified genetic alterations that were then integrated with the functionally important targets using the HitWalker algorithm to prioritize the mutant genes that most likely explain the observed drug sensitivity patterns. Through this process, we identified Tyrosine Kinase Non-receptor 2 (TNK2) point mutations that exhibited oncogenic capacity. Importantly, the integration of functional and genomic data using HitWalker allowed for prioritization of rare oncogenic mutations that may have been missed through genomic analysis alone. These mutations were sensitive to the multi-kinase inhibitor dasatinib, which antagonizes TNK2 kinase activity, as well as novel TNK2 inhibitors, XMD8-87 and XMD16-5, with greater target specificity. We also identified activating truncation mutations in other tumor types that were sensitive to XMD8-87 and XMD16-5, exemplifying the potential utility of these compounds across tumor types dependent on TNK2. Collectively, our findings highlight a more sensitive approach for identifying actionable genomic lesions that may be infrequently mutated or overlooked, and provide a new method for the prioritization of candidate genetic mutations.
Keywords: TNK2, kinase inhibitors, acute myeloid leukemia, chronic myelomonocytic leukemia
INTRODUCTION
A tremendous amount of information now exists detailing the genetic alterations in leukemia cells. Despite this wealth of genomic data, our understanding of the functional significance of these genetic events lags far behind. One major challenge is how to sort through all of the known mutations to find novel therapeutic targets. To this end, we have developed an algorithm called HitWalker (1), which can prioritize gene mutations based on functional data, detailing the underlying vulnerabilities of the leukemia cells. This functional data is derived from a kinase inhibitor screening platform (2). These kinase inhibitor screens are run on primary patient samples, and then the kinase targets of the effective drugs are calculated based on the known efficacy of the drugs against various targets (1). This study is the first example of the HitWalker algorithm being used to identify a novel therapeutic target, TNK2.
Tyrosine Kinase Non-receptor 2 (TNK2) is a cytoplasmic kinase also known as ACK1 (activated CDC42-associated kinase)(3). TNK2 is part of a family of cytoplasmic tyrosine kinases that also includes TNK1 (3). TNK2 was originally identified based on its binding to the cell cycle regulator, CDC42 (4). Together, TNK2 and CDC42 regulate cellular attachment and migration (5).
TNK2 is comprised of several functional domains including a sterile alpha motif (SAM) domain, a Tyrosine Kinase domain, a SH3 protein-protein interaction domain, a CDC42/RAC-interactive (CRIB) domain, and a region that is homologous to the EGFR binding domain of Mig6 and a ubiquitin association domain. In addition to phosphorylation in response to EGFR signaling, TNK2 can also be activated by other receptor tyrosine kinases (6), and phosphorylation of the TNK2 activation loop by SRC is required for its kinase activity (7).
Multiple mechanisms by which TNK2 contributes to solid tumors have been documented. TNK2 mutations have been found in renal cancer cells and also in lung, ovarian and gastric cancers (8,9). TNK2 genomic amplification has been associated with late stage or metastatic lung and prostate cancers (10). Overexpression of TNK2 promoted metastasis in a mouse model of breast cancer (10). Finally, TNK2 signaling is disrupted in prostate (11), breast (12) and gastrointestinal (13) tumors.
TNK2 can activate several pro-tumorigenic signaling pathways including modulation of the pro-survival AKT signaling pathway (14), phosphorylation of androgen receptor leading to androgen-independent prostate cancer growth (11,15), and negatively regulate the tumor suppressor Wwox (16). In myeloid malignancies, specifically chronic neutrophilic leukemia and atypical CML, non-mutated TNK2 has been shown to be a functional target in patients with CSF3R mutations (17). TNK2 levels increase in BaF3 cells harboring oncogenic CSF3R mutations, which have upregulated JAK kinase and SRC signaling promoting IL3-independent growth (17).
In this report we find a novel link between TNK2 mutations and leukemia using a combination of drug screening and deep sequencing of primary patient samples to identify novel mutations of TNK2 that are therapeutically targetable in leukemia. These mutations are oncogenic, and sensitive to previously developed TNK2 inhibitors. Additionally, we developed novel, potent TNK2 inhibitors that exhibit greater specificity for TNK2.
MATERIALS AND METHODS
Sequencing of Patient Samples
Genomic DNA was extracted from pellets of 5 × 106 purified primary patient sample mononuclear cells using the DNeasy Blood and Tissue Kit (Qiagen). Deep sequencing was performed on a panel of 1862 kinase-associated genes as previously described (17). The mutated regions of TNK2 were verified via Sanger sequencing (GeneWiz) and analyzed using Sequencher and DNASTAR software. All clinical samples were obtained with informed consent by the patients. The study was approved by the institutional review boards at Oregon Health and Science University (OHSU).
Hitwalker Prioritization
Genomic variants were prioritized based on the HitWalker variant prioritization for personalized functional cancer genomics algorithm as described previously (1). Functional screening of kinase inhibitors was performed using primary patient samples, and dependence on specific kinases was predicted based on an inhibitor sensitivity algorithm, as previously described (2). The gene mutations found within each patient sample were prioritized by HitWalker integration of mutant genes with the functionally predicted targets.
Plasmid Construction
TNK2 transcript variant 1 in pDONR was purchased from Genecopoeia (GC-Y4392). TNK2 mutations were made using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) and the following primers: TNK2-D163E-F gcctgaagcccgaggtcctgagccagc, TNK2-D163E-R gctggctcaggacctcgggcttcaggc, TNK2-R806Q-F cgctgccaccccagctctcaagctc, TNK2-R806Q-R gagcttgagagctggggtggcagcg, TNK2-P494fs-F gctcaggaggtcggggggtccatg, TNK2-P494fs-R catggaccccccgacctcctgagc, TNK2-P632fs-F ctgcccccccccgcccgcctatg, TNK2-P632fs-R cataggcgggcgggggggggcag, TNK2-S808*-F cccaggtgagctttagagccggggtgg, TNK2-S808*-R ccaccccggctctaaagctcacctggg, TNK2-Q831fs-F gcctggatcacctgggggtggcgtac, TNK2-Q831fs-R gtacgccacccccaggtgatccaggc. Wild type and mutated TNK2 were transferred into a Gateway converted version of pMXs-IRES-Puro (Cell Biolabs, Inc.) or MSCV-IRES-GFP using Gateway LR Clonase kit (Invitrogen). Plasmid sequences were confirmed via Sanger sequencing (Eurofins).
Cell Culture and Transfection
The human embryonic kidney, 293T17 cell line (ATCC) was grown in DMEM (Invitrogen) medium with 10% FBS (Atlanta Biologicals), L-glutamine, penicillin/streptomycin (Invitrogen), and amphotericin B (HyClone). Murine retrovirus was created by co-transfecting TNK2 plasmids and the EcoPac plasmid (provided by Dr. Rick Van Etten, Tufts University) into 293T17 using FuGENE 6 (ProMega). Supernatants were harvested 72 hours later. Ba/F3 cells were maintained in RPMI 1640 medium with 10% FBS, 15% WEHI conditioned medium (a source of IL3), L-glutamine, penicillin/streptomycin, and amphotericin B.
