Abstract
Background
Alzheimer's disease (AD) is a progressive brain disease, for which there is no effective drug therapy at present. Ginsenoside Rg1 (G-Rg1) and G-Rg2 have been reported to alleviate memory deterioration. However, the mechanism of their anti-AD effect has not yet been clearly elucidated.
Methods
Ultra performance liquid chromatography tandem MS (UPLC/MS)-based metabolomics was used to identify metabolites that are differentially expressed in the brains of AD mice with or without ginsenoside treatment. The cognitive function of mice and pathological changes in the brain were also assessed using the Morris water maze (MWM) and immunohistochemistry, respectively.
Results
The impaired cognitive function and increased hippocampal Aβ deposition in AD mice were ameliorated by G-Rg1 and G-Rg2. In addition, a total of 11 potential biomarkers that are associated with the metabolism of lysophosphatidylcholines (LPCs), hypoxanthine, and sphingolipids were identified in the brains of AD mice and their levels were partly restored after treatment with G-Rg1 and G-Rg2. G-Rg1 and G-Rg2 treatment influenced the levels of hypoxanthine, dihydrosphingosine, hexadecasphinganine, LPC C 16:0, and LPC C 18:0 in AD mice. Additionally, G-Rg1 treatment also influenced the levels of phytosphingosine, LPC C 13:0, LPC C 15:0, LPC C 18:1, and LPC C 18:3 in AD mice.
Conclusion
These results indicate that the improvements in cognitive function and morphological changes produced by G-Rg1 and G-Rg2 treatment are caused by regulation of related brain metabolic pathways. This will extend our understanding of the mechanisms involved in the effects of G-Rg1 and G-Rg2 on AD.
Keywords: Alzheimer's disease, ginseng, ginsenoside Rg1, ginsenoside Rg2, metabolomics
1. Introduction
Alzheimer's disease (AD) is an irreversible, progressive brain disease and one of the most debilitating neurodegenerative diseases in aging populations. As a type of dementia, it causes problems with memory, thinking, and behavior [1]. Worldwide, 36 million people suffer from dementia with numbers forecast to reach 80 million by 2040 [2]. Although AD is a major global public health problem, the actual biochemical basis for neurodegeneration is still poorly understood, and there are few treatments available to slow or stop the deterioration of brain cells [3]. Therefore, the development of potential therapeutic agents that can prevent or retard AD-related memory decline and studies of their effects on the brains of AD patients are urgently needed.
Ginseng, as a herbal medicine, has long been used to alleviate many ailments, particularly those associated with aging and memory deterioration [4], [5]. Ginseng produces its effects at multiple sites of action, which makes it an ideal candidate for developing multitarget drugs. This is most important in AD where multiple etiological and pathological factors work together to regulate the final pathophysiology of the disease [6], [7], [8]. Therefore, ginseng and ginsenosides are increasingly attracting attention because of their neuroprotective properties. Previous pathological studies have reported that Ginsenoside Rg1 (G-Rg1) displays promising effects by reducing cerebral Aβ levels and reversing certain neuropathological changes [9], [10]. G-Rg2 has been reported to exert a neuroprotective effect against glutamate-induced neurotoxicity in PC12 cells [11]. However, previous studies have mainly focused on the biochemical and pathological changes that occur in AD, and few studies have examined the changes in metabolite profiles that occur upon treatment with G-Rg1 and G-Rg2.
Metabolomics is defined as “a quantitative measurement of multi-parametric metabolic responses of multi-cellular systems to pathophysiological stimuli or genetic signaling” [12]. As an important component of systems biology, metabolomics can provide a comprehensive systems-level study of the relationships between metabolites, disease, and drugs [13]. Ultra performance liquid chromatography tandem MS (UPLC/MS), which is an established technology in metabolomics research, has main advantages including a wide dynamic range, reproducible quantitative analysis, and the ability to analyze biofluids with extreme molecular complexity [14], [15]. As a global biochemical approach to biomarker discovery, UPLC/MS has recently been applied to the study of a number of metabolic and neurodegenerative diseases [16], [17]. In our previous studies, UPLC/MS was used to determine changes in metabolite levels in the plasma of AD patients [18], [19]. Those studies provided preliminary basics for the study of drug effects on AD using metabolomics.
The aim of this study was to investigate the metabolic changes in the brains of G-Rg1 and G-Rg2 treated APP/PS1 transgenic mice. In addition, the Morris water maze (MWM) test and immunohistochemistry were used to assess the effects of G-Rg1 and G-Rg2 on the cognitive function and pathological changes in APP/PS1 mice. To our knowledge, this is the first report of the use of a UPLC/MS-based brain tissue metabolomics method to investigate the interventional effects of G-Rg1 and G-Rg2 on AD, which may facilitate our understanding of the anti-AD mechanism of these ginsenosides.
2. Materials and methods
2.1. Materials
Acetonitrile (HPLC grade) and methanol (HPLC grade) were purchased from Fisher Scientific (Fair Lawn, NJ, USA). Formic acid (HPLC grade) was obtained from Kermel Industrial (Tianjin, China). G-Rg1 and G-Rg2 (purity ≥ 98%) were purchased from Shanghai Yuanye Bio-Technology Co., Ltd (Shanghai, China). The reference standards (purity ≥ 98%) of hypoxanthine, hexadecasphinganine, dihydrosphingosine, phytosphingosine, lysophosphatidylcholine (LPC) C 13:0, LPC C 15:1, LPC C 15:0, LPC C 16:0, LPC C 18:3, LPC C 18:1, and LPC C 18:0 were obtained from the National Standard Substances Center (Beijing, China). Distilled water, prepared from demineralized water, was used throughout the experiments.
