Abstract
Rheumatoid arthritis (RA) is characterized by chronic joint inflammation and associates with HLA-DRB1*04. The Collagen IIp261-273-specific T cell repertoire in the peripheral blood of DR4 + patients at the onset of the disease shows a restricted TCR-beta chain usage among which the most frequent is TRBV25.
To define whether this group of DR4-restricted collagen-specific shared T cell could represent markers of active-severe disease and response to therapy, 90 subjects affected by early-RA were enrolled in the study; peripheral blood mononuclear cells were cultured with or without the human collagen II peptide p261-273 and were examined by immunoscope analysis for the usage of the previously identified shared TCR-beta chains.
We report that the presence of T cells carrying rearrangement TRBV25 associated with HLA-DR haplotype and disease activity. HLA-DRB1* haplotypes 04–04, 04–01 and 04–11 were significantly associated with usage of TRBV25, higher disease activity at the onset of disease and poor response to DMARDs.
Finally, the HLA-DRB1* haplotype appeared complementary with current serologic tools to predict good and poor responders in a treat to target strategy. The data reported here offer clues to predict the course of the disease and to foresee personalized treatments in RA patients.
Abbreviations: ERA, early rheumatoid arthritis; ACPA, anti-cyclic citrullinated peptide antibodies; RF, rheumatoid factor; ESR, erythrocyte sedimentation rate; CRP, C-reactive protein; TJC, tender joint count; SJC, swollen joint count; DAS, disease activity score; HAQ, Health Assessment Questionnaire; HLA, histocompatibility leucocyte antigen; GWAS, genome wide association studies; SNP, single nucleotide polymorphism; MHC, major histocompatibility complex; TCR, T cell receptor; TRBV, variable beta chain gene of TCR; TRBJ, junctional beta chain gene of TCR; Coll261-273, human collagen derived peptide; CDR3, complementarity-determining region 3; PBMC, peripheral blood mononuclear cells; RT-PCR, reverse transcription polymerase chain reaction; APCs, antigen presenting cells
Keywords: HLA-DRB1, Disease activity, ACPA, TRBV 25, Clonotypes
Highlights
-
•
In DR4 + RA patients disease activity is associated with detection of Collagen261-273-specific T cells carrying TRBV25.
-
•
HLA-DR 04/04, 04/01 and 04/11 alleles were associated with TRBV25, DAS at the onset, and poor response to DMARDs.
-
•
These findings could lead to tailor the treatment in the subgroup of patients with an active refractory disease.
In the era of costly medical care with monoclonal antibodies and new molecules, and of an increasing request of a personalized medicine, a relevant socio-economic problem in the management of Rheumatoid Arthritis patients is the possible identification of the subgroups of poor responders to treatment. Our study aimed to detect the refractory active patients using an HLA-DR test (available in most hospital centers) combined with a relatively new biomarker of active disease expressed on the cell surface of autoreactive T cells. These tests appear complementary tools to identify the best and the poor responders to a “treat to target strategy”.
1. Introduction
Rheumatoid Arthritis (RA) is an autoimmune disease and a systemic inflammatory disorder affecting joints and internal organs. It is one of the main causes of disability in the western world. The etiology and the triggers of autoimmunity and in particular of RA have been ascribed to different factors: genetic and environment. The humoral and cell-mediated immune responses in which self-reactivity plays an important role are key points in the pathophysiology of RA. The chronic synovial inflammation involved in joint destruction and disability is mediated by the infiltration of the activated immune cells: CD4 + T cells, B cells, dendritic cells and macrophages (Cooles and Isaacs, 2011, van der Woude et al., 2010, Willemze et al., 2012, Michelutti et al., 2011, Tolusso et al., 2009).
Family-based and twin-based studies have shown that the heritability of RA accounts for about 60% of the developmental risk, one third of which has been linked to two particular major histocompatibility antigens (human leukocyte antigens) namely HLA-DRB1*04 and 01 (Deighton et al., 1989). The genome-wide association studies (GWAS) and meta-analyses identified and combined a number of single nucleotide polymorphisms (SNPs) related to RA, both in HLA and non-HLA genes (Chatzikyriakidou et al., 2013, Mesko et al., 2010). Several studies about the amino-acids motifs at the position 70–74 of the third hyper-variable region of HLA-DRB1 molecules associated not only with RA susceptibility (RR-QK-QRRAA as in DRB1*0101, *0102, *0401, *0404, *0405, *0408, *0410, *1001 and *1402) or protection (DERAA as in *0103, *0402, *1102, *1103, *1301, *1302 and *1304) (Feitsma et al., 2008), but also to disease severity and progression (Gonzalez-Gay et al., 2002) with variations among different ethnicities and populations, also demonstrated in Kapitany et al. (Kapitany et al., 2005).
The response to (self or non-self) antigens restricted by similar class II MHC molecules leads to a usage of a limited number of rearrangements of the Beta chain of the TCR, shared among several patients, with common variable-diversity-junctional segments (V-D-J) and similar three-dimensional structures of the third complementarity-determining region (CDR3). Many studies have shown that in RA patients the TCR repertoire is skewed to certain BV genes, suggesting oligoclonal expansions (VanderBorght et al., 2000, Li et al., 1994, Wagner et al., 1998) and phenotype alterations responsible of autoreactivity, immunosenescence and a proinflammatory state of CD4 + T lymphocytes (Spurlock et al., 2015). Such TCR repertoire remains stable during the disease, in part due to central tolerance that reduces the potentially self-reactive repertoire, and in part for competition between memory and naïve T cells in the lymph nodes for the antigen presentation (Lanzavecchia and Sallusto, 2002).
Epitope encompassing residues 261–273 of type II human Collagen (CII261-273) is the dominant antigenic epitope of Type II Collagen restricted by HLA DRB1*04 and HLA DRB1*01, as supported also by the observation that it is the dominant epitope for the T cell response during collagen-induced arthritis (CIA) in mice transgenic for the human HLA DRB1*01 and DRB1*04 in mice (He et al., 2004). Other studies demonstrated that the use of an altered peptide CII261-273 inhibits T cell activation in CIA induced mice (Zhao et al., 2008, Li et al., 2006), and that T cell reactivity to the peptide 264–270 correlates with disease activity in a cohort of DRB1*04 RA patients (Snir et al., 2012). Even though it has not been clarified if CII-specific T cells are involved in the pathogenesis of early RA or if they are a consequence of joint damage, the status or the size of the autoreactive T cell repertoire appears to be modulated by the level of both unmodified and glycosylated Collagen II peptides (Backlund et al., 2002).
In a previous work (Ria et al., 2008) we performed a complete analysis of CDR3 length distribution of the TCR Vbeta-chain of T cells specifically expanded by Coll261-273 in HLA DRB1*04+ ERA patients. We analyzed TCR BV gene usage and clonality of circulating and infiltrating T cells in the synovial fluid of RA patients using semiquantitative PCRs and modified BV-BJ immunoscope technique, finding that the repertoires of Coll261–273-specific T cells in peripheral blood and synovial fluid show a restricted variety of rearrangements of the TCR-beta chain. The TCR-beta chains identified by immunoscope analysis were sequenced and showed that identical aa sequenced were enriched by antigen stimulation among patients, and therefore identified a group of “shared” clonotypic, collagen-specific T cells. T cells carrying TRBV25-TRBJ2.2 were most frequently used among DR4 + patients affected by ERA. Finally, the majority of the repertoire was shared between different patients affected by RA (Ria et al., 2008) with respect to HLA DRB1*04 + healthy controls. These results were confirmed through different methods in a small cohort of early and long standing RA patients, not selected on the basis of the HLA-DR(Zhou et al., 2014). When we investigated the mechanism of Coll261-273-HLA-DRB1*04 recognition by TRBV domain using molecular modeling, protein-protein docking and molecular dynamics simulations, we could demonstrate that the residues 70–74 shared by HLA-DRB1*01 and HLA-DRB1*04 directly contributes to the engagement of beta chain of a Coll216-273-specific TCR (De Rosa et al., 2010).
In the present work, we aimed to define the association of the antigen specific T cell receptor with DR alleles, with disease activity-severity and with response to conventional DMARDs. The results show that the TRBV25 rearrangement identifies, along with HLA-DRB1, patients with the most active and refractory RA.
2. Material and Methods
2.1. Patients
A total of 90 patients affected by rheumatoid arthritis (RA) were enrolled, with symptoms duration of less than 12 months at diagnosis (ERA patients). Overall we collected 111 samples, 25 of which from ERA patients with a high disease activity (DAS ≥ 3.7), 31 with a DAS comprised between 3.7 and 2.4, 17 with a DAS between 2.4 and 1.6, and 38 samples from patients during a remission phase (DAS < 1.6). All the recruited samples were prepared for the TCR analysis.
