Abstract
Background
Panax ginseng cannot be cultivated on the same land consecutively for an extended period, and the underlying mechanism regarding microorganisms is still being explored.
Methods
Polymerase chain reaction and denaturing gradient gel electrophoresis (PCR-DGGE) and BIOLOG methods were used to evaluate the microbial genetic and functional diversity associated with the P. ginseng rhizosphere soil in various cultivation ages and modes.
Results
The analysis of microbial diversity using PCR-DGGE showed that microbial communities were significantly variable in composition, of which six bacterial phyla and seven fungal classes were detected in P. ginseng soil. Among them, Proteobacteria and Hypocreales dominated. Fusarium oxysporum, a soilborne pathogen, was found in all P. ginseng soil samples except R0. The results from functional diversity suggested that the microbial metabolic diversity of fallow soil abandoned in 2003 was the maximum and transplanted soil was higher than direct-seeding soil and the forest soil uncultivated P. ginseng, whereas the increase in cultivation ages in the same mode led to decreases in microbial diversity in P. ginseng soil. Carbohydrates, amino acids, and polymers were the main carbon sources utilized. Furthermore, the microbial diversity index and multivariate comparisons indicated that the augmentation of P. ginseng cultivation ages resulted in decreased bacterial diversity and increased fungal diversity, whereas microbial diversity was improved strikingly in transplanted soil and fallow soil abandoned for at least one decade.
Conclusion
The key factors for discontinuous P. ginseng cultivation were the lack of balance in rhizosphere microbial communities and the outbreak of soilborne diseases caused by the accumulation of its root exudates.
Keywords: DGGE, discontinuous cultivation, metabolic function, microbial composition, Panax ginseng
1. Introduction
Panax ginseng Meyer (Araliaceae), one of the most well-known Chinese herbal medicines, was formerly a wild plant grown in the northeastern region of China. Generally, P. ginseng is mainly dependent on artificial cultivation in China, and it is also cultivated in Korea and Japan [1]. However, the yield is severely hindered by continuous cropping obstacles of P. ginseng. In brief, P. ginseng cannot be cultivated on the same plot of land consecutively for several years or even decades owing to high requirements for soil quality [2]. Continuous cropping obstacles of P. ginseng always make its roots turn rusty and rot on account of soilborne diseases [3]. A large-scale deforestation is increasingly sharpening owing to discontinuous cultivation that not only damages forest resources but is also a limiting bottleneck on the sustainable development of P. ginseng crops. Hence, the contradiction between P. ginseng industries and the forest industry has become a major technical problem that needs to be solved urgently. Accumulating lines of evidence indicate that four major factors—deterioration of soil physicochemical characteristics, outburst of soilborne diseases, imbalance of soil microbial community, and autotoxicity of P. ginseng—can result in discontinuous cultivation of P. ginseng [4], [5]. In the past several years, although a few researchers have carried out investigations on soil improvement and sterilization, the underlying mechanisms responsible for the relationship between microbial diversity and discontinuous cultivation are still poorly understood.
Increasing evidence indicates that rhizosphere microorganisms play a vital role in nutrient cycling, organic matter decomposition, and the maintenance of soil fertility [6]. In addition, the soil microbial community is also an important bioindicator of soil function [7]. Therefore, many investigations on discontinuous cultivation were focused on the evaluation of soil quality and the microbial community. Interestingly, several findings from previous studies have demonstrated that continuous farming resulted in an imbalance in soil ecology and alterations of microbial diversity in rhizosphere soil [8], [9]. Although a number of microbial strains (comprising < 1% in total) have been isolated from successive cultivation soil [10], [11], [12], most of the microbial communities and their composition in rhizosphere soil are still difficult to analyze. The availability of modern tools in microbial ecology have permitted the study of microbial communities related to plant growth and development, in situ localization of important forms, and the monitoring of microbes when their quantities change in the soil environment [13].
In the present study, molecular culture-independent methods based on 16S rDNA and 18S rDNA gene diversity [14], [15], polymerase chain reaction and denaturing gradient gel electrophoresis (PCR-DGGE), and random amplified polymorphic DNA were successfully used to examine the microbial community and dynamics of dominant microbial species in plant rhizosphere soil during its growth [4], [16]. Compared with the traditional analysis method, PCR-DGGE has been one of the most widely used tools to assess the structure of microbial communities in soil and to determine the community dynamics in response to soil and other variations both quickly and economically [17], [18]. Moreover, soil functional diversity was commonly used as an indicator for soil quality, and studies on the relationship between the rhizosphere microbial structure and function have been becoming a hot topic in the field of soil ecosystem. For a better understanding of the relationship between microbial diversity and soil function, BIOLOG EcoPlate was used to study the metabolic function variance of rhizosphere soil microbes [19]. It is known that BIOLOG is an effective method based on carbon substrate utilization by microbial communities to analyze data via multivariate statistics, including principal component analysis (PCA) and the dynamics of the microbial community, as it is revealed by BIOLOG metabolic variance [20].
Recently, a variety of culture-independent approaches—including random amplified polymorphic DNA, PCR-DGGE, and BIOLOG—have been applied for the investigation of bacterial diversity and metabolic function diversity of P. ginseng soil [4], [21], [22]. However, only a few trials have reported on the microbial community diversity of P. ginseng soil among different cultivation ages using both PCR-DGGE and BIOLOG. Therefore, it is of great interest to evaluate the influence of P. ginseng cultivation ages and modes on the bacterial and fungal genetic diversity and metabolic functional diversity with the combination of PCR-DGGE and BIOLOG, respectively. Findings from the present studies will help to elucidate variations in soil microbial community and to elucidate the status of microflora variation underlying the mechanisms of continuous P. ginseng farming, although further studies are required prior to practical application.
2. Materials and methods
2.1. Soil sampling and DNA extraction
Two fields—Yushu Village (Y; 42°32′N, 127°08′E, 537 m) and Funan Village (F or F′; 42°08′N, 127°32′E, 781 m), located in Jilin province, China—were selected for the experiments. A bioassay test was carried out, which comprised five cultivation ages—three different direct-seeding ages [direct seeding for 1 yr (R1), direct seeding for 2 yr (R2) and direct seeding for 4 yr (R4)] from Y, and two different transplanted ages [2-yr-old P. ginseng transplanted to another field for 2 yr (R2 + 2) and 3-yr-old P. ginseng transplanted to another field for 3 yr (R3 + 3)]; in addition, three cultivation modes—direct seeding mode, transplanted mode, and fallow soil of P. ginseng abandoned for a long period [fallow soil of P. ginseng abandoned in 2007 (RL-07) and in 2003 (RL-03) from F′ soil]—were compared with each other. Soil samples were collected from the rhizosphere soil of P. ginseng in growth stages (May 2014), and the forest soil of uncultivated P. ginseng (R0) was used as the control soil (Table 1).
Table 1.
