Abstract
Aggregation at the neuronal cell membrane’s lipid bilayer surface is implicated in amyloid-β (Aβ) toxicity associated with Alzheimer’s disease; however, structural and mechanistic insights into the process remain scarce. We have identified a conserved binding mode of Aβ40 on lipid bilayer surfaces with a conserved helix containing the self-recognition site (K16-E22).
Alzheimer’s disease (AD), one of the leading causes of death worldwide, is commonly considered a protein misfolding disease.1 While identified by the deposition of insoluble aggregates of amyloid-β (Aβ) peptides as plaques in the brains of diseased patients,2 it is presently thought that soluble, intermediate oligomers and the process of their interconversion are mostly responsible for neuronal death in AD.3 The interaction of these soluble Aβ species with the lipid bilayer of neurons is believed to be responsible for toxicity, through both the formation of ion-selective channels as well as more dramatic membrane permeabilization and disruption via a two-step mechanism.4 Preventing such toxic interactions and structural transitions could be an important strategy for blocking Aβ toxicity in AD. Unfortunately, high resolution mechanisms of peptide interaction with both itself and the cell membrane are unclear due, in large part, to the heterogeneous protein environments present during amyloid formation.5 To date, full length structural models of Aβ have only been generated for freshly dissolved species and the stable amyloid fibrils which result from aggregation.6–10 Greater understanding of in vitro intermediate structures adopted, specifically within the biological context of lipid bilayers, is necessary to fully elucidate the in vivo aggregation network and toxic mechanisms of Aβ.
High resolution structural insights into the membrane-associated Aβ species have been elusive to date. Aβ aggregation inherently generates a mixture of lowly populated, transient species which makes NMR the most amenable approach for characterization. NMR, however, is hindered by the large size of vesicle membrane models, which can broaden the lipid-bound protein signal beyond detection, and by the ability of membranes to accelerate the aggregation of Aβ, reducing the free monomer concentration and limiting its intensity while simultaneously generating solution NMR invisible proteinaceous species.11,12 It has been previously shown, however, that maintaining a static sample at lower temperature stabilizes the monomeric peptide13 while using 100 nm large unilamellar vesicles (LUVs) composed solely of zwitterionic phosphatidylcholine (PC), containing lipids limits membrane-mediated catalysis of amyloid formation.12 Herein, coupling these sample conditions with a suite of NMR experiments14,15 and other biophysical approaches, we are able to successfully investigate the effect of early membrane interactions on the structure of Aβ prior to its aggregation on the bilayer surface and identify a conserved, partially helical structure which is adopted upon binding to the lipid bilayer.
The interaction of Aβ40 on the bilayer surface was probed with LUVs composed entirely of dilauroyl phosphatidylcholine (DLPC), dioleoyl phosphatidylcholine (DOPC), or 1-palmitoyl-2-oleoyl phosphatidylcholine (POPC) (Fig. 1). All three lipids form a liquid crystalline, zwitterionic bilayer above 0 °C while having variations in their hydrophobic thickness.16,17 In so doing, we investigated the universality of binding events to fluid PC bilayers which have variations in degrees of unsaturation and hydrophobicity. Upon the addition of substoichiometric concentrations of LUVs to Aβ40, circular dichroism (CD) suggests that the majority of the peptide is disordered in the presence of all three different bilayer systems as has been previously identified.18 This predominantly disordered state remains stable for at least 24 h under quiescent conditions (Fig. S1). While the unchanged CD spectra show a bulk population which is unbound from the bilayer and disordered, observing the same conditions by 1H NMR reveals global broadening of all the peptide’s resonances following the addition of LUVs to the peptide (Fig. S2). Line broadening is suggestive of rapid exchange between free, NMR-visible peptide and some form(s) of lipid-associated, NMR-invisible peptide in the NMR time scale. The existence of this invisible state solely in the presence of LUVs indicates the formation of an Aβ40–bilayer complex and consequently suggests that there is a sub-population of the peptide which exists in a structured (or semi-structured), membrane-bound conformation which is overpowered by the bulk peptide’s random coil conformation in CD measurements.
