Abstract
The process of Epithelial-mesenchymal transition (EMT), in addition to being an initiating event for tumor metastasis, is implicated in conferring several clinically relevant properties to disseminating cancer cells. These include stem cell like properties, resistance to targeted therapies and ability to evade immune surveillance. Enrichment analysis of gene expression changes during TGF-β induced EMT in lung cancer cells identified complement cascade as one of the significantly enriched pathway. Further analysis of the genes in the complement pathway revealed an increase in the expression of complement inhibitors and a decrease in the expression of proteins essential for complement activity. In this study, we tested whether EMT confers resistance to complement-dependent cytotoxicity (CDC) in lung cancer cells and promotes tumor progression. CD59 is a potent inhibitor of membrane attack complex that mediates complement-dependent cell lysis. We observed a significant increase in the CD59 expression on the surface of cells after TGF-β-induced EMT. Furthermore, CD59 knock down restored susceptibility of cells undergoing EMT to Cetuximab-mediated CDC. TGF-β-induced CD59 expression during EMT is dependent on Smad3 but not Smad2. ChIP analysis confirmed that Smad3 directly binds to the CD59 promoter. Stable knock-down of CD59 in A549 cells inhibited experimental metastasis. These results demonstrate that TGF-β-induced EMT and CD59 expression confers an immune evasive mechanism to disseminating tumor cells facilitating tumor progression. Together, our data demonstrates that CD59 inhibition may serve as an adjuvant to enhance the efficacy of antibody-mediated therapies, as well as to inhibit metastasis in lung cancer.
Keywords: Complement regulatory proteins, CD59, metastasis, immune evasion, lung cancer
INTRODUCTION
The ability of cancer cells to disseminate from the primary tumor to distant sites through the complex process of metastasis is responsible for more than ninety percent of cancer-related mortality. Epithelial-mesenchymal transition (EMT) is a reversible cellular process by which epithelial cancer cells trans-differentiate into highly mobile and invasive mesenchymal-like cells, giving rise to disseminating or circulating tumor cells to initiate tumor metastasis (1,2). EMT involves dissolution of cell-cell adhesions, down-regulation of epithelial markers, induction of mesenchymal markers, and breach of the basement membrane to invade surrounding tissue (3,4). In addition to conferring an invasive phenotype, EMT also endows cancer cells stem cell-like properties, resistance to targeted therapies and the ability to evade host immune surveillance (5). Together, these properties enable cancer cells to successfully disseminate to distant sites and illustrate the biological relevance of EMT in tumor progression.
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that acts as a tumor suppressor in early stages, but promotes tumor progression in late stages through multiple mechanisms, including inducing EMT in cancer cells (6). Consistent with these tumor-promoting functions, expression of TGF-β is frequently up-regulated in non-small cell lung cancer (NSCLC) along with many other human cancers (7), and is correlated with enhanced invasion, metastasis and poor prognosis for patients with lung cancer (8). We have shown that TGF-β-induced EMT confers a migratory and invasive phenotype and promotes metastasis of lung cancer cells (9). Inhibition of EMT blocked ability of lung cancer cells to invade and metastasize (10,11). We also demonstrated that the EMT-associated secretory phenotype predicts survival of lung cancer patients (12). Here we demonstrate a new immune evasive mechanism conferred by TGF-β-induced EMT on lung cancer cells.
The complement pathway is recognized as a first line of defense in host immune surveillance against non-self microbial and tumor cells (13). Cancer cells, including that of lung, are known to develop multiple mechanisms to avoid complement mediated damage and impair the efficacy of monoclonal antibody-based anti-cancer therapies (14–16). This suggests a critical role for complement in host immune surveillance against cancer. Up regulation of complement regulatory proteins that can inhibit complement activation are often observed in cancer cells and are considered an important mechanism by which malignant cells may escape complement-mediated elimination (17). These proteins include CD59, decay accelerating factor (DAF) and membrane cofactor protein (MCP). They protect cells on which they are expressed by inhibiting different steps of complement activation. DAF and MCP interfere with formation and assembly of C3- and C5-convertases during complement activation, whereas, CD59 inhibits the transmembrane channel formation preventing the assembly of membrane attack complex and protect the target cell (18,19). Consistently, over expression of complement regulatory proteins, including CD59 is associated with a poor prognosis in lung and other tumor types (20–22). However, the context and the pathways that regulate these proteins in the tumor microenvironment are not known. In this study, we demonstrate up regulation of CD59 during TGF-β-induced EMT, conferring resistance to complement dependent cytotoxicity (CDC). Inhibition of CD59 expression restored susceptibility to CDC, enhanced efficacy of antibody therapy and inhibited EMT-mediated metastasis of lung cancer cells. In addition, we show the mechanism by which TGF-β regulates CD59 expression in lung cancer cells.
RESULTS
Pathway enrichment analysis identified complement as a major pathway modulated during EMT
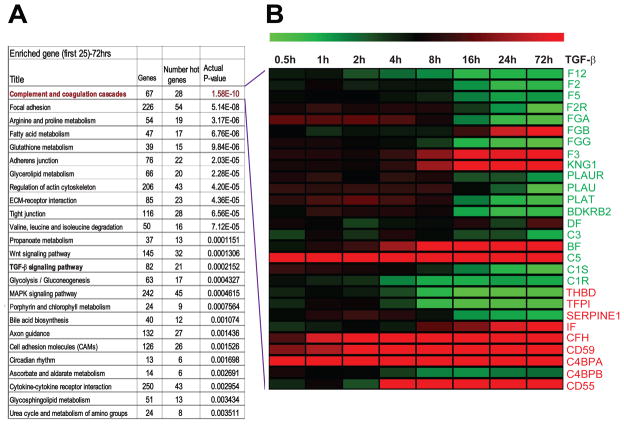
In an earlier study, we determined the time course transcriptome of cells undergoing EMT in response to TGF-β (GSE 17708) (23). Pathway enrichment analysis of the transcriptome data obtained after 72 h of TGF-β treatment was carried out using Metacore, a pathway mapping algorithm (GeneGo, St. Joseph, MI). This analysis identified multiple pathways that are consistent with biology of EMT and regulation by TGF-β. These pathways include regulation of actin cytoskeleton, adherent junction, tight junction, focal adhesion, ECM-receptor interaction, Wnt and TGF-β signalling pathways. Interestingly, the complement and coagulation cascade pathway was the top most statistically significant pathway enriched in this analysis (Fig 1A). Closer examination of this pathway revealed a modulation of 28 out of 67 total genes that constitute complement and coagulation cascade pathway in response to TGF-β. In general, we observed down regulation of genes that promote complement activation. Interestingly, C5 the precursor of anaphylatoxin C5a was up regulated. This is consistent with the recent observation that C5a promotes tumor progression in the lung tumor microenvironment (24). The majority of genes that inhibit complement function were up regulated, including CD59, CD55, and CFH (Fig. 1B).
Figure 1. List of top 25 biological pathways enriched and heat map of complement pathway genes differentially expressed during TGF-β-induced EMT.
