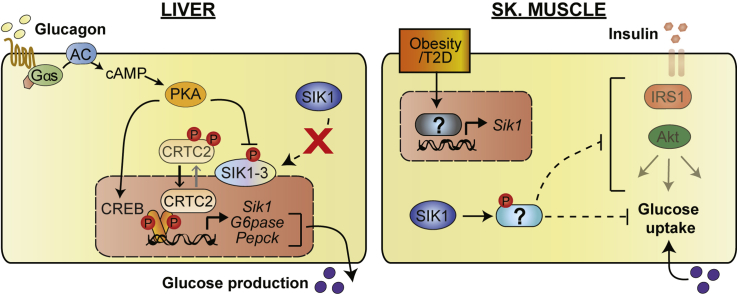
Abstract
Objective
Insulin resistance causes type 2 diabetes mellitus and hyperglycemia due to excessive hepatic glucose production and inadequate peripheral glucose uptake. Our objectives were to test the hypothesis that the proposed CREB/CRTC2 inhibitor salt inducible kinase 1 (SIK1) contributes to whole body glucose homeostasis in vivo by regulating hepatic transcription of gluconeogenic genes and also to identify novel SIK1 actions on glucose metabolism.
Methods
We created conditional (floxed) SIK1-knockout mice and studied glucose metabolism in animals with global, liver, adipose or skeletal muscle Sik1 deletion. We examined cAMP-dependent regulation of SIK1 and the consequences of SIK1 depletion on primary mouse hepatocytes. We probed metabolic phenotypes in tissue-specific SIK1 knockout mice fed high fat diet through hyperinsulinemic-euglycemic clamps and biochemical analysis of insulin signaling.
Results
SIK1 knockout mice are viable and largely normoglycemic on chow diet. On high fat diet, global SIK1 knockout animals are strikingly protected from glucose intolerance, with both increased plasma insulin and enhanced peripheral insulin sensitivity. Surprisingly, liver SIK1 is not required for regulation of CRTC2 and gluconeogenesis, despite contributions of SIK1 to hepatocyte CRTC2 and gluconeogenesis regulation ex vivo. Sik1 mRNA accumulates in skeletal muscle of obese high fat diet-fed mice, and knockout of SIK1 in skeletal muscle, but not liver or adipose tissue, improves insulin sensitivity and muscle glucose uptake on high fat diet.
Conclusions
SIK1 is dispensable for glycemic control on chow diet. SIK1 promotes insulin resistance on high fat diet by a cell-autonomous mechanism in skeletal muscle. Our study establishes SIK1 as a promising therapeutic target to improve skeletal muscle insulin sensitivity in obese individuals without deleterious effects on hepatic glucose production.
Keywords: SIK1, Salt inducible kinase, CRTC, Gluconeogenesis, Insulin resistance, CREB
Abbreviations: AKT, protein kinase B; AMPK, AMP-activated protein kinase; BAT, brown adipose tissue; cAMP, cyclic adenosine monophosphate; CHX, cycloheximide; CREB, cAMP response element-binding protein; CRTC, CREB regulated transcription coactivator; EndoRa, endogenous rate of glucose appearance; FGF21, fibroblast growth factor 21; FOXO1, forkhead box protein O1; FSK, forskolin; G6pase, glucose 6-phosphatase; GDR, glucose disposal rate; GIR, glucose infusion rate; Glgn, glucagon; GTT, glucose tolerance test; Glut, glucose transporter; HDAC, histone deacetylase; HFD, high fat diet; HSP, heat shock protein; IBMX, 3-isobutyl-1-methylxantine; ITT, insulin tolerance test; Pepck, phosphoenolpyruvate carboxykinase; Pgc, peroxisome proliferator-activated receptor gamma coactivator; PTT, pyruvate tolerance test; SIK, salt inducible kinase; WAT, white adipose tissue
Graphical abstract
Highlights
-
•
We report the first conditional SIK1 knockout mouse models.
-
•
Global and muscle-specific SIK1-KO mice are insulin sensitive on HFD.
-
•
Liver- and fat-specific SIK1-KO mice are normoglycemic.
-
•
SIK1 expression increases in skeletal muscle of obese mice.
-
•
SIK1 is a promising therapeutic target to improve insulin sensitivity in obesity.