Soil samples used for diversity analysis and main physicochemical characteristics (, n = 3)
| Soil samples | Age (y) | Growth model1) | pH | Organic matter (g/kg)2) |
|---|---|---|---|---|
| R0 (control) | 0 | Forest soil uncultivated Panax ginseng | 6.47 ± 0.04a | 28.71 ± 1.99a |
| R1 | 1 | 1 | 5.74 ± 0.05b | 27.48 ± 1.23a,b |
| R2 | 2 | 2 | 5.53 ± 0.04c | 24.42 ± 1.39b |
| R4 | 4 | 4 | 5.44 ± 0.03c | 14.39 ± 1.24c |
| R2 + 2 | 4 | 2 + 2 | 5.42 ± 0.02c | 13.75 ± 0.78c |
| R3 + 3 | 6 | 3 + 3 | 5.21 ± 0.04c | 10.96 ± 0.76d |
| RL-07 | / | Fallow soil abandoned for 6 y | 5.49 ± 0.03bc | 6.37 ± 0.38e |
| RL-03 | / | Fallow soil abandoned for 10 y | 5.62 ± 0.04b | 25.89 ± 0.79b |
In the growth model column, a + b means Panax ginseng growing at one place for a yr, then transplanted to another place and growing for b yr
The letters indicate the tested with Shortest Significant ranges (SSR) at P = 0.05 of different treatments. Different letters denote a significant difference at p < 0.05 level
At the beginning of the growing season, these fields were administered following good agricultural practice. Soil samples were collected from five replicate plots randomly distributed over the fields. The entire root system along with rhizosphere soil was collected by digging at a depth of 20 cm from five healthy plants, and sampling was performed as described previously [21], [22]. Soil samples of different cultivation ages and modes of P. ginseng were used directly for DNA extraction using the UltraClean Soil DNA Isolation Kit (Mo Bio Laboratories Inc., Bohemia, NY, USA) according to the manufacturer's instructions [23].
2.2. PCR amplification and electrophoresis
The universal DNA Purification Kit [DP214, TIANGEN Biotech (Beijing) Co., Ltd., Beijing, China] was used for the PCR Clean-Up. On the basis of DNA extracted from P. ginseng soil, the variable region V3 of the 16S rDNA was amplified using primers GC-338F and 518R, designed to be specific for most bacteria for the analysis of bacterial diversity [24]. For the analysis of fungal diversity, PCR amplification of the 18S rDNA gene was performed using the fungal universal primers NS1 and GC-Fung [25].
2.3. DGGE community fingerprints, DNA sequences, and phylogenetic analysis
The PCR products were analyzed with DGGE using a BioRad DCode Universal Mutation Detection System (Bio-Rad, Richmond, CA, USA). Samples were subjected to 8% (w/v) polyacrylamide gels in 1× Tris-acetate-EDTA solution. Optimal separation was achieved with a 35–55% urea-formamide denaturing gradient for the bacterial community and 15–35% for the fungal community [100% denaturant corresponds to 7M urea and 40% (v/v) formamide]. Bacterial gel runs were performed for 4 h, whereas fungal gel runs lasted 6 h at 150 V at 60°C. Subsequently, banding patterns of the DGGE profile were analyzed using the software QuantityOne-1-D (version 4.5; Bio-Rad Laboratories). Images were normalized using the markers, and the patterns were subsequently compared using clustering methods with CANOCO for Windows software (version 4.5; Microcomputer Power, Ithaca, NY, USA). Similarity matrices consisting of defined numbers within each gel were generated using Pearson's correlation coefficient (r). Additionally, computer-assisted analysis of DGGE fingerprints, DNA sequences, and phylogenetic analysis were used as described previously [24], [25], [26].
2.4. BIOLOG analysis
Functional diversity of the soil microbial community was characterized by community level physiological profiles using BIOLOG EcoPlate (BIOLOG, Hayward, CA, USA) [27]. All BIOLOG profiles were generated by a BIOLOG reader on ELx808BLG (BIO-TEK Instrument, Inc., Winooski, VT, USA) at 24-h intervals for 168 h [22].
The average well color development (AWCD), the metabolic profile of microbial communities, and PCA were used to analyze the metabolic variance of P. ginseng rhizosphere soil. AWCD was calculated according to the procedure described by Garland et al [28], and the total ability of the microbial community on carbon substrate utilization was evaluated. The metabolic profile of microbial communities included the Shannon index (H′), the evenness index (E), and Simpson index (Ds). All the community-level physiological profiles were calculated as described previously [22]. The AWCD value at 72 h was used to calculate the H′, and IBM SPSS Statistics software (version 19.0; IBM, Armonk, NY, USA) was used for PCA [27].
3. Results
3.1. Bacterial community structures in P. ginseng rhizosphere soil as assessed by PCR-DGGE
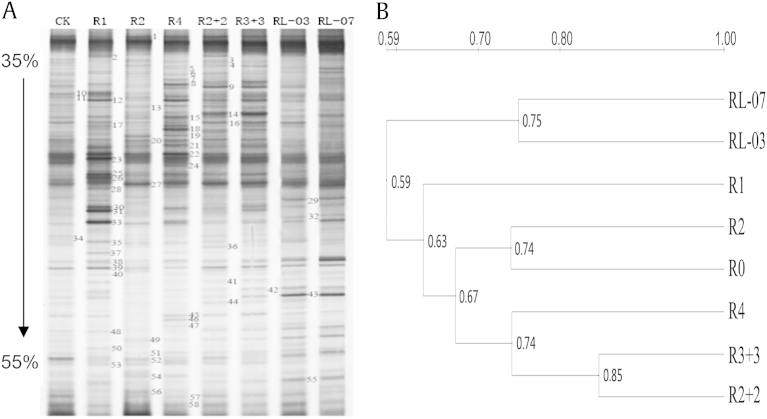
For eight soil samples of P. ginseng, bacterial diversity was evaluated using PCR-DGGE analysis of the amplified partial 16S rDNA genes (Fig. 1). Overall, the bacterial community structures were relatively complex between different cultivation ages and modes. The PCR-DGGE profile revealed 38–47 bands per lane in all samples, whereas some special bands were only found in individual soil samples (Fig. 1A). All patterns (using Unweighted Pair Group Method with Arithmetic Mean, a simple agglomerative (bottom-up) hierarchical clustering method, based on the PCR-DGGE profiles) derived from the same cultivation modes were generally similar with each other with an average similarity of 74–85%; however, patterns derived from lower cultivation ages were different from those derived in higher ages, such as R4, R2 + 2, and R3 + 3, which had similarity rates apart from R0 and R2 with an average similarity of 67% (Fig. 1B).
Fig. 1.
Denaturing gradient gel electrophoresis (DGGE) banding patterns of 16S rDNA fragment and the clustering of DGGE profiles in Panax ginseng soil between different cultivation ages and modes. Lanes corresponding to different soil samples were indicated by numbers at the top (R0, forest soil uncultivated P. ginseng; R1–R4, direct-seeding soil; R2 + 2 and R3 + 3, transplanted soil; RL-07 and RL-03, fallow soil abandoned in 2007 and in 2003, respectively). The bands of DGGE profiles indicated by the numbers were detected, and some were excised, reamplified and subjected to sequencing. The sequenced results are listed in Table 2. The arrow on the left indicates the direction of DGGE electrophoresis and the percentage number indicates % denaturant.
To evaluate the bacterial species in P. ginseng soil, 27 bands in the DGGE profiles were commonly sequenced, and the relative identification is reported in Table 2. The similarity of all band sequences was ≥ 96% compared with those available in the GenBank database. Six bacterial phyla (Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Acidobacteria, and uncultured bacteria) were detected, of which Proteobacteria dominated in P. ginseng soil. Among these groups, Sphingomonadales, Hyphomicrobiales, Rhodospirillales, and Rhizobiales, belonging to Proteobacteria, were dominant and were found to be present in all ages and modes of P. ginseng. For instance, uncultured actinobacterium (band-30) was evident in all soil samples, and there was a larger content in R1, indicating that it was the major bacterial group in R1. However, Alcaligenaceae and Comamonadaceae emerged in several soil samples. For example, Paralcaligenes ureilyticus (band-28) and uncultured Helicobacter sp. (band-31) were respectively evident in R1 and R3 + 3 with a high intensity signal, whereas uncultured Comamonadaceae bacterium was only found in R2 + 2 (band-36) with a faint band, indicating that it was not the major bacterial species, and uncultured actinobacterium (band-12) was only detected in P. ginseng soil. Furthermore, uncultured Fusobacterium sp. (band-9) from Fusobacteria only emerged in R2 + 2 and R3 + 3, as well as uncultured Prosthecobacter sp. (band-29) from Verrucomicrobia in R3 + 3, RL-07, and RL-03, suggesting that special groups, such as Fusobacteria and Verrucomicrobia, were the main groups to change the bacterial community structure of P. ginseng rhizosphere soil.