All the RA patients fulfilled the 1987 and 2010 American College of Rheumatology (ACR) criteria for RA (Arnett et al., 1988, Aletaha et al., 2010).
At baseline, ERA patients were treated with methotrexate (up to 20 mg weekly) and, when necessary, with a low steroid dosage for three months; a combination with a TNFα blocker (adalimumab 40 mg every two weeks, or etanercept 50 mg weekly) was started if patients did not reach at least a good response within three months from the onset of therapy, according to EULAR criteria (DAS44: DAS ≤ 2.4). ERA patients were followed every month up to three months and every three months thereafter. At each visit the ACR European League Against Rheumatism (EULAR) core data set (erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), swollen joint count (SJC), tender joint count (TJC), physician and patient global assessment, pain, health assessment questionnaire (HAQ)) was registered and DAS was calculated (Fransen and van Riel, 2005).
At each visit, clinical improvement and remission were evaluated according to DAS (van Gestel et al., 1996, Prevoo et al., 1996). Written informed consent was obtained from all study subjects. This study was performed according to the Declaration of Helsinki and was approved by the local ethics committee.
2.2. HLA Genotyping
We performed HLA-DRB1 genotyping and CDR3 length analysis in 90 ERA patients enrolled at different time points: 35 at diagnosis (T0), of which 20 with high disease activity (DAS ≥ 3.7), 12 with a moderate disease activity (3,7 > DAS ≥ 2.4) and 3 with low disease activity (DAS < 2.4); the other 55 patients also during the follow up of the disease (T ≠ 0) reaching a total number of 111 samples (for some patients more samples were collected at different time points). The mean disease duration since the enrollment of all the ERA patients was 10.7 months after diagnosis.
Genomic DNA for the HLA genotyping was obtained from whole blood of the 90 ERA patients using QIAamp DNA Mini kits for genomic DNA purification (Qiagen GmbH, Hilden, Germany) and 0.05–0.1 μg of purified gDNA were used for the PCR amplification of exon 2 of HLA-DRB1.
After PCR amplification molecular typing of HLA-DRB1 by a reverse hybridization method was performed using the INNO-LiPA HLA-DRB1 plus kit (Innogenetics N. V., Ghent, Belgium) and following manufacturer's instructions. Interpretation of hybridization of HLA-DRB1 probes was made by use of LiRAS software, to predict one digit HLA.
2.3. Cell Culture
Peripheral blood mononuclear cells (PBMCs) were purified from whole blood by Lympholite-H density gradient (CEDARLANE Laboratories Ltd., Burlington, Canada), as described in manufacturer's protocol.
Purified PBMCs were cultured in presence or absence of 20 μg/ml Coll261-273 for 72 h in RPMI 1640 medium (Sigma Aldrich, St. Louis, MO, USA) supplemented with 2 millimoles/l l-glutamine, 50 millimoles/l of 2-ME (mercaptoethanol), 50 μg/ml of gentamicin (Sigma-Aldrich, St. Louis, MO, USA), and 1% human AB serum (following the protocol described in Nicolò et al. (Nicolo et al., 2006) and Ria et al. (Ria et al., 2008)). Seventy-two hours later cells were harvested and antigen-specific cell proliferation was assessed by cell count after Trypan blue staining (Burker chamber). Cell death was shown not to be different between antigen-stimulated and unstimulated PBMC by staining for annexin V and Propide Iodide as described in our previous work (Ria et al., 2008) to avoid any bias.
2.4. Staining and Enrichment of IL-13 and IL-17-secreting T Cell
Coll261-273-specific T cells secreting IL-13 and IL-17 were stained and enriched from PBMCs obtained from ERA patients using MACS® secretion kit (Miltenyi Biotec, Germany) according to the manufacturer's instruction, following the protocols for enrichment of low-frequency secreting cells. Briefly, 2 × 107 to 4 × 107 cells obtained from PBMCs were stimulated in absence (background) or in presence of 10 μg/ml of Coll261-273 peptide (positive sample), in a 24-well plate at a concentration of 0.5 × 107 cells ml/well. Three hours (for IL13 staining) and 16 h (for IL17 staining), cells were harvested and submitted to the staining procedure for each cytokine. The enrichment for cytokine-secreting cells was checked by flow cytometry (Coulter Epics FACS, equipped with Lysis software) (Beckman Coulter Inc., Fullerton, CA, USA) analysis. In order to evaluate correctly the number of antigen-specific cells, we examined by FACS 5 × 105 cells both in the background and in positive samples. Total, negatively selected and positively selected cells were collected and prepared for mRNA isolation. As previously published (Nicolo et al., 2006), in order to prevent uncontrolled loss of mRNA due to scarcity of cells in the positively selected fraction (usually ~ 104 total cells were recovered in the cytokine-positive fraction), mRNA obtained from 106 TCR alpha-beta-BW cells (a mouse T cell line lacking the expression of alpha- and beta- chains of the TCR) was added to the positively selected cells before proceeding with mRNA isolation for the TCR repertoire analysis.
2.5. TCR Repertoire Analysis
Total mRNA was isolated from cell suspensions using RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany), in accordance with the manufacturer's instruction. cDNA was synthesized using an oligo-dT primer (dT15; Gibco BRL Life Technologies). From each cDNA, PCR reactions were then performed. Sequences of TRBV25, TRBC1alfa and TRBJ2.2 specific primers were deduced from the IMGT (ImMunoGeneTics, www.imgt.com) and described in Ria et al. (Ria et al., 2008).
Using 2 μl of this product as a template, run-off reactions were performed with a single internal fluorescent primer for each BJ tested. These products were then denatured in formamide and analyzed on an Applied Biosystem 3130 Prism using Gene-mapper version 4.0 software (Life Technologies, Amsterdam, The Netherlands). Results are also reported as RSI (rate stimulation index = normalized peak area obtained from cells stimulated with antigen/normalized peak area of non-stimulated cells). Analyzing the 90 ERA patients we looked for TCRs expansions, in particular TRBV25-TRBJ2.2 with the length of 139 bases (alias name vb11-jb2.2, hereafter TRBV25). The cut-off for positive specific expansions defined as rate stimulation index (RSI) was settled RSI ≥ 1.8) and the TCRs sequences, identifying common variable-diversity-junctional segments (V-D-J), were described in our previous work (Ria et al., 2008).
We found on a large collection of samples that peaks of length other than those previously identified as associated with the response to Coll peptide had a SI of 1 ± 0.34 when ag − versus ag + were compared. Thus, we chose the SI value of 1.8 (> average + 2SD) as a cut off value for a specific expansion.
2.6. Statistical Analysis
Data were analyzed using SPSS Statistics 20.0 (IBM Corp., Armonk, NY, USA) and Prism software 6.01 (Graph-Pad, San Diego, CA 92121-USA). Categorical and quantitative variables were respectively described as numbers, percentages (%) and mean ± standard deviation (SD). Mann–Whitney's test was used to compare continuous variable. Categorical variables were analyzed using χ2 test or Fisher's test, depending on sample size restrictions. Kaplan–Meier survival plots were created using Prism software 6.01 (Graph-Pad, San Diego, CA 92121-USA). Statistical significance of survival analyses and P-values were determined by applying the Log-rank statistical method (Mantel-Cox test) and the Gehan–Breslow method (Wilcoxon test) taking into account multiple comparisons. A value of P < 0.05 was considered statistically significant.
3. Funding
This work was partially supported by “linea D1” from Catholic University of Rome (Gianfranco Ferraccioli and Francesco Ria).
4. Results
4.1. HLA-DRB1 and Disease Activity at Diagnosis: Group Differences
We examined in our cohort whether DR4-4, DR4-11 and DR4-1 patients represented collectively a subgroup of patients with a more aggressive disease.
A total of 90 ERA patients treated with the treat-to-target strategy were enrolled. The mean DAS value in the whole ERA cohort was 3.8 ± 1.1 (51.1% with an active disease-DAS ≥ 3.7, 42.2% with a moderate disease-2.4 < DAS < 3.7 and 6.7% with a low disease activity-DAS ≤ 2.4). The demographic, immunological and clinical characteristics of RA patients are shown in Table 1.
Table 1.