Ba/F3 Cytokine Independent Growth Assays
Ba/F3 cells were infected with murine retrovirus obtained from 293T17 cells via addition of Polybrene and spinoculation at 30°C for 90 min at 2500 rpm. Infected cells were placed under puromycin selection for 72 hours. Cells were expanded after being washed three times and finally plated at 5 × 105 cells per mL in RPMI 1640 medium with 10% FBS, penicillin/streptomycin, L -glutamine, and amphotericin B. Viable cell counts were measured using propidium iodide exclusion with a Guava personal cell analysis system (Millipore).
pTNK2 Inhibition Assay
293T cells expressing TNK2 were plated in 6-well format at a density of 250,000 cells per well 48 hours prior to treatment. Cells were then treated with a 100 μl of XMD8-87 or XMD16-5 at 5 μM and with 9 1:1 serial dilutions down to ≈10 nM. Two additional samples were treated with DMSO only. Cells were then incubated for 6 hours at 37° C. Protein extraction was accomplished by adding 300 μl of lysis buffer (complete Lysis-M EDTA-free: Roche + complete protease inhibitors Roche + PhosSTOP: Roche) to cells after removing media. Plates were gently shaken for 5 minutes at room temperature. Lysates were collected and cleared of incompletely solubilized material by spinning for 10 minutes at maximum speed in a microcentrifuge. Samples were prepared for SDS-PAGE using the EPage loading buffer (Invitrogen) by diluting 10 μl of lysate with 0.3 μl BME and 3.5 μl 4X loading buffer. Samples were heated at 70 °C for 10 minutes than run on EPage 8 % gels (Invitrogen). For western blot detection, EPage gels were transferred to nitrocellulose using the Invitrogen iBlot transfer instrument. After transfer, blots were blocked for 45 minutes in 5% milk diluted in TBS-T (TRIS buffered saline containing 0.1% tween), then were washed 3 times 5 minutes with TBS-T. Primary antibody (Millipore ACK1-PO-Y284 #09-142) was diluted into blocking buffer and incubated for 2 hours at room temperature. HRP labeled anti-rabbit secondary was diluted into blocking buffer and incubated for 30 minutes. After 3 × 5 minutes washes in TBS-T, Pierce SuperSignal chemiluminescent substrate was added, and imaged using a Fugi chemiluminescence Imager.
TNK2 ELISA Assay
293T cells expressing TNK2 were plated in 96 well format at a density of 50,000 cells per well one day prior to inhibitor treatments. Cells were treated with TNK2 inhibitors for six hours, washed in DPBS, and lysed in ice-cold RIPA buffer (Boston Bioproducts) containing Halt protease and phosphatase inhibitor cocktail (Thermo Scientific). Plates were incubated on Ice for 10-20 minutes. 96 well ELISA plates were pre-coated with anti-ACK chicken antibody overnight, then washed, blocked in PBS with 1% BSA. Sample from lysed cells was incubated for two hours at room temperature, washed and detected with ACK1-PO-Y284 antibody or total ACK antibody (A11, Santa Cruz Biotechnology) for 1 hour. Plates were then washed and incubated with anti-Rabbit HRP (Cell Signaling Technology) and then detected using Glo Substrate (R&D) and a Pherastar Microplate Reader (BMG Labtech).
Kinase Profiling
KiNativ™ is based on biotinylated acyl phosphates of ATP and ADP that irreversibly react with protein kinases on conserved lysine residues in the ATP-binding pocket. The compounds were screened following the detailed procedures described in KiNativ methodology (18).
Enzymatic kinase assay
Kinase targets were tested with biochemical enzymatic kinase assays using the SelectScreen Kinase Profiling Service (Life Technologies Corporation) to determine IC50 values. The compounds were assayed at 10 concentrations (3-fold serial dilutions starting from 1 μM) at an ATP concentration equal to the ATP Km,app for the assay following the detailed procedures described in the SelectScreen Customer Protocol and Assay Conditions documents located at www.invitrogen.com/kinaseprofiling.
Inhibitor Studies
Infected Ba/F3 cell lines were distributed in 96-well plates with increasing concentrations of TNK2 inhibitors. Control lines (Parental Ba/F3s, pMX empty vector, and TNK2 wild-type-expressing) were plated in RPMI 1640 medium with 10% FBS, 15% WEHI conditioned medium, L-glutamine, penicillin/streptomycin, and amphotericin B, while post-transformation lines were plated in WEHI-deficient media. Cells were plated at a density of 6 × 104 cells per mL and treated with the following inhibitors for 72 hours: dasatinib (SellekChem), AIM-100(19) (CalBiochem), XMD8-87 and XMD16-5. All conditions were plated with three replicates. Cell viability was measured using a methanethiosulfonate (MTS)-based assay (CellTiter96 Aqueous One Solution, Promega) and absorbance (490 nm) was read at 1 and 3 hours after adding reagent, using a BioTek Synergy 2 plate reader. MTS-absorbances of inhibitor-treated wells were normalized to those of untreated cells. Regression curve fit analysis of the normalized data to determine IC50 values was performed using GraphPad Prism software.
Immunoblotting
Transfected 293T17 cells were treated with cell lysis buffer (Cell Signaling Technologies) with added complete mini protease inhibitor mixture tablets (Roche), phenylmethylsulfonyl fluoride and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich). Lysates were spun at 8000 rpm for 10 min at 4 °C to pellet cell debris, mixed 2:1 with 3X GS sample buffer with β-ME (75 mM Tris (pH 6.8), 3% sodium dodecyl sulfate, 15% glycerol, 8% β-mercaptoethanol, and 0.1% bromphenol blue) and heated at 95 °C for 5 min. Lysates were run on Criterion 4–15% Tris-HCl gradient gels (Bio Rad), transferred to a PVDF membrane, and blocked for 30 min in Tris-buffered saline with Tween (TBST) with 5% BSA. Blots were probed overnight at 4 °C with anti-ACK1 (TNK2) rabbit antibody (Abcam, catalog no. ab65108), anti-ACK1 Phospho Y284 rabbit antibody (Abcam, ab74091), anti-ACK N-terminal antibody (Abcam ab137506), anti-actin mouse antibody (Millipore), followed by anti-rabbit or anti-mouse IgG HRP conjugate secondary antibodies. Blots were developed using Clarity chemiluminescent substrate (BioRad) or SuperSignal West Dura Extended Duration Substrate (Life Technologies) and imaged using a BioRad ChemiDoc MP Imaging System or exposed to x-ray film.
