Abstract
Background
Korean Red Ginseng (KRG) is a herbal medicine used in Asian countries and is very popular for its beneficial biological properties. Diabetes mellitus (DM) and its complications are rapidly becoming a global public health concern. The literature on transcriptional changes induced by KRG in rat models of diabetic retinopathy is limited. Considering these facts, we designed this study to determine whether retinopathy-associated genes are altered in retinas of rats with DM and whether the induced changes are reversed by KRG.
Methods
Male Sprague–Dawley rats were intravenously injected with streptozotocin (50 mg/kg body weight) to induce DM, following which, KRG powder (200 mg/kg body weight) was orally administered to the KRG-treated DM rat group for 10 wks. The rats were then sacrificed, and their retinas were harvested for total RNA extraction. Microarray gene expression profiling was performed on the extracted RNA samples.
Results
From among > 31,000 genes investigated, the expression of 268 genes was observed to be upregulated and that of 58 genes was downregulated, with twofold altered expression levels in the DM group compared with those in the control group. Moreover, 39 genes were upregulated more than twofold and 84 genes were downregulated in the KRG-treated group compared to the DM group. The expression of the genes was significantly reversed by KRG treatment; some of these genes were analyzed further to verify the results of the microarray experiments.
Conclusion
Taken together, our data suggest that reversed changes in the gene expression may mediate alleviating activities of KRG in rats with diabetic retinopathy.
Keywords: diabetic retinopathy, gene profiling, microarray, Panax ginseng
1. Introduction
A dramatic increase in the total number of patients with diabetes worldwide shows that diabetes mellitus (DM) and its complications are rapidly becoming a major global public health concern. Every individual with DM is considered to be at risk of developing diabetic retinopathy (DR). DR is a diabetic complication that affects the blood vessels and causes death of retinal cells, ultimately leading to blindness [1], [2]. Indeed, DR is the most important cause of blindness occurring in working-age adults, and it is also the common microvascular complication of DM [3]. The retina consists of multiple layers of retinal cells. Loss of cells from the various retinal layers seems to be a prominent feature of DR. Previous researchers observed DM-induced cell death in all retinal layers, implying that several types of retinal cells may be affected by DM [4], [5], [6], [7]. Understanding the retinal changes occurring during DM and developing new drug candidates are important for designing effective therapies for DR treatment. The precise mechanism of DR is not completely understood. However, it is believed that various factors such as oxidative stress, inflammation, cell death, and genetic and metabolic alterations are involved in the development and progression of DM and DR.
Panax ginseng Mayer has been a popular herbal medicine in Eastern Asia for > 2,000 years. Recent studies have demonstrated various effects of ginseng, including antioxidant, antithrombotic, antihyperlipidemic, angiogenic, and anticancer effects [8], [9], [10], [11], [12]. Korean Red Ginseng (KRG), produced by repeated heating and drying of the root of ginseng, has strong potent pharmacological actions effective in the treatment of several human diseases. In DM, KRG not only improves glucose control but also enhances the renal, auditory, and immune functions [13], [14], [15], [16], [17].
Microarrays are high-throughput genomic tools that offer the fastest and most comprehensive molecular evaluations [18]. Over the past few years, microarrays have been used to compare the global changes in gene expression profiles to enable researchers investigate the molecular and functional alterations of genes in human diseases such as DM [19], [20], [21], [22].
In the current study, the systematic analysis of gene expression profiling was performed in the retinal tissues of rats with streptozotocin (STZ)-induced diabetes before and after KRG treatment to identify the mechanism for the mitigating effect of KRG on retinopathy.
2. Materials and methods
2.1. Animals
Seven-wk-old male Sprague–Dawley (SD) rats were housed under a 12 h:12 h light-dark cycle at room temperature. All the rats were allowed water ad libitum. The rats were divided into normal control, DM, and KRG-treated DM groups. The rats were injected intravenously with a single dose of STZ (50 mg/kg body weight; Sigma-Aldrich Corporation, St. Louis, MO, USA). STZ was dissolved in 0.01M of sodium citrate buffer (pH 4.5), kept on an ice bath, and used within 15 min of preparation. The control rats were injected with the vehicle only. Two days after STZ administration, only the rats with a blood glucose level of > 300 mg/dL were considered to have diabetes and included in further experiments. KRG powder was obtained from the Korea Ginseng Corporation (Daejeon, Korea). The general composition of the KRG powder was as follows: moisture 36%, solid volume 64%, ash 2.5%, total fat 0.05%, total crude saponin 70 mg/g, and total ginsenosides 20 mg/g. The rats in the KRG-treated group received KRG mixed into powdered food at a final concentration of 200 mg/kg/d for 10 wk. The control and DM groups received powdered food alone. In all the groups, > 95% of food was consumed, implying that the rats received the intended doses of KRG.
2.2. Sample preparation
Total RNA was extracted from rat retinal cells by using the miRNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The total RNA pellet was dissolved in diethylpyrocarbonate water, and its quality and quantity was analyzed using the Bioanalyzer 2100 System (Agilent Technologies, Palo Alto, CA, USA). Gene expression was analyzed using the GeneChip Rat Genome 230 2.0 Arrays (Affymetrix, Santa Clara, CA, USA), which comprises > 31,000 probe sets representing approximately 28,700 characterized rat genes. For each gene, 11 pairs of oligonucleotide probes were synthesized in situ on the arrays.
