Abstract
The temporal version of the pediatric sepsis biomarker risk model (tPERSEVERE) estimates the risk of a complicated course in children with septic shock based on biomarker changes from days 1 to 3 of septic shock. We validated tPERSEVERE performance in a prospective cohort, with an a priori plan to redesign tPERSEVERE if it did not perform well. Biomarkers were measured in the validation cohort (n = 168) and study subjects were classified according to tPERSEVERE. To redesign tPERSEVERE, the validation cohort and the original derivation cohort (n = 299) were combined and randomly allocated to training (n = 374) and test (n = 93) sets. tPERSEVERE was redesigned using the training set and CART methodology. tPERSEVERE performed poorly in the validation cohort, with an area under the curve (AUC) of 0.67 (95% CI: 0.58–0.75). Failure analysis revealed potential confounders related to clinical characteristics. The redesigned tPERSEVERE model had an AUC of 0.83 (0.79–0.87) and a sensitivity of 93% (68–97) for estimating the risk of a complicated course. Similar performance was seen in the test set. The classification tree segregated patients into two broad endotypes of septic shock characterized by either excessive inflammation or immune suppression.
Keywords: Sepsis, Biomarkers, Outcome, Prediction, Modeling, Inflammation, Immune Suppression, Endotype
Highlights
-
•
We prospectively tested the performance of the temporal version of the pediatric sepsis biomarker risk model (tPERSEVERE).
-
•
tPERSEVERE performed poorly in the test cohort, prompting a redesign.
-
•
The redesigned tPERSEVERE model performed well upon testing.
-
•
The redesigned tPERSEVERE provides information regarding septic shock endotypes.
Septic shock is characterized by individual heterogeneity and it is not known who is at greatest risk of poor outcome and would thus benefit from more aggressive treatment. We designed a biomarker-based model to estimate the risk of poor outcome in children with septic shock. The model measures biomarker concentrations over the early period of disease evolution, and estimates how the biomarker changes reflect changing risk for poor outcome. The model has potential to serve as a monitor to evaluate the effectiveness of therapy in children with septic shock and may provide information regarding the biological mechanisms of septic shock.
1. Introduction
Septic shock is a dynamic clinical and biological syndrome (Cohen et al., 2015). Patient outcomes are highly variable, reflecting a complex, time-dependent interplay between inflammation, immunity, pathogen-related factors, patient heterogeneity, and therapeutic interventions. We have attempted to navigate this complexity at the individual patient level by developing biomarker-based models to estimate the probability of poor outcomes in patients with septic shock (Alder et al., 2014, Wong et al., 2012, Wong et al., 2014a, Wong et al., 2014b, Kaplan and Wong, 2011).
Analogous to longstanding concepts in the oncology field, we contend that understanding baseline probability of poor outcome is fundamental to clinical practice and research in the field of septic shock. Prognostic models that reliably estimate the risk of poor outcome have the potential to serve as tools for enrichment of clinical trials, inform individual patient decision-making, to serve as a benchmark for quality improvement efforts, and to facilitate risk-stratified analyses of clinical data. Further, such models have the potential to provide insight regarding the pathogenesis of septic shock and how it varies among different patients.
The Pediatric Sepsis Biomarker Risk Model (PERSEVERE) incorporates a panel of biomarkers and age into a decision tree estimating the baseline risk of mortality in children with septic shock (Wong et al., 2012, Wong et al., 2014b). The PERSEVERE biomarkers are proteins measured in the blood compartment on day 1 of presentation to the intensive care unit with septic shock. To reflect change in risk over time, we developed a temporal version of the model (tPERSEVERE) (Wong et al., 2014c). tPERSEVERE considers how the PERSEVERE biomarker concentrations change from day 1 to day 3 of septic shock, and how these changes are associated with poor outcome.
We envisioned that a reliable temporal model could potentially serve as a monitor for therapeutic efficacy, in combination with traditional clinical parameters. For example, if the model signals a decreasing risk for poor outcome over time, this could be indicative of therapeutic efficacy. Alternatively, if the model signals increasing risk or unchanged risk from a high baseline risk, this could indicate lack of efficacy and could potentially trigger a reassessment of the therapeutic regimen.
Models such as tPERSEVERE require prospective testing in order to assess validity and generalizability. In the current study, we prospectively tested the performance of tPERSEVERE in an independent validation cohort, and use the results to explore how biological variation may be associated with the pathogenesis of poor outcomes in septic shock.