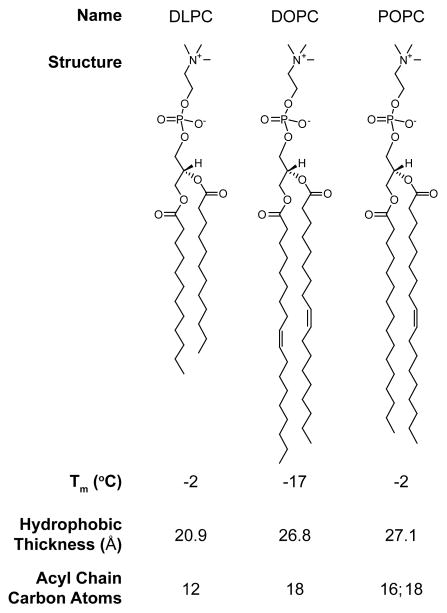
Figure 1.
Zwitterionic lipid structures. All lipids used in this study contain the zwitterionic phosphatidylcholine (PC) head group. They differ in their acyl chain length which determines the hydrophobic thickness (20.9–27.1 Å),16,17 but does not dramatically impact their transition temperature (Tm), ensuring that all bilayers are in the liquid crystalline phase under our experimental conditions.
Due to the evident fast exchange of Aβ40 between the free and membrane-associated states, transferred 1H–1H nuclear Overhauser effect spectroscopy (tr-NOESY) was applied to probe the conformational changes induced by bilayer interactions while filtering out structural information from the unbound population Aβ40.19–21 The tr-NOESY spectra of Aβ40 in the presence of all three vesicles present a large number of NOEs suggesting the existence of an at least partially folded structure when bound to each PC-containing LUV (Fig. 2). Similar NOEs originating from the peptide were observed with all three LUVs, implying that the partially folded conformation of Aβ40 bound to the lipid bilayer is conserved across DLPC, DOPC, and POPC bilayers (Fig. 3a). This demonstrates that initial interactions between Aβ40 and the lipid membrane may, therefore, be predominantly controlled by the surface characteristics of the membrane (charge and fluidity) rather than hydrophobic characteristics (acyl chain saturation and thickness). The N-terminal residues (D1-Q15) show few NOEs, signifying a greater degree of flexibility relative to the rest of the peptide for which additional NOEs are observable. It is likely, therefore, that the N-terminus of Aβ40 plays a minimal role in initial adsorption to the bilayer surface. Sequential NH–NH and Hα–NH NOEs are prominent throughout the central region (K16-G25) of the Aβ40 peptide, indicating enhanced rigidity and a propensity towards structure. This region has previously been found to adopt a partially folded structure in solution6 while also being proposed to instigate the formation of amyloid fibrils as a self-recognition sequence.1 A second string of sequential NH–NH and Hα–NH NOEs was seen for the more C-terminal hydrophobic region (G29-M35) which, similarly to the self-recognition sequence, has been implicated in fiber structure and formation.1 Along with these sequential NOEs, multiple Hα–Hβ i,i+4 NOEs were observed in the central sequence (L17-A21, V18-E22). These side chain NOEs are accompanied by an Hα–NH i,i+5 NOE between L17 and E22. This combination of i,i+4 i,i+5 NOEs is suggestive of the peptide folding into a loosely packed π-helix in the central region.
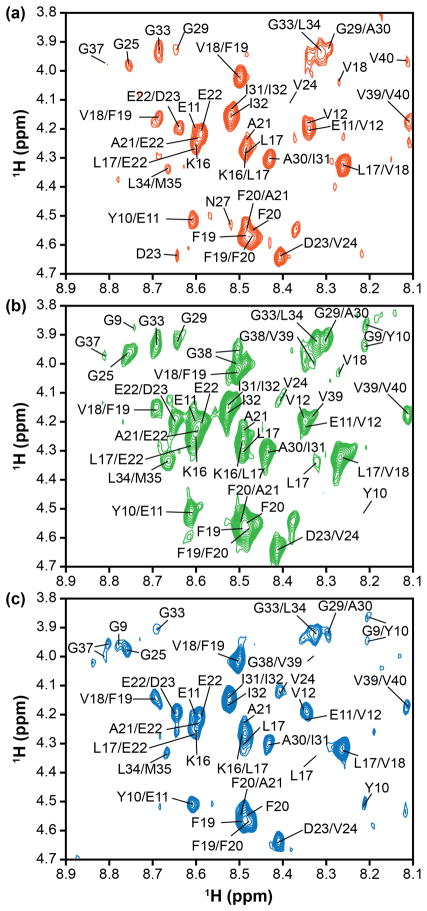
Figure 2.