RESULTS
Functional genomic prioritization identifies TNK2 as an important target in a patient with AML
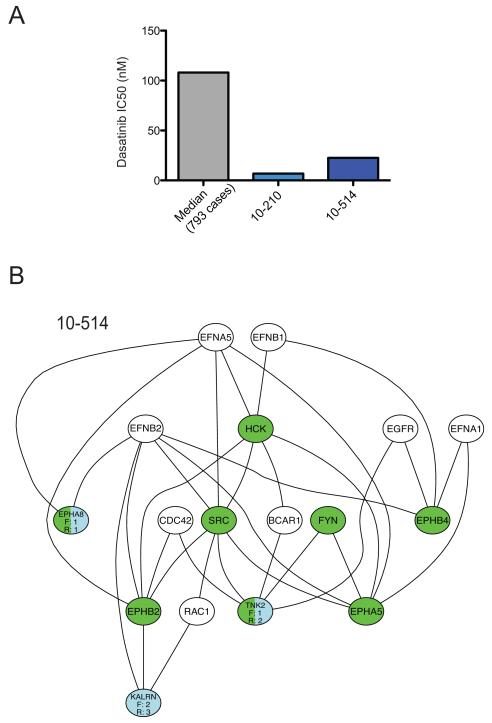
In order to identify the actionable genetic drivers in a patient with AML (10-514), we utilized functional inhibitor screen data to prioritize genomic mutations found by sequencing of 1862 kinase associated genes. The functional data was obtained by screening the primary patient sample against a panel of small-molecules comprised largely of kinase inhibitors. An IC50 value was then calculated for each inhibitor and compared to the median of the IC50s for the full patient sample cohort that has been screened to date. Inhibitors for which the IC50 value was less than 20% of the cohort median were considered effective. As an example, the cohort median IC50 observed for dasatinib to date has been 109 nM, and in comparison the two specimens analyzed for this study exhibited significantly lower IC50s of 6.8 and 22.6 nM (Figure 1A) The full list of effective drugs was used to predict specific kinase targets that were important for mediating the observed drug sensitivity profile, using a previously described algorithm (2). These functional targets were then integrated with mutant genes observed within this same specimen using the previously described HitWalker algorithm, which annotates functional and genomic targets onto the STRING database of protein-protein interaction networks to calculate the shortest path between mutated genes to functional gene targets (1). In this way, we prioritized mutant genes that were likely to explain the drug sensitivity pattern observed for AML patient specimen 10-514 (Figure 1B). In this specimen two genes were both highly ranked functional targets and were themselves mutated genes—the tyrosine kinases EPHA8 and TNK2 (Figure 1B and 2A, B).
Figure 1. Functional genomic prioritization identifies TNK2 as an important target in a patient with AML.
(A) Example of hypersensitive drugs in two patient specimens analyzed in this study. The calculated IC50s for dasatinib of the two individual primary patient samples are shown as well as the Median IC50 value for dasatinib for 793 cases. (B) The HitWalker variant prioritization for functional genomics algorithm was used to prioritize mutated genes found by deep sequencing based on the kinase inhibitor sensitivities observed in the same patient samples. The functional targets are shown in green and mutated genes are shown in blue. Each mutated gene is assigned a rank (ex. R:1 in the figure). The two most highly prioritized genes in this leukemia patient sample were EPHA8 and TNK2.
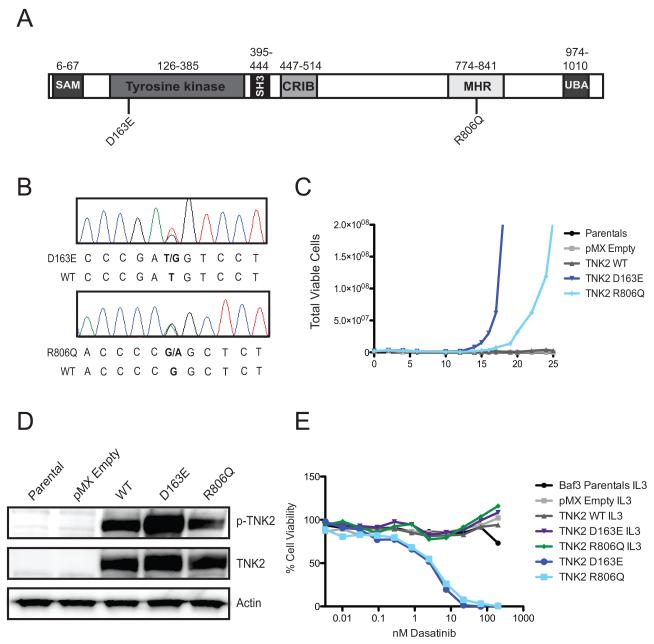
Figure 2. Prioritized TNK2 mutations exhibit transformative capacity and confer dasatinib sensitivity.
(A) Schematic of TNK2 gene and locations of mutations of interest. The D163E mutation was found in a patient with acute myeloid leukemia (AML) and the R806Q mutation was found in a patient with chronic myelomonocytic leukemia (CMML). The sterile alpha motif (SAM), Tyrosine kinase domain, SRC Homology 3 domain (SH3, a protein-protein interaction domain), CDC42/Rac interactive binding domain (CRIB), SRC, Mig6 homology region (MHR, aka the EGFR binding domain), and ubiquitin association domain (UBA) are shown. The amino acid positions of each domain is noted above (40). (B) Confirmation of TNK2 mutations found by deep sequencing. Sanger sequencing was performed on genomic DNA isolated from patient leukemia samples and identified heterozygous D163E and R806Q mutations in patient samples. Electropherogram peaks are shown and correspond to the following nucleotides A (green), T (red), C (blue), G (black). (C) Interleukin-3-dependent Ba/F3 cells were infected with murine retrovirus expressing mutated TNK2 constructs. TNK2 wild-type and empty vector expressing Ba/F3s in addition to uninfected parental Ba/F3 cells were used as controls. Both TNK2 mutations transformed cells to IL3-independent growth. (D) The TNK2 D163E mutation is hyperphosphorylated. 293T17 cells expressing TNK2 D163E and R806Q mutations were subjected to immunoblot analysis for TNK2, TNK2 pY284 and actin. Parental 293T17 cells as well as empty vector and TNK2 wild-type expressing cells were included as controls. (E) Ba/F3 cells pre and post-interleukin-3-independent transformation were treated with varying concentrations of dasatinib. Both post-transformation lines were sensitive to dasatinib at low nanomolar concentrations. The pre-IL3 transformation cell lines did not show sensitivity to dasatinib.
Prioritized TNK2 mutations are highly oncogenic
To determine whether or not the prioritized mutations were oncogenic, we tested them in a cytokine independent growth assay. The murine pro-B Ba/F3 cell line normally requires the cytokine IL3 for growth, but can grow in the absence of IL3 if a transforming oncogene is expressed. To test the oncogenic capacity of the EPHA8 and TNK2 mutations we introduced wild type and mutant versions of EPHA8 and TNK2 into Ba/F3 cells and withdrew IL3. The EPHA8 mutation exhibited no transforming capacity in this assay (data not shown), while the TNK2 D163E mutation allowed the cells to grow in the absence of IL3 (Figure 2C). Based on the oncogenic capacity of this novel TNK2 mutation, we looked for other patient samples in which a TNK2 mutation had been highly ranked by HitWalker. In a CMML patient sample (10-210), a TNK2 R806Q mutation was also highly prioritized (Supplemental Figure 1 and Figure 2A, B). This mutation also transformed Ba/F3 cells to cytokine independent growth. The D163E mutation occurs in the tyrosine kinase domain of TNK2, while that R806Q mutation is in the EGFR binding domain (EBD) of TNK2 (Figure 2A). Analysis of these TNK2 mutations by western blot revealed that the TNK2 D163E mutation results in increased phosphorylation of TNK2, while the R806Q mutation does not. This suggests that the D163E mutation may increase kinase activity of TNK2, while the R806Q mutation may lead to increased signaling through alternative mechanisms.