2.3. Microarray
Biotinylated complementary RNAs were prepared from 500 ng total RNA, according to the standard Affymetrix protocol. Briefly, following fragmentation, 15 μg of artificial RNA (aRNA) was hybridized for 16 h at 45°C on the GeneChip Rat Genome Array (Affymetrix). The GeneChips were washed and stained in the Fluidics Station 450 (Affymetrix) and then scanned by using the GeneChip Scanner 3000 7G (Affymetrix). The data obtained were analyzed by using the Robust Multichip Analysis (RMA) algorithm with Affymetrix default analysis settings and global scaling as the normalization method. The trimmed mean target intensity of each array was arbitrarily set to 100. The normalized and log-transformed intensity values were then analyzed using GeneSpring GX 12.6.1 (Agilent Technologies). Fold-change filters were set to retain only upregulated genes present in ≥ 200% of the controls and downregulated genes present in < 50% of the controls. Hierarchical clustering data were grouped according to similarity in behavior across the experiments by using GeneSpring GX 12.6.1. Clustering algorithm was based on the euclidean distance and average linkage.
2.4. Complementary DNA synthesis
The total amount of the extracted RNA was determined spectrophotometrically at 260/280 nm. The RNA quality was assessed as the absence of smear of 18S and 28S bands by using Bioanalyzer 2100. RNA samples were stored at −70°C until further use. Complementary DNAs (cDNAs) were synthesized from 1 μg of total RNA by using the High-Capacity RNA-to-cDNA Kit (Applied Biosystems), according to the manufacturer's protocol.
2.5. Real-time quantitative polymerase chain reaction
High-throughput real-time quantitative polymerase chain reaction (RT-qPCR) was performed using the Applied Biosystems Prism 7900 Sequence Detection System (PE Applied Biosystems, www.appliedbiosciences.com) in triplicates by using white-colored 384-well plates (ABgene, Hamburg, Germany) for intensifying the fluorescent signals by a factor of three. This system operates using a thermal cycler and a fiberoptics-directed laser to each of the 384 wells. The fluorescence emission from each well was collected using a charge-coupled device-camera, and the quantitative data were analyzed by using the Sequence Detection System Software (SDS version 2.2, PE Applied Biosystems). PCR primer sequences used in this study are given in Table 1. The reaction mixtures contained 10 pmol/μL of each primer and 2X SYBR Green PCR Master Mix (PE Applied Biosystems), which contains the HotStarTaq DNA-Polymerase in an optimized buffer, the Deoxynucleotide (dNTP) mix (with Deoxyuridine triphosphate, dUTP additive), the SYBRs Green I fluorescent dye, and the ROX dye as a passive reference. To each well of the 384-well RT-qPCR plates, serial dilutions (1, , , , and ) of cDNA were added, which were then used to generate relative standard curves for the genes. All the primers were amplified using the same conditions: thermal cycling at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 30 s, and finally at 72°C for 3 s. In order to exclude the presence of unspecific products, a melting curve analysis of the products was performed routinely after amplification via high-resolution data collection with a temperature increase from 60°C to 95°C, and with a ramp rate of 0.21°C/s. Then, the RT-qPCR cycle numbers were converted to the corresponding gene amounts (ng) on the basis of the equation.
Table 1.
Primer for real-time polymerase chain reaction
| Gene symbol | Primer sequence | Size |
|---|---|---|
| Crygb | F AATGTAATGGAGGGCTGCTG | 124 |
| R AGCCAACTTTGGCGTTTG | ||
| Gfap | F TTGGCTCTGAATCCTTGGAA | 135 |
| R GGGACTGAGCAACCAGGATA | ||
| Igf1r | F CCCTACCCAAACCCTTAACTG | 179 |
| R CACTGGGAAGCGGAGAAA | ||
| Gapdh | F ATGATTCTACCCACGGCAAG | 89 |
| R CTGGAAGATGGTGATGGGTT |
2.6. Statistical analysis
Statistical significance between each group was estimated by Student t test, and the results were expressed as mean ± standard deviation (GraphPad Prism version 4; GraphPad Software, San Diego, CA, USA).
3. Results and discussion
3.1. Establishment of diabetic rat model
During the experiment, rats with diabetes gradually lost their body mass and developed high blood glucose levels. Two days after DM onset, the average blood glucose level in the DM group was > 450 mg/dL; this level persisted with the progress of DM within 450–600 mg/dL range.
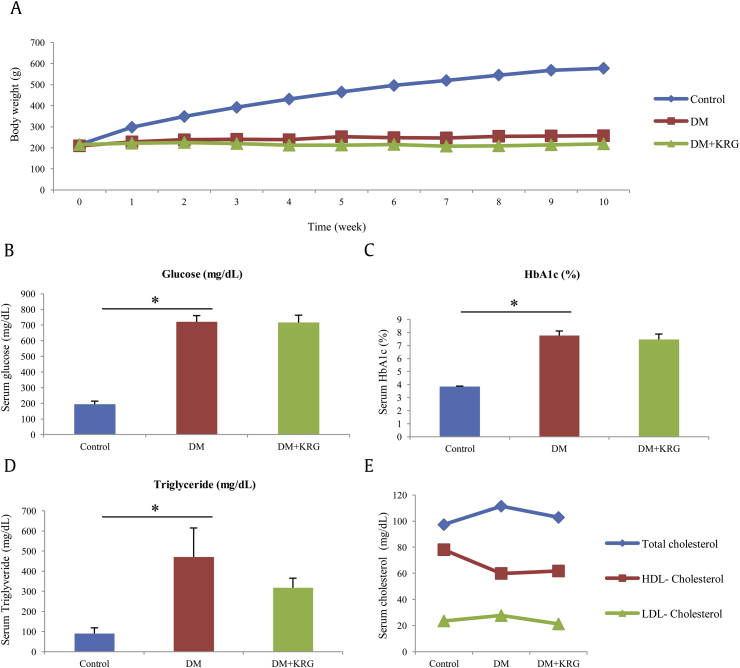
Prior to the microarray study, the effect of KRG on the biochemical indicators in the rats with STZ-induced DM was determined. The rats with KRG-treated DM did not show any change in body weight, plasma glucose, and hemoglobin A1c (HbA1c) levels as compared with those in rats with STZ-induced DM, suggesting that KRG has little effect on DM (Figs. 1A–C). The results, however, suggest that oral administration of KRG can lower the average levels of triglyceride and cholesterol in rats with KRG-treated DM than in rats with STZ-induced DM, although the difference in the levels is not statistically significant (Figs. 1D, 1E).