2.2. Animal handling and sample collection
Male APP/PS1 mice (No. 11401300006401), weighing 20 ± 2 g, and male C57BL/6J mice (No. 11401300006402), weighing 20 ± 2 g, were supplied by Beijing HFK Bio-Technology Co., Ltd (Beijing, China). The animals were genotyped by polymerase chain reaction analysis of the DNA extracted from tail biopsies. The animals, aged 7 mo, were maintained in an air-conditioned animal center at 23 ± 2°C and a relative humidity of 50 ± 10%, with a natural light-dark cycle. Food and water were available ad libitum. After acclimatization for 1 wk, the mice were divided into four groups (n = 10 in each group): the normal control group, the AD model group, the G-Rg1 group, and the G-Rg2 group. According to the concentration-response curves, the mice in the G-Rg1 and G-Rg2 groups were injected intraperitoneally once daily with G-Rg1 and G-Rg2 (30 mg/kg), respectively, dissolved in saline. The mice in the AD model group (APP/PS1 mice) and the normal control group (C57BL/6J nontransgenic littermates) were treated with isodose saline (0.9% w/v). All mice were treated for 1 mo before brain metabolite profiling. Experiments were conducted in accordance with the regulations for animal experimentation issued by the State Committee of Science and Technology of China.
2.3. MWM
Spatial learning and memory were tested with the MWM test [20], [21]. A circular water tank (diameter × height, 120 cm × 40 cm) was filled with water at 23 ± 2°C and divided into four equal quadrants. The water was made opaque by the addition of white nontoxic water-soluble paint. A submerged platform (diameter × height, 8 cm × 10 cm) painted black was centered in the fourth quadrant 1 cm below the water surface. A camera placed 2 m above the center of the tank recorded escape latencies during each trial. The chamber contained a number of fixed visual cues on the walls. The place navigation test was performed twice daily for 5 d. Mice were trained to find and escape onto the fixed platform from different starting positions within 60 s; when mice failed to locate the platform, they were gently guided and allowed to remain on it for 10 s before being returned to their cage. The average escape latency of each mouse was calculated. On the day after the place navigation test, the spatial exploration test was conducted for which the platform was removed. The time spent swimming in the target quadrant and the number of crossings of the previous platform site were determined for each mouse.
2.4. Immunohistochemistry
Immunohistochemistry was used to evaluate the histopathological changes in the brains of the mice [22]. Mice were deeply anesthetized with 7% choral hydrate and their brains were removed and fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned at a thickness of 4 μm. After deparaffinization in xylene and rehydration in a graded alcohol series, sections were incubated in 3% hydrogen peroxide at 37°C for 10 min to inactivate endogenous peroxidase. Antigen retrieval was performed by boiling sections in 0.01M citrate buffer (pH 6.0) for 5 min and blocking with goat serum. Aβ deposits were detected with mouse anti-Aβ antibody (6E10; Covance, Dedham, MA, USA) diluted 1:200 at 4°C overnight. Sections were washed with 0.01M phosphate-buffered saline, and then incubated with biotin-conjugated goat anti-mouse immunoglobulin G (1:500; Sigma-Aldrich, St. Louis, MO, USA) at 30°C for 30 min. After washing, horseradish peroxidase-labeled streptomycin-avidin working solution was added to the sections, which were incubated at 37°C for 30 min. The sections were washed with distilled water and immunoreactivity was detected with diaminobenzidine prior to counterstaining with hematoxylin.
2.5. Sample preparation
Mice were anesthetized with diethyl ether, and the brains were collected and weighed. Water (1 mL) was added to 0.1 g of brain tissue, and the mixtures were homogenized in an ice bath. An aliquot of 600 μL of ice-cold methanol was then added to 150 μL of the homogenate, and the mixture was vortexed for 5 min followed by centrifugation at 12,000g for 10 min at 4°C. The supernatant was transferred to another tube and evaporated to dryness at 30°C under a gentle stream of nitrogen. The dried residue was then reconstituted in 100 μL of acetonitrile-water (2:98, v/v) and a 10-μL aliquot was used in the UPLC/MS analysis.
2.6. Spectrum acquisition
Liquid chromatography was performed on a Waters Acquity UPLC system (Waters Corp., Milford, MA, USA). Separation was achieved with a Waters ethylene-bridged hybrid (BEH) C18 column (50 mm × 2.1 mm, 1.7 μm) held at 30°C. Gradient elution with a mobile phase composed of water and acetonitrile with 0.1% formic acid was performed at a flow rate of 0.3 mL/min (Table 1).
Table 1.
Ultra performance liquid chromatography-MS gradient elution program
| Time (min) | %A (0.1% FA-water) | %B (0.1% FA-acetonitrile) |
|---|---|---|
| Initial | 98 | 2 |
| 2 | 92 | 8 |
| 4 | 60 | 40 |
| 17 | 12 | 88 |
| 18 | 0 | 100 |
| 21 | 98 | 2 |
FA = formic acid.
Mass spectrometric detection was performed on a Micromass Quattro Micro API mass spectrometer (Waters Corp.) with an electrospray ionization interface and triple quadrupole mass analyzer. The electrospray ionization source was set in the positive mode. The following parameters were used: capillary voltage, 3.2 kV; cone voltage, 30 V; source temperature, 120°C; and desolvation temperature, 300°C. Nitrogen was used for desolvation and as the cone gas at flow rates of 600 L/h and 50 L/h, respectively. The full scan mode was used in a mass range of 100–1,000 amu. For MS/MS, argon was used as the collision gas, and the collision energy was set according to the metabolite composition. NaCsI was used for mass correction before the experiment was commenced. Data were collected in centroid mode.
2.7. Statistical analysis
SPSS version 19.0 software (SPSS Inc., Chicago, IL, USA) was used to analyze the data from the MWM test, which were evaluated by the analysis of variance, followed by Tukey's multiple comparison test. The results are expressed as mean ± standard deviation (SD), and differences are considered statistically significant at p < 0.05.