2. Methods
2.1. Study Subjects and Data Collection
The validation cohort consisted of 168 subjects prospectively enrolled since the initial derivation of tPERSEVERE. The protocol for collection and use of biological specimens and clinical data was approved by the Institutional Review Boards of each of 18 participating institutions. Children ≤ 18 years of age admitted to the pediatric intensive care unit (PICU) and meeting pediatric-specific consensus criteria for septic shock were eligible for enrollment (Goldstein et al., 2005, Wong et al., 2007). The only exclusion criterion was the inability to obtain informed consent, which was obtained from parents or legal guardians prior to any data or sample collection.
Serum samples were obtained within 24 h of first meeting the criteria for septic shock in the PICU, which was typically at presentation. These are referred to as “day 1” samples. “Day 3” samples were collected 48 h after the day 1 samples. Clinical and laboratory data were collected daily while in the PICU. Organ failure data were tracked up to day seven of septic shock using previously published criteria (Goldstein et al., 2005). Mortality was tracked for 28 days after enrollment. Complicated course was defined as the persistence of two or more organ failures at day seven of septic shock or 28-day mortality (Wong et al., 2015). Illness severity was estimated using PRISM scores (Pollack et al., 1997). Baseline mortality probability was estimated using PERSEVERE (Wong et al., 2012, Wong et al., 2014b).
2.2. PERSEVERE Biomarkers
PERSEVERE includes C–C chemokine ligand 3 (CCL3), interleukin 8 (IL8), heat shock protein 70 kDa 1B (HSPA1B), granzyme B (GZMB), and matrix metallopeptidase 8 (MMP8) (Wong et al., 2012). Serum concentrations of these biomarkers were measured using a multi-plex magnetic bead platform (MILLIPLEX™ MAP, EMD Millipore Corporation, Billerica, MA). Biomarker concentrations were measured in a Luminex® 100/200 System (Luminex Corporation, Austin, TX), according the manufacturers' specifications. Assay performance data were previously published (Wong et al., 2012).
2.3. Statistical Analysis and Validation of tPERSEVERE
Initially, data are described using medians, interquartile ranges, frequencies, and percentages. Comparisons between groups used the Mann–Whitney U-test, Chi-square, or Fisher's exact tests as appropriate. Descriptive statistics and comparisons used SigmaStat Software (Systat Software, Inc., San Jose, CA).
Each study subject was assigned a probability of a complicated course using the previously published tPERSEVERE model (Wong et al., 2014c). tPERSEVERE performance is reported using diagnostic test statistics with 95% confidence intervals computed using SPSS 23.0 (IBM Corporation, Armonk, NY), R (base version 3.1.1) and package epiR (version 0.9–62) (R Core Team, 2014, Stevenson and with contributions from Nunes T, Heuer C, Marsh).
2.4. Redesigning tPERSEVERE
A priori, we determined that if the area under the receiver operating curve was less than 0.7 in the validation cohort, we would redesign tPERSEVERE. Initially, we explored reasons for failure by comparing the validation cohort to the original cohort, and by characterizing false negatives and comparing them to true positives. Then, to redesign tPERSEVERE we combined the 168 prospectively enrolled subjects, and the 299 previously reported subjects. From this pooled cohort (n = 467) we randomly selected 80% of the subjects for a training cohort (n = 374) and the remaining 20% were retained as a test cohort (n = 93).
The modeling procedures for redesigning tPERSEVERE used CART methodology (Salford Predictive Modeler v6.6, Salford Systems, San Diego, CA) (Che et al., 2011, Muller and Mockel, 2008). The primary outcome variable for the modeling was a complicated course, as defined above (Wong et al., 2015). Using complicated course as the primary outcome variable allows for the exploration of association between temporal biomarker changes and nuances of sepsis severity beyond the dichotomy of “alive” vs. “dead”. Continuous, predictor variables for the modeling procedure included the PERSEVERE-based mortality probability, day 1 and day 3 PERSEVERE biomarker values, age, and a derived variable termed “delta”, which subtracted the day 1 value for a given biomarker from the respective day 3 value. Dichotomous predictor variables included gender, and the presence of any co-morbidity, malignancy, immune suppression, or previous bone marrow transplantation. Weighting of cases and the addition of cost for misclassification were not used in the modeling procedures. The code and data used to generate the model is available from the authors.
2.5. Funding
The study was funded by the National Institutes of Health, National Institute of General Medical Sciences. The funder played no other role in the study, in the writing of the manuscript, or in the decision to submit the manuscript.
3. Results
3.1. Prospective Validation of tPERSEVERE
Table 1 shows the demographic and clinical characteristics of the validation cohort (n = 168). Sixty-three subjects (38%) had a complicated course. Among these, 25 (40%) died by study day 28. Compared to subjects with a non-complicated course, those in the complicated course group had a higher PRISM score, a higher PERSEVERE mortality probability, and a greater proportion had immune suppression or had undergone previous bone marrow transplantation. No other differences were noted.