Select regions of tr-NOESY spectra. The NH–Hα region of the tr-NOESY spectrum in the presence of LUVs (a) DLPC, (b) DOPC, or (c) POPC (Bruker 900 MHz spectrometer, equipped with cryoprobe at 10 °C). Moderately broad peaks hinder complete peak assignment, but suggest a modest exchange rate between the lipid-bound and -free forms of the peptide.
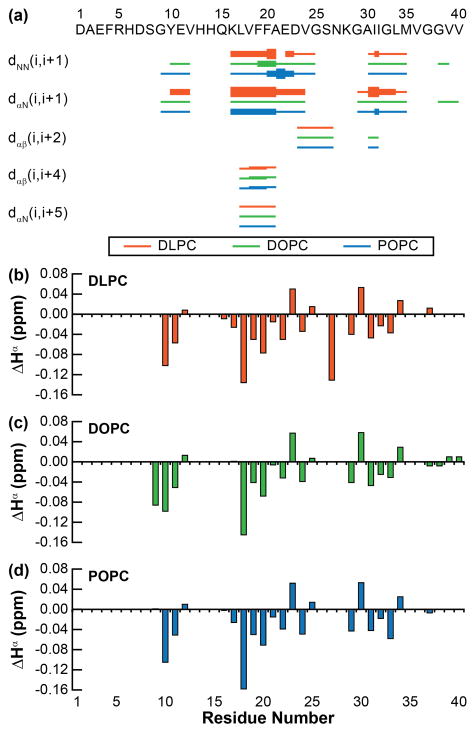
Figure 3.
tr-NOE derived structural constraints. (a) NOE connectivity plot shows a partially structured backbone conserved across all three bilayers investigated. The strengths of NOEs are indicated by the height of the bars, graded strong, medium, and weak. The Hα chemical shift for (b) DLPC, (c) DOPC, and (d) POPC was calculated for each resolved residue relative to the random coil chemical shift for each residue.
In conjunction with these long distance NOEs, the frequent negative ΔHα values (Fig. 3) are predictive of a helical propensity through the majority of the peptide (Fig. 3b–d). Sequential helical propensity is most pronounced between K16 and E22 region, which, along with the NOE connectivity observed above, further supporting that the central portion of Aβ40 adopts a helical conformation in the presence of all three PC bilayers upon binding. The presence of multiple, non-sequential negative ΔHα values in the more C-terminal region may demonstrate a propensity toward a more random coil and rigid membrane associated conformation. Finally, the N-terminus displays few resonances in the presence of all three bilayers, showing a lack of defined structure or rigidity.
In order to better define the boundaries of membrane-bound and -unbound regions of the peptide, a titration of paramagnetic MnCl2 was used to selectively quench the NMR signal from residues not associated with the hydrophobic lipid bilayer (Fig. S3). As was observed with tr-NOESY, all three lipid bilayers caused the Aβ40 peptide to react similarly in the presence of MnCl2. Substoichiometric concentrations of MnCl2 reduced the signal intensity of the N-terminus (D1-Q15) to ca. 50% of their original value while stoichiometric concentrations decreased the same regions to ca. 30% of their original intensity. The same concentrations of MnCl2 decreased the remainder of the peptide’s resonances to ca. 90% and 85%, respectively. This shows that the N-terminal residues, predicted to be flexible based on tr-NOESY signal, are, in fact, not bound to the bilayer surface and instead free and unstructured in solution. The remainder of the peptide is associated with the bilayer surface, though only adopting defined structure in selective hydrophobic regions identified by tr-NOESY (vide supra).