The UPLC/MS data from the brain samples were analyzed with Markerlynx within Masslynx software version 4.1 (Waters Corp.). The retention time and m/z were calculated for each peak. Principal components analysis (PCA) was used to select distinct variables, and Student t test was used to evaluate statistically significant differences between potential biomarkers (SPSS version19.0 software; SPSS Inc.). Statistical significance was accepted if p < 0.05.
2.8. Identification of biomarkers
In positive product ion scan spectra, samples were analyzed and low molecular weight metabolites were visible as chromatographic peaks in the total ion chromatograms. Fragmentation patterns of potential brain biomarkers were obtained from collision-induced dissociation experiments. The biomarkers were identified by comparing their retention time, mass value, and MS/MS fragmentation with the corresponding parameters of the reference standards and available references. In addition, full scan mass spectra of these metabolites were interpreted using biochemical databases including the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/), the Human Metabolome Database (http://www.hmdb.ca/), and the Lipidomics Gateway (http://www.lipidmaps.org/).
3. Results
3.1. G-Rg1 and G-Rg2 improve the performance of APP/PS1 mice in the MWM test
In the place navigation test, the average escape latency of the mice in searching for the hidden underwater platform decreased over the training days (Table 2). From Day 2 onwards, the escape latencies among the AD model group were significantly longer than those of the normal control group (p < 0.01). G-Rg1 and G-Rg2 reduced the escape latencies on the last two training days compared to the AD model group (p < 0.05). In the spatial exploration test, the total time spent in the target quadrant and the number of mice that exactly crossed the previous position of the platform were clearly shorter and lower, respectively, in the AD model group mice than in the normal control group mice (p < 0.01), a trend that was reversed by treatment with G-Rg1 and G-Rg2 (G-Rg1, p < 0.01; G-Rg2, p < 0.05). The effects of G-Rg1 and G-Rg2 treatment on the performance of the mice in the spatial probe trial are shown in Fig. 1.
Table 2.
Effects of ginsenoside Rg1 (G-Rg1) and G-Rg2 on the escape latency of Alzheimer's disease (AD) mice In the Morris water maze test
| Group (n = 10) | Training day |
||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
| Control | 59.9 ± 0.7 | 34.0 ± 6.1∗∗ | 21.3 ± 2.1∗∗ | 18.6 ± 3.3∗∗ | 18.9 ± 2.7∗∗ |
| AD model | 58.1 ± 2.8 | 59.9 ± 0.9 | 48.5 ± 5.6 | 50.4 ± 4.1 | 41.3 ± 6.3 |
| G-Rg1 | 59.9 ± 1.1 | 52.3 ± 6.0 | 39.2 ± 6.1 | 32.1 ± 5.0∗∗ | 27.1 ± 4.9∗∗ |
| G-Rg2 | 59.9 ± 1.2 | 42.2 ± 5.7 | 34.1 ± 5.2 | 30.6 ± 4.9∗∗ | 25.3 ± 3.7∗ |
* p < 0.05 versus AD model group (one-way analysis of variance).
** p < 0.01 versus AD model group (one-way analysis of variance).
AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
Fig. 1.
Effects of Ginsenoside Rg1 (G-Rg1) and G-Rg2 on the performance of Alzheimer's disease (AD) mice in the Morris water maze test. (A) Number of platform crossings within 60 s and (B) time in the target quadrant were measured. Values indicate mean ± standard deviation (SD) [one-way analysis of variance followed by Tukey's multiple comparison test (n = 10)]. *p < 0.05 versus AD model group, **p < 0.01 versus AD model group. AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
The MWM results show that the APP/PS1 mice were markedly deficient in cognitive function compared with age-matched C57BL/6J mice, which is consistent with previous studies [23]. Treatment with G-Rg1 and G-Rg2 effectively improved cognitive function of the mice that had declined due to AD.
3.2. G-Rg1 and G-Rg2 reduce Aβ1-42 accumulation in APP/PS1 mice
In the immunohistochemistry study, no clear Aβ depositions were observed in the hippocampi of the normal control mice (Fig. 2A). By contrast, marked morphological changes were observed in the AD model mice: neurons were loosely organized and darkly stained, and there were dark brown, scattered Aβ deposits (Fig. 2B). In the G-Rg1 and G-Rg2 treated mice, the pathological abnormalities observed in the APP/PS1 mice were gradually ameliorated. Clear nucleoli and light brown, sparsely scattered Aβ deposits were visible (Figs. 2C, 2D). These results, combined with the behavioral results discussed above, indicate that the impaired cognitive function and increased hippocampal Aβ depositions in the APP/PS1 mice were partly ameliorated by treatment with G-Rg1 and G-Rg2.
Fig. 2.