Structural consequences of the D163E and R806Q mutations
From a structural perspective the two activating mutations of TNK2, D163E and R806Q, are rather conservative and do not drastically change the electrostatic properties of the TNK2 domain in which they are located. Based on available structural data we tried to infer a possible mechanism as to how the mutants could influence kinase activity. For TNK2, structural data is available for the kinase domain in its active conformation (in the presence and absence of ATP-pocket ligands) as well as in its inactive conformation as part of a kinase-SH3 domain construct. Based on these structural studies, homodimerization has been proposed to allosterically activate TNK2 kinase activity similar to EGFR (20). The latter forms an asymmetric head-to-tail dimer with the N-lobe of one molecule interacting with the C-lobe of the second molecule (21). Another example of kinase domain dimerization is BRAF that forms a symmetric side-to-side dimer involving primarily residues of the N-lobe (22). In contrast to EGFR and BRAF, the structures of the TNK2 active state show a head-to-head symmetric dimer involving residues in proximity to the catalytically active αC-helix in the N-lobe (20).
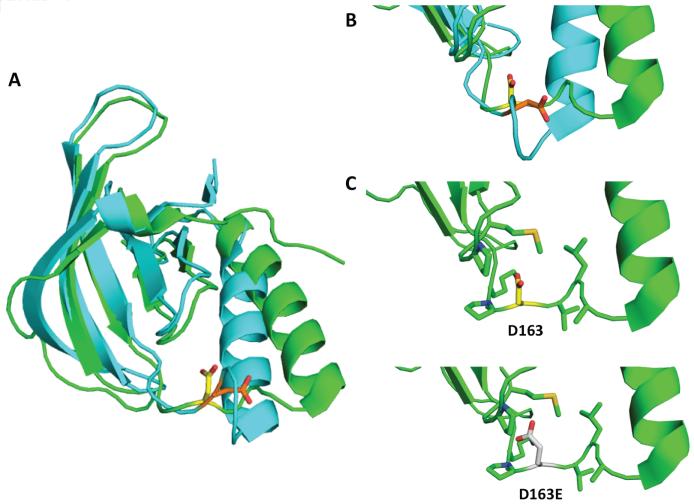
One of the two activating mutations, the D163E, resides in the N-lobe of the kinase domain and therefore existing structural data could be employed to build a hypothesis explaining the activating effect of the D to E mutation. The D163 residue is located in the loop connecting the β3 strand with the αC-helix (Figure 3A). This loop is not present in some of the structures deposited in the PDB database, which might reflect a certain degree of flexibility. Nonetheless, PDB entry 4ewh presents the active state of the kinase including this loop region (23), in which the side chain of D163 is solvent exposed and may be involved in the dimerization interface (Figure 3A). In the interface, however, no potentially interacting residue, that would engage the aspartate carboxyl group in either an ionic interaction (Lys/Arg) or a hydrogen bond (Ser/Thr), could be found in the near proximity (<5Å).
Figure 3. Structure of the kinase domain of TNK2 in the active and inactive state and position of D163.
(A) Structures of the N-lobe of the kinase domain in its active (PDB ID 4ewh, cyan) and inactive conformation (PDB ID 4hzs, green) are aligned and shown as cartoon representation. The side chain of D163 is presented in sticks illustrating the different orientation in the active (orange) and inactive state (yellow). A closer look of D163 is given in (B). (C) In the inactive conformation the D163 residue points to the inside of the N-lobe and the side chains of residues in proximity of 5Å are visualized as sticks. (D) Mutation of D163 into a glutamate prolongs the side chain by a methylene group that in the inactive conformation cannot easily accommodated in this inward-oriented position due to the proximity to P199 and M201.
In a different approach, comparison of the active and inactive conformation of the TNK2 kinase domain reveals the classical rearrangements within the N-lobe (Figure 3B) (20,23). Specifically, in the inactive state, the αC-helix undergoes an outward movement and also the N-terminal β-sheet is repositioned. These conformational changes also influence the position and orientation of the β3-αC loop that hosts the D163 residue. While in the active conformation the side chain of D163 points to the outside, it is buried between the C-terminal β-sheet and the αC-helix of the N-lobe in the inactive conformation (Figure 3B). Similar to the active state situation, there is no functional group in proximity of 5Å of D163 that would lock the residue in this position in the inactive state (Figure 3C). However, one could well imagine that upon a D163E mutation, the longer and thus bulkier glutamate side chain causes steric clashes within the core of the N-lobe (Figure 3D). As a consequence the β3-αC loop would undergo a conformational switch to a more outward-oriented position of the D163E residue thereby stabilizing the αC-helix in the active conformation.
A closer look at the sequence of this region of TNK1 and TNK2 across different species reveals additional interesting observations strengthening our above made hypothesis. While in TNK2, the KPD(161-163) segment in the β3-αC loop is only conserved in mouse and human, TNK1 possesses a Glu at position of D163. This glutamate, however, is either accommodated in a longer β3-αC loop or preceded by less bulky residues such as glycine. Furthermore, in other cytoplasmic tyrosine kinases this motif in the β3-αC loop is often composed of a KP[S/G] sequence which is also less bulky as compared to KP[D/E].
The importance of this β3-αC loop for the regulation of kinase activity is further corroborated by reports about other tyrosine kinases whose activity was influenced upon mutations in this region of the kinase domain. For example, targeted mutations in this region of SRC have shown to be activating (24,25) whereas alanine scanning in this loop of CSK have led to a loss of kinase activity due to disruption of the kinase-SH2 interface (25,26). Also for ABL kinase mutation of residue E275 has been shown to influence kinase activity (27).
In contrast to D163, the second mutation R806Q lies in a region of TNK2 that is structurally undefined. Based on the primary sequence this region has been identified as the EGFR binding domain (EBD) also known as MIG6 homology region (MHR). Secondary structure prediction by PsiPred (http://bioinf.cs.ucl.ac.uk/psipred/) indicates that this region is very likely to be unfolded. As the name implies and further shown by sequence alignments, this stretch of TNK2 possesses high sequence identity to a part of the MIG6 protein that regulates EGFR kinase activity upon interaction with its kinase domain (Supplementary Figure 2). Structural studies on a complex between EGFR and a MIG6 peptide have been published by Kuriyan and coworkers (28). MIG6 binds to the C-lobe of the kinase domain with one segment and likely extends towards the kinase active site with a segment C-terminal to the first one leading to kinase inhibition. Within the same publication the authors also speculate on a possible function of the EBD of TNK2 (28). Not only the EBD is highly homologous to MIG6 but also the C-lobes of the kinase domains of EGFR and TNK2 are very similar. This led to the hypothesis that the EBD of TNK2 could play a role in an autoinhibition mechanism of TNK2. The R806Q mutation lies in the segment of the EBD that would by homology bind to the C-lobe of the kinase (Figure 2) and this interaction is required as a docking interaction so that the second segment can be engaged in further interactions with either the activation loop or residues from the N-lobe of the kinase. If now this mutation weakens the binding to the kinase C-lobe, the autoinhibition of TNK2 would be lost resulting in an active kinase.
TNK2 mutations confer dasatinib sensitivity
In the TNK2 mutant leukemia samples, TNK2 was prioritized partly due to sensitivity of these samples to the multi-kinase inhibitor dasatinib (Figure 1A). Dasatinib has been shown to exhibit binding to TNK2 in the low nanomolar range (29), suggesting that dasatinib sensitivity in these cases could be mediated through targeting of TNK2. To test whether the TNK2 mutations directly confer sensitivity to dasatinib we treated the Ba/F3 cell lines expressing the TNK2 mutants with dasatinib and evaluated effects on cell growth and viability. Control or mutant cell lines grown in IL3 were completely insensitive to dasatinib, because the cytokine support renders their survival independent of TNK2. In contrast, both TNK2 mutant cell lines growing in the absence of IL3 were highly sensitive to dasatinib with IC50s of less than 10 nM.