Table 1.
Demographic and clinical characteristics of the validation cohort.
| Non-complicated course | Complicated course | |
|---|---|---|
| N (%) | 105 (63) | 63 (38) |
| Median age, years (IQR) | 5.3 (1.8–12.8) | 4.8 (1.1–14.9) |
| Males, # (%) | 60 (57) | 37 (59) |
| 28-day mortality, # (%) | 0 (0) | 25 (40) |
| Median PRISM score (IQR) | 10 (7–15) | 13 (8–21)1 |
| PERSEVERE mortality probability (95% C.I.) | 6.5 (4.6–8.4) | 12.9 (9.5–16.3)2 |
| # With gram negative bacteria (%) | 27 (26) | 15 (24) |
| # With gram positive bacteria (%) | 19 (18) | 14 (22) |
| # With other pathogen isolated (%) | 9 (9) | 5 (8) |
| # With no pathogen identified (%) | 50 (48) | 29 (46) |
| # With comorbidity (%) | 67 (64) | 47 (75) |
| # With malignancy (%) | 20 (19) | 16 (25) |
| # With immune suppression (%) | 20 (19) | 22 (35)3 |
| # With bone marrow transplantation (%) | 4 (4) | 10 (16)3 |
p < 0.05 vs. non-complicated course; Rank sum test.
p < 0.05 vs. non-complicated course; t-test.
p < 0.05 vs. non-complicated course; Chi-square.
The validation cohort subjects were classified according to the tPERSEVERE decision tree. For estimating the probability of a complicated course, tPERSEVERE had an area under the curve (AUC) of 0.67 (95% CI: 0.58–0.75). Table 2 shows the diagnostic test characteristics of tPERSEVERE in the validation cohort.
Table 2.
Test characteristics of tPERSEVERE for estimating the probability of a complicated course in the validation cohort.
| No. of false positives | 38 |
| No. of true positives | 42 |
| No. of true negatives | 67 |
| No. of false negatives | 21 |
| Sensitivity | 67% (54–78) |
| Specificity | 64% (54–73) |
| Positive predictive value | 52% (41–64) |
| Negative predictive value | 76% (66–84) |
| + Likelihood ratio | 1.84 (1.35–2.51) |
| − Likelihood ratio | 0.52 (0.36–0.76) |
| AUC | 0.67 (0.58–0.75) |
3.2. Failure Analysis
The diagnostic test characteristics of tPERSEVERE for estimating the probability of a complicated course in the validation cohort were poor. We therefore conducted analyses to determine what factors may have contributed to the poor performance of tPERSEVERE.
We first compared the clinical characteristics of the study subjects in the original derivation cohort (Wong et al., 2014c). to that of the validation cohort. The validation cohort had a higher prevalence of a complicated course (38%) than the original derivation cohort (23%, p < 0.001). This difference may have been due to a higher comorbidity burden in the validation cohort, compared to the derivation cohort (68% vs. 42%; p < 0.05). Of note, a higher proportion of subjects in the validation cohort had immune suppression (25% vs. 13%; p < 0.05) or malignancy (21% vs. 9%, p < 0.05), compared to the derivation cohort subjects.
Table 3 characterizes the false negative and true positive subjects within the validation cohort. The true positive subjects had a higher median PRISM score and a greater proportion of males, compared to the false negative subjects. No other differences were noted.
Table 3.
Comparison of the false negative and true positive subjects in the validation cohort.
| VARIABLE | False negatives | True positives | p Value |
|---|---|---|---|
| N | 21 | 42 | = |
| PRISM; median (IQR) | 10 (8–14) | 17 (8–22) | 0.031 |
| Age; median (IQR) | 4.9 (2.6–13.8) | 4.4 (0.8–15.2) | 0.370 |
| Deaths; # (%) | 5 (24) | 20 (48) | 0.069 |
| Males; # (%) | 8 (38) | 29 (69) | 0.019 |
| Malignancy; # (%) | 6 (29) | 10 (23) | 0.682 |
| Immune suppression; # (%) | 5 (24) | 17 (40) | 0.191 |
| Bone marrow transplantation; # (%) | 1 (5) | 9 (21) | 0.144 |
3.3. Redesigning tPERSEVERE
tPERSEVERE may have performed poorly in the validation cohort because of differences in the clinical characteristics between the derivation and the validation cohort subjects. Further, the difference in illness severity between the false negative and true positives subjects suggests that tPERSEVERE may not perform well across a wide spectrum of illness severity, as measured by PRISM. We therefore redesigned tPERSEVERE with the goals of improved performance across a diverse cohort of children with septic shock and providing biological insight regarding poor outcomes.