Demographic, immunological and clinical characteristics of ERA patients at diagnosis. Values are mean ± standard deviation unless otherwise indicated. ERA: early rheumatoid arthritis; ACPA: anti-citrullinated peptide antibodies; RF: rheumatoid factor; ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; TJC: tender joint count; SJC: swollen joint count; DAS: disease activity score; HAQ: Health Assessment Questionnaire. HLA: histocompatibility Leucocyte Antigen. ESR, CRP, TJC, SJC, HAQ, are missing in 5 patients.
| ERA patients | |
|---|---|
| N. | 90 |
| Age, (years) | 55.0 ± 13.6 |
| Sex, n° female, (%) | 70.0 (77.8) |
| ACPA > 5 U/ml n. (%) | 64 (71.1) |
| RF-IgM > 20 U/ml n. (%) | 46 (51.1) |
| ESR (mm/1 h) | 41.9 ± 26.4 |
| CRP (mg/l) | 24.9 ± 31.7 |
| TJC | 11.6 ± 8.0 |
| SJC | 14.7 ± 8.8 |
| DAS | 3.8 ± 1.1 |
| HAQ | 1.29 ± 0.77 |
| Symptoms' duration (months) | 6.27 ± 3.55 |
| HLA-DRB1*01 n. (%) | 21 (23.3) |
| HLA-DRB1*04 n. (%) | 27 (30.0) |
| HLA-DRB1*11 n. (%) | 32 (35.6) |
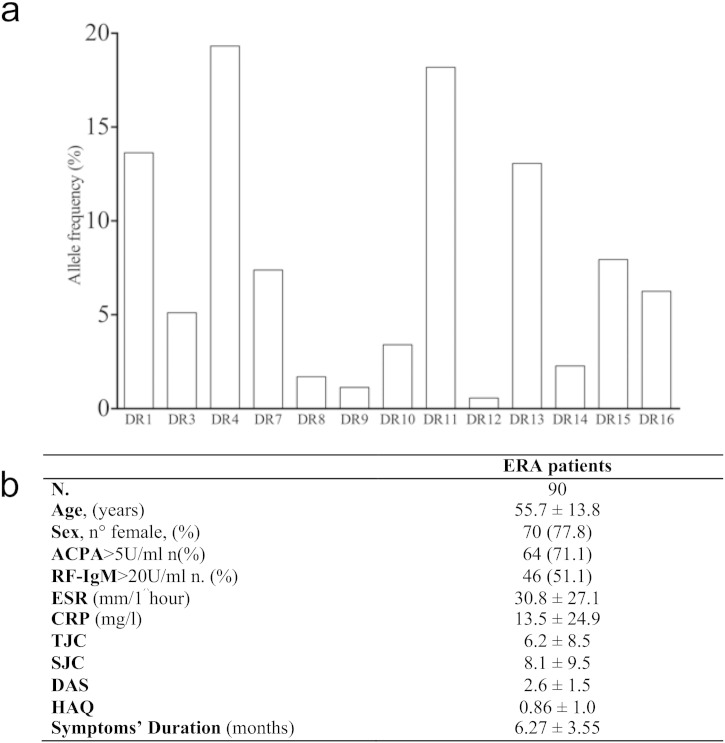
The HLA-DRB1 genotyping of ERA patients showed, as expected (Reveille, 1998, Rosloniec et al., 2002), a high prevalence of HLA DRB1*04 and *01 positive patients (Supplementary Fig. 1a). The allelic frequency of DR4 was 18.9% in a total of 27 out 90 ERA patients, of which 7 (7.8%) were homozygous; moreover, the allelic frequency of DR1 + was 13.3% in a total of 21 subjects, of which 3 (3.3%) were homozygous and 5 (5.6%) were DR4 + DR1 +. Finally, the allelic frequency of DR11 was 18.3% for a total of 32 out 90 ERA patients, of whom 2 (6.3%) were DR1 + DR11 + and 4 were DR4 + DR11 + (12.5%). (Table 2) Thus, DR11, despite its protective role in RA, appears to be the second most frequent allele in this cohort of patients (Supplementary Fig. 1a and Table 2), and for this reason we decided to examine in depth its association to RA.
Supplementary Fig. 1.
HLA-DRB1 frequency and demographic, immunological and clinical characteristics of ERA patients at the study entry. (a) The bars chart shows the allelic frequency of HLA-DRB1 alleles in the 90 ERA patients cohort. The corresponding population of reference is the central Italy where the allelic frequencies of the most represented alleles of the cohort are different (7.4% for DR1, 8.2% for DR4, 27% for DR11 and 10.5% for DR13). The source of Italian allelic frequency is the database www.allelefrequencies.net. (b) Values are mean ± standard deviation unless otherwise indicated. ERA: early rheumatoid arthritis; ACPA: anti-citrullinated peptide antibodies; RF: rheumatoid factor; ESR: erythrocyte sedimentation rate; CRP: C-reactive protein; TJC: tender joint count; SJC: swollen joint count; DAS: disease activity score; HAQ: Health Assessment Questionnaire. ACPA, RF, ESR, CRP, TJC, SJC and HAQ are missing in 6 patients.
Table 2.
HLA-DRB1*01, *04 and *11 alleles. Values report the absolute number of positive subjects and frequency (%) in the 90 ERA patients, for each indicated haplotype.
| ERA patients (N = 90) | |||||
|---|---|---|---|---|---|
| HLA-DR allele combinations | n. (%) | HLA-DRB1*01 | HLA-DRB1*04 | HLA-DRB1*11 | Other allele combinations |
| HLA-DRB1*01 | 21 (23.3) | 3 (3.3) | 5 (5.6) | 2 (2.2) | 11 (12.2) |
| HLA-DRB1*04 | 27 (30.0) | 5 (5.6) | 7 (7.8) | 4 (4.4) | 11 (12.2) |
| HLA-DRB1*11 | 32 (35.6) | 2 (2.2) | 4 (4.4) | - | 26 (28.9) |
| HLA-DRB1*04 + and (04 + or 01 + or 11 +) | 16 (17.8) | ||||
We did not consider the alleles with a frequency less than 10% because the low number of cases was not sufficient to do a correct power analysis. Nevertheless, in our cohort of ERA DR3, DR7, DR8, DR9, DR10, DR12, DR14, DR15 and DR16 seemed not to be involved in RA characteristics (severity or outcome). DR13 allele (13.5%) in our cohort of RA did not associate with the clinical parameters-data and was not considered into the analysis, also on the basis of its known protective role in RA (Lundstrom et al., 2009).
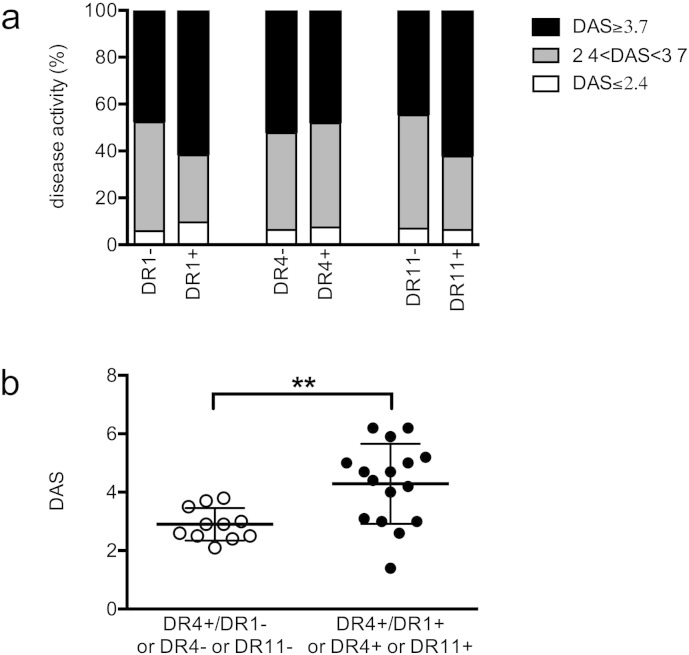
We examined whether the presence of isolated DR4, DR1 and DR11 had any effect on severity of the disease at diagnosis. Data reported in Fig. 1a show that the individual presence of each of these DR alleles per se did not associate with the severity of the disease at diagnosis. Next, we focused our analysis on the 27 DR4 + patients only, and we observed that patients homozygous for HLA-DRB1*04 or carrying HLA-DRB1*01 or HLA-DRB1*11 as the second allele showed a higher disease activity at diagnosis when compared to DR4+ patients carrying other HLA-DRB1 alleles, both when DAS was divided in classes (data not shown p = 0.03) and when the absolute value of DAS was considered (Fig. 1b, 4.3 ± 1.4 vs 2.9 ± 0.6, p = 0.002).
Fig. 1.