Pathway enrichment analysis was performed by Metacore algorithm (GeneGo.com) and Top 25 pathways we most significant p-values are listed (A). Heat map (green: down regulation, red: up regulation) representing time course of 28 differential expressed complement pathway genes during TGF-β-induced EMT. Gene names in green and red are positive and negative regulators of complement pathway respectively (B).
TGF-β-induced CD59 expression in lung adenocarcinoma cells
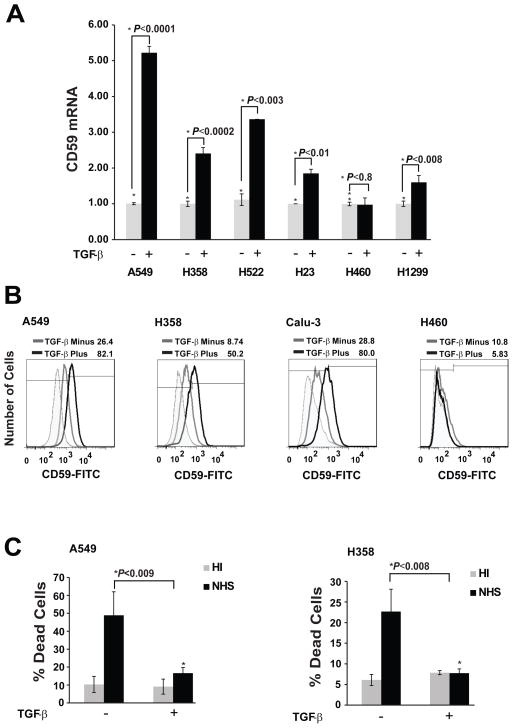
To validate the transcriptome analysis, we assessed TGF-β induced CD59 expression in a panel of human lung adenocarcinoma cell lines (A549, H358, Calu3, H522, H23, H460 and H1299) by qPCR. We observed TGF-β-induced CD59 mRNA expression in 5 out of 7 cell lines tested (Fig 2A). The magnitude of induction was in the range of 2 to 5 fold over untreated cells. Consistently, TGF-β stimulation also increased cell surface CD59 expression in cell lines (A549, H358 and Calu3) that showed mRNA induction, but none in the cell line (H460) that did not, as assessed by flow cytometry (Fig 2B). This suggests a transcriptional regulation of CD59 expression by TGF-β.
Figure 2. TGF-β-induced EMT increases CD59 expression and confers resistance to complement dependent cytotoxicity (CDC).
Indicated cell lines were serum starved for 24 h, and cultured with or without TGF-β (5 ng/ml) for 72 h. Total RNA was isolated to assess CD59 mRNA expression by qPCR. Fold changes relative to untreated control are plotted (A). To assess cell surface CD59 expression, cells were trypsinized, and fixed in paraformaldehyde before staining with FITC or PE conjugated anti-CD59 antibodies. CD59 expression was assessed by flow cytometry. FITC or PE conjugated IgG staining was used as controls and values are expressed as percentage of CD59 positive cells compared to IgG control. The cut-off bar for background fluorescence was set at 1%. Gray peak represents untreated samples; black peak indicates TGF-β treatment while shaded gray represents IgG controls. Numbers on top of plots indicate the percentage of CD59 positive cells (B). To assess complement dependent cytotoxicity, cells were incubated with normal human serum (NHS) or heat inactivated serum (HI), fixed in 70% alcohol before staining with 7-AAD. DNA fragmentation associated with complement dependent cell lysis was assessed by the percentage of 7-AAD positive cells in sub-G0 by flow cytometry (C).
TGF-β-induced EMT confers resistance to complement-dependent cytotoxicity (CDC)
We have previously shown that lung cancer cells undergo EMT in response to TGF-β stimulation (9,11). Here, we assessed the susceptibility of cells to CDC before and after TGF-β-induced EMT, in the presence of normal human serum (NHS) or heat-inactivated serum (HIS). CDC was determined by measuring number of cells in sub-G0, using 7-aminoactinomycin D (7-AAD) staining, by flow cytometry. Lung cancer cells (A549, and H358) were susceptible to CDC, triggered by NHS, as evident by the significant increase in the number of cells in sub-G0 phase compared to cells in the presence of HIS (Fig 2C). However, cells undergoing TGF-β-induced EMT were protected from CDC as indicated by a significant decrease in the number of cells in sub-G0 phase (Fig 2C). These findings indicate that TGF-β-induced EMT confers resistance to CDC and the induction of CD59 may be mediating this resistance.
CD59 inhibition restores CDC
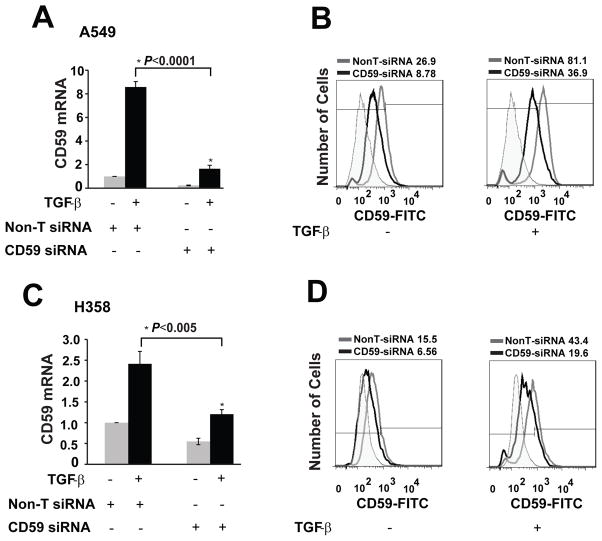
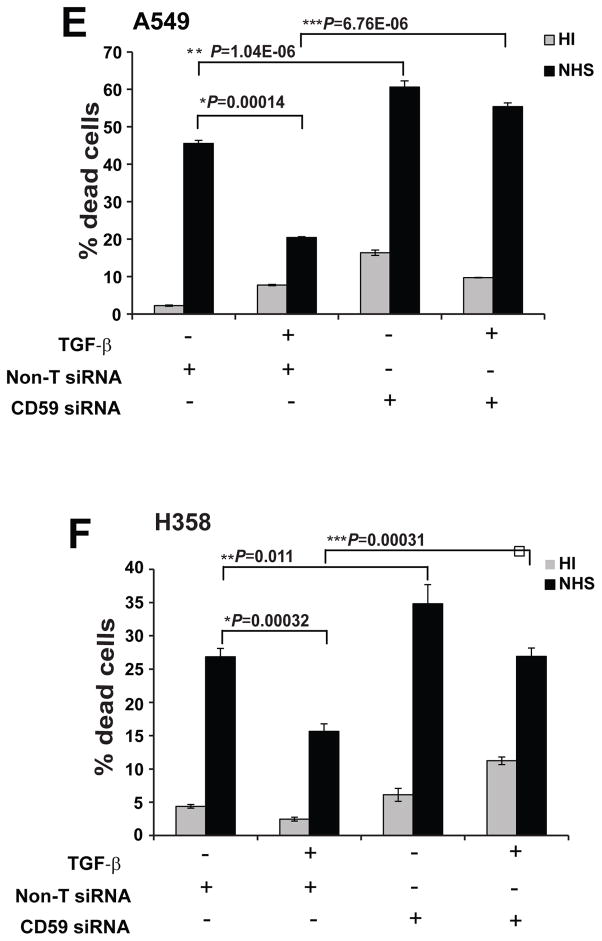
To test whether the induction of CD59 expression mediated the observed CDC resistance during EMT, we inhibited CD59 expression using Smart pool siRNA (Dharmacon) and assessed CDC before and after induction of EMT in two cell lines (A549 and H358). siRNA transfection achieved up to 80% knockdown of both base line and TGF-β-induced CD59 expression both at the mRNA (Fig. 3A & C) and protein levels (Fig. 3B & D). Inhibition of CD59 expression increased basal CDC of both cell lines without TGF-β treatment. CD59 inhibition also restored EMT-induced suppression of CDC in both A549 and H358 cells (Fig. 3E & F). This indicates that TGF-β-induced CD59 expression mediates the EMT-induced suppression of CDC.