Table 2.
Phylogenetic identification results of selected denaturing gradient gel electrophoresis (DGGE) bands from bacterial DGGE profiles in Fig. 11)
| Band no.2) | Similar strain (NCBI accession No.) | Similarity (%) | Classification3) |
|---|---|---|---|
| Band-6 | Uncultured bacterium (KF106741.1) | 96 | Bacteria; unknown species |
| Band-7 | Uncultured actinobacterium (EF073893.1) | 99 | Actinobacteria; unknown species |
| Band-8 | Uncultured Hyphomicrobiaceae bacterium (EF665789.1) | 99 | Proteobacteria; Hyphomicrobiaceae |
| Band-9 | Uncultured Fusobacterium sp. (KJ730169.1) | 100 | Fusobacteria; Fusobacteriaceae |
| Band-10 | Alpha proteobacterium LEMS (JF490046) | 96 | Proteobacteria; alpha proteobacterium |
| Band-12 | Uncultured actinobacterium (GU568941.1) | 98 | Actinobacteria; unknown species |
| Band-14 | Uncultured Firmicutes bacterium (EU300338.1) | 99 | Firmicutes; unknown species |
| Band-15 | Uncultured Rubrobacterales bacterium (GU983374.1) | 100 | Actinobacteria; Rubrobacteridae |
| Band-16 | Uncultured bacterium (KJ601351.1) | 100 | Bacteria |
| Band-18 | Sphingomonas astaxanthinifaciens (NR114037) | 100 | Proteobacteria; Sphingomonadaceae |
| Band-20 | Uncultured bacterium (KF411798.1) | 100 | Bacteria |
| Band-22 | Chitinophagaceae bacterium (AB850958) | 98 | Proteobacteria; Sphingobacteriales |
| Band-23 | Altererythrobacter xinjiangensis (NR108901) | 100 | Sphingomonadales; Erythrobacteraceae |
| Band-27 | Uncultured Acidobacteria bacterium (KJ191824.1) | 100 | Acidobacteriaceae; Acidobacteria |
| Band-28 | Paralcaligenes ureilyticus (NR116812) | 100 | Proteobacteria; Alcaligenaceae |
| Band-29 | Uncultured Prosthecobacter sp. (JX505079.1) | 99 | Verrucomicrobia; Verrucomicrobiaceae |
| Band-30 | Uncultured actinobacterium (EF019447.1) | 100 | Actinobacteria |
| Band-31 | Uncultured Helicobacter sp. (GQ375470.1) | 99 | Proteobacteria; Helicobacteraceae |
| Band-32 | Uncultured alpha proteobacterium (EF662861) | 99 | Proteobacteria; Alphaproteobacteria |
| Band-33 | Azospirillum sp. (KF318811.1) | 99 | Proteobacteria; Rhodospirillaceae |
| Band-36 | Uncultured Comamonadaceae bacterium (GU473122.1) | 97 | Proteobacteria; Comamonadaceae |
| Band-37 | Frigoribacterium sp. (KJ529092.1) | 99 | Actinobacteria; Microbacteriaceae |
| Band-39 | Nitrobacter winogradskyi (NR074324) | 99 | Bradyrhizobiaceae; Nitrobacter |
| Band-43 | Uncultured Acidobacteria bacterium (JX025749) | 100 | Acidobacteriales; Acidobacteriaceae |
| Band-46 | Uncultured actinobacterium (JX011472) | 100 | Actinobacteria |
| Band-52 | Bradyrhizobium sp. (AB121773.1) | 99 | Proteobacteria; Bradyrhizobiaceae |
| Band-55 | Novosphingobium nitrogenifigens (NR043857) | 99 | Proteobacteria; Sphingomonadaceae |
Only the highest homology matches are presented
Bands are numbered according to Fig. 1
Classification represents Phylum, order, and family to which a similar strain belongs
3.2. Fungal community structures in P. ginseng rhizosphere soil as assessed by PCR-DGGE
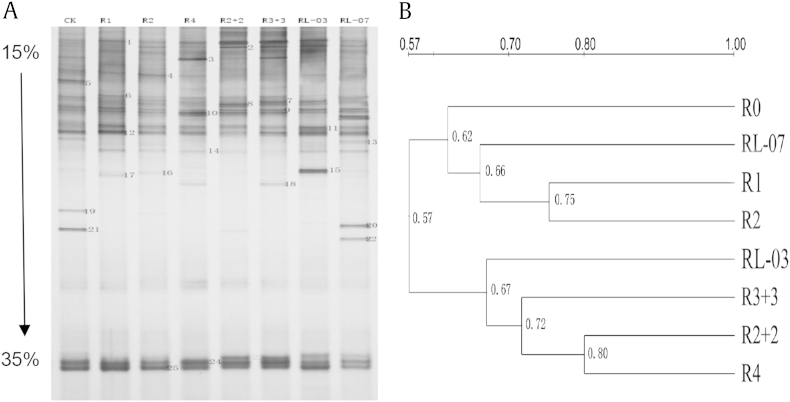
The fungal diversity of eight soil samples was evaluated using PCR-DGGE analysis, and the 18S rDNA gene analyses revealed that the fungal community structures in P. ginseng soil were similar to those from different cultivation ages and modes, but there were also some special species in individual soil samples (Fig. 2). As a whole, there were 16–20 bands per line in all soil samples. These differences indicated the fluctuation in fungal community structures between different cultivation ages and modes by comparing them with the number and intensity of the DNA bands in DGGE profiles (Fig. 2A). Patterns (using UPGMA based on the PCR-DGGE profiles) derived from higher cultivation ages were generally similar with each other with an average similarity of 72–80%. Moreover, patterns derived from lower cultivation ages were also similar to those from R0 and RL-07; R1, R2, and RL-07 were in the same group, with an average similarity of 66–75% (Fig. 2B).
Fig. 2.
Denaturing gradient gel electrophoresis (DGGE) banding patterns of 18S rDNA fragment and the clustering of DGGE profiles in Panax ginseng soil between different cultivation ages and modes. Lanes corresponding to different soil samples were indicated by numbers at the top (R0, forest soil uncultivated P. ginseng; R1–R4, direct-seeding soil; R2 + 2 and R3 + 3, transplanted soil; RL-07 and RL-03, fallow soil abandoned in 2007 and in 2003, respectively). The bands of DGGE profiles indicated by the numbers were detected and some of them were excised, re-amplified and subjected to sequencing. The sequenced results are listed in Table 3. The arrow on the left indicates the direction of DGGE electrophoresis and the percentage indicates % denaturant.