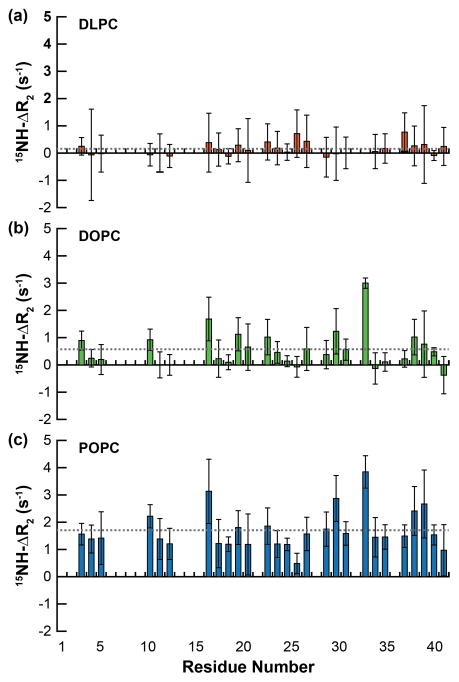
Given the two-state bilayer interaction (containing both a membrane-associated and -free regions), the relaxation and dynamics of the peptide in the presence of LUVs were measured; 15NH-ΔR2 values (the difference in 15NH-R2 in the presence of LUVs versus in the absence) measured the relative flexibility of the peptide backbone in the presence of PC bilayers. Unlike what was observed in the structural experiments above, the three lipids induced very different dynamic changes to the peptide backbone (Fig. 4). Instead of each bilayer selectively increasing the relaxation rate (R2) of the bound, structured region, there was no sequence specificity for the relaxation increases observed; instead the increases were sporadic throughout the peptide. Additionally, the average ΔR2 differs for each of the three lipid systems. This suggests that the relaxation changes are less due to specific structural alterations and are instead the result of differential binding propensities and partitioning of free versus bound peptide to the three different bilayers. Fluorescence polarization was used to measure the binding affinity (Kd) of the peptide for the three distinct LUVs (Fig. S4). There was an inverse correlation between bilayer thickness (Fig. 1) and the measured affinities (DLPC, 20.9 Å, Kd = 1188 ± 41 μM; DOPC, 26.8 Å, Kd = 801 ± 45 μM; DLPC, 27.1 Å, Kd = 366 ± 36 μM). An additional inverse correlation exists between the ΔR2 values and the binding affinity of Aβ40 for each bilayer (Fig. S5). Therefore, the increase in rigidity and subsequently accelerated relaxation rates observed for the peptide are the result of an increased population which binds to the bilayer surface, rather than a result of the structural changes induced by peptide binding. It is likely that the bound peptide is adopting the relaxation rate of the LUV itself and the higher the population bound with LUV the more the R2 value increases as the weighted average R2 value for all species of peptide shifts to faster rates.
Figure 4.
Peptide backbone dynamics in the presence of lipid bilayers. The ΔR2 value for each resolvable residue was calculated as the difference between the relaxation of Aβ40 in the presence LUVs containing either (a) DLPC, (b) DOPC, or (c) POPC and in the absence of lipid. The average ΔR2 of all residues of the peptide is represented by the dashed gray line.
It seems, therefore, that zwitterionic, liquid crystalline bilayers are capable of inducing a conserved fold in Aβ40, tuneable by small alterations in the bilayer structure (i.e., thickness and acyl chain saturation). In this common fold, the N-terminus is unstructured and unbound to membrane, while the more C-terminal region adopts a bound, more rigid coil structure and the central hydrophobic region become a more structured helix (Fig. 5). This general topology is similar to both a previously proposed structure of partially folded Aβ40 in solution6 and a model of Aβ40 bound to GM1 micelles.22 It is, then, possible that this conserved helical fold may extend beyond PC bilayers and be a more universal early step in Aβ40 folding and aggregation. General helical intermediates have previously been suggested, especially in the presence of heterogeneous bilayers, though the residue-specific topology is unclear in most cases.18,22,23 We believe that this helical, membrane bound intermediate represents the first step in membrane-mediated Aβ aggregate formation, preceding both transmembrane pore formation and surface-catalyzed fiber formation and subsequent membrane disruption (Fig. 5e).4 Our results suggest that the central region containing the self-recognition sequence of Aβ40 (K16-E22) commonly associated with the cross β-strand amyloid structure may also be essential for the formation of early helical intermediates.
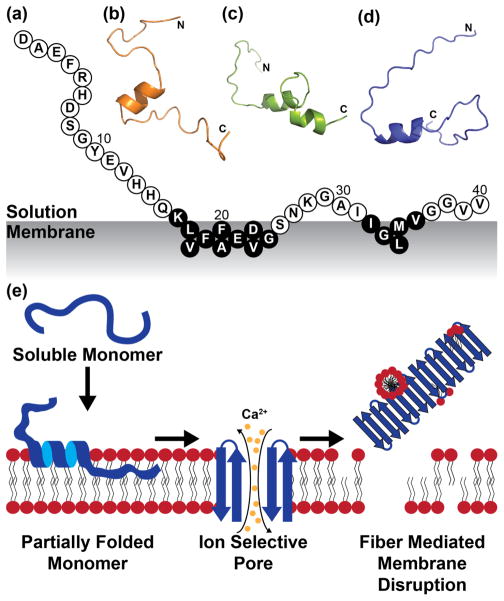
Figure 5.