Figure 3. CD59 inhibition restores CDC.
A549 and H358 cell lines transfected either with control or CD59 siRNA (50 nM) were serum starved for 24 h and stimulated with or without TGF-β (5 ng/ml) for 72 h. Total RNA was isolated, CD59 expression was measured by qPCR to assess knockdown efficiency of siRNA at mRNA level. Fold changes relative to non-targeting control siRNA are plotted (A & C). To assess knockdown efficiency at protein level, cell surface CD59 expression was measured. For this, cells were trypsinized, and fixed in paraformaldehyde before staining with FITC or PE conjugated anti-CD59 antibodies. CD59 expression was assessed by flow cytometry. FITC or PE conjugated IgG staining was used as controls and values are expressed as percentage of CD59 positive cells compared to IgG control. Gray peak represents non-target siRNA, black peak indicates CD-59 siRNA while shaded gray represents IgG controls. Numbers on top of plots indicate the percentage of CD59 positive cells (B & D). To assess complement dependent cytotoxicity, cells were incubated with normal human serum (NHS) or heat inactivated serum (HI), fixed in 70% alcohol before staining with 7-AAD. DNA fragmentation associated with complement dependent cell lysis was assessed by the percentage of 7-AAD positive cells by flow cytometry (E & F).
CD59 inhibition had no effect on TGF-β-induced EMT but resistance to CDC is dependent on EMT
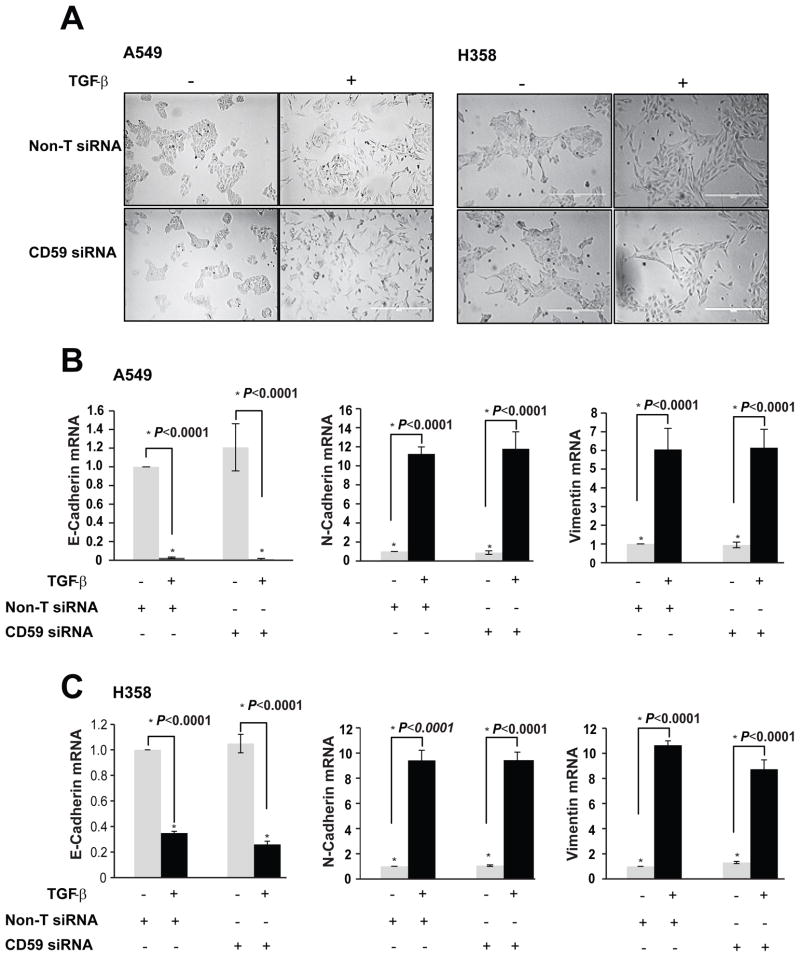
We determined the effect of CD59 inhibition on EMT. Interestingly, inhibition of CD59 expression had no effect on EMT induced by TGF-β as assessed by the effect on cellular morphology (Fig. 4A). In addition, treatment with CD59 siRNA did not block the down regulation of epithelial E-cadherin or it inhibit the induction of mesenchymal markers N-cadherin and vimentin in the two cell lines tested (Fig. 4B&C). This clearly demonstrates that CD59 induction does not regulate TGF-β-induced EMT.
Figure 4. CD59 inhibition had no effect on TGF-β-induced EMT but resistance to CDC is dependent on EMT.
A549 and H358 cell lines transfected either with control or CD59 siRNA (50 nM) were serum starved for 24 h and stimulated with or without TGF-β (5 ng/ml) for 72 h. Change in EMT-induced cellular morphology was monitored by real time imaging using IncuCyte. Representative images at 72 h time point are provided (A). Total RNA was isolated to assess mRNA expression of epithelial (E-cadherin) and mesenchymal (N-cadherin and Vimentin) markers by qPCR. Fold changes relative to untreated controls from non-targeted siRNA samples are plotted (B & C). A549 cells were plated at low (50% confluence) or high (<90% confluence) density. TGF-β stimulation induces EMT in low density cultures and minimally in high density cultures. To assess complement dependent cytotoxicity, cells were incubated with normal human serum (NHS) or heat inactivated serum (HI), fixed in 70% alcohol before staining with 7-AAD. DNA fragmentation associated with complement dependent cell lysis was assessed by the percentage of 7-AAD positive cells by flow cytometry (D). Degree of EMT was measured by assessing the expression of epithelial (E-cad) and mesenchymal (N-cad and Vimentin) markers. Activation of TGF-β signaling was demonstrated by assessing phospho-and total Smad3 expression by western immunoblotting (E).