Effects of Ginsenoside Rg1 (G-Rg1) and G-Rg2 on the histopathological changes in the hippocampus of APP/PS1 mice. (A) The normal control group; (B) the Alzheimer's disease (AD) model group; (C) G-Rg1 group; and (D) G-Rg2 group are shown at 100× magnification. Scale bars = 100 μm. AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
3.3. Metabolite profiles in AD
3.3.1. Method development and verification
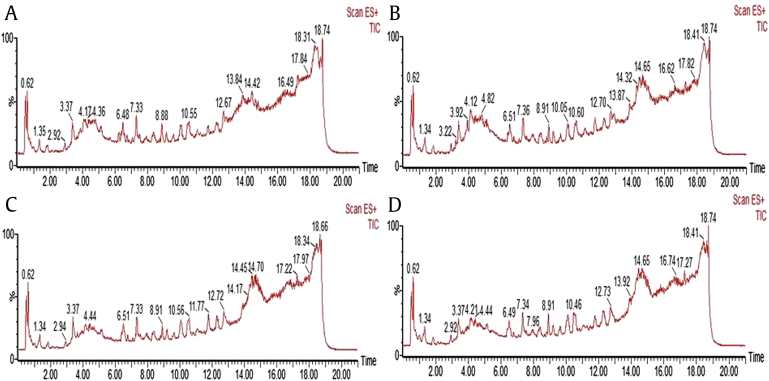
A metabolomics study was conducted to investigate the possible mechanisms of action of G-Rg1 and G-Rg2 in the APP/PS1 mice. The preparation of brain samples for metabolic profiling analysis with UPLC/MS involved a protein precipitation step to separate the low-molecular-weight compounds and remove large amounts of proteins that would otherwise interfere with the UPLC/MS analysis. Quantitative information was obtained in the positive ion mode rather than in the negative ion mode. Typical total ion chromatograms of brain samples from the normal control group (A), AD model group (B), G-Rg1 group (C), and G-Rg2 group (D) are shown in Fig. 3. The applied method was validated before the analysis of the experimental samples by determining the precision, within-day stability, and repeatability of sample preparation. The extracted ion chromatographic peaks of seven ions (m/z 132.2, 0.52 min; m/z 120.1, 1.90 min; 710.5, 3.37 min; 346.7, 8.41 min; 502.3, 8.93 min; 374.1, 10.6 min; and 402.3, 12.88 min, in positive ion mode) were distributed across different spectral regions, and the retention times were selected to validate the method. The relative SDs (RSDs) of the peak intensities and retention times for the selected ions in the pooled brain sample were calculated. The precision of injection was established with the continuous detection of five injections of the same sample. The RSDs ranged from 0.8% to 2.1% for the retention time and from 6.5% to 13.7% for the peak intensity. The post-preparation stability of the sample was tested by analyzing a sample left in the autosampler (maintained at 4°C) for 4 h, 8 h, 12 h, and 24 h. The RSDs ranged from 3.5% to 11.7% for the peak intensity and from 0.9% to 2.4% for the retention time. Method repeatability was evaluated with five replicate analyses of a brain sample. The RSDs of the peak intensities ranged from 3.5% to 13.7% and those for the retention time ranged from 0.9% to 3.4%.
Fig. 3.
Typical total ion chromatograms of brain samples in positive mode. (A) The normal control group; (B) the Alzheimer's disease (AD) model group; (C) Ginsenside Rg1 (G-Rg1) group; and (D) G-Rg2 group. AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
3.3.2. Metabolite profiling analysis
To determine whether the metabolites in brain differed between normal control and AD model mice, PCA, which is a classic unsupervised method (no prior knowledge concerning groups or tendencies within the data sets is necessary) of pattern recognition, was used. In the PCA score, each point represented an individual sample; the plot of PCA scores divided different brain samples into blocks, suggesting changes in the metabolic profiles. Samples from the normal control and AD model groups were clearly divided into two classes (Fig. 4A), indicating that AD was successfully reproduced by this model and that specific biomarkers could distinguish AD from normal control mice.
Fig. 4.
Principal components analysis (PCA) results derived from the ultra performance liquid chromatography tandem MS (UPLC/MS) of the Alzheimer's disease (AD) model and normal control mice brains in positive mode (n = 10). (A) PCA score plot indicating the separation between two groups and (B) PCA loading plot indicating the potential biomarkers. The variables are labeled with retention time and m/z. AD, Alzheimer's disease.
3.3.3. Biomarker identification
In the PCA loading plots for the AD model and normal control mice, the distance of an ion from the origin represents its influence on PCA components (Fig. 4B). Ions in the plot were selected as putative biomarkers (Table 3), and were those with retention time–m/z pairs of 0.82_136.8, 4.7_302.0, 6.4_274.2, 6.5_318.1, 10.0_454.0, 12.3_482.1, 10.2_480.1, 9.6_496.1, 10.1_518.1, 10.6_522.1, and 12.4_524.1. These biomarkers were compared with the data in the literature [18], [24], [25] and biochemical databases, and further identified by comparing with corresponding standards according to their retention times, m/z, and product ion scan spectra in positive mode. For instance, the biomarker at m/z 496.1 contained four major fragment ions, which are the same as those of LPC C 16:0 standard. The high-abundance fragment ions at m/z 104.1 and m/z 183.9 represent [HOCH2CH2N(CH3)3]+ and [H2O3PO-CH2CH2N(CH3)3]+, respectively, which identify the head group of the phosphatidylcholine class of biomarkers. Another two major fragment ions at m/z 258.1 (M-COC15H31+H)+ and m/z 478.5 (M-H2O + H)+ indicated that this biomarker is an LPC. The retention time, mass value, and MS/MS fragmentation of this biomarker are the same as those of LPC C 16:0 standard proved the metabolite is LPC C 16:0. To evaluate changes in biomarker profiles, peak intensities of putative biomarkers in the brains of AD and control mice were compared (Fig. 5). The peak intensities of LPCs, dihydrosphingosine, hexadecasphinganine, and phytosphingosine were reduced and that of hypoxanthine was elevated in the brains of AD mice compared to normal control mice (p < 0.05).
Table 3.
Potential biomarkers and change trends of them in different groups
| Retention time (min) | m/z | Scan mode | Quasi-molecular ion | Metabolites | AD mice vs. Controls | G-Rg1 treated mice vs. AD mice | G-Rg2 treated mice vs. AD mice |
|---|---|---|---|---|---|---|---|
| 0.82 | 136.8 | + | [M+H]+ | Hypoxanthine | ↑ | ↓ | ↓ |
| 6.4 | 274.2 | + | [M+H]+ | Hexadecasphinganine | ↓ | ↑ | ↑ |
| 4.7 | 302.0 | + | [M+H]+ | Dihydrosphingosine | ↓ | ↑ | ↑ |
| 6.5 | 318.1 | + | [M+H]+ | Phytosphingosine | ↓ | ↑ | |
| 10.0 | 454.0 | + | [M+H]+ | LPC C 13:0 | ↓ | ↑ | |
| 10.2 | 480.1 | + | [M+H]+ | LPC C 15:1 | ↓ | ||
| 12.3 | 482.1 | + | [M+H]+ | LPC C 15:0 | ↓ | ↑ | |
| 9.6 | 496.1 | + | [M+H]+ | LPC C 16:0 | ↓ | ↑ | ↑ |
| 10.1 | 518.1 | + | [M+H]+ | LPC C 18:3 | ↓ | ↑ | |
| 10.6 | 522.1 | + | [M+H]+ | LPC C 18:1 | ↓ | ↑ | |
| 12.4 | 524.1 | + | [M+H]+ | LPC C 18:0 | ↓ | ↑ | ↑ |
AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2; LPC, lysophosphatidylcholine
Fig. 5.