HLA DRB1 is associated with disease severity at the onset in DR4 + patients. (A) The six groups of patients positive or negative for HLA-DRB1* allele 01, 04 and 11 are divided in 3 classes, based on DAS at onset: DAS ≤ 2.4 (white), 2.4 < DAS < 3.7 (gray) and DAS ≥ 3.7 (black). (B) DR4 + patients (N = 27) were divided in two groups on the basis of the second DRB1* allele: all the DR4 + patients are shown individually on the basis of DAS at the onset and on the basis of the presence (black circles) or the absence (white circles) of DR4 or DR1 or DR11 as second allele. Each group is further subdivided, as described for panel A.
4.2. DR Haplotype and Disease Activity Determine the Presence of Circulating Coll263-271-specific TRBV25 + T Cells
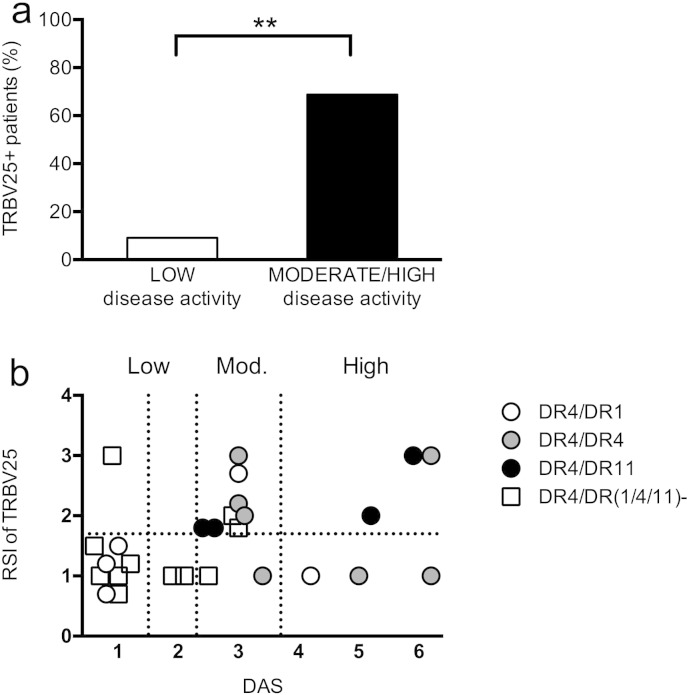
Next, we examined the possible relationship between disease activity score and the presence and/or level of stimulation index of T cells carrying the shared TRBV25 rearrangement in DR4 + patients. Patients were divided in low (n = 11) or moderate-high activity (n = 16) on the basis of their DAS at the moment of the test. We found expansions of the huColl261-273 specific TCR rearrangement of interest (TRBV25-TRBJ2.2 with a length of 139 base pairs) more frequently in patients with a moderate-high disease activity score than in patients with a DAS < 2.4. In DR4 + ERA patients, DAS seemed to be directly proportional to the absolute value of the RSI (data not shown, p = 0.01) and this was confirmed by the comparison of the two groups of patients (low versus moderately-highly active disease) on the basis of positivity (RSI ≥ 1.8) versus negativity (RSI < 1.8) of TRBV25; eleven out of sixteen (68.8%) patients with a moderate-high disease activity were positive for TRBV25 + cells versus one out of eleven (9.1%) patients with a DAS < 2.4 (p = 0.0047, exact Fischer's test, Fig. 2a).
Fig. 2.
TRBV25 usage and DR4 and DR11 are markers of disease activity and of response to “treat to target” strategy. (a) PBMC from 27 consecutive HLA DRB1* 04 + ERA patients at different phases of disease activity were stimulated with Coll261-273 and considered positively expanded when rate stimulation index (RSI) of TRBV25 was at least 1.8 or superior. The DR4 + patients, samples positive for the usage of shared TCR (here represented in percentage) are significantly more frequent in subjects with higher DAS. P value is evaluated with unpaired non-parametric Mann Whitney's test comparing patients on the basis of TRBV25 positivity and negativity (p = 0,04). (b) Associations between DAS, RSI for TRBV25 + cells and the second HLADRB1* allele in the 27 DR4 + ERA patients. Vertical dashed lines indicate DAS of 1.6, 2.4 and 3.7. Symbols indicate patients homozygous for DR4 (gray circles), or DR4/1 (white circles), DR4/11 (black circles) and finally those in which DR4 was associated with any other HLA allele (white squares).
Next we examined if the second HLA allele had an influence on the presence of cells carrying the TRBV25 TCR within the DR4 + group of patients. Fig. 2b reports the RSI of the TRBV25 rearrangement and the DAS value for each of the DR4 + patients examined (n = 27), with the different symbols representing the second DR allele. Thus, on a total of 11 DR4 + ERA patients that used the collagen-specific TRBV25 rearrangement, we observed that 4 were DR4 + DR11 +, 4 were homozygous for DR4, and 1 was DR4 + DR1 +. All these patients were examined when in high or moderate disease activity. Intriguingly, all DR4 + DR11 + patients grouped within the samples obtained during moderate-high disease activity and positive for TRBV25 usage, while some patients homozygous for DR4 or having DR1 as the second DR allele fell in the group of DR4 + ERA patients with high or moderate disease activity, not using the collagen-specific TRBV25 rearrangement. Thus, it appears that DR11 allele is able to promote the usage of TRBV25 by DR4 + subjects at least as well as (and possibly even better than) a second DR4 allele. Finally, the vast majority (8/11) of DR4 + patients with a low disease activity at the moment of the test were heterozygous, but having as a second DR allele DRs different from DR1 or DR11 (Fig. 2b).
Overall, these results, despite the low number of patients, suggest that the presence of peripheral blood T cells carrying the TRBV25 TCR specific for Coll261-273 represent a marker of moderate-high disease activity in RA patients carrying DR4 allele. Moreover, the homozygosity for DR4 or the combination of DR4 with DR1 or, surprisingly, with DR11 seems to be associated with the usage of Coll261-273 specific T cells, carrying the TRBV25 TCR, and with increased disease severity and resistance to therapy.
4.3. HLA-DRB1 Haplotype Predicts Response to Therapy in ERA Patients
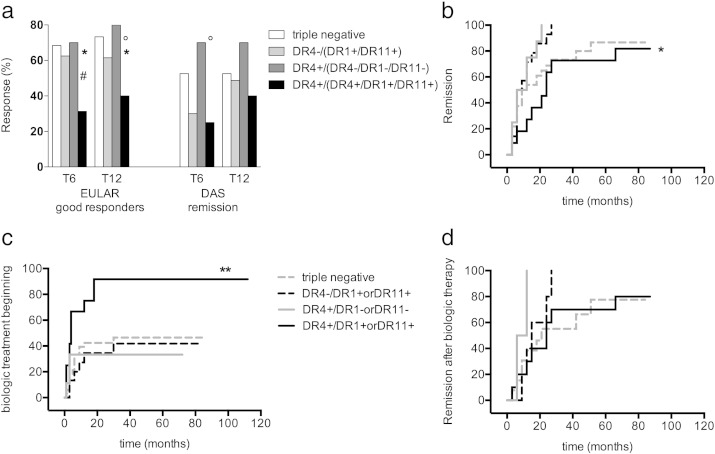
Since we had observed that HLA was associated with disease activity at the onset and with selection of a TCR repertoire in the DR4 + group of patients, next we examined the influence of HLA-DR on the therapeutic outcome of the patients. Overall, 60 (58.8%) ERA patients were classified as good EULAR responders, while 38.8% and 50.6% were in DAS remission (DAS < 1.6) at 6 and 12 months follow-up visit, respectively. Five ERA patients were lost during follow-up. Results are reported in Fig. 3a and b. Following the results reported in the above section, we divided patients in 4 groups: Group 1: DR4, DR1 and DR11 triple negative; Group 2: DR4- and DR1 + or DR11 +; Group 3: DR4 + and DR1- or DR11-; Group 4: DR4 + in combination with another DR4 or a DR1 or DR11. Patients belonging to Group 1 and 3 were more frequently good responders if compared to patients of Group 2 and of Group 4.
Fig. 3.
DR1, DR4 and DR11 allele combinations and the outcome of disease activity. (a) ERA patients are evaluated after six (T6) and twelve (T12) months of follow up during treat to target therapy and are displayed in 4 groups on the basis of the allelic combinations: 1) DR4, DR1 and DR11 triple negative (white bars), 2) DR4- and DR1 + or DR11 + (light gray bars), 3) DR4 + and DR1- or DR11- (dark gray bars) and 4) DR4 allele in combination with another DR4 or DR1 or DR11 (black bars). Patients' outcome is reported using EULAR criteria for good response (DAS reduction ≤ 1.2 left bars) or for the achievement of a DAS value < 1.6 (right bars). Patients in group 4 show a significant poorer response to treatment (unpaired non-parametric Mann Whitney's test, T6 p = 0.04 (* 4 versus 1, ° 4 versus 2, # 4 versus 3): T12 * p = 0.03 and ° p = 0.05. (b/c/d) Cumulative Kaplan Meyer plots of the same groups of patients described in (a) (Group 1, dashed gray; Group 2, dashed black; Group 3, continuous gray; Group 4, continuous black) indicating (b) the % of patients not in remission (DAS < 1,6 and DAS variance > 1.2) of the disease, (c) the % of patients not adding biologic treatments and (d) % of patients not in remission after addition of biologic therapy. The p values were evaluated with Log Rank (Mantel Cox's test) and ERA patients of group 4 showed a behavior significantly different for time to remission (b) p = 0.03 and for time to addition of biologic treatment (c) p = 0.002, but not for time of remission after addition of biologic therapy (d) p = 0.2.