It is well known that higher density impairs the efficiency of cancer cells to undergo EMT in cell cultures which is consistent with the detection of EMT at the invasive front of a tumor in clinical samples (25). To determine whether observed resistance to CDC is EMT dependent or not, we cultured A549 cells at low (50% confluence) and high (<90% confluence) density in the presence and absence of TGF-β. As expected we observed robust induction of EMT in low density cultures with down regulation of E-cadherin and up regulation of mesenchymal markers N-cadherin and vimentin (Fig. 4E). Consistent with previous studies there was less EMT in higher density cultures with very little modulation of epithelial and mesenchymal markers (Fig. 4E). In contrast, both cultures showed equally robust expression of phospho-Smad3 in response to TGF-β, demonstrating that activation of TGF-β signalling was not different between two densities. Interestingly, consistent with impaired EMT, TGF-β-treated cells in high density cultures were significantly less resistant to CDC compared to TGF-β-treated cells in low density cultures (Fig. 4D). This demonstrates that observed CDC resistance is most likely EMT dependent than only a TGF-β effect.
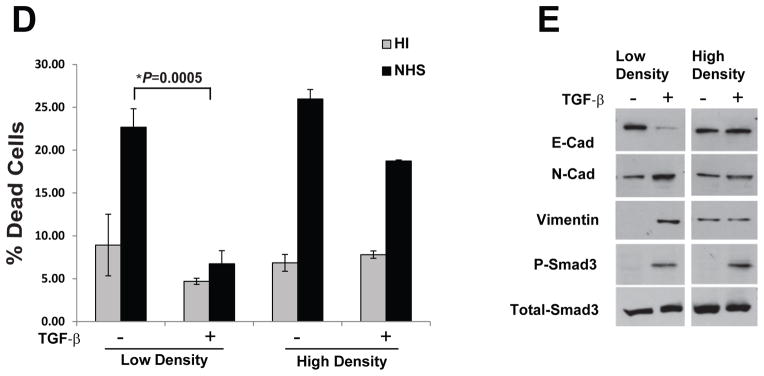
The inhibition of CD59 sensitizes cancer cells that undergo EMT to cetuximab-mediated CDC
Therapeutic monoclonal antibodies against the epidermal growth factor receptor (EGFR) have been in the clinic for the treatment of multiple solid tumors, including lung adenocarcinomas. In addition to the inhibition of receptor signalling, these anti-EGFR antibodies were been shown to trigger complement-dependent tumor cell lysis including cetuximab (26). Interestingly, resistance to EGFR-targeted therapies have been associated with acquisition of EMT phenotype (27). Here we tested whether inhibition of EMT-induced CD59 expression can sensitize lung cancer cells to cetuximab, an anti-EGFR antibody. Cetuximab treatment further enhanced NHS induced CDC and TGF-b treatment inhibited CDC both in the presence and absence of cetuximab (Fig. 5A & B). As anticipated, CD59 siRNA significantly restored TGF-b-induced inhibition of CDC both in the presence and absence of cetuximab (Fig. 5A & B). This suggests that inhibition of CD59 may be an important strategy to overcome resistance to anti-EGFR antibody therapy.
Figure 5. CD59 inhibition sensitizes cancer cells that undergo EMT to cetuximab mediated CDC.
A549 and H358 cell lines transfected either with control or CD59 siRNA (50 nM) were serum starved for 24 h and stimulated with or without TGF-β (5 ng/ml) for 72 h. To assess complement dependent cytotoxicity, cells were incubated with normal human serum (NHS) or heat inactivated serum (HI) in the presence or absence of cetuximab, fixed in 70% alcohol before staining with 7-AAD. Cituximab alone or IgG are used a controls. DNA fragmentation associated with complement dependent cell lysis was assessed by the percentage of 7-AAD positive cells in sub-G0 by flow cytometry (A & B).
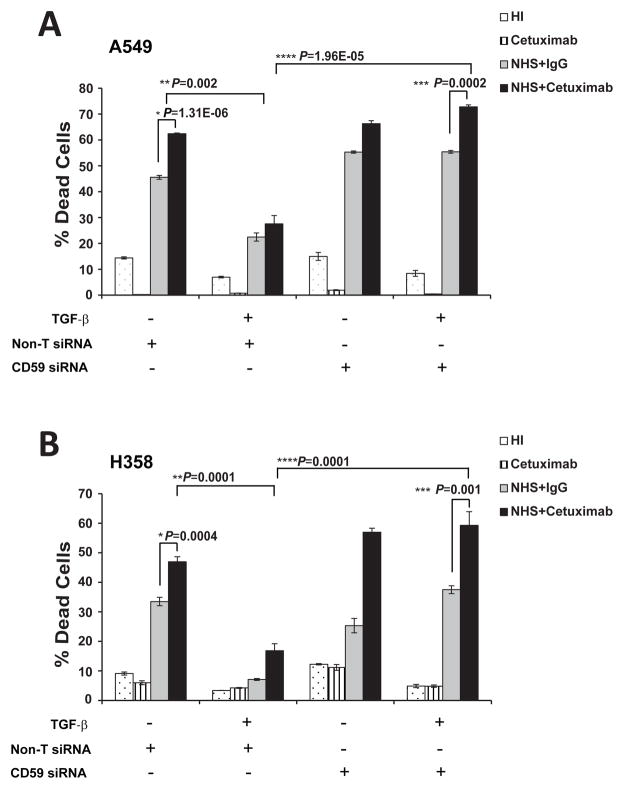
Stable knockdown of CD59 inhibits experimental metastasis of lung cancer cells
To achieve sustained inhibition of CD59 required for in-vivo experiments, we stably expressed CD59 shRNA in A549 cells by lentiviral-mediated delivery. We achieved approximately 80% knock-down in A549-CD59shRNA cells as evaluated by qPCR and flow cytometry (Fig. 6A). In an earlier study, we demonstrated that TGF-β-induced EMT promotes experimental metastasis of A549 cells in SCID mice when injected through the tail vein (11). Here, we observed a significant inhibition of EMT-induced experimental metastasis of A549-CD59shRNA cells, as compared to the corresponding vector control cells (Fig. 5B). Interestingly, inhibition of CD59 had no effect on the in-vitro growth of A549-CD59shRNA cells compared to corresponding vector control cells, as assessed by monitoring cellular confluence over time (Fig. 5C). However, in-vivo CD59 knockdown resulted in more than 50% inhibition of the primary tumor growth of subcutaneously implanted A549-CD59shRNA xenografts, compared to corresponding vector control xenografts (Fig. 5D). The effects of CD59 inhibition on primary tumor growth and metastasis establish the significance of CD59 expression in lung tumor progression.
Figure 6. Stable knockdown of CD59 inhibits experimental metastasis.
Parental cells with vector control or CD59-shRNA expressing A549 cells were serum starved for 24 h and stimulated with TGF-β (5 ng/ml) for 72 h. Total RNA was isolated, CD59 expression was measured by qPCR to assess knockdown efficiency of shRNA at mRNA level. Fold changes relative to non-targeting control shRNA are plotted (A). To assess knockdown efficiency at protein level, cell surface CD59 expression was measured. For this, cells were trypsinized, and fixed in paraformaldehyde before staining with FITC or PE conjugated anti-CD59 antibodies. CD59 expression was assessed by flow cytometry. FITC or PE conjugated IgG staining was used as controls and values are expressed as percentage of CD59 positive cells compared to IgG control. Gray peak represents vector control; black peak indicates CD-59 shRNA while shaded gray represents IgG controls. Numbers on top of plots indicate the percentage of CD59 positive cells (A). To assess experimental metastasis, luciferase expressing parental or CD59-shRNA clones of A549 cells were stimulated with TGF-β (5 ng/ml) for 72 h in-vitro and injected through tail vein into CB17/SCID-beige mice (n = 6). After 3 weeks, whole animals were imaged for bioluminescence in the lungs. The tumor burden in the lungs was further validated by ex-vivo bioluminescence imaging of the harvested lungs. Representative images from 4 animals with signal intensity scale are presented (B). The growth rate of the established CD59-shRNA clones and parental A549 cells were assessed by monitoring culture confluence over time using IncuCyte (C).