Peak intensities of putative potential biomarkers in the brains of the Alzheimer's disease (AD) model and normal control groups. Identification: 136.8, hypoxanthine; 274.2, hexadecasphinganine; 302.0, dihydrosphingosine; 318.1, phytosphingosine; 454.0, lysophosphatidylcholine (LPC) C 13:0; 480.1, LPC C 15:1; 482.1, LPC C 15:0; 496.1, LPC C 16:0; 518.1, LPC C 18:3; 522.1, LPC C 18:1; and 524.1, LPC C 18:0. Values indicate mean ± standard deviation (SD) [Student t test (n = 10)]. * p < 0.05 versus normal control group. AD, Alzheimer's disease.
3.4. G-Rg1 and G-Rg2 treatment alters metabolite profiles of AD mice
The PCA model was also used to determine whether G-Rg1 and G-Rg2 affect the metabolite profiles of AD mice. According to the PCA score plots (Fig. 6A) from processing dates of the normal control, AD model, and G-Rg1 and G-Rg2 treated groups, samples from the ginsenosides treated groups were separated from the AD model group and were closer to the controls, suggesting that G-Rg1 and G-Rg2 treatment partly restored brain metabolites in AD mice to near normal levels. Metabolites with altered levels were identified in the PCA loading plot (Fig. 6B); the peak intensity of hypoxanthine was lower, while those of dihydrosphingosine, hexadecasphinganine, phytosphingosine, LPC C 13:0, LPC C 15:0, LPC C 16:0, LPC C 18:0, LPC C 18:1, and LPC C 18:3 were higher in the G-Rg1 group compared to the AD group (p < 0.05). G-Rg2 treatment yielded similar results; the peak intensity of hypoxanthine was lower, while those of dihydrosphingosine, hexadecasphinganine, LPC C 16:0, and LPC C 18:0 were higher in the G-Rg2 compared to the AD group (p < 0.05; Fig. 7). These results show that G-Rg1 and G-Rg2 can partly correct brain metabolic alterations in AD mice.
Fig. 6.
Principal components analysis (PCA) results derived from the ultra performance liquid chromatography tandem MS (UPLC/MS) of the Alzheimer's disease (AD) model, normal control and ginsenosides treatment mice brains in positive mode (n = 10). (A) PCA score plot indicating the separation among all the groups; (B) PCA loading plot indicating the potential biomarkers. The variables are labeled with retention time and m/z. AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
Fig. 7.
Peak intensities of potential biomarkers in the brains of Alzheimer's disease (AD) model, normal control and ginsenosides treatment groups. Identification: 136.8, hypoxanthine; 274.2, hexadecasphinganine; 302.0, dihydrosphingosine; 318.1, phytosphingosine; 454.0, lysophosphatidylcholine (LPC) C 13:0; 480.1, LPC C 15:1; 482.1, LPC C 15:0; 496.1, LPC C 16:0; 518.1, LPC C 18:3; 522.1, LPC C 18:1; and 524.1, LPC C 18:0. Values indicate mean ± standard deviation (SD) [Student t test (n = 10)]. * p < 0.05 versus AD model group. AD, Alzheimer's disease; G-Rg1, Ginsenside Rg1; G-Rg2, Ginsenside Rg2.
4. Discussion
Ginseng is one of the most widely used herbal medicines in humans. In our study, a UPLC/MS-based metabolomics approach was applied to the characterization of AD, and the protective effects of G-Rg1 and G-Rg2 were investigated in APP/PS1 transgenic mice. APP/PS1 mice are a reliable animal model for studies of AD prevention and treatment because they exhibit neurodegenerative changes in behavioral, biochemical, and histopathological aspects that are similar to those observed in humans with AD [26], [27]. The cognitive dysfunctions and pathophysiological changes of APP/PS1 mice were alleviated by treatment with G-Rg1 and G-Rg2, which provides evidence for the protective effects of G-Rg1 and G-Rg2 against AD. Moreover, 11 potential biomarkers related to AD were changed in the brains of G-Rg1 and G-Rg2 treated mice. These metabolites were associated with hypoxanthine, LPCs, and sphingolipid metabolism, indicating the protective effects of G-Rg1 and G-Rg2 in AD are exerted through the modulation of these pathways.
Hypoxanthine is a purine compound and is the principal purine nucleobase involved in the salvage purine pathway in the brain [28]. Purinergic signaling plays an important role in the development of neural diseases. Numerous studies have provided evidence for the involvement of purine metabolites in the brains of patients with AD [29], [30], and this was also proved in the present study. In addition, a deficiency in the activity of hypoxanthine-guanine phosphoribosyltransferase, a hypoxanthine metabolic enzyme, has been found in patients with neurodegenerative diseases. This could lead to the accumulation of hypoxanthine [31]. It was reported that hypoxanthine enhanced acetylcholinesterase (AChE) activity in the hippocampus and striatum of 15- and 30-d-old mice, when added to the incubation medium [32]. AChE is associated with the pathophysiology of AD. Constant stimulation of AChE activity might decrease acetylcholine levels, in turn interfering with cholinergic transmission. This is an essential neurotransmitter in the central nervous system [33]. Therefore, AChE-related hypoxanthine metabolism disorder is associated with memory deficits. Moreover, hypoxanthine has also been reported to inhibit Na+, K+-ATPase activity and induces oxidative stress in rat striatum. Oxidative stress is a primary event in the pathogenesis of AD and has been found to impair brain memory of mice [34]. From the above, abnormal brain hypoxanthine concentration could be one of the factors leading to AD, and G-Rg1 and G-Rg2 may improve the metabolism of hypoxanthine and decrease the concentration of hypoxanthine, which is associated with AChE and oxidative stress, thus alleviating symptoms of AD.