DR4 + ERA patients having DR1, DR4 or DR11 as a second allele received a combination therapy with biologic agents within 12 months follow-up (73.3%). This finding was significantly higher compared to RA patients of other groups at the same time point of follow up (nearly 30.0%; p = 0.002) (Fig. 3c and Supplementary Fig. 2). However, in terms of outcome of the combination therapy of patients of group 4, compared to that of patients belonging to the other groups submitted to the same therapeutic regimen, no differences could be observed (Fig. 3d).
Supplementary Fig. 2.
Response to anti-TNF therapy. ERA patients are evaluated twelve months (T12) after study entry during treat to target therapy, in terms of percentage of subjects who were under a biologic treatment; they are displayed in 4 groups on the basis of the allelic combination: 1) DR4, DR1, DR11 triple negative (white bars), 2) DR4-and DR1 + or DR11 + (bright gray bars), 3) DR4 + and DR1- or DR11- (dark gray bars) and 4) DR4 allele in combination with another DR4 or a DR1 or DR11 (black bars). Patients of the group 4 were treated significantly more often with anti-TNF therapy than group 1 (*p = 0,05) and than group 3 (°p = 0,03).
Taken together, these data indicate that a DR4-4, DR4-1 or DR4-11 genotype marks a group of patients suffering a more active and severe disease, which appears resistant to DMARDs but responsive to biologic therapy.
4.4. HLA-DRB1 and Seropositivity for ACPA are Complementary in Identifying the Subsets of Good and Poor Responders
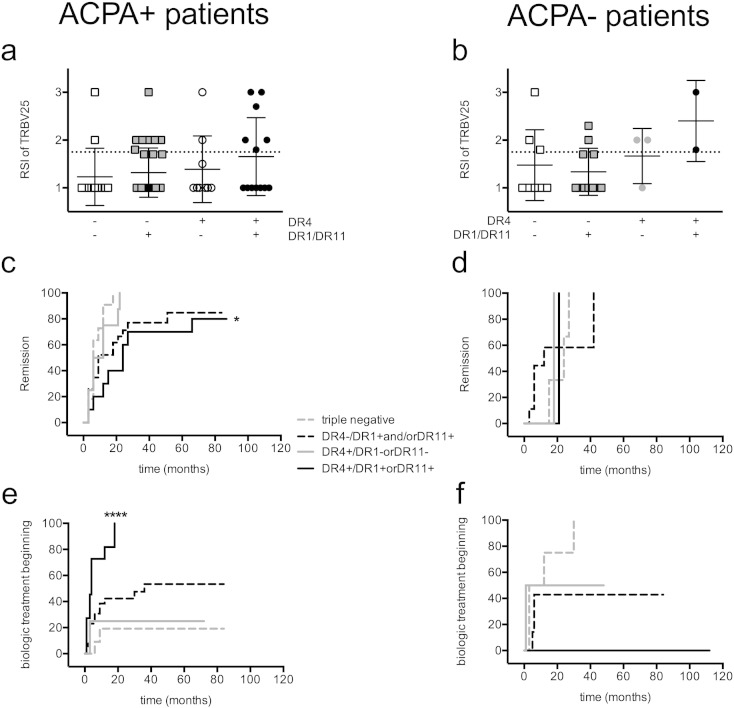
We wondered whether the combination of HLA DR alleles associated with severity of disease was simply highlighting the same population of RA patients showing presence of anti-cyclic citrullinated peptide antibodies. Thus, we separated our ERA patients cohort into ACPA + (seropositive) and ACPA- (seronegative) and, as done in Fig. 4, divided them into 4 subsets: Group 1: DR4, DR1 and DR11 triple negative; Group 2: DR4- and DR1 + or DR11 +; Group 3: DR4 + and DR1- or DR11-; Group 4: DR4 + in combination with another DR4 or a DR1 or DR11.
Fig. 4.
DR1, DR4 and DR11 allele combinations and the outcome of disease activity in ACPA positive patients. ERA patients are divided on the basis of the positivity to ACPA (ACPA +, a, c, e) and ACPA- (b, d, f) and are divided in the 4 groups described in Fig. 3, on the basis of the allelic combinations. ACPA statuses are missing in 5 patients. (a and b) Correlation between ACPA positivity, DR and RSI of the Coll261-273-specific TRBV25 + T cells (number of patients per group: ACPA + = 66 distributed in 1A = 13, 2A = 31, 3A = 9, 4A = 13; ACPA- = 24 divided in 1B = 8, 2B = 11, 3B = 3, 4B = 2). (c to f) Cumulative Kaplan Meyer plots of the same groups of patients divided according to DR as already described in Fig. 3 (BD) (Group 1, dashed gray; Group 2, dashed black; Group 3, continuous gray; Group 4, continuous black) indicating (c, d) the % of patients not in remission (DAS < 1,6 and DAS variance > 1.2) of the disease, (e, f) the % of patients not yet adding biologic treatments. Missing patients for follow up data are 14 for C, 9 for D and 10 for E and F, P values of Kaplan Meyer plots are evaluated with Log Rank (mantel Cox's test) and patients of Group 4 had a significantly worse outcome than the other groups in the ACPA + patients: p = 0.01 for c, p < 0.0001 for e.
First, we noticed that ACPA + ERA patients (n = 66) with expanded TRBV25 + T cells are more frequent in the group 4 (6 out 13, black circles, Fig. 4a and b). When we compared the response to therapy in seropositive patients, groups 1 (n = 13), 2 (n = 31) and 3 (n = 9) were frequently good responders; they could gain an early DAS reduction of at least 1.2 and a DAS < 1.6. ACPA + DR4 + ERA patients with DR4, DR1 or DR11 as second DR allele (group 4, black line), on the other hand, presented a poor response to a treat to target strategy with remission significantly delayed (Fig. 4c) and they were treated significantly more often with biologic DMARDs therapy (Fig. 4e) than the other groups. Thus, having a DR4-4, DR4-1 and DR4-11 allele combination represents a marker of poor response to therapy also when examined within the ACPA + group of ERA patients.
ACPA- ERA patients (n = 24) seem to have a different response to a treat to target strategy, but numbers are too small to predict a significantly different outcome (Fig. 4d and f). Overall DR and ACPA appear complementary tools to identify good and poor responders.
4.5. Cells Carrying TRBV25 can Produce IL-17
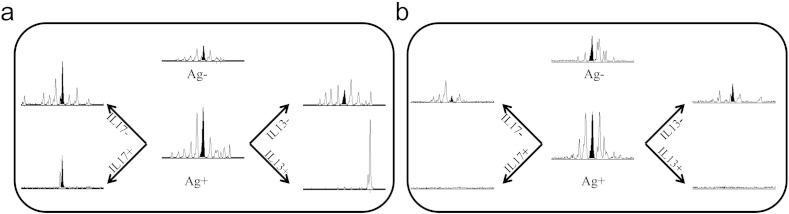
We have shown above that the same HLA-DRB1 haplotype predicts severity of the disease and resistance to treatment and in parallel usage of TRBV25 collagen-specific T cells, which are also associated with a more severe outcome of the disease. This observation leads to the hypothesis that an effect of HLA-DRB1 haplotype is the selection of collagen specific T cells characterized by higher aggressive behavior. An important goal to better investigate the role of T cells in RA is the characterization of their cytokine production, in particular IL17 and IL13. As a proof of concept here we report two case-reports where we explored the hypothesis of this connection. We stimulated PBMC from 2 DR4 + ERA active patients with Coll261-273 and sorted the T cells secreting IL17 and IL13. We examined the presence of T cells specific for huColl261-273 using the VB-JB pairs previously shown to host shared TCR-beta chain rearrangements. The RA patients presented in the PBMC 7 TCR-beta chains that were expanded by the stimulation with huColl261-273, among which there were the shared TRBV25 T cells (CDR3 length of 139 bases and Rate Stimulation Index of 1.8). When we examined the presence of the same 7 TCR-beta chain rearrangements found in the blood in the IL17-secreting cell population, we could detect only the TRBV25 TCR chains (Fig. 5a). Thus, the antigen specific T cells able to secrete IL17 seem to represent a restricted subpopulation within the total T cell repertoire responding to the self-antigen during the acute presentation of the disease (Fig. 5b). The TRBV25 rearrangement was not enriched in the IL-13 secreting population from the same patients, nor from PBMC IL17+ or IL13+ T cell populations in two more patients in low disease activity (data not shown) further suggesting that its presence may be involved in the activity of the disease.