TGF-β-induced CD59 is Smad3 dependent and a direct transcriptional target of Smad3
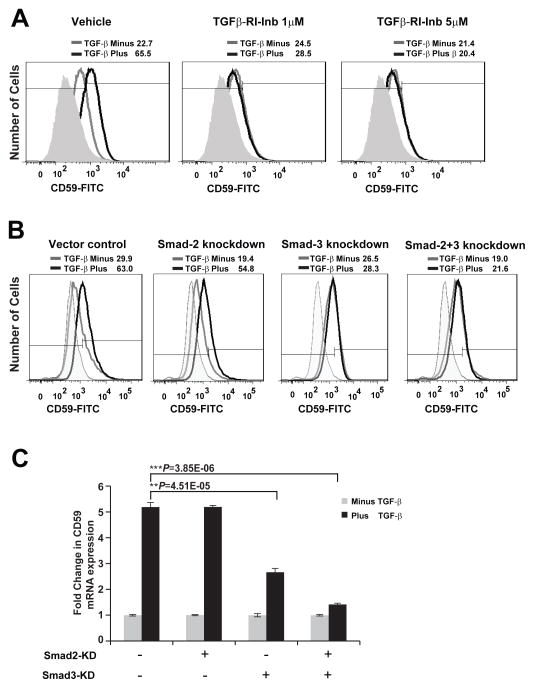
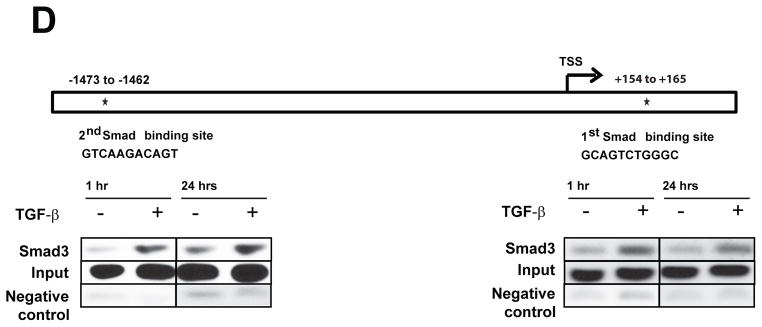
To date, it’s not known what regulates the expression of complement regulatory proteins, including CD59, on tumor cells. We observed a robust induction of CD59 expression by TGF-β at the mRNA and protein levels. Inhibition of TGF-β-receptor kinase completely abrogated CD59 expression, suggesting transcriptional regulation by TGF-β (Fig. 7A). TGF-b is known to signal through canonical Smad-dependent and non-canonical Smad-independent pathways including activation of p38 MAPK pathways. Utilizing previously established stable A549 cells expressing shRNA against Smad2, Smad3 and Smad2/3 (11), we found that TGF-β-induced CD59 expression is exclusively Smad3 dependent as demonstrated by robust inhibition of CD59 expression, both at protein (Fig. 7B) and mRNA level (Fig. 7C), in cells expressing Smad3-shRNA and Smad2&3-shRNA. Smad2-shRNA had no effect. This strongly implicates a canonical Smad3 dependent pathway in TGF-β-induced transcriptional regulation of CD59. Bioinformatics analysis of CD59 promoter for transcription factor binding sites identified two Smad3 binding sites within −2.0 to +0.5 kb region from transcription start site (Fig. 7C). To determine whether Smad3 directly binds to above identified regions of CD59 promoter we performed chromatin immunoprecipitation (ChIP) analysis, using anti-human Smad3 antibody. ChIP analyses revealed that stimulation with TGF-β increased Smad3 occupancy on both Smad3 sites identified, within 1 h and was sustained up to 24 h (Fig. 7C). These findings further suggest that CD59 is a direct transcriptional target of Smad3.
Figure 7. TGF-β-induced CD59 is Smad3 dependent and a direct transcriptional target of Smad3.
Parental A549 cells were serum starved for 24 h and stimulated with TGF-β (5 ng/ml) for 72 h in the presence or absence of TGF-β-RI inhibitor (SB-431542) (A). Stable A549 cells expressing shRNA for Vector control, Smad2, Smad3 and Smad2/3 were serum starved for 24 h and stimulated with or without TGF-β (5 ng/ml) for 72 h (B). To assess cell surface CD59 expression, cells were trypsinized, and fixed in paraformaldehyde before staining with FITC or PE conjugated anti-CD59 antibodies. CD59 expression was assessed by flow cytometry. FITC or PE conjugated IgG staining was used as controls and values are expressed as percentage CD59 positive cells compared to IgG control. Gray peak represents samples without TGF-β; black peak indicates TGF-β treatment while shaded gray represents IgG controls. Numbers on top of plots indicate the percentage of CD59 positive cells. To assess CD59 mRNA expression total RNA was isolated, CD59 expression was measured by qPCR. Fold changes relative to samples without TGF-β stimulation are plotted (C). Representation of CD59 promoter showing two putative Smad binding sites within −2kb to +0.5 kb of transcription start site (TSS) (D). Binding of Smad3 to CD59 promoter was assessed by ChIP using anti-human Smad3 antibody. Input genomic DNA and sample without antibody were used as positive and negative controls respectively. 2–4 μl of precipitated DNA was used as template for PCR reactions. Primers flanking 1st and 2nd Smad binding sites were designed to amplify 198 and 156 bases respectively (D).
DISCUSSION
Lung cancer is a leading cause of cancer related deaths worldwide. A large proportion of lung cancer patients are known to present with metastatic disease at the time of diagnosis leading to the dismal 5 year survival of less than 15% (28). EMT is considered an important initiating event in promoting tumor cell dissemination leading to metastasis (29). In the tumor microenvironment, a number of studies have implicated both innate and adaptive immune cell infiltrates as drivers of EMT through multiple cytokines and growth factors secreted by these cells (1). However, few studies have investigated the immunomodulatory consequences of EMT. Studies to date have shown evasion of T-cell responses and suppression of dendritic cell functions by cells undergoing EMT, suggesting an important role for EMT in immune editing in the tumor microenvironment (30). In this study, we demonstrate a new immune evasive consequence of EMT by which tumor cells escape CDC by modulating the complement regulatory protein, CD59.