Novel TNK2 Inhibitors
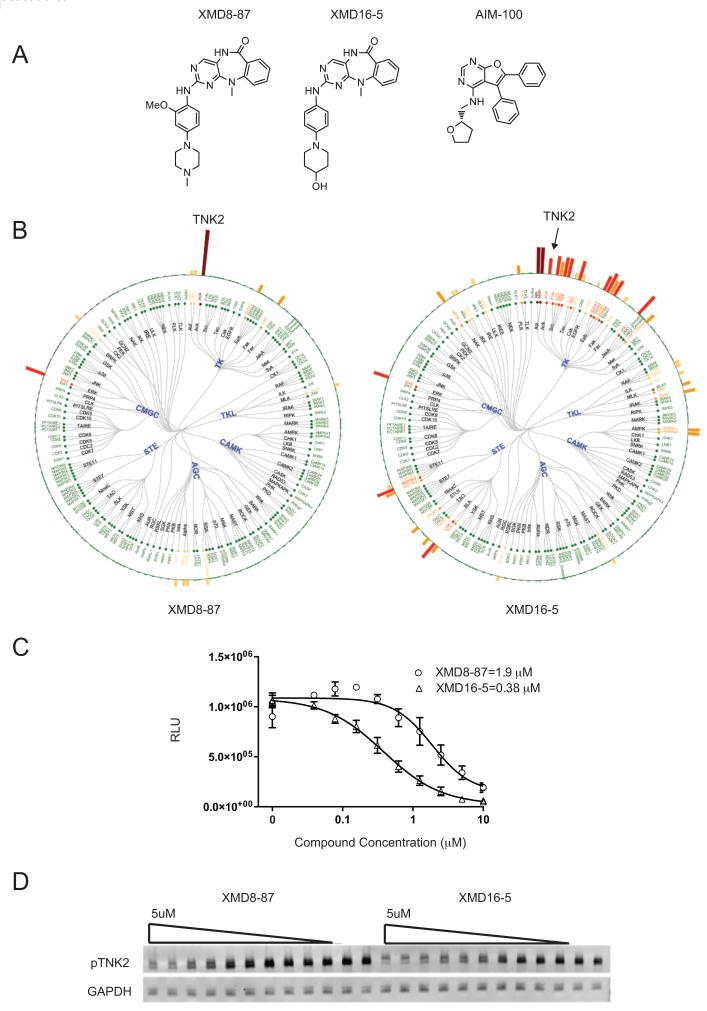
Despite dasatinib’s ability to potently inhibit TNK2, its promiscuity as a kinase inhibitor (30) makes it difficult to conclude that its cellular potency is derived primarily from TNK2 inhibition. Therefore we sought to identify structurally distinct TNK2 inhibitors with improved kinase selectivity that could serve as more precise mechanistic probes. We searched our in-house database for inhibitors that exhibited potent and selective binding to TNK2, which resulted in the identification of XMD8-87 (31) and XMD16-5 (32) (Figure 4A). The kinase selectivities of these two compounds were evaluated by screening against a diverse panel of 241 kinases using a chemical proteomic approach, KiNativ (33) (Figure 4B). The selectivity of XMD8-87 was also assessed using KinomeScan™ (29) (Ambit Biosciences, San Diego, CA) across a panel of 402 human kinases at concentration of 10 μM (see Supporting Information for complete profiling data). The top hits from both screening tests (kinases exhibit binding inhibition of 80% or more in Kinativ or 90% or more in KinomeScan) were further investigated by enzymatic kinase assay (SelectScreen Kinase Profiling Services) (Supplemental Table 1). This analysis revealed that both of these compounds are much more selective inhibitors of TNK2 than dasatinib. Activity of XMD8-87 and XMD16-5 against TNK2 was confirmed by ELISA (Figure 4C). To confirm that these compounds could inhibit TNK2 in a cellular context, we evaluated their ability to inhibit TNK2 autophosphorylation by immunoblot analysis in 293T cells (Figure 4D). Both compounds were found to be potent in the inhibition of TNK2 phosphorylation.
Figure 4. Novel TNK2 inhibitors.
(A) Structures of TNK2 inhibitors XMD8-87, XMD16-5 and AIM-100. (B) Selectivity of XMD8-87 and XMD16-5 were analyzed by KiNativ. Percent inhibitions were represented by bar height. Brown bar indicates >90% inhibition; red bar, 75%–90% inhibition; orange highlight, 50%–75% inhibition; and yellow bar, 35-50%. (C) ELISA of TNK2 inhibition by XMD8-87 and XMD16-5. Assay was performed as described in the methods with drug concentrations between 0 and 10 uM. (D) Inhibition of TNK2 phosphorylation was measured by western blot analysis. Cells were then treated with XMD8-87 or XMD16-5 at 5 μM and with 9 1:1 serial dilutions down to ≈10 nM. Two additional samples were treated with DMSO only. GAPDH is used as a loading control.
Selective TNK2 inhibitors potently block proliferation of TNK2 mutant cell lines
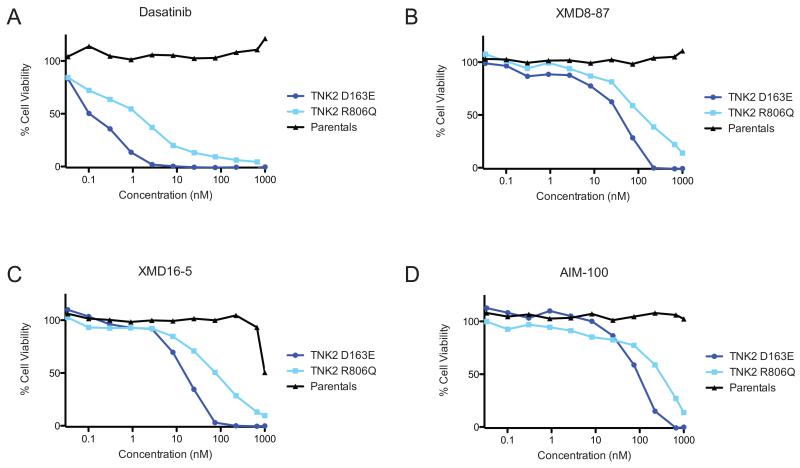
Dasatinib is a potent multi-kinase inhibitor and has many targets other than TNK2, such as ABL and SRC kinases. As such, we next wanted to test whether inhibitors with greater specificity to TNK2 are also effective against Ba/F3 cells that are dependent on mutant TNK2. In addition, to XMD8-87 and XMD16-5, we also evaluated a structurally distinct third previously reported TNK2 inhibitor, AIM-100 (15). All three of the inhibitors exhibited potent inhibition of growth of the TNK2 mutant expressing cell lines while having little or no effect on the control cells out to the highest tested concentrations (1,000 nM) (Figure 4B-D). The compound XMD8-87 had IC50s of 38 nM and 113 nM for the D163E and R806Q mutations (Figure 4B), and the compound XMD16-5 had IC50s of 16 nM and 77 nM for the D163E and R806Q mutations (Figure 4C). Both compounds show lower IC50 compared with the previously reported TNK2 inhibitor AIM-10021, which had IC50s of 91 nM for the D163E cells and 320 nM for the R806Q cells (Figure 4D).