LPCs are associated with the integrity of the neuronal membrane structure and exert a diverse array of effects on cellular functional activities. It is recognized that they can take part in multiple neuronal pathways [35], [36]. LPCs are related to lecithin metabolism. Lecithin, an important component of cell membranes, participates in the synthesis of the neurotransmitter acetylcholine [37]. Moreover, impaired lecithin metabolism could result in lesions to the cell, which are involved in numerous human diseases, particularly in the central nervous system because of its high lipid content [38]. Thus, lower levels of LPCs were also observed in the brains of AD mice compared to those of normal control mice. This is consistent with previously published reports [39]. The brain is the most lipid-rich tissue in mammals [40]. A disorder of lipid metabolism in the brain is one of the factors leading to AD according to the results. However, it was partly restored by treatment with G-Rg1 and G-Rg2, indicating that the therapeutic effects of G-Rg1 and G-Rg2 are also connected with the modulation of impaired LPCs metabolism, which is related to the neuronal membrane structure and multiple neuronal pathways.
Dihydrosphingosine, hexadecasphinganine, and phytosphingosine are classified as sphingolipids [41]. The central nervous system contains large amounts of sphingolipids; their metabolites play not only a structural role in biomembranes, but also act as second messengers involved in the transduction of numerous cell signals [42]. There is also strong evidence that cholesterol and sphingomyelin preferentially interact with each other in neuronal membranes, and that this interaction has a direct effect on the structure and permeability of the cell membrane and second-messenger systems [43]. In the brain, imbalances in the contents of various classes of sphingolipids can result in neuronal dysfunction and apoptosis of brain cells, ultimately leading to AD [44], [45]. In our previous study, we found changes in the sphingolipid levels in the plasma of AD patients [19]. The results of this study proved that disorders of sphingolipid levels are also observed in the brains of AD mice, and indicated that G-Rg1 and G-Rg2 also take part in sphingolipid metabolic pathways. G-Rg1 treatment increased the levels of dihydrosphingosine, hexadecasphinganine, and phytosphingosine, whereas G-Rg2 treatment just increased the levels of dihydrosphingosine and hexadecasphinganine. This difference, combined with LPCs metabolism results, shows that G-Rg1 restored more AD-related metabolites to near normal levels than G-Rg2.
In summary, G-Rg1 and G-Rg2 treatment can partly restore hypoxanthine, LPCs, and sphingolipid metabolism in the brains of AD mice based on metabolomics results. To our knowledge, this is the first retrospective study using UPLC/MS to biochemically profile the brain metabolic pathways affected by G-Rg1 and G-Rg2 in AD mice. It may contribute to our understanding of the biochemical mechanisms involved in this effect.
5. Conclusion
A brain metabolomics approach, combined with MWM and immunohistochemistry, was used to identify metabolic changes in the brains of ginsenoside treated mice and to evaluate the effects of G-Rg1 and G-Rg2 on AD. The cognitive dysfunctions and pathophysiological changes in APP/PS1 mice were alleviated by treatment with G-Rg1 or G-Rg2. A total of 11 metabolites related to the metabolism of sphingolipids, hypoxanthine, or lecithin were identified in the brains of AD mice. Treatment with G-Rg1 or G-Rg2 both influenced sphingolipid, hypoxanthine, and lecithin metabolism pathways. These results provide molecular evidence for the efficacy of these ginsenosides in AD treatment.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
Thanks to Yingge Gong, Pengyi Hou, and Can Xu for technical assistance. This study was supported by the National Science Foundation of China grant number 81203002 and Shenyang Science and Technology Program number F12-193-9-22.
Footnotes
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc-nd/4.0) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
References
- 1.Inoue K., Tsutsui H., Akatsu H., Hashizume Y., Matsukawa N., Yamamoto T., Toyo'oka T. Metabolic profiling of Alzheimer's disease brains. Sci Rep. 2013;3:2364–2372. doi: 10.1038/srep02364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan W., Chen H., Li Y. The potential mechanisms of Abeta-receptor for advanced glycation end-products interaction disrupting tight junctions of the blood-brain barrier in Alzheimer's disease. Int J Neurosci. 2014;124:75–81. doi: 10.3109/00207454.2013.825258. [DOI] [PubMed] [Google Scholar]