The deposition and activation of complement component proteins in tumor tissues, coupled with increased expression of inhibitory complement regulatory proteins on tumor cells illustrate the importance of complement pathway in host immune surveillance against cancer (14–17). Consistently, increased expression of complement regulatory proteins, CD59 and CD55 has been highly correlated with the histology and patient prognosis in multiple solid tumors (20–22). Despite such potential clinical implications, it is not clear what regulates the expression of proteins like CD59 in the tumor microenvironment. In this study we demonstrate that TGF-β, a cytokine rich in the tumor microenvironment, is a potent inducer of CD59. This is consistent with the earlier implication of TGF-β in activated monocytes-induced CD59 expression in normal human keratinocytes. However, anti-TGF-β neutralizing antibodies did not block CD59 expression suggesting involvement of additional factors (31). In contrast, we show that TGF-β-receptor kinase inhibitor or shRNA against Smad3 completely abrogated TGF-β-induced CD59 expression. Furthermore, we also established that TGF-β-induced CD59 expression is Smad3, but not Smad2, dependent and a direct transcriptional target of Smad3. Together, our data strongly implicates canonical Smad-dependent TGF-β pathway in the regulation of CD59 expression.
Lung cancer cells that develop resistance to EGFR targeted therapies were associated with the acquisition of mesenchymal phenotype through EMT (27). Interestingly, the inhibition of CD59 sensitized cells undergoing EMT to cetuximab (ant-EGFR) mediated CDC. This is consistent with earlier studies that showed antibody-based neutralization of CD59 increased the efficacy of anti-Her2 antibodies in lung and breast cancer cells (32,33) and rituximab (anti-CD21) efficacy in lymphomas (34). More importantly, knockdown of CD59 blocked primary tumor growth as well as EMT-induced metastasis of lung cancer cells in-vivo. However, inhibition of CD59 had no direct effect on the proliferation of tumor cells in-vitro, suggesting immune mediated inhibition of tumor growth and metastasis, potentially through complement, rather than a cell autonomous effect. Given its significant contribution to immune surveillance and antibody therapy, a number of strategies have been employed to neutralize CD59 in cancer therapy. One particularly promising approach has been the development of rILYd4, a novel recombinant protein inhibitor of CD59 (35). rILYd4 is a potent and specific human CD59 inhibitor. This molecule has been shown to significantly enhance the efficacy of rituximab in lymphomas and anti-CD24 Ab therapy in osteosarcoma (34) demonstrating that rILYd4 could be a viable platform for developing therapeutic CD59 inhibitors.
In addition to the strong evidence supporting a role for complement in host immune surveillance and improving the efficacy of antibody therapy, there is equally strong data supporting the tumor promoting functions of complement activating components (36). Recent studies including in lung cancer, showed that tumor cell derived C5a contributes to the immunosuppressive tumor microenvironment by recruiting myeloid derived suppressor cells. Neutralizing C5a inhibited tumor growth in-vivo and reduced immune suppression (24,37). Interestingly, along with CD59 we also observed increase in the gene expression of C5, precursor of C5a, during EMT (Fig. 1B). One possibility for such a paradoxical effect is that the tumor promoting effects of complement components might be a late stage consequence of immune editing, which initially renders cancer cells resistance to CDC through processes like EMT. In other words, once tumors become resistant to CDC they may begin to accumulate complement components and utilize these molecules for other tumor promoting functions. Therefore, to successfully exploit complement pathway for the control of cancer, it is necessary to identify pathways that modulate complement components and their regulators, and to establish the context under which they operate.
MATERIALS AND METHODS
Cell culture
All the human lung cancer cell lines (A549, H358, Calu3, H522, H23, H460 and H1299) were obtained from ATCC and were cultured in RPMI-1640 (plus glutamine) medium supplemented with 10% FBS and penicillin-streptomycin (Complete medium). For EMT experiments, cells were grown in 6-well plate in 30–40% confluency (50,000 cells/well) in complete medium. They were serum starved for 24hrs and then treated with TGF-β (5ng/ml) for 72hrs. Pharmacological inhibitors when used were added to cells 30 minutes prior to addition of TGF-β. shRNA stable clones were selected in presence of complete medium containing 2 μg/ml puromycin and maintained in complete medium additionally supplemented with 1 μg/ml puromycin. Where indicated, change in cellular morphology was monitored over time by real time imaging using IncuCyte (Essen biosciences, Inc., Ann Arbor, MI), a real time imaging system to monitor cell cultures.
siRNA transfection
CD59-specific siRNA included a pool of 4 synthetic duplexes (Dharmacon’s SMART pool). A scrambled sequence from the same company was used as control. Cells at 30–40% confluent (50,000 cells/well) were transfected with siRNA using Lipofectamine 2000 and OptiMEM medium. After 6 hours of transfection, cells were allowed to recover overnight from transfection in RPMI-1640 medium with 10% FBS before inducing EMT or further assessments as indicated.
Establishing stable shRNA clones
Lentivirus-based plasmids expressing CD59 shRNA (clone # V2LHS_226894 target sequence: “ATTCTTTCCCAACAGGATC”) were purchased from OpenBiosystems, Lafayette, CO. Lentiviral particles were prepared at vector core facility, University of Michigan (http://www.umms.med.umich.edu/mcore/pub). A549 cells were transduced with lentiviral particles; stable transfectants were isolated by puromycin (1 μg/ml) resistance and assessed for CD59 knock-down by both mRNA and protein levels. A549 clones expressing shRNA against Smads were developed and reported earlier (11). The growth rate of the established clones was assessed by monitoring culture confluence over time using IncuCyte.
Quantitative PCR analysis
Total RNA was isolated using an RNA isolation kit following manufacturer’s protocol (OmegaBiotek, Norcross, GA). Required PCR primers and probes (Human CD59, E-Cadherin, N-Cadherin and Vimentin) were purchased from IDT Inc, Coralville, IA. qPCR was performed using ABI Prism (Applied Biosystems, Carlsbad, CA) normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression in each sample. Reaction conditions were −30 second at 94° C, 1 minute at 50° C, 45 seconds at 68° C for 40 cycle amplification. Fold changes relative to control were plotted after normalization to GAPDH expression.
Western blot analysis
Cells were washed with PBS after treatment and lysed in radio immunoprecipitation assay buffer (RIPA) containing NaF, Na3Vo4 and protease inhibitor. Samples containing 20μg of total protein were electrophoresed on SDS-polyacrylamide gels and transferred onto a polyvinyldifluoride membrane by electro blotting. Membranes were probed with primary antibodies (E-cadherin, N-cadherin, P-Smad3 and Total Smad3 [Cell signalling Technologies, Beverly, MA,] Vimentin [Sigma, St. Louis, MO]) with overnight incubation at 4° C, followed by horseradish peroxidase-conjugated secondary antbodies. Finally, the immunoblots were visualized using enhanced chemiluminescence (ECL) reagents.