Fig. 1.
Changes in body weight and the serum biochemical parameters in different groups of rats.
Abnormalities of plasma lipid and lipoprotein concentrations are common in DM. Patients with DM showed reduced plasma high-density lipoprotein (HDL) cholesterol and increased plasma triglyceride and low-density lipoprotein (LDL) cholesterol levels [23], which are strongly correlated with cardiovascular complications [24], [25]. Furthermore, it has been reported that abnormal serum lipid levels in patients with DM can be restored by administration of antidiabetic agents to the patients [26], [27]. These results suggest that KRG exerts hypolipidemic effects in rats with established STZ-induced DM.
3.2. Identification of global gene expression changes
Findings from a previous study have suggested the ameliorating effect of alcoholic ginseng extract on diabetic complications, which demonstrated biochemical and functional changes in mouse retina and heart [28]. Accordingly, we explored global changes in gene expression to attempt systemic analysis of the effect of KRG on diabetic retinopathy.
Total RNAs extracted from rat retinal cells were analyzed using the whole rat genome microarray. To obtain a rough estimate of the number of patterns between DM and KRG+DM groups, we performed hierarchical clustering. In Fig. 2, red color indicates overexpression and green color indicates underexpression of genes. A total of 326 genes were differentially expressed in the DM rat retinal cells than in the control cells, of which 268 genes showed upregulated expression and 58 genes showed more than twofold downregulated expression. In the retina of KRG-treated DM rats, the expressions of 39 genes were upregulated and of 84 genes were downregulated as compared with those in the retina of rats with DM (Table 2). Thus, we observed a reversal of the pattern of the number of differentially expressed genes in the DM rat retinal cells by KRG treatment.
Fig. 2.
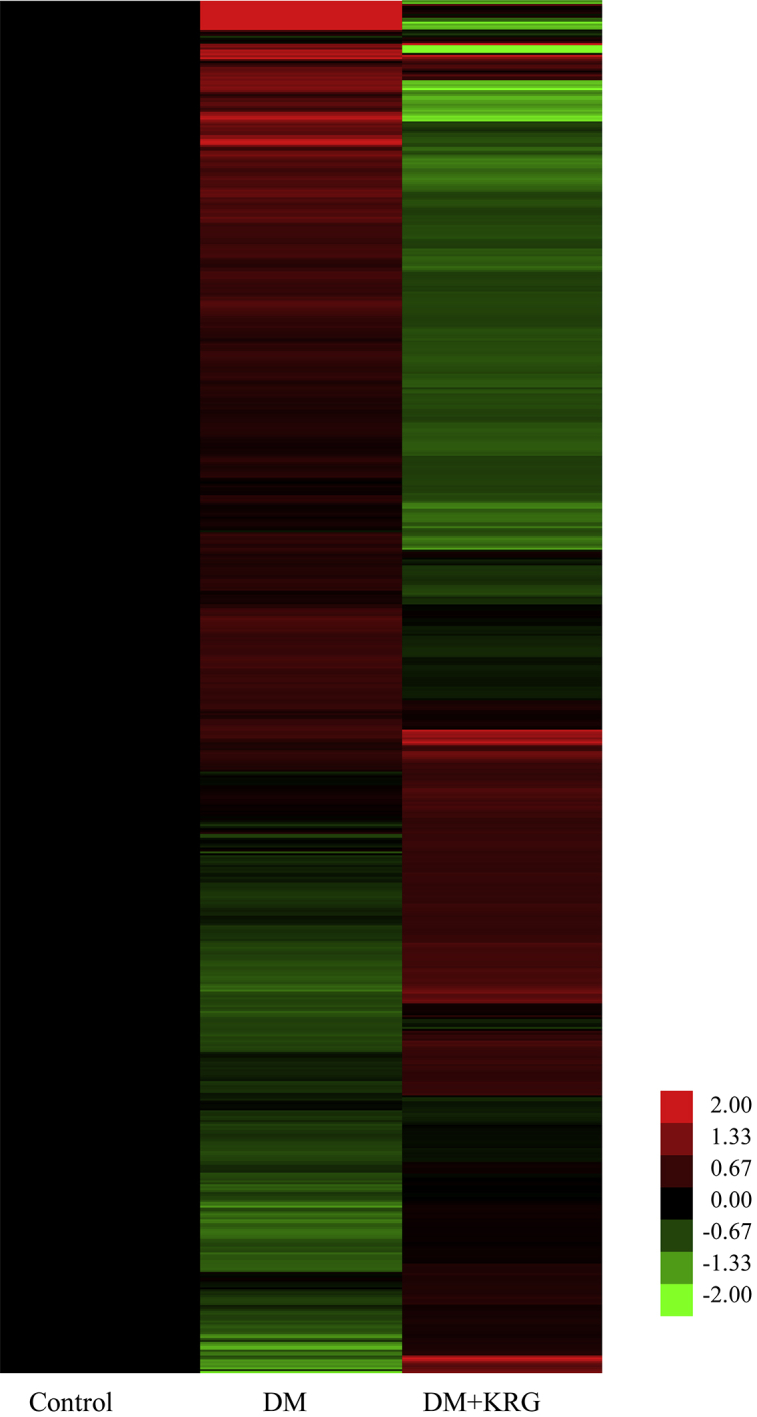
Hierarchical cluster image showing the differential gene expression profiles in DM and DM+KRG group. Gene expression levels of normal control rats are shown in black for base-lines; upregulated genes are shown in red color, whereas downregulated genes are shown in green color. DM, diabetes mellitus; KRG, Korean Red Ginseng.
Table 2.