To evaluate the fungal species in P. ginseng soil, 25 selected bands in the DGGE profiles were elucidated as indicated in Table 3. Comparison of the sequences from the excised bands with those available in the GenBank database revealed that all were ≥ 93% similar to 18S rDNA fragments already in the database. The results of DGGE and direct sequencing revealed that Hypocreales dominated, and showed the presence of representatives of four classes (Sordariomycetea, Pezizomycetes, Dothideomycetes, and Leotiomycetes) from Ascomycota and three classes (Urediniomycetes, Hymenomycetes, and Zygomycetes) belonging to Basidiomycota and Zygomycota, respectively. Among these groups, some species, such as Phialocephala repens (band-1), Mortierella alpine (band-7), Rhizomucor endophyticus (band-9), Geomyces destructans (band-10), Acremonium recifei (band-11), Phacidium lacerum (band-12), Hymenoscyphus scutula (band-24), and uncultured Lecythophora sp. (band-25), were present in all soil samples with high intensity signal, indicating that these fungi were the most common species in P. ginseng soil. Creolimax fragrantissima (band-5), Clonostachys rosea (band-13), and Rheomorpha neiswestonovae (band-19) were only found in R0, whereas other species, such as Cryptococcus humicolus (band-6) and Fusarium oxysporum (band-8), were detected only in P. ginseng cultivation soil. Moreover, some special species were also observed or deleted because of the changes in cultivation ages or modes. For instance, Trichosporon pullulans (band-3) cannot be found in lower cultivation ages (R1, R2), and Botryosphaeria mamane (band-23) was only found in transplanted soil and fallow soil of P. ginseng. This illustrated that fungal diversity could be changed with variations in cultivation ages and modes of P. ginseng.
Table 3.
Phylogenetic identification results of selected denaturing gradient gel electrophoresis (DGGE) bands from fungal DGGE profiles in Fig. 21)
| Band no.2) | Similar strain (NCBI accession no.) | Similarity (%) | Classification3) |
|---|---|---|---|
| Band-1 | Phialocephala repens (EU434874) | 98 | Pezizomycotina |
| Band-2 | Uncultured fungus (JN166410.1) | 99 | Fungi |
| Band-3 | Trichosporon pullulans (AB001766) | 100 | Cystofilobasidiaceae; Guehomyces |
| Band-4 | Fusarium fujikuroi (HF679024) | 99 | Hypocreales; Nectriaceae; Fusarium |
| Band-5 | Creolimax fragrantissima (EU124916) | 93 | Ichthyophonida; Creolimax |
| Band-6 | Cryptococcus humicolus (AB035587) | 100 | Tremellaceae; Asterotremella |
| Band-7 | Mortierella alpina (AY546097) | 99 | Mortierellaceae; Mortierella |
| Band-8 | Fusarium oxysporum (HM210090) | 98 | Hypocreales; Nectriaceae; Fusarium |
| Band-9 | Rhizomucor endophyticus (HM623313) | 97 | Mucorineae; Mucoraceae; Mucor |
| Band-10 | Hymenoscyphus sp. (KC164672.1) | 99 | Helotiales; Helotiaceae |
| Band-11 | Phacidium lacerum (DQ471028) | 99 | Phacidiaceae; Phacidium |
| Band-12 | Acremonium recifei (HQ232206) | 97 | Hypocreales; Acremonium |
| Band-13 | Clonostachys rosea (GU112755) | 99 | Bionectriaceae; Clonostachys |
| Band-14 | Tetrachaetum elegans (AY357280) | 98 | Ascomycota; Tetrachaetum |
| Band-15 | Alicorhagia sp. (KF650049.1) | 99 | Metazoa; Alicorhagiidae |
| Band-16 | Uncultured Hygrophoraceae (EU300937.1) | 99 | Dikarya; Hygrophoraceae |
| Band-17 | Kionochaeta sp. (AB521038) | 100 | Sordariomycetidae; Kionochaeta |
| Band-18 | Tullbergia yosii (DQ016556) | 97 | Tullbergiinae; Tullbergia |
| Band-19 | Rheomorpha neiswestonovae (AY527049) | 95 | Lumbricina; Aeolosomatidae |
| Band-20 | Hemienchytraeus sp.(GU901905.1) | 99 | Metazoa; Enchytraeidae |
| Band-21 | Tardigrada sp. (AJ617459.2) | 99 | Metazoa; Tardigrada |
| Band-22 | Odontolaimus sp. (FJ969131.1) | 93 | Metazoa; Oxystominidae |
| Band-23 | Botryosphaeria mamane (KF531821) | 97 | Botryosphaeriaceae; Botryosphaeria |
| Band-24 | Hymenoscyphus scutula (AY789430) | 95 | Helotiales; Helotiaceae |
| Band-25 | Uncultured Lecythophora sp. (HQ424852.1) | 99 | Hypocreales; Clavicipitaccae |
Only the highest homology matches are presented
Bands are numbered according to Fig. 2
Classification represents phylum, class, order, and family to which a similar strain belongs
3.3. Carbon substrate metabolic profiles of soil microbial communities
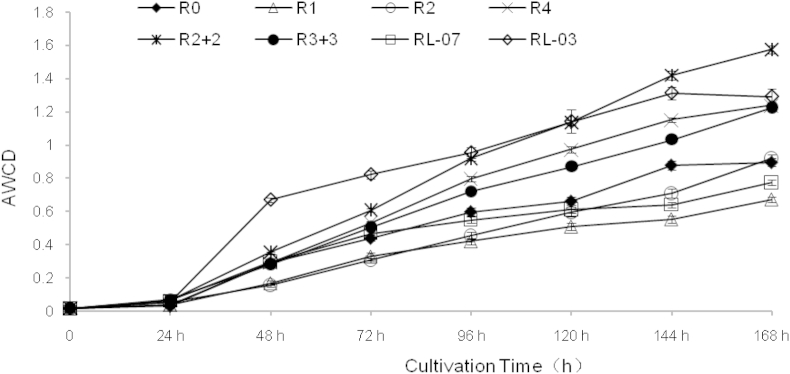
Microbial community functional diversity reflected the soil microbial ecological function. AWCD is one of the most important indexes to determine the capacity of the soil microbial community utilizing carbon sources, and it is an important indicator of the activity of microbial communities [29]. The dynamics of AWCD were investigated with the P. ginseng soil cultivated at intervals of 24 h later, as shown in Fig. 3. As a whole, AWCD was gradually increased with the cultivation time prolonged. The carbon substrates utilized by microbes were nonsignificant during the first 24 h, which showed that the carbon source was not used very well at the beginning of soil cultivation. After that, the growth of soil microorganisms entered logarithmic growth until 144 h later and the AWCD of all soil samples increased rapidly to approximately 1.0, whereas it was almost slow and steady after 144 h because the soil microbes had to gradually adapt to the environment of BIOLOG microplate after the lag phase. The increasing rate and the final value of AWCD depend on the abundance and activity of the microbial community [28]. Moreover, in comparing the AWCD from different P. ginseng soil samples in 72 h, it was observed that the AWCD values of P. ginseng soil were between 0.304 and 0.821. The AWCD of RL-03 was the maximum and significantly higher than that in R0 and transplanted soil, which demonstrated that the ability to use a single carbon source from the soil microbial community was significantly increased when the fallow soil of P. ginseng was abandoned for 10 yr. Meanwhile, the AWCD of R1 was the minimum, whereas the AWCD of the transplanted soil was significantly higher than that of R0 and the direct-seeding soil of P. ginseng. As a whole, on account of the increase in cultivation ages, the metabolic activity of the soil microbial communities was affected to a large extent; however, both the change in cultivation modes and fallow soil abandoned for 10 yr significantly improved microbial metabolic activity in P. ginseng soil.
Fig. 3.
Average well color development (AWCD) development of soil microbial communities in Panax ginseng soil between different cultivation ages and modes.