Structural modelling of membrane-bound Aβ40. (a) Aβ40 adopts a conserved binding mode with PC lipid bilayers. Those residues in black circles are believed to have an especially high propensity for structure based on observed tr-NOEs. 3D cartoon models were generated based on tr-NOE and paramagnetic quenching constraints for (b) DLPC, (c) DOPC, and (d) POPC lipid bilayers. (e) The folding of Aβ40 on the bilayer into this conserved helix likely precedes the formation of other membrane-associated aggregate species related to membrane disruption.4
Supplementary Material
Acknowledgments
This study was supported by the Protein Folding Initiative at The University of Michigan (to A.R. and M.H.L.) and partly by funds from the NIH (to A.R.); the National Research Foundation of Korea (NRF) grant funded by the Korean government [NRF-2014S1A2A2028270 (to M.H.L. and A.R.). We thank Dr. Subramanian Vivekanandan for his assistance with acquiring NMR relaxation data.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
References
- 1.Savelieff MG, Lee S, Liu Y, Lim MH. ACS Chem Biol. 2013;8:856–865. doi: 10.1021/cb400080f. [DOI] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Cold Spring Harb Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 4.Sciacca MF, Kotler SA, Brender JR, Chen J, Lee DK, Ramamoorthy A. Biophys J. 2012;103:702–710. doi: 10.1016/j.bpj.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotler SA, Walsh P, Brender JR, Ramamoorthy A. Chem Soc Rev. 2014;43:6692–6700. doi: 10.1039/c3cs60431d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A. Biochem Biophys Res Commun. 2011;411:312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milojevic J, Esposito V, Das R, Melacini G. J Am Chem Soc. 2007;129:4282–4290. doi: 10.1021/ja067367+. [DOI] [PubMed] [Google Scholar]
- 8.Narayanan S, Reif B. Biochemistry. 2005;44:1444–1452. doi: 10.1021/bi048264b. [DOI] [PubMed] [Google Scholar]
- 9.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Cell. 2013;154:1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schutz AK, Vagt T, Huber M, Ovchinnikova OY, Cadalbert R, Wall J, Guntert P, Bockmann A, Glockshuber R, Meier BH. Angew Chem Int Ed Engl. 2015;54:331–335. doi: 10.1002/anie.201408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodner CR, Dobson CM, Bax A. J Mol Biol. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terakawa MS, Yagi H, Adachi M, Lee YH, Goto Y. J Biol Chem. 2015;290:815–826. doi: 10.1074/jbc.M114.592527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki Y, Brender JR, Soper MT, Krishnamoorthy J, Zhou Y, Ruotolo BT, Kotov NA, Ramamoorthy A, Marsh EN. Biochemistry. 2013;52:1903–1912. doi: 10.1021/bi400027y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karamanos TK, Kalverda AP, Thompson GS, Radford SE. Prog Nucl Magn Reson Spectrosc. 2015;88–89:86–104. doi: 10.1016/j.pnmrs.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthis NJ, Clore GM. Q Rev Biophys. 2015;48:35–116. doi: 10.1017/S0033583514000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kucerka N, Liu Y, Chu N, Petrache HI, Tristram-Nagle S, Nagle JF. Biophys J. 2005;88:2626–2637. doi: 10.1529/biophysj.104.056606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kucerka N, Tristram-Nagle S, Nagle JF. J Membr Biol. 2005;208:193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 18.Wong PT, Schauerte JA, Wisser KC, Ding H, Lee EL, Steel DG, Gafni A. J Mol Biol. 2009;386:81–96. doi: 10.1016/j.jmb.2008.11.060. [DOI] [PubMed] [Google Scholar]
- 19.Clore GM, Gronenborn AM. J Magn Reson. 1982;48:402–417. [Google Scholar]
- 20.Bhunia A, Mohanram H, Bhattacharjya S. Biopolymers. 2009;92:9–22. doi: 10.1002/bip.21104. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Jones JD, Rizo J, Gierasch LM. Biochemistry. 1993;32:13991–13999. doi: 10.1021/bi00213a032. [DOI] [PubMed] [Google Scholar]
- 22.Utsumi M, Yamaguchi Y, Sasakawa H, Yamamoto N, Yanagisawa K, Kato K. Glycoconj J. 2009;26:999–1006. doi: 10.1007/s10719-008-9216-7. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda K, Yamaguchi T, Fukunaga S, Hoshino M, Matsuzaki K. Biochemistry. 2011;50:6433–6440. doi: 10.1021/bi200771m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.