The number of differentially expressed genes in retina of rats with diabetes before and after treatments of Korean Red Ginseng
| Control vs. DM | DM vs. DM+KRG | |
|---|---|---|
| Upregulated genes | 268 | 39 |
| Downregulated genes | 58 | 84 |
DM, diabetes mellitus; KRG, Korean Red Ginseng.
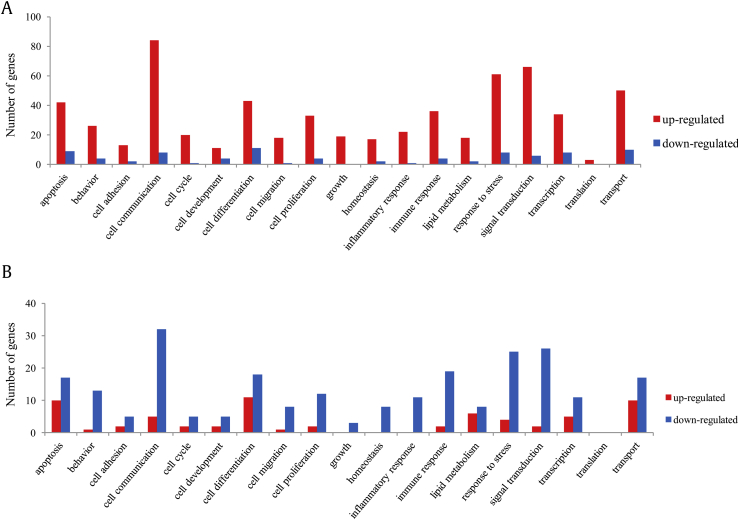
3.3. Functional classification of genes
The differentially expressed genes were divided into the following 19 functional groups: apoptosis, behavior, cell adhesion, cell communication, cell cycle, cell development, cell differentiation, cell migration, cell proliferation, growth, homeostasis, inflammatory response, immune response, lipid metabolism, response to stress, signal transduction, transcription, translation, and transport. The number of genes of DM and DM+KRG rat retina in each category is shown in Fig. 3A and 3B, respectively. As Fig. 3B displays, the number of genes altered in Fig. 3A showed a reversed pattern after KRG treatment, which indicates that the change in the expression of genes induced by DM may be restored by KRG treatment. These data revealed that KRG may play roles in different biological pathways in DR and that some of the genes involved in these pathways are fairly responsive to KRG treatment.
Fig. 3.
Functional classification of differentially expressed genes. Differentially expressed genes in (A) DM group compared with the control group or (B) DM+KRG group compared with DM group were classified into 19 groups and the numbers of genes in each group were shown as the graph. Red and blue bars indicate the number of upregulated (≥ 2.0) and downregulated (≤ 0.5) genes, respectively. DM, diabetes mellitus; KRG, Korean Red Ginseng.
3.4. Effect of KRG in restoration of gene expression altered in DM
Ginseng is currently one of the most famous herbal medicines in the world, which is well known to be beneficial in the treatment of various diseases such as cancer, cardiovascular diseases, and DM. KRG, prepared by the traditional method of steaming and sun drying ginseng, has better pharmacological activities such as antioxidant, anti-inflammatory, anticarcinogenic, and ameliorative effects on blood circulation as compared to white ginseng prepared by only sun drying fresh ginseng [29]. On using high pressure and temperature during KRG preparation, the total phenolic and flavonoid content increases and some ginsenosides (the major active ingredients of KRG) are newly produced [30], [31]. These components of ginseng have been reported to modulate the gene expression [32], [33].
In our experiment, numerous genes differentially expressed in DM rat retinal cells showed significant reversals by KRG treatment (Table 3). Altered genes in DM raise the possibility that these changes contribute to the pathogenesis of retinopathy. Indeed, the results involved several genes related to the development and progression of retinopathy, such as Cd47 (or integrin-associated protein; IAP), Cyp26a1, Igf1r, Litaf, and Mmp14. Moreover, the markers of retinopathy, such as Gfap, Crystallin, and Hmox1, were also identified. Hmox1 is known as an antioxidant gene mediating an adaptive response under pathophysiologic conditions, such as vascular disease [34]. Although blood glucose level was unaffected by KRG administration in this experiment, a number of genes showed changes in their expression, which suggests more mechanisms, such as a direct modulation by components in KRG on gene expression.
Table 3.