3.4. Specific substrate utilization of soil microbial communities
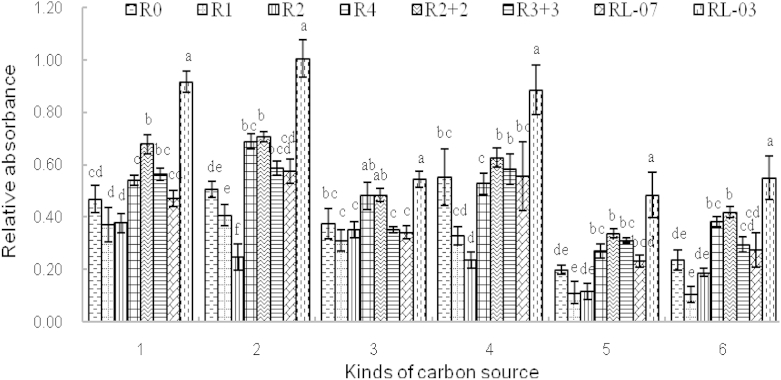
There are 31 types of different carbon sources in the BIOLOG EcoPlate, i.e., 12 types of carbohydrates, six types of amino acids, four types of polymers, five types of carboxylic acids, two types of phenolic acids, and two types of amines/amides. The utilization ability of carbon sources was researched (and shown in Fig. 4), which was conducive to a more comprehensive understanding of the metabolic characteristics of the soil microbial community. The results showed that carbohydrates, amino acids, and polymers were the main carbon sources utilized in cultivated and abandoned soil of P. ginseng, whereas the utilization abilities of carboxylic acids, phenolic acids, and amines/amides were weaker than those of the others. The utilization ratios of six types of carbon sources in RL-03 were the highest, of which the utilization of carbohydrates, amino acids, and polymers was higher than that in R0, and determined as 48.89%, 49.77%, and 37.85%, respectively. It was indicated that the metabolic function of soil microbial communities was increased significantly in fallow soil of P. ginseng abandoned for 10 yr. Higher utilization ratios of polymers were detected in R2 + 2, and significant differences were found compared with those in R3 + 3 and R0 (respectively 17.06% and 28.60% higher). Utilization ratios of carbohydrates, amino acids, and polymers were the lowest in R1, and no significant differences were found compared with R0. It can be observed that the utilization ratios of carbon sources were enhanced with increases of P. ginseng cultivation ages, and the catabolic function of the soil microbial community was changed in different cultivation modes.
Fig. 4.
Utilization abilities of six types of carbon sources in the BIOLOG EcoPlate by soil microbial communities in different cultivation ages and modes. 1, Miscellaneous carbohydrates; 2, amino acids; 3, carboxylic acids; 4, polymers; 5, phenolic acids; and 6, amines/amides.
3.5. Diversity index of soil microbial communities of PCR-DGGE and BIOLOG profiles
The overall species richness and catabolic diversity of microbial communities in P. ginseng soil were characterized by the microbial diversity index as shown in Table 4 (as evaluated by the number of major bands present in the PCR-DGGE and the AWCD value in 72-h BIOLOG profiles). It was clearly indicated that the metabolic diversity of fallow soil (RL-07, RL-03), transplanted soil (R2 + 2, R3 + 3), and R4 was higher compared with R0, R1, and R2 as calculated by H′ and Ds. By contrast, the overall bacterial H′ in lower cultivation ages of direct-seeding and transplanted soils appeared to have a higher microbial diversity (3.73 < H′ < 3.84) compared with that in higher cultivation ages (3.62 < H′ < 3.69). Remarkably, the variation was opposite in the fungal community diversity, which infers that with the increase in cultivation ages, there was a greater diversity of fungal community in P. ginseng soil. E and Ds effectively distinguished the cultivation ages and modes from each other and showed that bacterial diversity was highest in R0, R1, and RL-03 soil samples (Ds = 0.98–0.99). However, fungal diversity was highest in R0, R1, and RL-07 (Ds = 0.93–0.95), which indicates the presence of more homogeneous and stable ecosystems in R0 and R1. The increase in cultivation ages resulted in disequilibrium in microbial diversity, whereas microbial diversity was improved strikingly in transplanted soil and fallow soil abandoned for at least one decade.
Table 4.
Microbial diversity index of Panax ginseng soil calculated from DGGE fingerprints and BIOLOG data
| Soil samples | Analysis of microbial diversity1) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Analysis by PCR-DGGE method |
Analysis by BIOLOG method |
||||||||
| Bacterial diversity |
Fungal diversity |
Shannon index (H′) | Substrate evenness (E) | Simpson's index (Ds) | |||||
| Shannon index (H′) | Substrate evenness (E) | Simpson's index (Ds) | Shannon index (H′) | Substrate evenness (E) | Simpson's index (Ds) | ||||
| R0 | 3.82 ± 0.009a,b | 0.98 ± 0.001b | 0.98 ± 0.001a,b | 2.63 ± 0.009c | 0.99 ± 0.000a,b | 0.94 ± 0.001a,b | 3.06 ± 0.02c | 0.98 ± 0.004b,c | 0.927 ± 0.012b |
| R1 | 3.84 ± 0.009a | 0.97 ± 0.000d | 0.98 ± 0.000a,b | 2.27 ± 0.006e | 0.99 ± 0.000e | 0.93 ± 0.002a,b | 2.89 ± 0.02d | 0.99 ± 0.008a,b | 0.928 ± 0.002b |
| R2 | 3.80 ± 0.007b | 0.96 ± 0.000f | 0.97 ± 0.000d | 2.61 ± 0.012c | 0.99 ± 0.000c,d | 0.94 ± 0.001d | 2.92 ± 0.03d | 1.00 ± 0.012a | 0.926 ± 0.003b |
| R4 | 3.62 ± 0.015e | 0.97 ± 0.000e | 0.98 ± 0.001b | 2.68 ± 0.009b | 0.99 ± 0.001d,e | 0.94 ± 0.002b | 3.15 ± 0.01b | 0.98 ± 0.004b | 0.951 ± 0.001a |
| R2 + 2 | 3.73 ± 0.015c | 0.97 ± 0.001c | 0.97 ± 0.000c | 2.36 ± 0.006d | 0.99 ± 0.001e | 0.93 ± 0.000c | 3.21 ± 0.01a | 0.96 ± 0.003c | 0.954 ± 0.001a |
| R3 + 3 | 3.69 ± 0.009d | 0.97 ± 0.001d | 0.97 ± 0.000c | 2.37 ± 0.009d | 0.99 ± 0.001c | 0.94 ± 0.001c | 3.13 ± 0.01b | 0.98 ± 0.003b,c | 0.949 ± 0.001a |
| RL-07 | 3.68 ± 0.009d | 0.98 ± 0.000a | 0.97 ± 0.001c | 2.91 ± 0.003a | 0.10 ± 0.001a | 0.95 ± 0.001a | 3.16 ± 0.02b | 0.98 ± 0.005b,c | 0.949 ± 0.001a |
| RL-03 | 3.81 ± 0.003a,b | 0.99 ± 0.001a | 0.98 ± 0.000a | 2.92 ± 0.009b | 0.99 ± 0.001b | 0.94 ± 0.001c | 3.25 ± 0.01a | 0.97 ± 0.002b,c | 0.957 ± 0.001a |
LSR; PCR-DGGE, polymerase chain reaction and denaturing gradient gel electrophoresis
The letters indicate tested with Shortest Significant ranges (SSR) at p = 0.05 of different treatments. Different letters indicate a significant difference at p < 0.05 level
3.6. Multivariate analysis of DGGE fingerprints and BIOLOG data
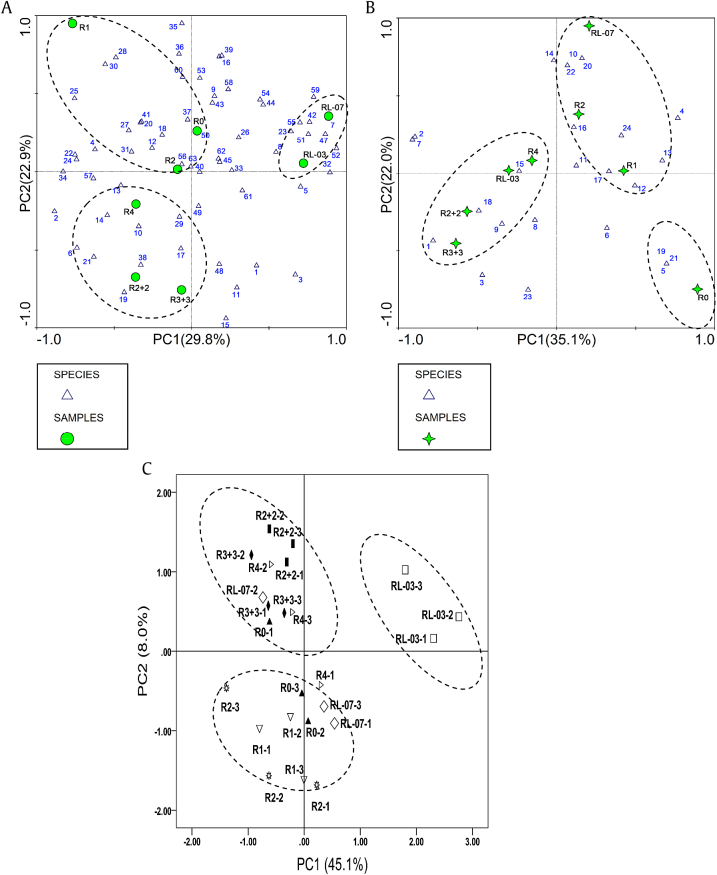
Taking into account the relative intensity of the bands in DGGE gel profiles and utilization of carbon substrate in BIOLOG EcoPlate, PCA analyses were used to investigate the possible correlation between cultivation ages and modes of P. ginseng (Fig. 5). The results obtained from multivariate comparisons showed that the complex microbial communities were different between cultivation ages and modes of P. ginseng (Figs. 5A and 5B). However, no significant effect of cultivation ages on carbon substrate consumption was found in any of the P. ginseng soil samples (Fig. 5C).