Mutations of gatekeeper residue inhibit drug sensitivity
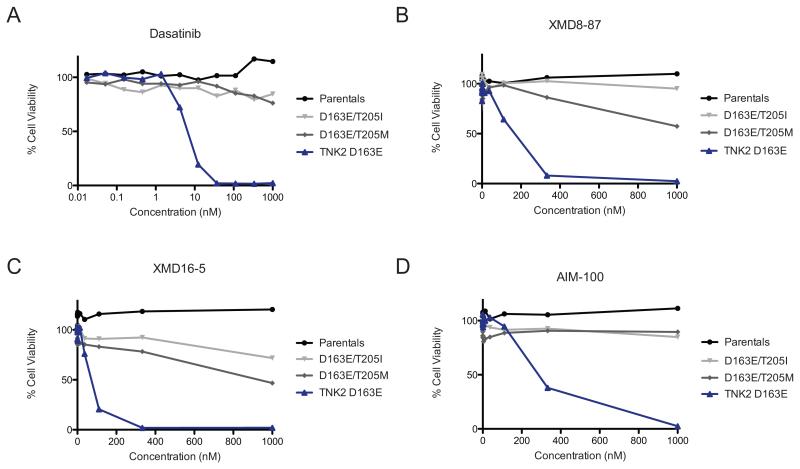
In order to confirm that the death of cells harboring TNK2 mutations was specifically due to inhibition of TNK2 we designed potentially inhibitor resistant alleles by mutating the ‘gatekeeper’ threonine (T205) to a methionine (T205M) or an isoleucine (T205I). This residue was selected for mutation as it is well characterized as a hot-spot that can confer resistance in many of the therapeutically important kinases such as BCR-ABL, c-KIT, PDGFR and EGFR (34-37). In addition, from crystallography and molecular modeling, dasatinib, XMD8-87 and XMD16-5 are all predicted to engage in hydrogen bonding interaction with the hydroxyl of T205. We engineered these gatekeeper mutations in the context of the most potent oncogenic of the TNK2 mutations, D163E. We then generated IL3 independent Ba/F3 lines and tested the cell lines for sensitivity to dasatinib, AIM-100, XMD8-87 and XMD16-5 (Figure 5). The gatekeeper mutations conferred near complete resistance of the D163E mutant cell lines to all of the compounds tested, indicating that the effects of these inhibitors on TNK2 cell lines are largely due to on-target effects on TNK2. It is important to note, however, that the gatekeeper mutations are in and of themselves activating, and thus increased growth rate of the cells could affect drug sensitivity.
Figure 5. TNK2 inhibitors block proliferation of TNK2 mutant cell lines.
TNK2 D163E or R806Q IL3-independent cell lines were tested against TNK2 inhibitors. Parental Ba/F3 cells grown in IL3 containing medium were used as a control. Percent viability as compared to untreated cells was determined using a tetrazolamine based viability assay. (A) Dasatinib inhibits TNK2 D163E cells with an IC50 of approximately 0.1 nM and TNK2 R806Q cells with an IC50 of 1.3 nM. (B) XMD8-87 inhibits TNK2 D163E cells with an IC50 of 38 nM and TNK2 R806Q cells with an IC50 of 113 nM. None of the inhibitors reached an IC50 in the parental Ba/F3 cells. (C) XMD16-5 inhibits TNK2 D163E cells with an IC50 of 16 nM and TNK2 R806Q cells with an IC50 of 77 nM. (D) AIM-100 inhibits TNK2 D163E cells with an IC50 of 91 nM and TNK2 R806Q cells with an IC50 of 320 nM.
TNK2 mutations in other tumor types are sensitive to the novel TNK2 inhibitors
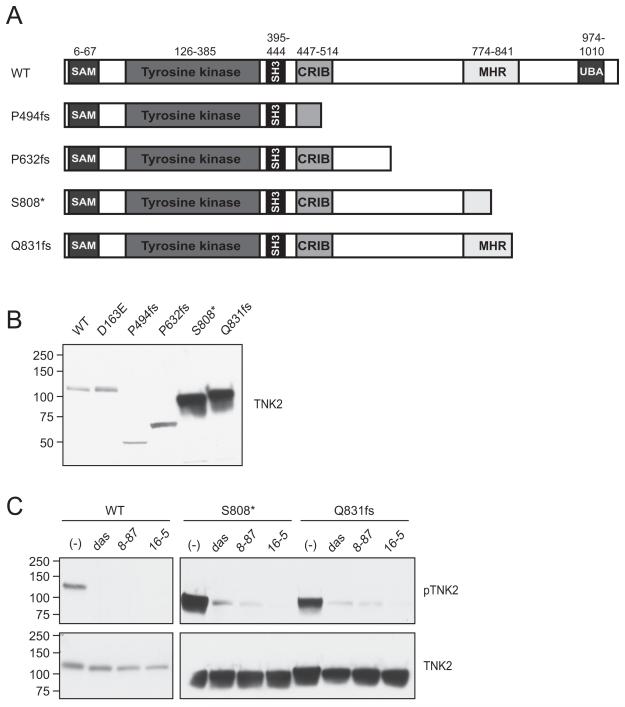
Analysis of the TCGA data using cBioPortal(38) revealed that many tumor types harbor TNK2 mutations. Of particular interest, were several recurrent truncation mutations were found in TNK2. Since these truncation mutations resulted in complete or partial loss of the portion of TNK2 that is homologous to MIG6, which in addition to binding EGFR also participates in negative regulation of the kinase domain, we hypothesized that they could be activating. We generated 4 of these truncation mutations: P494fs (found in several cell lines in the Cancer Cell Line Encyclopedia (39) (CCLE), P632fs (CCLE, glioblastoma, pancreatic, and stomach cancers), S808* (bladder cancer) and Q831fs (stomach cancer) (Figure 7A). Of particular interest the S808* and Q831fs mutations were markedly overexpressed when expressed in 293T17 cells (Figure 7B) with a corresponding increase in phosphorylation (Figure 7C). To determine if auto-phosphorylation of these overexpressed TNK2 mutants could be blocked with TNK2 inhibitors we treated the cells with dasatinib, XMD8-87 or XMD16-5 and indeed found that the vast majority of phosphorylation was blocked. This data suggests that these novel TNK2 inhibitors may be useful therapeutically in other tumor types harboring activating mutations in TNK2.
Figure 7. TNK2 truncation mutations found in solid tumors are sensitive to TNK2 inhibitors.
(A) Schematic of a subset of TNK2 truncation mutations found in solid tumors. (B) TNK2 S808* and Q831fs mutations result in protein overexpression. TNK2 mutations were transiently transfected into 293T17 cells and then total TNK2 levels were measured using an antibody to the TNK2 N-terminus. The TNK2 S808* and Q831fs were noticeably overexpressed compared to WT TNK2 or the other TNK2 truncations. (C) Phosphorylation of TNK2 mutants is inhibited by dasatinib, XMD8-87 and XMD16-5. 293T17 cells were transiently transfected with WT TNK2, TNK2 S808* or TNK2 Q831fs. After 48 hours the cells were treated with 1 μM dasatinib (das), 2.5 μM XMD8-87 or 2.5 μM XMD16-5 for 3.5 hours, and then harvested and subjected to immunoblot analysis for phosphorylated TNK2 (pTNK2) or total N-terminal TNK2. Drug concentrations were chosen based on the inhibition of WT TNK2 phosphorylation shown in figure 4D. Blots for WT and mutant TNK2 were run simultaneously, under the same conditions and exposed for identical time. These experiments were repeated with similar results.