List of genes showing reversed expression by Korean Red Ginseng in retina of rats with diabetes
| Description | Gene symbol | Fold change(DM/Con) | Fold change(DM+KRG/DM) |
|---|---|---|---|
| Adamts4 | ADAM metallopeptidase with thrombospondin type 1 motif, 4 | 5.704 | 0.336 |
| Adfp | Adipose differentiation related protein | 3.888 | 0.284 |
| Aldh3a1 | Aldehyde dehydrogenase 3 family, member A1 | 10.476 | 0.421 |
| Alox5ap | Arachidonate 5-lipoxygenase activating protein | 2.694 | 0.334 |
| Bcl3 | B-cell CLL/lymphoma 3 | 3.228 | 0.412 |
| Bfsp1 | Beaded filament structural protein 1 | 47.469 | 0.481 |
| Birc7 | Baculoviral IAP repeat-containing 7 | 4.274 | 0.312 |
| C1qb | Complement component 1, q subcomponent, B chain | 2.234 | 0.486 |
| C1qc | Complement component 1, q subcomponent, C chain | 3.839 | 0.455 |
| Ccnd2 | Cyclin D2 | 2.713 | 0.388 |
| Cd47 | Cd47 molecule | 2.248 | 0.495 |
| Cidea | Cell death-inducing DFFA-like effector a | 10.025 | 0.483 |
| Copg2 | Coatomer protein complex, subunit gamma 2 | 3.392 | 0.322 |
| Crip | Cysteine-rich intestinal protein | 2.590 | 0.499 |
| Cryga | Crystallin, gamma A | 4.765 | 0.368 |
| Crygb | Crystallin, gamma B | 62.504 | 0.397 |
| Crygc | Crystallin, gamma C | 78.570 | 0.483 |
| Crygd | Crystallin, gamma D | 83.967 | 0.415 |
| Cryge | Crystallin, gamma E | 5.100 | 0.301 |
| Cyp26a1 | Cytochrome P450, family 26, subfamily a, polypeptide 1 | 2.749 | 0.483 |
| Dclk1 | Doublecortin-like kinase 1 | 4.700 | 0.330 |
| Dennd4a | DENN/MADD domain containing 4A | 2.161 | 0.445 |
| Edn2 | Endothelin 2 | 10.581 | 0.208 |
| Emp1 | Epithelial membrane protein 1 | 4.698 | 0.357 |
| Fancb | Fanconi anemia, complementation group B | 4.804 | 0.386 |
| Fcgr2a | Fc fragment of IgG, low affinity IIa, receptor | 2.190 | 0.468 |
| Fgl2 | Fibrinogen-like 2 | 4.665 | 0.492 |
| Gadd45g | Growth arrest and DNA-damage-inducible, gamma | 3.838 | 0.345 |
| Gal | Galanin prepropeptide | 3.789 | 0.205 |
| Gfap | Glial fibrillary acidic protein | 12.043 | 0.482 |
| Gnb3 | Guanine nucleotide binding protein (G protein), beta polypeptide 3 | 2.735 | 0.382 |
| Gpnmb | Glycoprotein (transmembrane) nmb | 10.825 | 0.483 |
| Hmox1 | Heme oxygenase (decycling) 1 | 4.906 | 0.500 |
| Igf1r | Insulin-like growth factor 1 receptor | 2.000 | 0.490 |
| Il4ra | Interleukin 4 receptor, alpha | 2.750 | 0.252 |
| Inmt | Indolethylamine N-methyltransferase | 2.487 | 0.442 |
| Irak2 | Interleukin-1 receptor-associated kinase 2 | 2.669 | 0.210 |
| Kif11 | Kinesin family member 11 | 2.152 | 0.339 |
| Klf10 | Kruppel-like factor 10 | 2.202 | 0.378 |
| Krt12 | Keratin 12 | 41.472 | 0.263 |
| Krt18 | Keratin 18 | 3.840 | 0.343 |
| Krt6a | Similar to keratin complex 2, basic, gene 6a | 3.399 | 0.351 |
| Lcn2 | Lipocalin 2 | 16.957 | 0.434 |
| Lgals3 | Lectin, galactoside-binding, soluble, 3 | 40.062 | 0.372 |
| Litaf | Lipopolysaccharide-induced TNF factor | 4.198 | 0.441 |
| Lypd2 | Ly6/Plaur domain containing 2 | 7.488 | 0.297 |
| Matn2 | Matrilin 2 | 3.221 | 0.468 |
| Mlf1 | Myeloid leukemia factor 1 | 3.459 | 0.124 |
| Mmp14 | Matrix metallopeptidase 14 (membrane-inserted) | 2.255 | 0.487 |
| Mt1a | Metallothionein 1a | 10.301 | 0.493 |
| Mt2A | Metallothionein 2A | 18.029 | 0.433 |
| Muc3a | Mucin 3A, cell surface associated | 2.430 | 0.131 |
| Pdpn | Podoplanin | 3.687 | 0.430 |
| Plekha8 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 | 2.313 | 0.396 |
| Postn | Periostin, osteoblast specific factor | 2.090 | 0.387 |
| Prtg | Protogenin homolog (Gallus gallus) | 5.292 | 0.377 |
| S100a3 | S100 calcium binding protein A3 | 3.271 | 0.269 |
| Samd4a | Sterile alpha motif domain containing 4A | 4.179 | 0.487 |
| Sec16b | SEC16 homolog B (Saccharomyces cerevisiae) | 2.733 | 0.457 |
| Serpina3n | Serine (or cysteine) peptidase inhibitor, clade A, member 3N | 7.249 | 0.368 |
| Slc25a30 | Solute carrier family 25, member 30 | 2.251 | 0.348 |
| Slc26a8 | Solute carrier family 26, member 8 | 3.966 | 0.273 |
| Stat3 | Signal transducer and activator of transcription 3 | 3.115 | 0.397 |
| Stc1 | Stanniocalcin 1 | 2.472 | 0.497 |
| Tagln2 | Transgelin 2 | 3.708 | 0.461 |
| Tbata | Thymus, brain and testes associated | 2.623 | 0.468 |
| Timp1 | TIMP metallopeptidase inhibitor 1 | 11.742 | 0.282 |
| Tmbim1 | Transmembrane BAX inhibitor motif containing 1 | 5.420 | 0.388 |
| Tmod1 | Tropomodulin 1 | 3.865 | 0.407 |
| Cdr2 | Cerebellar degeneration-related 2 | 0.325 | 2.300 |
| Grk1 | G protein-coupled receptor kinase 1 | 0.409 | 2.307 |
| Herc3 | Hect domain and RLD 3 | 0.413 | 2.341 |
| Id3 | Inhibitor of DNA binding 3 | 0.574 | 2.263 |
| Lamb3 | Laminin, beta 3 | 0.200 | 2.333 |
| LOC689064 | Beta-globin | 0.264 | 2.262 |
| Pax4 | Paired box 4 | 0.436 | 4.471 |
| Pla2r1 | Phospholipase A2 receptor 1 | 0.408 | 2.520 |
| Prc1 | Protein regulator of cytokinesis 1 | 0.317 | 3.163 |
| RT1-S3 | RT1 class Ib, locus S3 | 0.449 | 2.086 |
| Sntg2 | Syntrophin, gamma 2 | 0.449 | 2.489 |
| Tmem116 | Transmembrane protein 116 | 0.439 | 2.533 |
| Trove2 | TROVE domain family, member 2 | 0.473 | 2.066 |
We employed the Database for Annotation, Visualization, and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) tools to analyze the gene ontology (GO) of selected genes, which allows enrichment of functional genes. We observed a range of degrees of GO term enrichments in a selected gene set (Table 4). Among the identified categories, most genes belonged to the intracellular signaling group. Several other selected genes were also related to cell death and apoptosis. These results indicate that the reversed expression of genes mediate alleviating effect of KRG by influencing various cell signals and retinal cell death during DM.