Fig. 5.
TRBV25 + cells are enriched among T cells secreting IL17 after stimulation with Coll261-273 during active presentation of disease. PBMC from DR4 + patients during moderate disease activity phase (a, DAS = 2.6) or low disease activity (b, DAS = 1.3) were cultured in presence of Coll261-273 and magnetically sorted for secretion of IL17 (left panels) or IL13 (right panels). For each patient, the presence of a Coll261-273-driven expansion of T cells was examined after 72 h of culture in the absence or presence of the peptide antigen (middle panels). All panels report the distribution of CDR3-beta peaks obtained by Immunoscope analysis of rearrangement TRBV25-TRBJ2.2. The black peak corresponds to the length of 139 bases that identifies the shared Coll261-273-specific rearrangement. This peak appears enriched only in IL-17 secreting cells obtained from the patient in a moderate disease activity phase (a).
5. Discussion
RA has long been considered an autoimmune disease, in which immune complexes mediate tissue injury (Vaughan, 1973, Johnson et al., 1973). The failure to correlate disease activity to the levels of autoantibodies and the realization that RA exists in individuals not expressing RF, suggest that T cell very likely play a major role through T cell-dependent effector mechanisms and T cell-derived cytokines, released upon T cell recognition of Ag in the synovial membrane (Ferraccioli and Zizzo, 2011, Zizzo et al., 2011). Activation of macrophages and synovial fibroblasts causes the formation of a tissue-invasive pannus.
HLA-D, expressed by antigen presenting cells (APCs), hosts in the peptide-binding groove the antigenic fragments to present to T cell receptor (TCR) on cognate T cells. The polymorphisms of HLA-D molecules can change the amino-acid side chains of the binding pockets, altering the TCR-peptide-HLA-D interface. For example the residues at positions 11, 71 and 74 of the HLA-DRB1 chain are associated with the higher RA susceptibility in HLA-DRB1*0401 and *0404, while the *0402 allele has a protective role (Gregersen et al., 1987, Taneja et al., 2008, Luckey et al., 2014).
Our data confirm the link between HLA-DRB1 and RA and in particular in our cohort of 90 Italian patients as expected a 44 present DR4 and/or a DR1 allele, but the second most frequent allele is DR11, despite in literature it is described as having a protective role in RA. It is not the first observation of an HLA allele with controversial role: in the case of HLA-DRB1*13 allele, it has been described an association with increasing risk of RA development when in combination with DRB1*03 and in ACPA negative patients, and at same time a protecting role in ACPA positive patients (Lundstrom et al., 2009); similarly DR11 seems to have a “janus” effect in RA, in particular modulated by the association with DR4.
A modification of the selection of self-epitopes by simultaneous presence of two HLA alleles may be one mechanism that explains this “janus” effects. As demonstrated in other autoimmune models, the modification of peptide presentation can result in the promotion of positive selection of pathogenic autoreactive thymocytes and/or in the failure to trigger their subsequent deletion; moreover protective class II molecules would select a repertoire of T-cells endowed with decreased pathogenicity (Luhder et al., 1998). The disease predisposing class II MHC molecules have also been suggested to contribute to autoimmune inflammation by modulating the cytokine production of T lymphocytes in the peripheral tolerance homeostasis (Mangalam et al., 2013). Protective class II MHCs have been proposed to compete against or to interfere with self-antigen presentation for capture of determinants (Deng et al., 1993, Ge et al., 2011). In this context it has been demonstrated that pathogenic autoreactive TCRs are “MHC-promiscuous” (Schmidt et al., 1997), evidence derived from the fact that individual TCRs have the inherent potential to recognize a wide set of peptides embedded in a single MHC molecule or multiple MHC molecules presenting the same or different peptides (Wooldridge et al., 2012, Brock et al., 1996).
A further role of the protective class II molecules in autoimmune disease resistance could be related to the modulation of T regulatory cells against the self-reactions (Tsai and Santamaria, 2013).
The restriction of the TCR and the sharing of T cells among different Germinal Centers strongly support a role for few antigens driving the formation of tertiary lymphoid tissue and subsequent tissue destruction in the rheumatoid joint (Tsai and Santamaria, 2013).
These antigens are obviously shared among patients because the follicular CD4 T cell clones undergo stimulation in the autologous as well as in the heterologous tissues. The restriction to HLA-DRB1 molecules is in line with the recognition of classical peptide antigens.
In the present report, we demonstrate that the already known biomarker of severity DRB1 (it associates with seropositivity and erosions) also associates with circulating T cells specific for Coll261-273. These cells using the shared TCR-beta chains TRBV25-TRBJ2.2 (alias VB11-BJ2.2) are significantly associated with disease activity in the group of DR4 + RA patients, and with a worse outcome. The relevance of the antigen-specific T helper cells has been recently confirmed and their analysis could represent a new possible approach in the evaluation of RA (Shoda et al., 2015). Furthermore it has been demonstrated that HLA-DRB1*04 contributes to RA risk, specifically in CD4 + T cells, with a cognate and continuous stimulation of TCR-CD3 complexes that culminates in intracellular signaling able to increase the NF-κB activity and to promote a proinflammatory state (Spurlock et al., 2015). Thus, we propose that the usage of this T cell receptor is able to predict the outcome of the disease in DR4 + patients (Ferraccioli and Zizzo, 2011, Raza et al., 2005, Annunziato et al., 2009) and even if it seems to involve a portion of RA patients, we show here that this subgroup presents a delayed remission. Probably one of the main reasons is that DMARDs therapy after diagnosis, as indicated by the treat to target strategy, is insufficient and only biological treatment is useful for them; in this sense a TCR specificity in combination with HLA appears to lead to a more aggressive therapy since the beginning.
Previous works from other groups have demonstrated that homozygosity for alleles of the DR predisposes to RA severity (Lundstrom et al., 2009). Here we confirm this observation and report for the first time that also heterozygosity for DR4-DR1 and DR4-DR11 associate with a more severe disease. Interestingly, this role of DR11 as a predictor of worse disease is limited to the DR4 + and ACPA positive RA patients, while no effect of DR11 per se could be observed in the whole cohort of RA patients.
Heterozygosity of DR4-DR11 and, to a lesser extent, DR4-DR1 and homozygosity for DR4, are associated with the presence of TRBV25-TRBJ2.2 +, collagen-specific T cells during the acute presentation of the disease. As a proof of concept of our hypothesis we show here that TRBV25-TRBJ2.2 + T cells likely secrete IL-17, thus contributing to the aggressiveness of the disease (Leipe et al., 2010, Hot and Miossec, 2011). Thus, these data suggest that these allelic combinations may promote the selection of T cells primed for the collagen-specific self-reactive response and with possible polarization to a milieu much more inflammatory (Nicolo et al., 2006).
6. Conclusions
In the present work, we hypothesize that autoreactive T cells, could be selected on the background of DR4-DR1-DR11 as second alleles in the DR4 + RA patients (enhancing the differentiation of autoreactive T cells) leading to detect the subgroup of patients with a more severe presentation of RA and a marked resistance to DMARDs. The analysis of the HLA-DRB1 (determination of the HLA DRB1* is a test available in most hospital centers) and of T cells specific for Coll261-273 carrying the shared TRBV25-TRBJ2.2 rearrangement can be proposed as a possible biomarker matrix that identify those RA patients who will not satisfactorily respond to treatment following a treat to target strategy with TNFα blockers.
Prospective studies stratifying the patients at diagnosis will be necessary. We are aware of the limits of our cohort, diverse numbers in different subsets, heterogeneity of the genetic background, different therapies at follow-up, yet all the data move into the same direction: our study shows that the analysis of collagen specific the T cell repertoire, of HLA-DR and seropositivity appear complementary tools to identify the best and the poorest responders to DMARD therapy in a treat to target strategy. Therefore the study offers some clues to predict the course in terms of a personalized treatment in RA patients (Isaacs and Ferraccioli, 2011).
The following are the supplementary data related to this article.
Supplementary material.