In-vivo mouse experiments
Stable CMV-luciferase (A549-Luc) or CD59-shRNA (A549-CD59KD-Luc) expressing clones of A549 cells were used in this study. Cells were serum starved for 24 h then stimulated with TGF-β (5 ng/ml) for 72h to induce EMT. Care of animals and experimental procedures were according to University of Michigan, UCUCA (University committee for use and care of animals) approved protocol # PRO00004116. To assess experimental metastasis, 0.5 ×106 cells in 100 μL of serum free medium were injected into 8 week old, male, CB17/SCID-beige mice (n= 6 in each group) (Harlan laboratories, Indianapolis, USA) through the tail vein. The number of animals is each group is estimated to detect 35% difference between the groups with 80% power and 95% significance. After 3 weeks, mice were anesthetized with isoflurane (2%) and injected with 100 μl of an aqueous solution of luciferin (5 mg/mL, intraperitoneally) 10 min prior to imaging. The animals were placed into the light-tight chamber of the bioluminescence imaging system (Xenogen, Alameda, CA) and the photons emitted from the luciferase-expressing cells were captured by CCD camera and quantitated using the software program Living Image (Xenogen) as an overlay on Igor (Wavemetrics, Seattle, WA). The tumor burden in the lungs were further validated by re- imaging in the Xenogen system after harvesting from the animal, as well as by counting the number of visible colonies in lung.
CD59 expression by flow cytometry
Cells were detached using trypsin-EDTA (Invitrogen) and re-suspended in serum free RPMI. After washing, cells were fixed in final concentration of 2% paraformaldehyde. Subsequently, cells were washed and labelled with FITC or PE anti-human CD59 antibodies (BD Biosciences, San Jose, CA and Molecular probes, Life Technologies, Grand Island, NY) and evaluated for staining with flow cytometry (Attune, Applied Biosystems, Carlsbad, CA) in BL-1 (488 nm) or BL2 (565 nm) channels. Mouse anti-human IgG-FITC and IgG-PE were used as controls for normalization. 1% was normally used as the background fluorescence and fixed as the cut-off for staining positive cells.
Complement Dependent Cytotoxicity (CDC) Assay
Pooled normal human serum from healthy volunteers was used as source of complement. Serum was allowed to coagulate at 37° C for 30 mins, spun and supernatant (complement) was snap-frozen at −80° C. Heat Inactivated serum (HI) was obtained by incubation of the serum at 56° C for 30 minutes. Complement-dependent cytotoxicity is associated with DNA fragmentation and cell death (26,38). DNA fragmentation was evaluated using 7-aminoactinomycin D (7-AAD) staining. Briefly, approximately, 2×105 cells were incubated with 200–400 μl of NHS or HI (diluted 1:5) in triplicates, in the presence or absence of Cetuximab, an anti-EGFR antibody (40–100 μg/ml) for 1 h. After complement treatment, cells were washed in PBS and pelleted in 1ml of cold 70% alcohol for at least 60 mins. Cells were then washed twice with PBS, re-suspended in PBS containing 1mg/ml of 7-AAD for 45 minutes in dark at room temperature. Labelled cells were analysed by flow cytometry in BL-3 channel (Attune, Applied Biosystems, Carlsbad, CA). The percentage of cells in sub-G0 phase were taken as measure of DNA fragmentation and therefore cell death.
Chromatin Immunoprecipitation analysis (ChIP)
Analysis of CD59 promoter using Genomatix software (www.Genomatix.de) predicted presence of 2 putative Smad binding sites within −2 kb to + 0.5 kb region from transcription start site. ChIP analysis was done on control and TGF-β treated cells using ChiP kit (USB) following manufacturer’s protocol. Briefly, 150 mm dish of control and TGF-β treated cells were cross-linked using 2% PFA. Cross-linked DNA was fragmented, reverse cross-linked and immunoprecipitation was carried out using anti-human Smad3 antibody (Cell Signaling Tech, Danvers, MA). Input genomic DNA and sample without antibody were used as positive and negative controls respectively. 2–4 μl of precipitated DNA was used as template for PCR reactions. Primers flanking 1st and 2nd Smad binding sites were designed to amplify 198 and 156 bases respectively. PCR was carried out using Hotstart Taq PCR mix (Qiagen, Valencia, CA) under the following conditions 95° C–15mins, denaturation at 95° C for 30 seconds, annealing at 58° C for 30 seconds, extension 72° C for 40 seconds with 35 cycles. The amplification product was loaded in 2% gel with ethidium bromide for analysis.
Statistical analysis
Data are represented as mean ± SD from at least three independent biological replicates. Groups were compared using Student’s t-test to evaluate differences. P values < 0.05 were considered significant and indicated in figures using asterisk.
Supplementary Material
Acknowledgments
This research is funded by, NIH/NCI (CA132571-01) grant, and the Elizabeth A. Crary Fund to V.G.K. NIH/NHLBI HL097564 to T.J.S.
Footnotes
Conflict of interest
The authors declare no conflict of interest
References
- 1.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews Molecular cell biology. 2014;15(3):178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339(6119):580–4. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 4.Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer Res. 2005;65(14):5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. discussion 00-1. [DOI] [PubMed] [Google Scholar]
- 5.Zavadil J, Haley J, Kalluri R, Muthuswamy SK, Thompson E. Epithelial-mesenchymal transition. Cancer Res. 2008;68(23):9574–7. doi: 10.1158/0008-5472.CAN-08-2316. [DOI] [PubMed] [Google Scholar]
- 6.Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–74. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 7.Kim WS, Park C, Jung YS, Kim HS, Han J, Park CH, et al. Reduced transforming growth factor-beta type II receptor (TGF-beta RII) expression in adenocarcinoma of the lung. Anticancer Res. 1999;19(1A):301–6. [PubMed] [Google Scholar]
- 8.Kong F, Jirtle RL, Huang DH, Clough RW, Anscher MS. Plasma transforming growth factor-beta1 level before radiotherapy correlates with long term outcome of patients with lung carcinoma. Cancer. 1999;86(9):1712–9. [PubMed] [Google Scholar]
- 9.Keshamouni VG, Michailidis G, Grasso CS, Anthwal S, Strahler JR, Walker A, et al. Differential protein expression profiling by iTRAQ-2DLC-MS/MS of lung cancer cells undergoing epithelial-mesenchymal transition reveals a migratory/invasive phenotype. Journal of proteome research. 2006;5(5):1143–54. doi: 10.1021/pr050455t. [DOI] [PubMed] [Google Scholar]