Fig. 5.
Principal component analysis (PCA) of the microbial composition of Panax ginseng soil samples in different cultivation ages and modes. (A) PCA of the bacterial composition of P. ginseng soil samples by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR-DGGE) analysis. (B) PCA of the fungal composition of P. ginseng soil samples by PCR-DGGE analysis. (C) PCA of the microbial composition of P. ginseng soil samples by BIOLOG analysis.
Using the PCA plot for the bacterial communities in P. ginseng soil, the first two components accounted for 29.8% and 22.9% of the variance, respectively. Positions of the different cultivation ages and modes in the PCA plot were classified into three groups, in which one group contained R0, R1, and R2, suggesting that the bacterial community structure in these soil samples were similar, and the other two groups contained RL-03 and RL-07, and R2 + 2, R3 + 3, and R4, respectively (Fig. 5A). As for the fungi analysis, the second two principal components accounted for 35.1% and 22.0% of the variance, respectively. The PCA plot analysis of fungal DGGE patterns indicated that the fungal community profiles were clustered into the following three groups: Group 1 (R0), Group 2 (R1, R2, and RL-07), and Group 3 (R4, R2 + 2, R3 + 3, and RL-03) (Fig. 5B). Through the PCA plot for the utilization of the single carbon substrate of P. ginseng soil, the accumulative contribution of PC was 86.01%, including the first component, which accounted for 67.83%, and the second component, which accounted for 10.78%. The PCA plot showed that the carbon substrate utilizing profiles were clearly separated following three groups: Group 1 (RL-03), Group 2 (R1, R2, and RL-07), and Group 3 (R0, R4, R2 + 2, and R3 + 3) (Fig. 5C). Collectively, the results of the PCA indicated that metabolic functional diversity was similar to soil fungal community diversity, whereas bacterial community diversity exerted a significant influence, in which the dominant factor of functional variation in the soil bacterial community was the alteration in P. ginseng cultivation ages.
4. Discussion
The aim of the present study was to investigate the effects of cultivation ages and modes on microbial genetic and functional diversity in P. ginseng soil using 16S rDNA and 18S rDNA gene profiles generated by PCR-DGGE and metabolic functional analyses generated with the BIOLOG EcoPlate. The combination of these two methods was found to be useful in systematically understanding microbial communities in P. ginseng soil. The reasons for using the combined approach were that soil functionality was thought to be dependent not only on the microbial species present but also on the potential metabolic activity of the microbiota in P. ginseng soil. The BIOLOG profiles, which could not separately represent the activity of bacterial or fungal communities, were the total values of microbial communities. The main finding in this study was that the cultivation of P. ginseng, especially cultivation ages, exerted the most profound influence on the genetic and functional diversity in P. ginseng rhizosphere soil, which is consistent with the results of previous studies [21], [30], [31].
The bacterial community structure analyzed by PCR-DGGE profile in P. ginseng soil was relatively complex, with significant diversity observed between different cultivation ages and modes. Six bacterial phyla (Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia, Acidobacteriaceae, and uncultured bacteria) were detected. Most of the DGGE bands (48.15%) were identified as Proteobacteria, suggesting that it was the major bacterial group in P. ginseng soil. All members of Proteobacteria are gram negative [32]. In the rhizosphere soil of plants, a selective effect favored Proteobacteria over Acidobacterium and gram-positive bacteria, leading to the prevalence of the Pseudomonas bacterial group, which caused diseases or interfered negatively with plant development [33], [34]. Therefore, Proteobacteria of the bacterial community played an important role during the growth of P. ginseng. Rubrobacterales and some uncultured actinobacterium belonging to Actinobacteria were also found in P. ginseng soil. Similar results were found in Regupathy Thamizh Vendan through DGGE at varying age levels of P. ginseng, which indicated that α-Proteobacteria and Actinobacteria were predominant in P. ginseng soil [21]. In the present study, Actinobacteria represented an important component of the microbial population in soil [35], which was used as a biocontrolling agent to control soil- and seed-borne diseases of plants [36]. Interestingly, some Actinomyces species gradually appeared or disappeared with the cultivation of P. ginseng, suggesting that the environment of P. ginseng soil significantly affected the Actinobacteria community.
As for the fungal community, the 18S rDNA gene analyses using PCR-DGGE revealed that the fungal community structures were similar to those from different cultivation ages and modes in P. ginseng soil, but there were also some special species in individual soil samples. Most of the DGGE bands (48.00%) were identified as representatives of four classes (Sordariomycetea, Pezizomycetes, Dothideomycetes, and Leotiomycetes) from Ascomycota and three classes (Urediniomycetes, Hymenomycetes, and Zygomycetes) belonging to Basidiomycota and Zygomycota, respectively. Fusarium sp., belonging to Hypocreales, Sordariomycetea, was the most predominant species detected in the cultivated soil of P. ginseng. F. oxysporum was found in the cultivated soil of P. ginseng except in R0, which was in higher cultivation ages with a significantly larger amount. F. oxysporum is a ubiquitous soilborne pathogen that occurs worldwide across various soil types and causes severe damage and yield losses across a wide range of plant families [37], [38]. Several studies have shown that F. oxysporum, which caused diseases in vegetables [39], wheat [40], maize [41], and cotton [42], seriously threatened commercial crop production worldwide. F. oxysporum infected vascular bundles in the plant host, leading to wilt symptoms [43]. Based on accumulating data, it was speculated that the germination of dormant spores of F. oxysporum was able to result in diseases of P. ginseng roots in cultivated soil, whereas F. oxysporum was also detected in RL-03, which showed that the accumulation of soilborne diseases played an important role in discontinuous cultivation of P. ginseng. Fusarium sp. could be used to assess the recuperative degree of the microbial community directly from continuous farming ground using PCR-DGGE.