Table 4 shows the demographic and clinical characteristics of the training and test sets for the modeling procedures. Subjects in the test set were older and had a higher mortality rate, compared to subjects in the training set. No other differences were noted.
Table 4.
Demographic and clinical characteristics of the training and test sets for re-designing tPERSEVERE.
| Training set | Test set | |
|---|---|---|
| N | 374 | 93 |
| Median age, years (IQR) | 3.0 (1.0–7.5) | 5.6 (1.6–9.9)1 |
| Males, # (%) | 220 (58) | 55 (59) |
| 28-day mortality, # (%) | 31 (8) | 15 (16)2 |
| # with complicated course, (%) | 105 (28) | 26 (28) |
| Median PRISM score (IQR) | 12 (8–20) | 11 (8–17) |
| PERSEVERE mortality probability (95% C.I.) | 9.1 (7.6–10.6) | 9.9 (6.9–12.9) |
| # With gram negative bacteria (%) | 91 (24) | 22 (24) |
| # With gram positive bacteria (%) | 85 (23) | 20 (22) |
| # With other pathogen isolated (%) | 41 (11) | 10 (11) |
| # With no pathogen identified (%) | 157 (42) | 41 (44) |
| # With comorbidity (%) | 194 (52) | 46 (49) |
| # With malignancy (%) | 50 (13) | 14 (15) |
| # With immune suppression (%) | 66 (18) | 14 (15) |
| # With bone marrow transplantation (%) | 16 (4) | 8 (9) |
p < 0.05 vs. training set; Rank sum test.
p < 0.05 vs. training set; Chi-square.
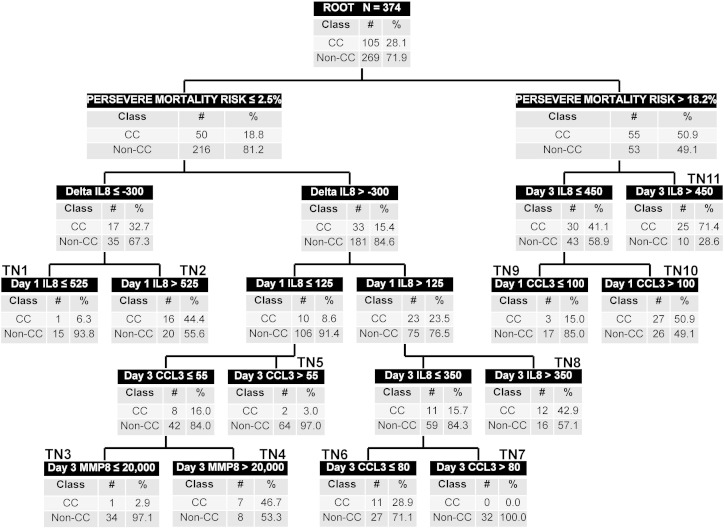
Fig. 1 shows tPERSEVERE based on the training set. Baseline PERSEVERE mortality risk occupied the first level decision rule. Day 1 and day 3 IL8 values, delta IL8, day 1 and day 3 CCL3 values, and day 3 MMP8 values also contributed to the predictive capacity of the redesigned tPERSEVERE model. None of the other predictor variables considered in the modeling process contributed to predictive capacity.
Fig. 1.
The redesigned tPERSEVERE classification tree. The classification tree includes the PERSEVERE-based mortality probability, day 1 and day 3 interleukin 8 (IL8) concentrations, the delta IL8 value, day 1 and day 3 C–C chemokine ligand 3 (CCL3) concentrations, and the day 3 matrix metallopeptidase 8 (MMP8) concentration. The biomarker concentrations are shown in ng/ml. The root node provides the total number of subjects in the training set, and the number of subjects with and without a complicated course, with the respective rates. Each daughter node provides the respective decision rule criterion and the number of subjects with and without a complicated course, with the respective rates. A negative value for a “delta” occurs when the biomarker level is, on average, decreasing. Terminal nodes (TN) TN1, TN3, TN5, TN7, and TN9 are low risk terminal nodes (0.0% to 15.0% risk of complicated course). TN2, TN4, TN6, and TN8 are intermediate risk terminal nodes (28.9% to 46.7% risk of complicated course). TN10 and TN11 are high risk terminal nodes (≥ 50% risk of complicated course).