- 3.Pratico D. Oxidative stress hypothesis in Alzheimer's disease: a reappraisal. Trends Pharmacol Sci. 2008;29:609–615. doi: 10.1016/j.tips.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Fang F., Chen X., Huang T., Lue L.F., Luddy J.S., Yan S.S. Multi-faced neuroprotective effects of Ginsenoside Rg1 in an Alzheimer mouse model. Biochim Biophys Acta. 2012;1822:286–292. doi: 10.1016/j.bbadis.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan X., Gu J., Zhao B., Wang S., Yuan J., Wang C., Chen J., Liu J., Feng L., Jia X. Ginseng improves cognitive deficit via the RAGE/NF-kB pathway in advanced glycation end product-induced rats. J Ginseng Res. 2014;9:1–9. doi: 10.1016/j.jgr.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L., Huang J., Hu X., Li K., Sun C. Simultaneous determination of ginsenoside (G-Re, G-Rg1, G-Rg2, G-F1, G-Rh1) and protopanaxatriol in human plasma and urine by LC–MS/MS and its application in a pharmacokinetics study of G-Re in volunteers. J Chromatogr B. 2011;879:2011–2017. doi: 10.1016/j.jchromb.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Li N., Liu B., Dluzen D.E., Jin Y. Protective effects of ginsenoside Rg2 against glutamate-induced neurotoxicity in PC12 cells. J Ethnopharmacol. 2007;111:458–463. doi: 10.1016/j.jep.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.H. Cardiovascular diseases and Panax ginseng: a review on molecular mechanisms and medical applications. J Ginseng Res. 2012;36:16–26. doi: 10.5142/jgr.2012.36.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen F., Eckman E.A., Eckman C.B. Reductions in levels of the Alzheimer's amyloid peptide after oral administration of ginsenosides. Faseb J. 2006;20:1269–1271. doi: 10.1096/fj.05-5530fje. [DOI] [PubMed] [Google Scholar]
- 10.Shi C., Zheng D.D., Fang L., Wu F., Kwong W.H., Xu J. Ginsenoside Rg1 promotes nonamyloidgenic cleavage of APP via estrogen receptor signaling to MAPK/ERK and PI3K/Akt. Biochim Biophys Acta. 2012;1820:453–460. doi: 10.1016/j.bbagen.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Kim H.J., Kim P., Shin C.Y. A comprehensive review of the therapeutic and pharmacological effects of ginseng and ginsenosides in central nervous system. J Ginseng Res. 2013;37:8–29. doi: 10.5142/jgr.2013.37.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholson J.K., Lindon J.C., Holmes E. 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 13.Lan M.J., McLoughlin G.A., Griffin J.L., Tsang T.M., Huang J.T.J., Yuan P., Manji H., Holmes E., Bahn S. Metabonomic analysis identifies molecular changes associated with the pathophysiology and drug treatment of bipolar disorder. Mol Psychiatr. 2009;14:269–279. doi: 10.1038/sj.mp.4002130. [DOI] [PubMed] [Google Scholar]
- 14.Want E.J., Nordströ m A., Morita H., Siuzdak G. From exogenous to endogenous. the inevitable imprint of mass spectrometry in metabolomics. J Proteome Res. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- 15.Zheng X., Kang A., Dai C., Liang Y., Xie T., Xie L., Peng Y., Wang G., Hao H. Quantitative analysis of neurochemical panel in rat brain and plasma by liquid chromatography-tandem mass spectrometry. Anal Chem. 2012;84:10044–10051. doi: 10.1021/ac3025202. [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Yang B., Sun H., Zhang A. Pattern recognition approaches and computational systems tools for ultra performance liquid chromatography-mass spectrometry-based comprehensive metabolomic profiling and pathways analysis of biological data sets. Anal Chem. 2012;84:428–439. doi: 10.1021/ac202828r. [DOI] [PubMed] [Google Scholar]
- 17.Dai W., Wei C., Kong H., Jia Z., Han J., Zhang F., Wu Z., Gu Y., Chen S., Gu Q. Effect of the traditional Chinese medicine tongxinluo on endothelial dysfunction rats studied by using urinary metabonomics based on liquid chromatography-mass spectrometry. J Pharmaceut Biomed. 2011;56:86–92. doi: 10.1016/j.jpba.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 18.Li N.J., Liu W.T., Li W., Li S.Q., Chen X.H., Bi K.S., He P. Plasma metabolic profiling of Alzheimer's disease by liquid chromatography/mass spectrometry. Clin Biochem. 2010;43:992–997. doi: 10.1016/j.clinbiochem.2010.04.072. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Li N.J., Zhou L., Li Q., Li W. Plasma metabolic profiling of mild cognitive impairment and Alzheimer's disease using liquid chromatography/mass spectrometry. Cent Nerv Syst Agents Med Chem. 2014;14:113–120. doi: 10.2174/1871524915666141216161246. [DOI] [PubMed] [Google Scholar]
- 20.Li X., Zhao X., Xu X., Mao X., Liu Z., Li H., Guo L., Bi K., Jia Y. Schisantherin A recovers Abeta-induced neurodegeneration with cognitive decline in mice. Physiol Behav. 2014;132:10–16. doi: 10.1016/j.physbeh.2014.04.046. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z., Zhao X., Liu B., Liu A.J., Li H., Mao X., Wu B., Bi K.S., Jia Y., Jujuboside A. a neuroprotective agent from semen Ziziphi Spinosae ameliorates behavioral disorders of the dementia mouse model induced by Abeta 1-42. Eur J Pharmacol. 2014;738:206–213. doi: 10.1016/j.ejphar.2014.05.041. [DOI] [PubMed] [Google Scholar]
- 22.Li Y., Ma Y., Zong L.-X., Xing X.-N., Guo R., Jiang T.-Z., Sha S., Liu L., Cao Y.-P. Intranasal inoculation with an adenovirus vaccine encoding ten repeats of Aβ3-10 reduces AD-like pathology and cognitive impairment in Tg-APPswe/PSEN1dE9 mice. J Neuroimmunol. 2012;249:16–26. doi: 10.1016/j.jneuroim.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 23.Ma Y., Li Y., Zong L.X., Xing X.N., Zhang W.G., Cao Y.P. Improving memory and decreasing cognitive impairment in Tg-APPswe/PSEN1dE9 mice with Abeta3-10 repeat fragment plasmid by reducing Abeta deposition and inflammatory response. Brain Res. 2011;1400:112–124. doi: 10.1016/j.brainres.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 24.Yin P., Mohemaiti P., Chen J., Zhao X., Lu X., Yimiti A., Upur H., Xu G. Serum metabolic profiling of abnormal savda by liquid chromatography/mass spectrometry. J Chromatogr B. 2008;871:322–327. doi: 10.1016/j.jchromb.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 25.Huo T., Cai S., Lu X., Sha Y., Yu M., Li F. Metabonomic study of biochemical changes in the serum of type 2 diabetes mellitus patients after the treatment of metformin hydrochloride. J Pharmaceut Biomed. 2009;49:976–982. doi: 10.1016/j.jpba.