Authors' Contributions and Acknowledgments
GDS made substantial contributions to the design of the project, acquisition of data, analysis and interpretation of data and writing and revising the manuscript. BT supervised the field activities, designed the study's analytic strategy and prepared the Material and Methods, the Results and the figures sections of the text. ALF and SA allowed their clinical experience to be included in the management and follow–up of enrolled patients. EG and CN contributed to the discussion, reviewed the preparation of the manuscript and conducted the literature review. FR designed the study and participated in drafting the article and revising it critically for important intellectual content. GF directed the implementation of the study, including quality assurance and control, and gave final approval of the version to be submitted and any revised version.
Conflict of Interest
The authors declare that they have no competing interest.
Contributor Information
Gabriele Di Sante, Email: gabriele.disante@hotmail.it.
Barbara Tolusso, Email: barbara.tolusso@unicatt.it.
Anna Laura Fedele, Email: annalaurafedele@gmail.com.
Elisa Gremese, Email: elisa.gremese@rm.unicatt.it.
Stefano Alivernini, Email: stefano.alivernini@hotmail.it.
Chiara Nicolò, Email: nicolochiara@yahoo.it.
Francesco Ria, Email: fria@rm.unicatt.it.
Gianfranco Ferraccioli, Email: gf.ferraccioli@rm.unicatt.it.
References
- Aletaha D., Neogi T., Silman A.J., Funovits J., Felson D.T., Bingham C.O., 3RD 2010 Rheumatoid Arthritis Classification Criteria: an American College of Rheumatology/European League Against Rheumatism Collaborative Initiative. Arthritis Rheum. 2010;62(9):2569–2581. doi: 10.1002/art.27584. [DOI] [PubMed] [Google Scholar]
- Annunziato F., Cosmi L., Liotta F., Maggi E., Romagnani S. Type 17T helper cells-origins, features and possible roles in rheumatic disease. Nat. Rev. Rheumatol. 2009;5(6):325–331. doi: 10.1038/nrrheum.2009.80. [DOI] [PubMed] [Google Scholar]
- Arnett F.C., Edworthy S.M., Bloch D.A., McShane D.J., Fries J.F., Cooper N.S. The American Rheumatism Association 1987 Revised Criteria for the Classification of Rheumatoid Arthritis. Arthritis Rheum. 1988;31(3):315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- Backlund J., Carlsen S., Hoger T., Holm B., Fugger L., Kihlberg J. Predominant selection of T cells specific for the glycosylated collagen type II epitope (263–270) in humanized transgenic mice and in rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 2002;99(15):9960–9965. doi: 10.1073/pnas.132254199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock R., Wiesmuller K.H., Jung G., Walden P. Molecular basis for the recognition of two structurally different Major histocompatibility complex/peptide complexes by a single T-cell receptor. Proc. Natl. Acad. Sci. U. S. A. 1996;93(23):13108–13113. doi: 10.1073/pnas.93.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzikyriakidou A., Voulgari P.V., Lambropoulos A., Drosos A.A. Genetics in rheumatoid arthritis beyond HLA genes: what meta-analyses have shown? Semin. Arthritis Rheum. 2013;43(1):29–38. doi: 10.1016/j.semarthrit.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Cooles F.A., Isaacs J.D. Pathophysiology of rheumatoid arthritis. Curr. Opin. Rheumatol. 2011;23(3):233–240. doi: 10.1097/BOR.0b013e32834518a3. [DOI] [PubMed] [Google Scholar]
- De Rosa M.C., Giardina B., Bianchi C., Carelli Alinovi C., Pirolli D., Ferraccioli G. Modeling the ternary complex TCR-Vbeta/COLLAGENII(261–273)/HLA-DR4 associated with rheumatoid arthritis. PLoS One. 2010;5(7) doi: 10.1371/journal.pone.0011550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton C.M., Walker D.J., Griffiths I.D., Roberts D.F. The contribution of HLA to rheumatoid arthritis. Clin. Genet. 1989;36(3):178–182. doi: 10.1111/j.1399-0004.1989.tb03185.x. [DOI] [PubMed] [Google Scholar]
- Deng H., Apple R., Clare-Salzler M., Trembleau S., Mathis D., Adorini L. Determinant capture as a possible mechanism of protection afforded by major histocompatibility complex class II molecules in autoimmune disease. J. Exp. Med. 1993;178(5):1675–1680. doi: 10.1084/jem.178.5.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitsma A.L., van der Helm-van Mil A.H., Huizinga T.W., de Vries R.R., Toes R.E. Protection against rheumatoid arthritis by HLA: nature and nurture. Ann. Rheum. Dis. 2008;67(Suppl. 3):iii61–iii63. doi: 10.1136/ard.2008.098509. [DOI] [PubMed] [Google Scholar]
- Ferraccioli G., Zizzo G. The potential role of Th17 in mediating the transition from acute to chronic autoimmune inflammation: rheumatoid arthritis as a model. Discov. Med. 2011;11(60):413–424. [PubMed] [Google Scholar]
- Fransen J., van Riel P.L. The disease activity score and the EULAR response criteria. Clin. Exp. Rheumatol. 2005;23(5 Suppl. 39):S93–S99. [PubMed] [Google Scholar]
- Ge X., James E.A., Reijonen H., Kwok W.W. Differences in self-peptide binding between T1D-related susceptible and protective DR4 subtypes. J. Autoimmun. 2011;36(2):155–160. doi: 10.1016/j.jaut.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gay M.A., Garcia-Porrua C., Hajeer A.H. Influence of human leukocyte antigen-DRB1 on the susceptibility and severity of rheumatoid arthritis. Semin. Arthritis Rheum. 2002;31(6):355–360. doi: 10.1053/sarh.2002.32552. [DOI] [PubMed] [Google Scholar]
- Gregersen P.K., Silver J., Winchester R.J. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30(11):1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- He X., Rosloniec E.F., Myers L.K., McColgan W.L., III, Gumanovskaya M., Kang A.H. T cell receptors recognizing type II collagen in HLA-DR-transgenic mice characterized by highly restricted V beta usage. Arthritis Rheum. 2004;50(6):1996–2004. doi: 10.1002/art.20289. [DOI] [PubMed] [Google Scholar]
- Hot A., Miossec P. Effects of interleukin (IL)-17A and IL-17F in human rheumatoid arthritis synoviocytes. Ann. Rheum. Dis. 2011;70(5):727–732. doi: 10.1136/ard.2010.143768. [DOI] [PubMed] [Google Scholar]
- Isaacs J.D., Ferraccioli G. The need for personalised medicine for rheumatoid arthritis. Ann. Rheum. Dis. 2011;70(1):4–7. doi: 10.1136/ard.2010.135376. [DOI] [PubMed] [Google Scholar]
- Johnson J.S., Vaughan J.H., Hench P.K., Blomgren S.E. Rheumatoid arthritis, 1970–1972. Ann. Intern. Med. 1973;78(6):937–953. doi: 10.7326/0003-4819-78-6-937. [DOI] [PubMed] [Google Scholar]
- Kapitany A., Zilahi E., Szanto S., Szucs G., Szabo Z., Vegvari A. Association of rheumatoid arthritis with HLA-DR1 and HLA-DR4 in Hungary. Ann. N. Y. Acad. Sci. 2005;1051:263–270. doi: 10.1196/annals.1361.067. [DOI] [PubMed] [Google Scholar]
- Lanzavecchia A., Sallusto F. Progressive differentiation and selection of the fittest in the immune response. Nat. Rev. Immunol. 2002;2(12):982–987. doi: 10.1038/nri959. [DOI] [PubMed] [Google Scholar]
- Leipe J., Grunke M., Dechant C., Reindl C., Kerzendorf U., Schulze-Koops H. Role of Th17 cells in human autoimmune arthritis. Arthritis Rheum. 2010;62(10):2876–2885. doi: 10.1002/art.27622. [DOI] [PubMed] [Google Scholar]
- Li Y., Sun G.R., Tumang J.R., Crow M.K., Friedman S.M. CDR3 sequence motifs shared by oligoclonal rheumatoid arthritis synovial T cells. Evidence for an antigen-driven response. J. Clin. Invest. 1994;94(6):2525–2531. doi: 10.1172/JCI117624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Li X., Li Z. Altered collagen II peptides inhibited T-cell activation in rheumatoid arthritis. Clin. Immunol. 2006;118(2–3):317–323. doi: 10.1016/j.clim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Luckey D., Behrens M., Smart M., Luthra H., David C.S., Taneja V. DRB1*0402 may influence arthritis by promoting naive CD4 + T-cell differentiation in to regulatory T cells. Eur. J. Immunol. 2014;44(11):3429–3438. doi: 10.1002/eji.201344424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhder F., Katz J., Benoist C., Mathis D. Major histocompatibility complex class II molecules can protect from diabetes by positively selecting T cells with additional specificities. J. Exp. Med. 1998;187(3):379–387. doi: 10.1084/jem.187.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom E., Kallberg H., Smolnikova M., Ding B., Ronnelid J., Alfredsson L. Opposing effects of HLA-DRB1*13 alleles on the risk of developing anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis. Arthritis Rheum. 2009;60(4):924–930. doi: 10.1002/art.24410. [DOI] [PubMed] [Google Scholar]