The redesigned tPERSEVERE model had five low risk terminal nodes (0.0% to 15.0% risk of complicated course; nodes 1, 3, 5, 7, and 9), four intermediate risk terminal nodes (28.9% to 46.7% risk of complicated course; nodes 2, 4, 6, and 8), and two high risk terminal nodes (> 50% risk of complicated course; nodes 10 and 11). Among the 169 subjects classified as low risk, 162 (96%) did not have a complicated course. Among the 117 subjects classified as intermediate risk, 46 (39%) had a complicated course. Among the 88 subjects classified as high risk, 52 (59%) had a complicated course. Table 5 shows the diagnostic test characteristics of the redesigned tPERSEVERE model for estimating the risk of a complicated course in the training set.
Table 5.
Test characteristics of the redesigned tPERSEVERE model for estimating the probability of a complicated course in the training and test sets.
| Training set | Test set | |
|---|---|---|
| No. of false positives | 107 | 24 |
| No. of true positives | 98 | 23 |
| No. of true negatives | 162 | 43 |
| No. of false negatives | 7 | 3 |
| Sensitivity | 93% (86–97) | 88% (69–97) |
| Specificity | 60% (54–66) | 64% (51–75) |
| Positive predictive value | 48% (41–55) | 49% (34–64) |
| Negative predictive value | 96% (92–98) | 93% (82–99) |
| + Likelihood ratio | 2.35 (2.01–2.74) | 2.47 (1.74–3.50) |
| − Likelihood ratio | 0.11 (0.05–0.23) | 0.18 (0.06–0.53) |
| AUC | 0.83 (0.79–0.87) | 0.84 (0.75–0.92) |
3.4. Testing the Redesigned tPERSEVERE Model
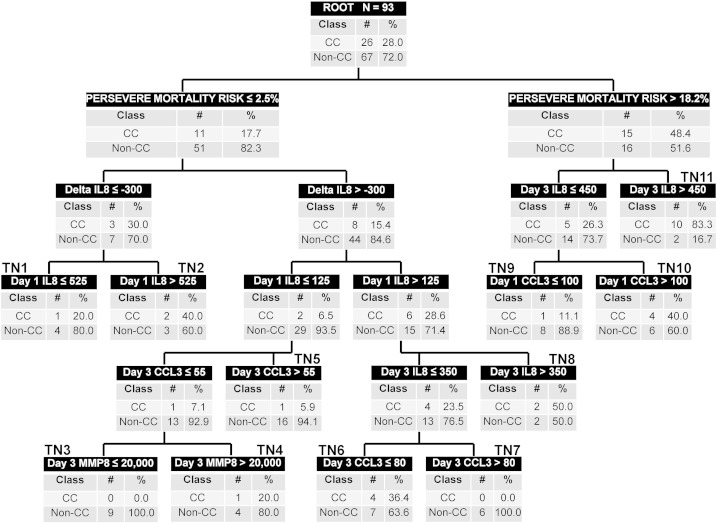
Fig. 2 shows the classification of the 93 subjects in the test set, according to the redesigned tPERSEVERE model. Among the 46 subjects classified as low risk, 43 (93%) did not have a complicated course. Among the 25 subjects classified as intermediate risk, 25 (36%) had a complicated course. Among the 22 subjects classified as high risk, 14 (64%) had a complicated course. Table 5 shows the diagnostic test characteristics of the redesigned tPERSEVERE model for estimating the risk of a complicated course in the test set. Based on the performance of the redesigned tPERSEVERE model, we estimate that if 350 subjects were included in a prospective evaluation and the accuracy remained at 60% with a sensitivity of 93%, the confidence intervals would be sufficiently narrow to conclude validity at about ± 5%.
Fig. 2.
Classification of the test set according to the redesigned tPERSEVERE model. The test set subjects (n = 93) were classified according to the redesigned tPERSEVERE model without any modifications. The same conventions apply to the decision tree as described for Fig. 1.
4. Discussion
Prospective testing of clinical risk models is imperative for assessing validity. We tested the performance of tPERSEVERE prospectively and found that it performed poorly. One possible reason for failure is that the originally derived model was mathematically over fit, without providing any biological insight (Wong et al., 2014c). Another possible reason is that clinical characteristics having the potential to affect the risk of poor outcome differed between the derivation and validation cohorts, including the prevalence of a complicated course. Finally, our failure analysis suggested that tPERSEVERE performed poorly in those subjects with lower PRISM-based illness severity who went on to develop a complicated course. Accordingly, we redesigned tPERSEVERE using a modeling approach that included these potential confounders.