DISCUSSION
In this study we have identified two oncogenic mutations in TNK2 from leukemia patient samples using the HitWalker algorithm, which uses functional data to prioritize mutations that may be functionally relevant in the patient sample. This method allowed us to rapidly prioritize TNK2 as a novel therapeutic target in leukemia. It also has the advantage of allowing for the identification of mutations that are of functional importance, but may be overlooked due to their rarity in genomic data alone. The first mutation, D163E is located in the tyrosine kinase domain of TNK2. A previous report also identified a mutation in the kinase domain (E346K) from a patient with ovarian endometroid carcinoma (9). The E346K mutation had the highest levels of phosphorylation at tyrosine 284, of all the mutations screened in that study (9). This is consistent with our finding that the D163E mutant, also in the kinase domain, results in high levels of TNK2 phosphorylation (Figure 2D). Interestingly, although the D163E and E346K mutations are in different lobes of the kinase domain they result in similar phenotypes. Molecular modeling suggests that although a fairly conservative substitution, the D163E mutation may stabilize the αC-helix of the N-lobe in an active conformation (Figure 3).
The second mutation is present in the Mig6 homology domain (MHC, aka EGFR binding domain, EBD) of TNK2. This mutation R806Q, is very close to a synthetic mutation F820A designed to block interaction of the MHD with the kinase domain (9). This interaction of the kinase domain with the MHD is thought to inhibit the kinase activity of TNK2 (9). Interestingly, the F820A mutation was hyperphosphorylated (9), while the R806Q mutation is not (Figure 2D), suggesting that different regions of the MHD may have different functional importance. The R806Q mutation resides within a segment that is highly homologous to the C-lobe binding peptide of Mig6, and therefore may be involved in kinase auto-inhibition. In summary, based on TNK2 kinase domain structures as well as sequence alignments and homology, it is possible to derive hypotheses giving a possible explanation for the two activating mutations in this kinase. To further elucidate the molecular mechanism of kinase activation and a possible autoinhibition mechanism of the MHD domain further experimental evidence is needed.
Mutated TNK2 may also represent a therapeutic target in other malignancies. In this study we identified two truncation mutations found in solid tumors that led to receptor overexpression and a correspondingly high level of phosphorylation (Figure 7). Interestingly these two mutations both lead to a loss of a portion of the MHD, which may negatively regulate kinase activity, but also a loss of the ubiquitin association domain (UBD) (Figure 7A). The UBD is the region that interacts with ubiquitin ligases, leading to protein degradation and turnover, and therefore the loss of the UBD could contribute to protein overexpression (40).
Dasatinib is a multi-targeted kinase inhibitor initially developed against ABL and SRC kinases (30). Dasatinib sensitivity in samples from patients with TNK2 mutations (Figure 1B) could result from either direct inhibition of TNK2 kinase activity itself, or indirectly through inhibition of SRC kinases by dasatinib, since SRC can activate TNK2 by directly phosphorylating the TNK2 activation loop (7). Alternatively any number of other targets of dasatinib could be relevant. Dasatinib has been shown to exhibit low nanomolar binding affinity for both TNK2 and SRC with Kds of 5.6 and 0.5 nM respectively (30). Other TNK2 inhibitors, such as AIM-100 have been shown to be effective in cell culture models of prostate cancer (15).
In this study we newly describe two kinase inhibitors that exhibit greater selectivity for TNK2 compared with dasatinib. XMD8-87 in particular demonstrated a high degree of selectivity for TNK2 (Figure 4B), and is also more potent than the previously reported TNK2 inhibitor, AIM-100 (Figure 4). Furthermore, XMD8-87 and XMD16-5.potently inhibit phosphorylation of TNK2 truncation mutations found in solid tumor types (Figure 7C). These compounds represent promising new lead chemical candidates for further development of clinically applicable, selective TNK2 inhibitors across multiple tumor types.
Here we prioritized TNK2 mutations as important functional targets using the HitWalker algorithm (1). This highlights the utility of the HitWalker algorithm in identifying actionable genomic lesions, including those that are infrequently mutated and may otherwise be overlooked in the analysis of genomic data alone. It is important to note that other genes not sequenced as part of the panel of kinase associated genes used in this study, could also contribute to the kinase sensitivity observed in these samples, and it will therefore be important in the future to perform a more comprehensive genomic analysis through whole exome sequencing. Given the large number of mutations present in a wide variety of tumors, the ability to prioritize genetic lesions greatly reduces the time and resources necessary to validate candidate mutations. Furthermore, this study highlights the utility of drug screening data for understanding the underlying vulnerabilities of leukemia cells and their accompanying gene mutations.
Supplementary Material
Figure 6. Mutations of gatekeeper residue blocks sensitivity to TNK2 inhibitors.
Addition of the T205I and T205M gatekeeper mutations to the D163E TNK2 mutations causes resistance to Dasatinib (A), XMD8-87 (B), XMD16-5 (C) and AIM-100 (D). Inhibitor studies were performed on IL-3 independent Ba/F3 lines as described in Figure 5. Parental Ba/F3 cells grown in IL3 containing medium were used as a control.
Acknowledgments
Financial Support: This work was supported by Howard Hughes Medical Institute and The Leukemia & Lymphoma Society. J.M. is supported by a Leukemia & Lymphoma Society Fellow Award and a Medical Research Foundation Early Clinical Investigator Award, and NCI K99 CA190605-01. J.W.T. is supported by grants from the V Foundation for Cancer Research, the Gabrielle’s Angel Foundation for Cancer Research, and the National Cancer Institute (5R00CA151457-04; 1R01CA183947-01). N.S.G, X.M.D and J.W. are supported by NIH grant (5 R01 CA173469-02).
Footnotes
Conflicts of Interest: JEM, MLA, SBL, HS, JG, REM, OH, SR, and NSG have no competing financial interests to disclose. JWT receives research funding from Array BioPharma, Constellation Pharmaceuticals, Aptose Biosciences, and Janssen Pharmaceutical Companies. Dr. Druker is or has been within the last year a consultant to Gilead Sciences, AstraZeneca, Cylene Pharmaceuticals, Aptose Biosciences, and Roche TCRC. He has equity interest in Gilead, Cylene, and Aptose. Dr. Druker is currently principal investigator or co-investigator on Novartis, Bristol-Myers Squibb, and ARIAD clinical trials. His institution has contracts with these companies to pay for patient costs, nurse and data manager salaries, and institutional overhead. He does not derive salary, nor does his lab receive funds from these contracts. OHSU and Dr. Druker have a financial interest in MolecularMD. This potential individual and institutional conflict of interest has been reviewed and managed by OHSU.