Table 4.
Overrepresented gene ontology biological process annotations associated with differential expression of selected messenger RNAs using the DAVID tool
| Term | Count | Fold | p |
|---|---|---|---|
| GO0035556: intracellular signaling cascade | 10 | 2.3 | 0.0091 |
| GO0042981: regulation of apoptosis | 9 | 2.9 | 0.0036 |
| GO0043067: regulation of programmed cell death | 9 | 2.8 | 0.0039 |
| GO0001654: eye development | 8 | 12.3 | <0.001 |
| GO0007423: sensory organ development | 8 | 7 | <0.001 |
| GO0009888: tissue development | 8 | 2.8 | 0.0076 |
| GO0043065: positive regulation of apoptosis | 6 | 3.9 | 0.004 |
| GO0043068: positive regulation of programmed cell death | 6 | 3.9 | 0.0042 |
| GO0010942: positive regulation of cell death | 6 | 3.9 | 0.0043 |
| GO0043627: response to estrogen stimulus | 5 | 7.3 | <0.001 |
Database for Annotation, Visualization and Integrated Discovery (DAVID) tools were used for gene ontology (GO) analysis. Results show the top 10 GO terms that are significantly overrepresented (fold change > 2 and p < 0.01, Fisher's exact test) in the selected messenger RNAs of retinas of rats with diabetes.
Taken together, our results demonstrate that the expression levels of numerous genes are significantly altered by KRG treatment in retinal cells of rats with DM; thus, they can be considered to be directly or indirectly associated with the alleviating activity of KRG toward diabetic complications.
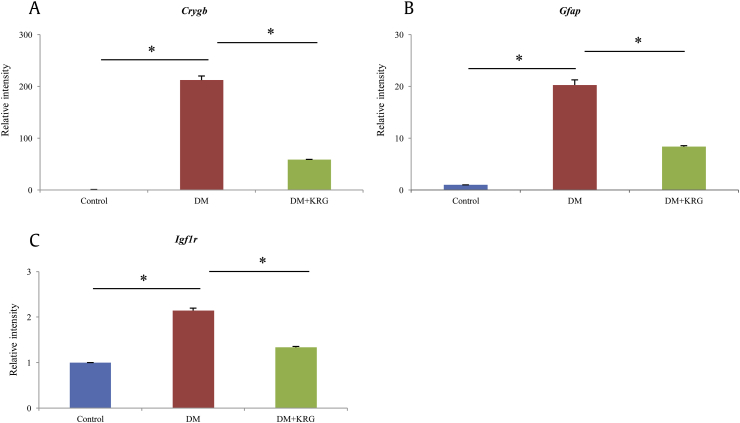
3.5. Validation of microarray data
To confirm the reliability of the microarray data, the expression of three differentially expressed genes, Gfap, Igf1r, and Crygb, related to retinopathy was analyzed by performing RT-qPCR. The expression levels of these three genes from retinal cells of rats with DM were upregulated as compared to those from control rat retinal cells, whereas they were decreased in the retinal cells of rats with KRG-treated DM (Fig. 4).
Fig. 4.
Analysis of the relative expression of differentially expressed genes by real-time quantitative polymerase chain reaction (PCR). Retina tissues were analyzed for Gfap, Igf1r, and Crygb messenger RNA levels by real-time quantitative PCR, as described in the Materials and methods section. Data are expressed as mean ± standard deviation of three independent experiments. *p > 0.005.
Glial fibrillary acidic protein (GFAP) is a representative marker associated with polyol pathway in diabetic retinopathy [7]. In diabetic retina, GFAP is upregulated, indicating dysfunction and structural abnormalities of Müller cells [35]. Insulin-like growth factor 1 receptor (IGF1R) is also increased in retinas of rats with DM in a time-dependent manner; moreover, Kummer et al [36] and Kuang et al [37] reported that reduced IGF1R immunoreactivity by using IGF-1 analogue can prevent predegenerative changes associated with the progression of DR, despite existence of sustained hyperglycemia. In our study, several subunits of crystallin showed greatly altered expression between DM and DM+KRG rat groups (Table 3). Crystallin acts as a small heat shock protein in the retina in response to stress, rather than as a structural protein [38]. It is reported that crystallins at both the protein and messenger RNA (mRNA) levels were highly upregulated in SD rats with STZ-induced diabetes [39], [40], which is a hallmark of DM in the retina [41].
The consistency of RT-qPCR results with the microarray data confirmed the influence of KRG on the expression of the selected genes. Furthermore, the data were consistent with the suggestion that KRG may contribute to the mitigation of retinopathy by altering the mRNA levels of retinopathy-related genes. Reversal of gene expression after KRG treatment may provide a validation of drug efficacy. From this perspective, reversal of transcriptional changes in KRG-treated rat retina implies that KRG can have therapeutic effects on DR by modulating the expression levels of relevant genes.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. 2011-0030072).