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H., Gao Y., Qiao P.F., Zhao F.L., Yan Y. Fenofibrate reduces amyloidogenic processing of APP in APP/PS1transgenic mice via PPAR- α/PI3-K pathway. Int J Dev Neurosci. 2014;38:223–231. doi: 10.1016/j.ijdevneu.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Kim H.Y., Kim H.V., Yoon J.H., Kang B.R., Cho S.M., Lee S., Kim J.Y., Kim J.W., Cho Y., Woo J. Taurine in drinking water recovers learning and memory in the adult APP/PS1 mouse model of Alzheimer’s disease. Sci Rep. 2014;4:7467–7473. doi: 10.1038/srep07467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bavaresco C.S., Chiarani F., Matte C., Wajner M., Netto C.A., de Souza Wyse A.T. Effect of hypoxanthine on Na+, K+-ATPase activity and some parameters of oxidative stress in rat striatum. Brain Res. 2005;1041:198–204. doi: 10.1016/j.brainres.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 29.González-Domínguez R., García-Barrera T., Vitorica J., Gómez-Ariza J.L. Metabolomic screening of regional brain alterations in the APP/PS1 transgenic model of Alzheimer’s disease by direct infusion mass spectrometry. J Pharmaceut Biomed. 2015;102:425–435. doi: 10.1016/j.jpba.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Xiang Z., Xu M., Liao M., Jiang Y., Jiang Q., Feng R., Zhang L., Ma G., Wang G., Chen Z. Integrating genome-wide association study and brain expression data highlights cell adhesion molecules and purine metabolism in Alzheimer's disease. Mol Neurobiol. 2014;09:1–8. doi: 10.1007/s12035-014-8884-5. [DOI] [PubMed] [Google Scholar]
- 31.Bavaresco C.S., Chiarani F., Kolling J., Netto C.A., Souza Wyse A.T.D. Biochemical effects of pretreatment with vitamins E and C in rats submitted to intrastriatal hypoxanthine administration. Neurochem Int. 2008;52:1276–1283. doi: 10.1016/j.neuint.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Wamser M.N., Leite E.F., Ferreira V.V., Delwing-de Lima D., da Cruz J.G.P., Wyse A.T.S., Delwing-Dal Magro D. Effect of hypoxanthine, antioxidants and allopurinol on cholinesterase activities in rats. J Neural Transm. 2013;120:1359–1367. doi: 10.1007/s00702-013-0989-x. [DOI] [PubMed] [Google Scholar]
- 33.Melo J.B., Agostinho P., Oliveira C.R. Involvement of oxidative stress in the enhancement of acetylcholinesterase activity induced by amyloid beta-peptide. Neurosci Res. 2003;45:117–127. doi: 10.1016/s0168-0102(02)00201-8. [DOI] [PubMed] [Google Scholar]
- 34.Bavaresco C.S., Chiarani F., Duringon E., Ferro M.M., Cunha C.D., Netto C.A., Wyse A.T. Intrastriatal injection of hypoxanthine reduces striatal serotonin content and impairs spatial memory performance in rats. Metab Brain Dis. 2007;22:67–76. doi: 10.1007/s11011-006-9038-x. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez-Dominguez R., García-Barrera T., Gomez-Ariza J.L. Combination of metabolomic and phospholipid-profiling approaches for the study of Alzheimer's disease. J Proteomics. 2014;104:37–47. doi: 10.1016/j.jprot.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 36.Frisardi V., Panza F., Seripa D., Farooqui T., Farooqui A.A. Glycerophospholipids and glycerophospholipid-derived lipid mediators: a complex meshwork in Alzheimer’s disease pathology. Prog Lipid Res. 2011;50:313–330. doi: 10.1016/j.plipres.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Klein J. Membrane breakdown in acute and chronic neurodegeneration: focus on choline-containing phospholipids. J Neural Transm. 2000;107:1027–1063. doi: 10.1007/s007020070051. [DOI] [PubMed] [Google Scholar]
- 38.Vestergaard M.C., Morita M., Hamada T., Takagi M. Membrane fusion and vesicular transformation induced by Alzheimer's amyloid beta. Biochim Biophys Acta. 2013;1828:1314–1321. doi: 10.1016/j.bbamem.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Chan R.B., Oliveira T.G., Cortes E.P., Honig L.S., Duff K.E., Small S.A., Wenk M.R., Shui G., Paolo G.D. Comparative lipidomic analysis of mouse and human brain with Alzheimer disease. J Biol Chem. 2012;287:2678–2688. doi: 10.1074/jbc.M111.274142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axelsen P.H., Murphy R.C. Quantitative analysis of phospholipids containing arachidonate and docosahexaenoate chains in microdissected regions of mouse brain. J Lipid Res. 2010;51:660–671. doi: 10.1194/jlr.D001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haughey N.J., Bandaru V.V., Bae M., Mattson M.P. Roles for dysfunctional sphingolipid metabolism in Alzheimer's disease neuropathogenesis. Biochim Biophys Acta. 2010;1801:878–886. doi: 10.1016/j.bbalip.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alessenko A.V. The potential role for sphingolipids in neuropathogenesis of Alzheimer's disease. Biomed Khim. 2013;59:25–50. [PubMed] [Google Scholar]
- 43.Kitatani K., Idkowiak-Baldys J., Hannun Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell Signal. 2008;20:1010–1018. doi: 10.1016/j.cellsig.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mielke M.M., Bandaru V.V., Haughey N.J., Rabins P.V., Lyketsos C.G., Carlson M.C. Serum sphingomyelins and ceramides are early predictors of memory impairment. Neurobiol Aging. 2010;31:17–24. doi: 10.1016/j.neurobiolaging.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mielke M.M., Lyketsos C.G. Alterations of the sphingolipid pathway in Alzheimer's disease: new biomarkers and treatment targets? Neuromol Med. 2010;12:331–340. doi: 10.1007/s12017-010-8121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]