REFERENCES
- 1.Bottomly D, Wilmot B, Tyner JW, Eide CA, Loriaux MM, Druker BJ, et al. HitWalker: variant prioritization for personalized functional cancer genomics. Bioinformatics. 2013;29:509–10. doi: 10.1093/bioinformatics/btt003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyner JW, Yang WF, Bankhead A, 3rd, Fan G, Fletcher LB, Bryant J, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer research. 2013;73:285–96. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prieto-Echague V, Miller WT. Regulation of ack-family nonreceptor tyrosine kinases. Journal of signal transduction. 2011;2011:742372. doi: 10.1155/2011/742372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manser E, Leung T, Salihuddin H, Tan L, Lim L. A non-receptor tyrosine kinase that inhibits the GTPase activity of p21cdc42. Nature. 1993;363:364–7. doi: 10.1038/363364a0. [DOI] [PubMed] [Google Scholar]
- 5.Modzelewska K, Newman LP, Desai R, Keely PJ. Ack1 mediates Cdc42-dependent cell migration and signaling to p130Cas. The Journal of biological chemistry. 2006;281:37527–35. doi: 10.1074/jbc.M604342200. [DOI] [PubMed] [Google Scholar]
- 6.Galisteo ML, Yang Y, Urena J, Schlessinger J. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9796–801. doi: 10.1073/pnas.0603714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan W, Sit ST, Manser E. The Cdc42-associated kinase ACK1 is not autoinhibited but requires SRC for activation. The Biochemical journal. 2011;435:355–64. doi: 10.1042/BJ20102156. [DOI] [PubMed] [Google Scholar]
- 8.Chua BT, Lim SJ, Tham SC, Poh WJ, Ullrich A. Somatic mutation in the ACK1 ubiquitin association domain enhances oncogenic signaling through EGFR regulation in renal cancer derived cells. Mol Oncol. 2010;4:323–34. doi: 10.1016/j.molonc.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prieto-Echague V, Gucwa A, Craddock BP, Brown DA, Miller WT. Cancer-associated mutations activate the nonreceptor tyrosine kinase Ack1. The Journal of biological chemistry. 2010;285:10605–15. doi: 10.1074/jbc.M109.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Horst EH, Degenhardt YY, Strelow A, Slavin A, Chinn L, Orf J, et al. Metastatic properties and genomic amplification of the tyrosine kinase gene ACK1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15901–6. doi: 10.1073/pnas.0508014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahajan NP, Liu Y, Majumder S, Warren MR, Parker CE, Mohler JL, et al. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8438–43. doi: 10.1073/pnas.0700420104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grovdal LM, Johannessen LE, Rodland MS, Madshus IH, Stang E. Dysregulation of Ack1 inhibits down-regulation of the EGF receptor. Experimental cell research. 2008;314:1292–300. doi: 10.1016/j.yexcr.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Wang L, Zhu JS, Song MQ, Chen GQ, Chen JL. Comparison of gene expression profiles between primary tumor and metastatic lesions in gastric cancer patients using laser microdissection and cDNA microarray. World J Gastroenterol. 2006;12:6949–54. doi: 10.3748/wjg.v12.i43.6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahajan K, Coppola D, Challa S, Fang B, Chen YA, Zhu W, et al. Ack1 mediated AKT/PKB tyrosine 176 phosphorylation regulates its activation. PLoS One. 2010;5:e9646. doi: 10.1371/journal.pone.0009646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahajan K, Challa S, Coppola D, Lawrence H, Luo Y, Gevariya H, et al. Effect of Ack1 tyrosine kinase inhibitor on ligand-independent androgen receptor activity. The Prostate. 2010;70:1274–85. doi: 10.1002/pros.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer research. 2005;65:10514–23. doi: 10.1158/0008-5472.CAN-05-1127. [DOI] [PubMed] [Google Scholar]
- 17.Maxson JE, Gotlib J, Pollyea DA, Fleischman AG, Agarwal A, Eide CA, et al. Oncogenic CSF3R mutations in chronic neutrophilic leukemia and atypical CML. The New England journal of medicine. 2013;368:1781–90. doi: 10.1056/NEJMoa1214514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chemistry & biology. 2011;18:699–710. doi: 10.1016/j.chembiol.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahajan K, Coppola D, Rawal B, Chen YA, Lawrence HR, Engelman RW, et al. Ack1-mediated androgen receptor phosphorylation modulates radiation resistance in castration-resistant prostate cancer. The Journal of biological chemistry. 2012;287:22112–22. doi: 10.1074/jbc.M112.357384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gajiwala KS, Maegley K, Ferre R, He YA, Yu X. Ack1: activation and regulation by allostery. PloS one. 2013;8:e53994. doi: 10.1371/journal.pone.0053994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Endres NF, Engel K, Das R, Kovacs E, Kuriyan J. Regulation of the catalytic activity of the EGF receptor. Current opinion in structural biology. 2011;21:777–84. doi: 10.1016/j.sbi.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–5. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 23.Jiao X, Kopecky DJ, Liu J, Liu J, Jaen JC, Cardozo MG, et al. Synthesis and optimization of substituted furo[2,3-d]-pyrimidin-4-amines and 7H-pyrrolo[2,3-d]pyrimidin-4-amines as ACK1 inhibitors. Bioorganic & medicinal chemistry letters. 2012;22:6212–7. doi: 10.1016/j.bmcl.2012.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Gonfloni S, Williams JC, Hattula K, Weijland A, Wierenga RK, Superti-Furga G. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of SRC. The EMBO journal. 1997;16:7261–71. doi: 10.1093/emboj/16.24.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang K, Wang YH, Brown A, Sun G. Identification of N-terminal lobe motifs that determine the kinase activity of the catalytic domains and regulatory strategies of SRC and CSK protein tyrosine kinases. Journal of molecular biology. 2009;386:1066–77. doi: 10.1016/j.jmb.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mikkola ET, Gahmberg CG. Hydrophobic interaction between the SH2 domain and the kinase domain is required for the activation of CSK. Journal of molecular biology. 2010;399:618–27. doi: 10.1016/j.jmb.2010.04.045. [DOI] [PubMed] [Google Scholar]
- 27.Sherbenou DW, Hantschel O, Kaupe I, Willis S, Bumm T, Turaga LP, et al. BCR-ABL SH3-SH2 domain mutations in chronic myeloid leukemia patients on imatinib. Blood. 2010;116:3278–85. doi: 10.1182/blood-2008-10-183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–4. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, et al. Comprehensive analysis of kinase inhibitor selectivity. Nature biotechnology. 2011;29:1046–51. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 30.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual SRC/ABL kinase inhibitor with potent antitumor activity in preclinical assays. Journal of medicinal chemistry. 2004;47:6658–61. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 31.Kwiatkowski N, Deng X, Wang J, Tan L, Villa F, Santaguida S, et al. Selective aurora kinase inhibitors identified using a taxol-induced checkpoint sensitivity screen. ACS chemical biology. 2012;7:185–96. doi: 10.1021/cb200305u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miduturu CV, Deng X, Kwiatkowski N, Yang W, Brault L, Filippakopoulos P, et al. High-throughput kinase profiling: a more efficient approach toward the discovery of new kinase inhibitors. Chemistry & biology. 2011;18:868–79. doi: 10.1016/j.chembiol.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patricelli MP, Szardenings AK, Liyanage M, Nomanbhoy TK, Wu M, Weissig H, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry. 2007;46:350–8. doi: 10.1021/bi062142x. [DOI] [PubMed] [Google Scholar]
- 34.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 35.Tamborini E, Bonadiman L, Greco A, Albertini V, Negri T, Gronchi A, et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–9. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Cools J, Stover EH, Boulton CL, Gotlib J, Legare RD, Amaral SM, et al. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFRalpha-induced myeloproliferative disease. Cancer cell. 2003;3:459–69. doi: 10.1016/s1535-6108(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 38.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahajan K, Mahajan NP. ACK1/TNK2 tyrosine kinase: molecular signaling and evolving role in cancers. Oncogene. 2014 doi: 10.1038/onc.2014.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.