Moreover, to improve our understanding of the effect of cultivation ages and modes on microbial communities in P. ginseng soil, we applied the PCR-DGGE and BIOLOG methods to evaluate the metabolic activity and diversity indexes. Wang et al [44] also used these two methods to investigate the effects of fertilization on bacterial community structure and function in black soil. The microbial metabolic activity in P. ginseng soil was described by AWCD of substrates arranged on the BIOLOG EcoPlate [45]. BIOLOG analysis indicated that the microbial activity of RL-03 was the strongest with better fungal diversity, whereas transplanted soil was significantly stronger than direct-seeding soil and R0. The increase in P. ginseng cultivation ages in the same mode led to decreases in microbial activities in P. ginseng soil (Fig. 3). Carbohydrates, amino acids, and polymers were the main carbon sources utilized in P. ginseng soil (Fig. 4). Furthermore, the microbial diversity index and multivariate comparisons revealed that the increase in P. ginseng cultivation ages led to decreases in bacterial diversity and increases in fungal diversity, whereas microbial diversity was improved strikingly in transplanted soil and fallow soil abandoned for at least one decade (Table 4 and Fig. 5). In previous studies, the cultivation age was assumed to be an important factor that influenced microbial activity and diversity in the rhizosphere of plants [9], [46], [47]. In addition, soil characteristics had a significant influence on soil microbial communities [48], of which soil pH was thought to exert primary domination on the composition of soil bacterial communities [49]. In the present study, evidence supporting this controversial concept came from the fact that decline in soil pH occurred with the increase in P. ginseng cultivation (Table 1). Prior to our investigation, it was also shown that the rhizospheric microbial community and soil pH were influenced to a certain extent by the accumulation of root exudates [50]. Therefore, root exudates might be a key factor influencing the microbial diversity in the rhizosphere of P. ginseng with cultivation age increasing, although the links between the microbial community and soil functions were unclear [51]. Moreover, variation in the microbial community may not result in the alteration of soil function [52], and thus the definite reasons for this need to be investigated in a further experiment.
In this study, our results showed that the microbial structure of the rhizosphere, particularly the outbreak of soilborne pathogenic fungus, was colonized by the cultivation ages of P. ginseng. Moreover, the transplantation mode was effective in improving microbial diversity in order to avoid discontinuous cultivation of P. ginseng. Although the microbial metabolic diversity of fallow soil abandoned for a long time was at a higher level, the microbial genetic diversity was still unbalanced. Therefore, we speculated that the key factors for discontinuous P. ginseng cultivation were the changes in the microbial community structure and the microecological imbalance of rhizosphere soil caused by the accumulation of P. ginseng root exudates. Further study with a more detailed examination of the correlation between a certain soilborne disease and the relevant compounds of P. ginseng root exudates and the elimination of P. ginseng root exudates utilizing metabolic decomposition of particular microbe or addition of adsorption material is necessary.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
The work was financially supported by grants from the Science and Technology Supporting Project of China (no. 2011BAI03B01-02) and the Nature Science Foundation of China (no. 31270371).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc-nd/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Jung J., Kim K.H., Yang K., Bang K.H., Yang T.J. Practical application of DNA markers for high-throughput authentication of Panax ginseng and Panax quinquefolius from commercial ginseng products. J Ginseng Res. 2014;38:123–129. doi: 10.1016/j.jgr.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang H.Y., Xue Q.H. Research progress in control of continuous cropping obstacle in Panax ginseng. Acta Agriculturae Jiangxi. 2010;22:68–71. [Google Scholar]
- 3.Zhang L.X., Chen C.B., Wang Y.P., Xu S.Q., Chang C. Study on discontinuous cultivating of Panax ginseng and its workable solution. J Jilin Agric Univ. 2008;30:481–485. [Google Scholar]
- 4.Li Y., Ying Y.X., Zhao D.Y., Ding W.L. Microbial community diversity analysis of Panax ginseng rhizosphere and non-rhizosphere soil using randomly amplified ‵ method. Open J Genet. 2012;2:95–102. [Google Scholar]
- 5.Yang L.M., Chen C.B., Wang X.Q., Zhang L.X., Tian Y.X. Ecological restoration and reused modes of old ginseng land in the Changbai mountainous area and its existing problems. J Jilin Agric Univ. 2004;26:546–549. [Google Scholar]
- 6.Larkin R.P. Characterization of soil microbial communities under different potato cropping systems by microbial population dynamics, substrate utilization, and fatty acid profiles. Soil Biol Biochem. 2003;35:1451–1466. [Google Scholar]
- 7.Zuppinger-Dingley D., Schmid B., Petermann J.S., Yadav V., De Deyn G.B., Flynn D.F. Selection for niche differentiation in plant communities increases biodiversity effects. Nature. 2014;515:108–111. doi: 10.1038/nature13869. [DOI] [PubMed] [Google Scholar]
- 8.Wu F.Z., Wang X.Z., Xue C.Y. Effect of cinnamic acid on soil microbial characteristics in the cucumber rhizosphere. Eur J Soil Biol. 2009;45:356–362. [Google Scholar]
- 9.Urashima Y., Sonoda T., Fujita Y., Uragami A. Application of PCR-denaturing-gradient gel electrophoresis (DGGE) method to examine microbial community structure in asparagus fields with growth inhibition due to continuous cropping. Microbes Environ. 2012;27:43–48. doi: 10.1264/jsme2.ME11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amann R.I., Ludwig W., Schleifer K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivated. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sang M.K., Chun S.C., Kim K.D. Biological control of Phytophthora blight of pepper by antagonistic rhizobacteria selected from a sequential screening procedure. Biol Control. 2008;46:424–433. [Google Scholar]
- 12.Hoang V.A., Kim Y.J., Nguyen N.L., Yang D.C. Brachybacterium ginsengisoli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol. 2014;64:3063–3068. doi: 10.1099/ijs.0.058388-0. [DOI] [PubMed] [Google Scholar]
- 13.Johri B.N., Sharma A., Virdi J.S. Rhizobacterial diversity in India and its influence on soil and plant health. Adv Biochem Eng Biot. 2003;84:49–89. doi: 10.1007/3-540-36488-9_2. [DOI] [PubMed] [Google Scholar]
- 14.Lv X.C., Weng X., Zhang W., Rao P.F., Ni L. Microbial diversity of traditional fermentation starters for Hong Qu glutinous rice wine as determined by PCR-mediated DGGE. Food Control. 2012;28:426–434. [Google Scholar]
- 15.Ma W.K., Siciliano S.D., Germida J.J. A PCR-DGGE method for detecting arbuscular mycorrhizal fungi in cultivated soil. Soil Biol Biochem. 2005;37:1589–1597. [Google Scholar]
- 16.Matsuyama T., Nakajima Y., Matsuya K., Ikenaga M., Asakawa S., Kimura M. Bacterial community in plant residues in a Japanese paddy field estimated by RFLP and DGGE analyses. Soil Biol Biochem. 2007;39:463–472. [Google Scholar]