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc-nd/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.El-Asrar A.M. Role of inflammation in the pathogenesis of diabetic retinopathy. Middle East Afr J Ophthalmol. 2012;19:70–74. doi: 10.4103/0974-9233.92118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldebasi Y.H., Rahmani A.H., Khan A.A., Aly S.M. The effect of vascular endothelial growth factor in the progression of bladder cancer and diabetic retinopathy. Int J Clin Exp Med. 2013;6:239–251. [PMC free article] [PubMed] [Google Scholar]
- 3.Mohamed Q., Gillies M.C., Wong T.Y. Management of diabetic retinopathy: a systematic review. JAMA. 2007;298:902–916. doi: 10.1001/jama.298.8.902. [DOI] [PubMed] [Google Scholar]
- 4.Mizutani M., Kern T.S., Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest. 1996;97:2883–2890. doi: 10.1172/JCI118746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joussen A.M., Murata T., Tsujikawa A., Kirchhof B., Bursell S.E., Adamis A.P. Leukocyte-mediated endothelial cell injury and death in the diabetic retina. Am J Pathol. 2001;158:147–152. doi: 10.1016/S0002-9440(10)63952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber A.J., Lieth E., Khin S.A., Antonetti D.A., Buchanan A.G., Gardner T.W. Neural apoptosis in the retina during experimental and human diabetes. Early onset and effect of insulin. J Clin Invest. 1998;102:783–791. doi: 10.1172/JCI2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asnaghi V., Gerhardinger C., Hoehn T., Adeboje A., Lorenzi M. A role for the polyol pathway in the early neuroretinal apoptosis and glial changes induced by diabetes in the rat. Diabetes. 2003;52:506–511. doi: 10.2337/diabetes.52.2.506. [DOI] [PubMed] [Google Scholar]
- 8.Park B.J., Lim Y.S., Lee H.J., Eum W.S., Park J., Han K.H., Choi S.Y., Lee K.S. Anti-oxidative effects of Phellinus linteus and red ginseng extracts on oxidative stress-induced DNA damage. BMB Rep. 2009;42:500–505. doi: 10.5483/bmbrep.2009.42.8.500. [DOI] [PubMed] [Google Scholar]
- 9.Jin Y.R., Yu J.Y., Lee J.J., You S.H., Chung J.H., Noh J.Y., Im J.H., Han X.H., Kim T.J., Shin K.S. Antithrombotic and antiplatelet activities of Korean red ginseng extract. Basic Clin Pharmacol Toxicol. 2007;100:170–175. doi: 10.1111/j.1742-7843.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 10.Kwak Y.S., Kyung J.S., Kim J.S., Cho J.Y., Rhee M.H. Anti-hyperlipidemic effects of red ginseng acidic polysaccharide from Korean red ginseng. Biol Pharm Bull. 2010;33:468–472. doi: 10.1248/bpb.33.468. [DOI] [PubMed] [Google Scholar]
- 11.Wong V.K., Cheung S.S., Li T., Jiang Z.H., Wang J.R., Dong H., Yi X.Q., Zhou H., Liu L. Asian ginseng extract inhibits in vitro and in vivo growth of mouse Lewis lung carcinoma via modulation of ERK-p53 and NF-kappaB signaling. J Cell Biochem. 2010;111:899–910. doi: 10.1002/jcb.22778. [DOI] [PubMed] [Google Scholar]
- 12.Morisaki N., Watanabe S., Tezuka M., Zenibayashi M., Shiina R., Koyama N., Kanzaki T., Saito Y. Mechanism of angiogenic effects of saponin from ginseng Radix rubra in human umbilical vein endothelial cells. Br J Pharmacol. 1995;115:1188–1193. doi: 10.1111/j.1476-5381.1995.tb15023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quan H.Y., Kim do Y., Chung S.H. Korean red ginseng extract alleviates advanced glycation end product-mediated renal injury. J Ginseng Res. 2013;37:187–193. doi: 10.5142/jgr.2013.37.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong B.N., Ji M.G., Kang T.H. The efficacy of red ginseng in type 1 and type 2 diabetes in animals. Evid Based Complement Alternat Med. 2013;2013:593181. doi: 10.1155/2013/593181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bang H., Kwak J.H., Ahn H.Y., Shin D.Y., Lee J.H. Korean red ginseng improves glucose control in subjects with impaired fasting glucose, impaired glucose tolerance, or newly diagnosed type 2 diabetes mellitus. J Med Food. 2014;17:128–134. doi: 10.1089/jmf.2013.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee Y.K., Chin Y.W., Choi Y.H. Effects of Korean red ginseng extract on acute renal failure induced by gentamicin and pharmacokinetic changes by metformin in rats. Food Chem Toxicol. 2013;59:153–159. doi: 10.1016/j.fct.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 17.Hong Y.J., Kim N., Lee K., Hee Sonn C., Eun Lee J., Tae Kim S., Ho Baeg I., Lee K.M. Korean red ginseng (Panax ginseng) ameliorates type 1 diabetes and restores immune cell compartments. J Ethnopharmacol. 2012;144:225–233. doi: 10.1016/j.jep.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Trevino V., Falciani F., Barrera-Saldana H.A. DNA microarrays: a powerful genomic tool for biomedical and clinical research. Mol Med. 2007;13:527–541. doi: 10.2119/2006-00107.Trevino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang H., Lee S.E., Kim G.D., Park C.S., Jin Y.H., Park Y.S. An integrated analysis of microRNA and mRNA expression in salvianolic acid B-treated human umbilical vein endothelial cells. Mol Cell Toxicol. 2013;9:1–7. [Google Scholar]
- 20.Dunbar D.R. Gene expression mining in type 2 diabetes research. Methods Mol Biol. 2009;560:263–271. doi: 10.1007/978-1-59745-448-3_17. [DOI] [PubMed] [Google Scholar]