We attempted to account for differences in disease severity by combining the derivation and validation cohorts, and by randomly selecting subjects for training and test sets. This generated two groups of subjects with similar complicated course prevalence. The two groups were also well matched for the majority of clinical variables examined, except for age and mortality. To further account for differences in clinical phenotype, we considered comorbidity variables, age, and gender in the modeling procedures. None of these clinical variables added to the predictive capacity of the redesigned tPERSEVERE model.
The original version of tPERSEVERE contains only the absolute day 1 and day 3 PERSEVERE biomarker values. To redesign a better performing model that could also provide biological insight regarding the development of poor outcome in children with septic shock, we considered two additional biomarker-based variables. First, we considered the PERSEVERE-based mortality probability. This is a composite variable reflecting day 1 biomarker values for CCL3, IL8, HSPA1B, GZMB, and MMP8, and age (Wong et al., 2012, Wong et al., 2014b). Second, we considered the possibility that a derived variable reflecting the changes in biomarker values from day 1 to day 3, could provide biological information beyond the absolute day 1 and day 3 biomarker values.
The redesigned tPERSEVERE model performed well in the test set. Beyond risk prediction, we suggest that the redesigned model provides useful biological information regarding the risk of a complicated course in children with septic shock. Current paradigms for understanding the pathophysiology of sepsis place patients into two broad endotypes (Hotchkiss and Sherwood, 2015, Hutchins et al., 2014, Skrupky et al., 2011). One endotype involves patients with excessive inflammation. In these patients, inflammatory mechanisms intended for the eradication of infection become excessive and thereby cause collateral damage to host tissues, which in turn leads to organ injury and higher risk for poor outcome. The other endotype involves patients with ineffective inflammation and a state of relative immune suppression. These patients are also at risk for poor outcome because they are unable to effectively clear infections or are at risk for secondary infections. These two endotypes represent extremes of a spectrum; it is likely that many patients with sepsis reside somewhere within this spectrum. In addition, many patients move along this spectrum during the course of illness.
IL8 and CCL3 are the major chemokines responsible for recruitment and activation of white blood cells and are thus key mediators of inflammation (Aziz et al., 2013). In murine models of experimental sepsis, the respective homologs for IL8 and CCL3 increase in proportion to sepsis severity (Ebong et al., 1999). Subjects occupying the right side of the redesigned decision tree (Fig. 1) had an intermediate to high PERSEVERE-based mortality risk. Among these subjects, if IL8 levels remained relatively high on day 3 (> 450 ng/ml, terminal node 11) or if the day 1 CCL3 level was greater than 100 ng/ml (terminal node 10), there was a greater than 50% probability of a complicated course. Conversely, subjects with a high PERSEVERE-based mortality probability, but a relatively low day 3 IL8 concentration (≤ 450 ng/ml) and a day 1 CCL3 concentration less than 100 ng/ml (terminal node 9), had a much lower probability (15%) of a complicated course. Thus, excessive inflammation in the presence of high baseline risk portends poor outcome, while lesser inflammation despite high baseline risk is associated with diminishing risk. These observations are consistent with the sepsis endotype characterized by excessive inflammation.
Subjects occupying the left side of the redesigned decision tree had a low PERSEVERE-based mortality risk. In these subjects, if IL8 decreased by at least 300 ng/ml from day 1 to day 3, the probability of a complicated course was 6.3% in the context of a day 1 IL8 concentration ≤ 525 ng/ml (terminal node 1), but 44.4% in the context of a relatively high day 1 IL8 concentration (> 525 ng/ml, terminal node 2). In other words, relatively lower baseline IL8 concentrations that are decreasing portend lower risk than relatively higher baseline IL8 concentrations that are decreasing. This suggests those subjects whose inflammatory state is excessive at baseline do poorly, even though inflammation may be diminishing over the first few days. In these same low-baseline-risk subjects, subjects with a day 1 IL8 concentration > 125 ng/ml and a day 3 IL8 concentration of > 350 ng/ml had a 42.8% probability of a complicated course (terminal node 8). These observations are also consistent with the sepsis endotype characterized by excessive inflammation and, in particular, suggest evolving excessive inflammation.
In contrast, subjects occupying terminal nodes 5, 6, and 7 may represent the sepsis endotype characterized by immune suppression. In these subjects higher CCL3 concentrations on day 3 were associated with a low risk of a complicated course (terminal nodes 5 and 7), whereas subjects with relatively lower CCL3 concentrations on day 3 had a higher risk of a complicated course (terminal node 6). It may be that these subjects have insufficient recruitment of white blood cells and therefore had a state of relative immune suppression.