- 10.Reka AK, Kuick R, Kurapati H, Standiford TJ, Omenn GS, Keshamouni VG. Identifying inhibitors of epithelial-mesenchymal transition by connectivity map-based systems approach. J Thorac Oncol. 2011;6(11):1784–92. doi: 10.1097/JTO.0b013e31822adfb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reka AK, Kurapati H, Narala VR, Bommer G, Chen J, Standiford TJ, et al. Peroxisome proliferator-activated receptor-gamma activation inhibits tumor metastasis by antagonizing Smad3-mediated epithelial-mesenchymal transition. Mol Cancer Ther. 2010;9(12):3221–32. doi: 10.1158/1535-7163.MCT-10-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reka AK, Chen G, Jones RC, Amunugama R, Kim S, Karnovsky A, et al. Epithelial-mesenchymal transition-associated secretory phenotype predicts survival in lung cancer patients. Carcinogenesis. 2014;35(6):1292–300. doi: 10.1093/carcin/bgu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walport MJ. Complement. First of two parts. The New England journal of medicine. 2001;344(14):1058–66. doi: 10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 14.Fishelson Z, Donin N, Zell S, Schultz S, Kirschfink M. Obstacles to cancer immunotherapy: expression of membrane complement regulatory proteins (mCRPs) in tumors. Molecular immunology. 2003;40(2–4):109–23. doi: 10.1016/s0161-5890(03)00112-3. [DOI] [PubMed] [Google Scholar]
- 15.Jurianz K, Ziegler S, Garcia-Schuler H, Kraus S, Bohana-Kashtan O, Fishelson Z, et al. Complement resistance of tumor cells: basal and induced mechanisms. Molecular immunology. 1999;36(13–14):929–39. doi: 10.1016/s0161-5890(99)00115-7. [DOI] [PubMed] [Google Scholar]
- 16.Varsano S, Rashkovsky L, Shapiro H, Ophir D, Mark-Bentankur T. Human lung cancer cell lines express cell membrane complement inhibitory proteins and are extremely resistant to complement-mediated lysis; a comparison with normal human respiratory epithelium in vitro, and an insight into mechanism(s) of resistance. Clinical and experimental immunology. 1998;113(2):173–82. doi: 10.1046/j.1365-2249.1998.00581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donin N, Jurianz K, Ziporen L, Schultz S, Kirschfink M, Fishelson Z. Complement resistance of human carcinoma cells depends on membrane regulatory proteins, protein kinases and sialic acid. Clinical and experimental immunology. 2003;131(2):254–63. doi: 10.1046/j.1365-2249.2003.02066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DD, Song WC. Membrane complement regulatory proteins. Clinical immunology. 2006;118(2–3):127–36. doi: 10.1016/j.clim.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Yan J, Allendorf DJ, Li B, Yan R, Hansen R, Donev R. The role of membrane complement regulatory proteins in cancer immunotherapy. Adv Exp Med Biol. 2008;632:159–74. [PubMed] [Google Scholar]
- 20.Watson NF, Durrant LG, Madjd Z, Ellis IO, Scholefield JH, Spendlove I. Expression of the membrane complement regulatory protein CD59 (protectin) is associated with reduced survival in colorectal cancer patients. Cancer Immunol Immunother. 2006;55(8):973–80. doi: 10.1007/s00262-005-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu C, Jung M, Burkhardt M, Stephan C, Schnorr D, Loening S, et al. Increased CD59 protein expression predicts a PSA relapse in patients after radical prostatectomy. Prostate. 2005;62(3):224–32. doi: 10.1002/pros.20134. [DOI] [PubMed] [Google Scholar]
- 22.Nishioka K, Kawamura K, Hirayama T, Kawashima T, Shimada K. The complement system in tumor immunity: significance of elevated levels of complement in tumor bearing hosts. Annals of the New York Academy of Sciences. 1976;276:303–15. doi: 10.1111/j.1749-6632.1976.tb41656.x. [DOI] [PubMed] [Google Scholar]
- 23.Keshamouni VG, Jagtap P, Michailidis G, Strahler JR, Kuick R, Reka AK, et al. Temporal quantitative proteomics by iTRAQ 2D-LC-MS/MS and corresponding mRNA expression analysis identify post-transcriptional modulation of actin-cytoskeleton regulators during TGF-beta-Induced epithelial-mesenchymal transition. Journal of proteome research. 2009;8(1):35–47. doi: 10.1021/pr8006478. [DOI] [PubMed] [Google Scholar]
- 24.Corrales L, Ajona D, Rafail S, Lasarte JJ, Riezu-Boj JI, Lambris JD, et al. Anaphylatoxin C5a creates a favorable microenvironment for lung cancer progression. Journal of immunology. 2012;189(9):4674–83. doi: 10.4049/jimmunol.1201654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masszi A, Fan L, Rosivall L, McCulloch CA, Rotstein OD, Mucsi I, et al. Integrity of cell-cell contacts is a critical regulator of TGF-beta 1-induced epithelial-to-myofibroblast transition: role for beta-catenin. The American journal of pathology. 2004;165(6):1955–67. doi: 10.1016/s0002-9440(10)63247-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu YF, Ajona D, Corrales L, Lopez-Picazo JM, Gurpide A, Montuenga LM, et al. Complement activation mediates cetuximab inhibition of non-small cell lung cancer tumor growth in vivo. Mol Cancer. 2010;9:139. doi: 10.1186/1476-4598-9-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, et al. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer research. 2005;65(20):9455–62. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- 28.Kohler BA, Ward E, McCarthy BJ, Schymura MJ, Ries LA, Eheman C, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. Journal of the National Cancer Institute. 2011;103(9):714–36. doi: 10.1093/jnci/djr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalluri R. EMT: when epithelial cells decide to become mesenchymal-like cells. The Journal of clinical investigation. 2009;119(6):1417–9. doi: 10.1172/JCI39675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasch MC, Bos JD, Daha MR, Asghar SS. Transforming growth factor-beta isoforms regulate the surface expression of membrane cofactor protein (CD46) and CD59 on human keratinocytes [corrected] European journal of immunology. 1999;29(1):100–8. doi: 10.1002/(SICI)1521-4141(199901)29:01<100::AID-IMMU100>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 32.Jurianz K, Maslak S, Garcia-Schuler H, Fishelson Z, Kirschfink M. Neutralization of complement regulatory proteins augments lysis of breast carcinoma cells targeted with rhumAb anti-HER2. Immunopharmacology. 1999;42(1–3):209–18. doi: 10.1016/s0162-3109(99)00006-5. [DOI] [PubMed] [Google Scholar]
- 33.Zhao WP, Zhu B, Duan YZ, Chen ZT. Neutralization of complement regulatory proteins CD55 and CD59 augments therapeutic effect of herceptin against lung carcinoma cells. Oncology reports. 2009;21(6):1405–11. doi: 10.3892/or_00000368. [DOI] [PubMed] [Google Scholar]
- 34.Hu W, Ge X, You T, Xu T, Zhang J, Wu G, et al. Human CD59 inhibitor sensitizes rituximab-resistant lymphoma cells to complement-mediated cytolysis. Cancer research. 2011;71(6):2298–307. doi: 10.1158/0008-5472.CAN-10-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ge X, Wu L, Hu W, Fernandes S, Wang C, Li X, et al. rILYd4, a human CD59 inhibitor, enhances complement-dependent cytotoxicity of ofatumumab against rituximab-resistant B-cell lymphoma cells and chronic lymphocytic leukemia. Clin Cancer Res. 2011;17(21):6702–11. doi: 10.1158/1078-0432.CCR-11-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Markiewski MM, Lambris JD. Is complement good or bad for cancer patients? A new perspective on an old dilemma. Trends in immunology. 2009;30(6):286–92. doi: 10.1016/j.it.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, et al. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9(11):1225–35. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cragg MS, Howatt WJ, Bloodworth L, Anderson VA, Morgan BP, Glennie MJ. Complement mediated cell death is associated with DNA fragmentation. Cell Death Differ. 2000;7(1):48–58. doi: 10.1038/sj.cdd.4400627. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.