- Mangalam A.K., Taneja V., David C.S. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J. Immunol. 2013;190(2):513–518. doi: 10.4049/jimmunol.1201891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesko B., Poliska S., Szegedi A., Szekanecz Z., Palatka K., Papp M. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med. Genet. 2010;3:15. doi: 10.1186/1755-8794-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelutti A., Gremese E., Morassi F., Petricca L., Arena V., Tolusso B. B-cell subsets in the joint compartments of seropositive and seronegative rheumatoid arthritis (RA) and no-RA arthritides express memory markers and ZAP70 and characterize the aggregate pattern irrespectively of the autoantibody status. Mol. Med. 2011;17(9–10):901–909. doi: 10.2119/molmed.2011.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolo C., Di Sante G., Orsini M., Rolla S., Columba-Cabezas S., Romano Spica V. Mycobacterium tuberculosis in the adjuvant modulates the balance of Th immune response to self-antigen of the CNS without influencing a “Core” repertoire of specific T cells. Int. Immunol. 2006;18(2):363–374. doi: 10.1093/intimm/dxh376. [DOI] [PubMed] [Google Scholar]
- Prevoo M.L., van Gestel A.M., van T.H.M.A., van Rijswijk M.H., van de Putte L.B., van Riel P.L. Remission in a prospective study of patients with rheumatoid arthritis. American Rheumatism Association Preliminary Remission Criteria in Relation to the Disease Activity Score. Br. J. Rheumatol. 1996;35(11):1101–1105. doi: 10.1093/rheumatology/35.11.1101. [DOI] [PubMed] [Google Scholar]
- Raza K., Falciani F., Curnow S.J., Ross E.J., Lee C.Y., Akbar A.N. Early rheumatoid arthritis is characterized by a distinct and transient synovial fluid cytokine profile of T cell and stromal cell origin. Arthritis Res. Ther. 2005;7(4):R784–R795. doi: 10.1186/ar1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveille J.D. The genetic contribution to the pathogenesis of rheumatoid arthritis. Curr. Opin. Rheumatol. 1998;10(3):187–200. doi: 10.1097/00002281-199805000-00007. [DOI] [PubMed] [Google Scholar]
- Ria F., Penitente R., De Santis M., Nicolo C., Di Sante G., Orsini M. Collagen-specific T-cell repertoire in blood and synovial fluid varies with disease activity in early rheumatoid arthritis. Arthritis Res. Ther. 2008;10(6):R135. doi: 10.1186/ar2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosloniec E.F., Whittington K.B., Zaller D.M., Kang A.H. HLA-DR1 (DRB1*0101) and DR4 (DRB1*0401) use the same anchor residues for binding an immunodominant peptide derived from human type II collagen. J. Immunol. 2002;168(1):253–259. doi: 10.4049/jimmunol.168.1.253. [DOI] [PubMed] [Google Scholar]
- Schmidt D., Verdaguer J., Averill N., Santamaria P. A mechanism for the major histocompatibility complex-linked resistance to autoimmunity. J. Exp. Med. 1997;186(7):1059–1075. doi: 10.1084/jem.186.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoda H., Fujio K., Sakurai K., Ishigaki K., Nagafuchi Y., Shibuya M. Autoantigen BiP-derived HLA-DR4 epitopes differentially recognized by effector and regulatory T cells in rheumatoid arthritis. Arthritis Rheumatol. 2015;67(5):1171–1181. doi: 10.1002/art.39054. [DOI] [PubMed] [Google Scholar]
- Snir O., Backlund J., Bostrom J., Andersson I., Kihlberg J., Buckner J.H. Multifunctional T cell reactivity with native and glycosylated type II collagen in rheumatoid arthritis. Arthritis Rheum. 2012;64(8):2482–2488. doi: 10.1002/art.34459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock C.F., 3RD, Tossberg J.T., Olsen N.J., Aune T.M. Cutting edge: chronic NF-kappaB activation in CD4 + T cells in rheumatoid arthritis is genetically determined by HLA risk alleles. J. Immunol. 2015;195(3):791–795. doi: 10.4049/jimmunol.1500267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja V., Behrens M., Basal E., Sparks J., Griffiths M.M., Luthra H. Delineating the role of the HLA-DR4 “shared epitope” in susceptibility versus resistance to develop arthritis. J. Immunol. 2008;181(4):2869–2877. doi: 10.4049/jimmunol.181.4.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolusso B., De Santis M., Bosello S., Gremese E., Gobessi S., Cuoghi I. Synovial B cells of rheumatoid arthritis express ZAP-70 which increases the survival and correlates with the inflammatory and autoimmune phenotype. Clin. Immunol. 2009;131(1):98–108. doi: 10.1016/j.clim.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Tsai S., Santamaria P. MHC class II polymorphisms, autoreactive T-cells, and autoimmunity. Front. Immunol. 2013;4:321. doi: 10.3389/fimmu.2013.00321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude D., Alemayehu W.G., Verduijn W., de Vries R.R., Houwing-Duistermaat J.J., Huizinga T.W. Gene-environment interaction influences the reactivity of autoantibodies to citrullinated antigens in rheumatoid arthritis. Nat. Genet. 2010;42(10):814–816. doi: 10.1038/ng1010-814. author reply 6. [DOI] [PubMed] [Google Scholar]
- van Gestel A.M., Prevoo M.L., van’t Hof M.A., van Rijswijk M.H., van de Putte L.B., van Riel P.L. Development and validation of the European league against rheumatism response criteria for rheumatoid arthritis. Comparison with the Preliminary American College of Rheumatology and the World Health Organization/International League Against Rheumatism Criteria. Arthritis Rheum. 1996;39(1):34–40. doi: 10.1002/art.1780390105. [DOI] [PubMed] [Google Scholar]
- VanderBorght A., Geusens P., Vandevyver C., Raus J., Stinissen P. Skewed T-cell receptor variable gene usage in the synovium of early and chronic rheumatoid arthritis patients and persistence of clonally expanded T cells in a chronic patient. Rheumatology (Oxford) 2000;39(11):1189–1201. doi: 10.1093/rheumatology/39.11.1189. [DOI] [PubMed] [Google Scholar]
- Vaughan J.H. Rheumatologic disorders due to immune complexes. Postgrad. Med. 1973;54(5):129–136. doi: 10.1080/00325481.1973.11713616. [DOI] [PubMed] [Google Scholar]
- Wagner U.G., Koetz K., Weyand C.M., Goronzy J.J. Perturbation of the T cell repertoire in rheumatoid arthritis. Proc. Natl. Acad. Sci. U. S. A. 1998;95(24):14447–14452. doi: 10.1073/pnas.95.24.14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemze A., Toes R.E., Huizinga T.W., Trouw L.A. New biomarkers in rheumatoid arthritis. Neth. J. Med. 2012;70(9):392–399. [PubMed] [Google Scholar]
- Wooldridge L., Ekeruche-Makinde J., van den Berg H.A., Skowera A., Miles J.J., Tan M.P. A single autoimmune T cell receptor recognizes more than a million different peptides. J. Biol. Chem. 2012;287(2):1168–1177. doi: 10.1074/jbc.M111.289488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li R., He J., Shi J., Long L., Li Z. Mucosal administration of an altered CII263-272 peptide inhibits collagen-induced arthritis by suppression of Th1/Th17 cells and expansion of regulatory T cells. Rheumatol. Int. 2008;29(1):9–16. doi: 10.1007/s00296-008-0634-4. [DOI] [PubMed] [Google Scholar]
- Zhou J., Kong C., Yu J., Dong H., Jin C., Song Q. Skewness of TCR Vbeta of peripheral blood and synovial fluid of patients with rheumatoid arthritis. J. Immunoass. Immunochem. 2014;35(2):207–219. doi: 10.1080/15321819.2013.841192. [DOI] [PubMed] [Google Scholar]
- Zizzo G., De Santis M., Bosello S.L., Fedele A.L., Peluso G., Gremese E. Synovial fluid-derived T helper 17 cells correlate with inflammatory activity in arthritis, irrespectively of diagnosis. Clin. Immunol. 2011;138(1):107–116. doi: 10.1016/j.clim.2010.10.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.