Finally, subjects in terminal nodes 3 and 4 may represent patients along the spectrum of excessive inflammation and immune suppression. These patients are defined initially by a low PERSEVERE-based mortality risk, a relatively low starting IL8 and a low day 3 CCL3 concentration. In these subjects, day 3 MMP8 differentiates between those at low risk and those at intermediate risk of a poor outcome. This is consistent with data from murine models exploring the role of MMP8 in sepsis. In these studies, genetic ablation or pharmacological inhibition of MMP8 conferred a survival advantage in mice subjected to sepsis, and this effect was associated with decreased activation of the pro-inflammatory transcription factor, NF-κB, and other indices of inflammation (Solan et al., 2012, Atkinson et al., 2015).
We note the limitations of the study. The process of redesigning and testing the tPERSEVERE model was based on pooling of all available subjects, random selection, and subsequent allocation into training and test sets. This process could have introduced unknown or unintended biases. It is therefore possible that the redesigned model is over fit, despite its good performance in the test set. The redesigned model requires testing with a fully independent validation cohort. Our interpretation of the biological information that the model might be providing is based on existing principles of potential sepsis endotypes and data from murine models of sepsis, but is speculative nonetheless. It will be important in future studies to directly determine the relative states of inflammation and immune suppression in subjects allocated to the respective terminal nodes potentially associated with excessive inflammation or immune suppression.
In conclusion, tPERSEVERE performed poorly upon prospective testing. We identified potential confounders leading to failure, and this prompted a redesign of the tPERSEVERE model using randomly selected training and test sets. The redesigned model performed well in the test set, and is consistent with current paradigms of sepsis endotypes involving excessive inflammation and immune suppression. The redesigned model requires prospective testing in a validation cohort and functional studies to confirm the validity of our interpretations of the biological information provided by the model. Successful validation could position tPERSEVERE as a monitor of therapeutic efficacy in patients with septic shock and as a tool for understanding septic shock endotypes predicated on patterns of inflammation and immunity.
Author Contributions
Hector R. Wong: Conceived and developed the study, obtained funding for the study, directly took part in the analyses, and wrote the manuscript.
Natalie Z. Cvijanovich, Nick Anas, Geoffrey L. Allen, Neal J. Thomas, Michael T. Bigham, Scott L. Weiss, Julie Fitzgerald, Paul A. Checchia, Keith Meyer, Michael Quasney, Mark Hall, Rainer Gedeit, Robert J. Freishtat, Jeffrey Nowak, Shekhar S. Raj, and Shira Gertz: Enrolled patients, provided biological samples and clinical data for the database, and edited the manuscript.
Kelli Howard and Erin Frank: Maintained the clinical database and coordinated all inter-institutional research activity.
Kelli Harmon: Maintained the biological repository and processed all biological samples.
Patrick Lahni: Conducted all biomarker assays.
Kimberly Hart: Assisted with statistical analysis.
Christopher J. Lindsell: Developed the study, assisted with analysis, and edited the manuscript.
Funding Source
Supported by National Institutes of Health Grants RO1GM064619, RO1GM099773, and R01GM108025. Supported in part by an Institutional Clinical and Translational Science Award, NIH/NCRR 8UL1 TR000077.
Author Competing Interests
Dr. Wong and the Cincinnati Children's Hospital Research Foundation have submitted a provisional patent application for tPERSEVERE.
Dr. Lindsell is named as a co-inventor in the above patent application.
The other authors have no competing interests to report.
Acknowledgments
We thank the following clinical research coordinators for enrolling patients at the various study sites: Debra Spear, Jenny Bush, Mary Ann De Liberto, Trisha Williams, Amber Hughes, Michelle Goldsworthy, Christi Rider, Mary Ellen Riodan, Tiffany Patterson, Ofelia Vargas, Monica Weber, Lauren Hoadley, Heather Anthony, Lisa Steele, Angela Doucette, Katherine Woods, and Claudia Rodriquez Paez.