- 21.White P., Kaestner K.H. Gene expression analysis in diabetes research. Methods Mol Biol. 2009;560:239–261. doi: 10.1007/978-1-59745-448-3_16. [DOI] [PubMed] [Google Scholar]
- 22.Park H.R., Yang H., Kim G.D., Son G.W., Park Y.S. Microarray analysis of gene expression in 3-methylcholanthrene-treated human endothelial cells. Mol Cell Toxicol. 2014;10:19–27. [Google Scholar]
- 23.Ginsberg H.N. Diabetic dyslipidemia: basic mechanisms underlying the common hypertriglyceridemia and low HDL cholesterol levels. Diabetes. 1996;45:S27–S30. doi: 10.2337/diab.45.3.s27. [DOI] [PubMed] [Google Scholar]
- 24.Albrink M.J., Lavietes P.H., Man E.B. Vascular disease and serum lipids in diabetes mellitus. Observations over thirty years (1931-1961) Ann Intern Med. 1963;58:305–323. doi: 10.7326/0003-4819-58-2-305. [DOI] [PubMed] [Google Scholar]
- 25.Ginsberg H.N. Lipoprotein physiology in nondiabetic and diabetic states. Relationship to atherogenesis. Diabetes Care. 1991;14:839–855. doi: 10.2337/diacare.14.9.839. [DOI] [PubMed] [Google Scholar]
- 26.Pyorala K., Pedersen T.R., Kjekshus J., Faergeman O., Olsson A.G., Thorgeirsson G. Cholesterol lowering with simvastatin improves prognosis of diabetic patients with coronary heart disease. A subgroup analysis of the Scandinavian Simvastatin Survival Study (4S) Diabetes Care. 1997;20:614–620. doi: 10.2337/diacare.20.4.614. [DOI] [PubMed] [Google Scholar]
- 27.Wulffele M.G., Kooy A., de Zeeuw D., Stehouwer C.D., Gansevoort R.T. The effect of metformin on blood pressure, plasma cholesterol and triglycerides in type 2 diabetes mellitus: a systematic review. J Intern Med. 2004;256:1–14. doi: 10.1111/j.1365-2796.2004.01328.x. [DOI] [PubMed] [Google Scholar]
- 28.Sen S., Chen S., Wu Y., Feng B., Lui E.K., Chakrabarti S. Preventive effects of North American ginseng (Panax quinquefolius) on diabetic retinopathy and cardiomyopathy. Phytother Res. 2013;27:290–298. doi: 10.1002/ptr.4719. [DOI] [PubMed] [Google Scholar]
- 29.Nam K.Y. The Comparative understanding between Red Ginseng and White Ginsengs, processed Ginsengs (Panax ginseng C. A. Meyer) J Ginseng Res. 2005;29:1–18. [Google Scholar]
- 30.Yang S.J., Woo K.S., Yoo J.S., Kang T.S., Noh Y.H., Lee J.S., Jeong H.S. Change of Korean Ginseng components with high temperature and pressure treatment. Korean J Food Sci Technol. 2006;38:521–525. [Google Scholar]
- 31.Hwang I.G., Kim H.Y., Joung E.M., Woo K.S., Jeong J.H., Yu K.W., Lee J., Jeong H.S. Changes in ginsenosides and antioxidant activity of Korean Ginseng (Panax ginseng CA Meyer) with heating temperature and pressure. Food Sci Biotechnol. 2010;19:941–949. [Google Scholar]
- 32.Shang W., Yang Y., Jiang B., Jin H., Zhou L., Liu S., Chen M. Ginsenoside Rb1 promotes adipogenesis in 3T3-L1 cells by enhancing PPARgamma2 and C/EBPalpha gene expression. Life Sci. 2007;80:618–625. doi: 10.1016/j.lfs.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 33.Salim K.N., McEwen B.S., Chao H.M. Ginsenoside Rb1 regulates ChAT, NGF and trkA mRNA expression in the rat brain. Brain Res Mol Brain Res. 1997;47:177–182. doi: 10.1016/s0169-328x(97)00042-9. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.E., Park Y.S. The role of antioxidant enzymes in adaptive responses to environmental toxicants in vascular disease. Mol Cell Toxicol. 2013;9:95–101. [Google Scholar]
- 35.Li Q., Puro D.G. Diabetes-induced dysfunction of the glutamate transporter in retinal Muller cells. Invest Ophthalmol Vis Sci. 2002;43:3109–3116. [PubMed] [Google Scholar]
- 36.Kummer A., Pulford B.E., Ishii D.N., Seigel G.M. Des(1-3)IGF-1 treatment normalizes type 1 IGF receptor and phospho-Akt (Thr 308) immunoreactivity in predegenerative retina of diabetic rats. Int J Exp Diabesity Res. 2003;4:45–57. doi: 10.1080/15438600303729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuang H., Zou W., Liu D., Shi R., Cheng L., Yin H., Liu X. The potential role of IGF-I receptor mRNA in rats with diabetic retinopathy. Chin Med J (Engl) 2003;116:478–480. [PubMed] [Google Scholar]
- 38.Piatigorsky J. Crystallin genes: specialization by changes in gene regulation may precede gene duplication. J Struct Funct Genomics. 2003;3:131–137. [PubMed] [Google Scholar]
- 39.Fort P.E., Freeman W.M., Losiewicz M.K., Singh R.S., Gardner T.W. The retinal proteome in experimental diabetic retinopathy: up-regulation of crystallins and reversal by systemic and periocular insulin. Mol Cell Proteomics. 2009;8:767–779. doi: 10.1074/mcp.M800326-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P.A., Haseeb A., Suryanarayana P., Ehtesham N.Z., Reddy G.B. Elevated expression of alphaA- and alphaB-crystallins in streptozotocin-induced diabetic rat. Arch Biochem Biophys. 2005;444:77–83. doi: 10.1016/j.abb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 41.Heise E.A., Marozas L.M., Grafton S.A., Green K.M., Kirwin S.J., Fort P.E. Strain-independent increases of crystallin proteins in the retina of type 1 diabetic rats. PLoS One. 2013;8:e82520. doi: 10.1371/journal.pone.0082520. [DOI] [PMC free article] [PubMed] [Google Scholar]