- 17.Giraffa G. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol Rev. 2004;28:251–260. doi: 10.1016/j.femsre.2003.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Ercolini D. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Meth. 2004;56:297–314. doi: 10.1016/j.mimet.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Garland J.L., Mills A.L., Young J.S. Relative effectiveness of kinetic analysis vs single point readings for classifying environmental samples based on community-level physiological profiles (CLPP) Soil Biol Biochem. 2001;33:1059–1066. doi: 10.1016/s0038-0717(01)00011-6. [DOI] [PubMed] [Google Scholar]
- 20.Rogers B.F., Tate R.L., III Temporal analysis of the soil microbial community along a toposequence in Pineland soils. Soil Biol Biochem. 2001;33:1389–1401. [Google Scholar]
- 21.Vendan R.T., Lee S.H., Yu Y.J., Rhee Y.H. Analysis of bacterial community in the ginseng soil using denaturing gradient gel electrophoresis (DGGE) Indian J Microbiol. 2012;52:286–288. doi: 10.1007/s12088-011-0193-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ying Y.X., Ding W.L., Zhou Y.Q., Li Y. Influence of Panax ginseng continuous cropping on metabolic function of soil microbial communities. Chin Herb Med. 2012;4:329–334. [Google Scholar]
- 23.Inceoğlu O., Hoogwout E.F., Hill P., van Elsas J.D. Effect of DNA extraction method on the apparent microbial diversity of soil. Appl Environ Microbiol. 2010;76:3378–3382. doi: 10.1128/AEM.02715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inceoğlu O., Salles J.F., van Overbeek L., van Elsas J.D. Effect of plant genotype and growth stage on the β-proteobacterial community associated with different potato cultivars in two fields. Appl Environ Microbiol. 2010;76:3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo J., Ran W., Hu J., Yang X.M., Xu Y.C., Shen Q.R. Application of bio-organic fertilizer significantly affected fungal diversity of soil. Soil Biol Biochem. 2010;74:2039–2048. [Google Scholar]
- 26.Lyautey E., Lacoste B., Hage L.T., Rols J.L., Garabetian F. Analysis of bacterial diversity in river biofilms using16S rDNA PCR-DGGE: methodological settings and fingerprints interpretation. Water Res. 2005;39:380–388. doi: 10.1016/j.watres.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 27.Schutter M., Dick R. Shift in substrate utilization potential and structure of soil microbial communities in response to carbon substrates. Soil Biol Biochem. 2001;33:1481–1491. [Google Scholar]
- 28.Garland J.L., Mills A.L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zabinski C.A., Gannon J.E. Effects of recreational impacts on soil microbial communities. Environ Manage. 1997;21:233–238. doi: 10.1007/s002679900022. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths B.S., Ritz K., Ebblewhite N., Dobson G. Soil microbial community structure: effects of substrate loading rates. Soil Biol Biochem. 1999;31:145–153. [Google Scholar]
- 31.Ferreira E., Dusi A.N., Xavier G.R., Rumjanek N.G. Rhizosphere bacterial communities of potato cultivars evaluated through PCR-DGGE profiles. Pesqui Agropecu Bras. 2008;43:605–612. [Google Scholar]
- 32.Berman J.J. 1st ed. Academic Press; Salt Lake City, UT: 2012. Taxonomic guide to infectious diseases: understanding the biologic classes of pathogenic organisms; pp. 25–31. [Google Scholar]
- 33.Marilley L., Aragno M. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl Soil Ecol. 1999;13:127–136. [Google Scholar]
- 34.Berggren I., Alstrőm S., van Vuurde J.W.L., Mårtensson A.M. Rhizoplane colonisation of peas by Rhizobium leguminosarum bv. viceae and a deleterious Pseudomonas putida. Fems Microbiol Ecol. 2005;52:71–78. doi: 10.1016/j.femsec.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Poltia M.A., Aparicioa J.D., Benimelic C.S., Amorosoa M.J. Role of Actinobacteria in bioremediation. In: Surajit D., editor. Microbial biodegradation and bioremediation. Elsevier; London: 2014. pp. 269–286. [Google Scholar]
- 36.Priyadharsini P., Dhanasekaran D. Diversity of soil allelopathic Actinobacteria in Tiruchirappalli district, Tamilnadu, India. J Saudi Soc Agric Sci. 2015;14:54–60. [Google Scholar]
- 37.Van der Does H.C., Lievens B., Claes L., Houterman P.M., Cornelissen B.J.C., Rep M. The presence of a virulence locus discriminates Fusarium oxysporum isolates causing tomato wilt from other isolates. Environ Microbiol. 2008;10:1475–1485. doi: 10.1111/j.1462-2920.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- 38.Michielse C.B., Rep M. Pathogen profile update: Fusarium oxysporum. Mol Plant Pathol. 2009;10:311–324. doi: 10.1111/j.1364-3703.2009.00538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang S., Park G., Atamian H.S., Han C.S., Stajich J.E., Kaloshian I., Borkovich K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014;10:e1004464. doi: 10.1371/journal.ppat.1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Y., Dong K., Zheng Y., Tang L., Yang Z.X. Faba bean fusarium wilt (Fusarium oxysporum) control and its mechanism in different wheat varieties and faba bean intercropping system. Chin J of Appl Ecol. 2014;25:1979–1987. [PubMed] [Google Scholar]
- 41.Fu M., Li R., Guo C., Pang M., Liu Y., Dong J. Natural incidence of Fusarium species and fumonisins B1 and B2 associated with maize kernels from nine provinces in China in 2012. Food Addit Contam A. 2014;6:1–9. doi: 10.1080/19440049.2014.976846. [DOI] [PubMed] [Google Scholar]
- 42.Gaspar Y.M., McKenna J.A., McGinness B.S., Hinch J., Poon S., Connelly A.A., Anderson M.A., Heath R.L. Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J Exp Bot. 2014;65:1541–1550. doi: 10.1093/jxb/eru021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leslie J.F., Summerell B.A. Techniques for recovering Fusarium. In: Leslie J.F., Summerell B.A., editors. The Fusarium laboratory manual. Wiley-Blackwell; Ames, IA: 2006. pp. 15–20. [Google Scholar]
- 44.Wang G.H., Liu J.J., Qi X.N., Jin J., Wang Y., Liu X.B. Effects of fertilization on bacterial community structure and function in a black soil of Dehui region estimated by Biolog and PCR-DGGE methods. Acta Ecol Sin. 2008;28:220–226. [Google Scholar]
- 45.Wang Y., Ou Z.Y., Zheng H., Wang X.K., Chen F.L., Zeng J. Carbon metabolism of soil microbial communities of restores forests in Southern China. J Soil Sediment. 2011;11:789–799. [Google Scholar]
- 46.Yue B.B., Li X., Zhang H.H., Jin W.W., Xu N., Zhu W.X., Sun G.Y. Soil microbial diversity and community structure under continuous Tobacco cropping. Soils. 2013;45:116–119. [Google Scholar]
- 47.Ma K., Zhang L., Du Q., Song N.P. Effect of potato continuous cropping on soil microorganism community structure and function. J Soil Water Conserv. 2010;24:229–233. [Google Scholar]
- 48.Girvan M.S., Bullimore J., Pretty J.N., Osborn A.M., Ball A.S. Soil type is the primary determinant of the composition of the total and active bacterial communities in arable soil. Appl Environ Microbiol. 2003;69:1800–1809. doi: 10.1128/AEM.69.3.1800-1809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Landesman W.J., Nelson D.M., Fitzpatrick M.C. Soil properties and tree species drive β-diversity of soil bacterial communities. Soil Biol Biochem. 2014;76:201–209. [Google Scholar]
- 50.Wu L.K., Lin X.M., Lin W.X. Advances and perspective in research on plant–soil–microbe interactions mediated by root exudates. Chin J Plant Ecol. 2014;38:298–310. [Google Scholar]
- 51.Anglet W.R., Gattin I.T., Laurent F.M., Ajzenberg E.L., Norini M.P., Latour X., Laval K. Soil microbial community structure and function relationships: a heat stress experiment. Appl Soil Ecol. 2014;86:121–130. [Google Scholar]
- 52.Chapin F.S., III, Walker B.H., Hobbs R.J., Hooper D.U., Lawton J.H., Sala O.E., Tilman D. Biotic control over the functioning of ecosystems. Science. 1997;277:500–504. [Google Scholar]