References
- Alder M.N., Lindsell C.J., Wong H.R. The pediatric sepsis biomarker risk model: potential implications for sepsis therapy and biology. Expert Rev. Anti-Infect. Ther. 2014;12(7):809–816. doi: 10.1586/14787210.2014.912131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson S.J., Nolan M., Klingbeil L., Harmon K., Lahni P., Zingarelli B. Intestine-derive matrix metalloproteinase-8 is a critical mediator of polymicrobial peritonitis. Crit. Care Med. 2015 doi: 10.1097/CCM.0000000000001374. (Epub ahead of print). (doi: 10.1097/CCM.0000000000001374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz M., Jacob A., Yang W.L., Matsuda A., Wang P. Current trends in inflammatory and immunomodulatory mediators in sepsis. J. Leukoc. Biol. 2013;93(3):329–342. doi: 10.1189/jlb.0912437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che D., Liu Q., Rasheed K., Tao X. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv. Exp. Med. Biol. 2011;696:191–199. doi: 10.1007/978-1-4419-7046-6_19. [DOI] [PubMed] [Google Scholar]
- Cohen J., Vincent J.L., Adhikari N.K., Machado F.R., Angus D.C., Calandra T. Sepsis: A roadmap for future research. Lancet Infect. Dis. 2015;15(5):581–614. doi: 10.1016/S1473-3099(15)70112-X. [DOI] [PubMed] [Google Scholar]
- Ebong S., Call D., Nemzek J., Bolgos G., Newcomb D., Remick D. Immunopathologic alterations in murine models of sepsis of increasing severity. Infect. Immun. 1999;67(12):6603–6610. doi: 10.1128/iai.67.12.6603-6610.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B., Giroir B., Randolph A. International pediatric sepsis consensus conference: definitions for sepsis and organ dysfunction in pediatrics. Pediatr. Crit. Care Med. 2005;6(1):2–8. doi: 10.1097/01.PCC.0000149131.72248.E6. [DOI] [PubMed] [Google Scholar]
- Hotchkiss R.S., Sherwood E.R. Immunology. getting sepsis therapy right. Science. 2015;347(6227):1201–1202. doi: 10.1126/science.aaa8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins N.A., Unsinger J., Hotchkiss R.S., Ayala A. The new normal: immunomodulatory agents against sepsis immune suppression. Trends Mol. Med. 2014;20(4):224–233. doi: 10.1016/j.molmed.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan J.M., Wong H.R. Biomarker discovery and development in pediatric critical care medicine. Pediatr. Crit. Care Med. 2011;12(2):165–173. doi: 10.1097/PCC.0b013e3181e28876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R., Mockel M. Logistic regression and CART in the analysis of multimarker studies. Clin. Chim. Acta. 2008;394(1–2):1–6. doi: 10.1016/j.cca.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Pollack M.M., Patel K.M., Ruttimann U.E. The pediatric risk of mortality III—acute physiology score (PRISM III-APS): a method of assessing physiologic instability for pediatric intensive care unit patients. J. Pediatr. 1997;131(4):575–581. doi: 10.1016/s0022-3476(97)70065-9. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: a language and environment for statistical computing. (URL http://www.R-project.org/) [Google Scholar]
- Skrupky L.P., Kerby P.W., Hotchkiss R.S. Advances in the management of sepsis and the understanding of key immunologic defects. Anesthesiology. 2011;115(6):1349–1362. doi: 10.1097/ALN.0b013e31823422e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solan P.D., Dunsmore K.E., Denenberg A.G., Odoms K., Zingarelli B., Wong H.R. A novel role for matrix metalloproteinase-8 in sepsis. Crit. Care Med. 2012;40(2):379–387. doi: 10.1097/CCM.0b013e318232e404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson M with contributions from Nunes T, Heuer C, Marshall J, Sanchez J, Thornton R, Reiczigel J, Robison-Cox J, Sebastiani P, Solymos P, Yoshida K, and Firestone S (2015). epiR: Tools for the Analysis of Epidemiological Data. R package version 0.9–62. (http://CRAN.R-project.org/package=epiR).
- Wong H.R., Shanley T.P., Sakthivel B., Cvijanovich N., Lin R., Allen G.L. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol. Genomics. 2007;30(2):146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.R., Salisbury S., Xiao Q., Cvijanovich N.Z., Hall M., Allen G.L. The pediatric sepsis biomarker risk model. Crit. Care. 2012;16(5):R174. doi: 10.1186/cc11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.R., Lindsell C.J., Pettila V., Meyer N.J., Thair S.A., Karlsson S. A multibiomarker-based outcome risk stratification model for adult septic shock. Crit. Care Med. 2014;42(4):781–789. doi: 10.1097/CCM.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.R., Weiss S.L., Giuliano J.S., Jr., Wainwright M.S., Cvijanovich N.Z., Thomas N.J. Testing the prognostic accuracy of the updated pediatric sepsis biomarker risk model. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0086242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.R., Weiss S.L., Giuliano J.S., Jr., Wainwright M.S., Cvijanovich N.Z., Thomas N.J. The temporal version of the pediatric sepsis biomarker risk model. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0092121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.R., Cvijanovich N.Z., Anas N., Allen G.L., Thomas N.J., Bigham M.T. Developing a Clinically feasible personalized medicine approach to pediatric septic shock. Am. J. Respir. Crit. Care Med. 2015;191(3):309–315. doi: 10.1164/rccm.201410-1864OC. [DOI] [PMC free article] [PubMed] [Google Scholar]