1. Introduction
Maintenance of glucose homeostasis is fundamental to mammalian survival. Multiple molecular mechanisms driven by endocrine hormones contribute to appropriate timing of glucose liberation and synthesis when food is scarce and glucose uptake and storage when food is plentiful. The liver is the primary source of endogenous glucose production during fasting, when the catabolic hormone glucagon stimulates hepatic glycogen breakdown as well as gluconeogenesis by activating transcription of rate-limiting gluconeogenic enzymes and transcriptional regulators including Pepck, G6pase and Pgc1α [1]. After feeding, insulin stimulates glucose uptake and storage primarily in skeletal muscle and suppresses hepatic glucose production [2]. When animals become obese and insulin resistant, blood glucose becomes poorly controlled due to the combination of excess hepatic glucagon action and multi-tissue insulin resistance. The increasing prevalence of pre-diabetes and type 2 diabetes has resulted in an urgent need to identify new therapeutic strategies to improve insulin sensitivity.
The cAMP response element binding protein (CREB) directly stimulates expression of hepatic gluconeogenesis genes and is required to maintain fasting blood glucose levels in mice [3]. cAMP-regulated transcriptional co-activator 2 (CRTC2) is a key component of activated CREB complexes on Pepck, G6pase and Pgc1α promoters and contributes to the hepatic fasting gluconeogenic response [4], [5], [6], [7]. In insulin resistant rodents, in which hepatic CREB/CRTC activity becomes elevated [8], acute knock-down of CREB or CRTC2 in liver normalizes blood glucose [9], [10]. Global CRTC2 knockout mice fed high fat diet have improved insulin sensitivity, possibly secondary to reduced hepatic glucose production [6]. CRTC subcellular localization and subsequently activity are regulated by AMPK-related kinases. In particular, salt-inducible kinases (SIK1-3) directly phosphorylate CRTCs on 14-3-3 binding sites and thereby cause cytosolic retention [11]. In addition to SIKs, MARK2 and AMPK itself also phosphorylate CRTCs on the same sites [4], [12], [13].
Among CRTC kinases, SIK1 is unique in that it is a direct CREB target gene in liver, skeletal muscle, adrenal cortical cells and neurons [14]. SIK1 is therefore thought to constitute an intrinsic inhibitory feedback circuit to efficiently stop CREB target gene expression after the original stimulus ends, such as after re-feeding when glucagon and catecholamine signaling decline. Indeed, adenoviral knockdown of Sik1 in liver was sufficient to increase CREB activity and blood glucose levels [4]. However, in obese db/db mice, Sik1 mRNA increases in several tissues including liver [15], yet hepatic CRTC2 activity remains abnormally high [8]. It is therefore clear that, at least in some settings, SIK1 is not required to regulate CRTC2. Indeed, a recently reported global SIK1 knockout mouse strain did not exhibit hyperglycemia but rather improved insulin secretion. SIK1 was found to catalyze activating phosphorylation of PDE4D in beta cells; SIK1 knockout increased intracellular cAMP in beta cells and potentiated glucose-stimulated insulin secretion [16]. However, these studies were conducted on global SIK1 knockout mice, so tissue specific effects may not have been apparent.
Sik1 is broadly expressed, indicating that it may have multiple roles in physiology. We previously showed that in skeletal muscle, SIK1 maintains MEF2 activity by catalyzing inhibitory phosphorylation on class II HDAC kinases [17]. This pathway is also operant during myoblast differentiation, when SIK1 accumulates by transcriptional and post-translational mechanisms [18]. In adult muscle, Sik1 expression is acutely induced by strenuous exercise training [19], as well as by over-nutrition (obese db/db) [15] and acute muscle injury [20]. However, nothing is known about how skeletal muscle SIK1 contributes to metabolic homeostasis or how SIK1 may exert distinct functions in other metabolic tissues because to date no conditional Sik1 knockout model has been available.
To analyze cell-autonomous and cell non-autonomous roles of Sik1 in glucose homeostasis in vivo, we generated conditional Sik1 knockout mice lacking exons encoding the catalytic kinase domain. Here we show that genetic deletion of Sik1 in all tissues does not result in hyperglycemia or increased hepatic gluconeogenesis in vivo, but rather a marked improvement in glucose tolerance, peripheral insulin sensitivity and skeletal muscle glucose uptake on high fat diet. Liver Sik1 deletion alone did not de-repress gluconeogenesis, despite the fact that isolated hepatocytes lacking SIK1 showed elevated transcription of gluconeogenic genes and glucose output. Sik1 mRNA is elevated in skeletal muscle of HFD-fed mice, and skeletal muscle-specific SIK1-KO mice, but not liver or adipose tissue SIK1-KO, have enhanced insulin sensitivity after HFD feeding. We therefore identify skeletal muscle as the site of SIK1 action required for development of full insulin resistance in obesity and provide the first evidence that SIK1 is a promising therapeutic target to improve peripheral insulin sensitivity in obese individuals.
2. Methods
2.1. Mice
Generation of Sik1 conditional KO mice (MGI accession nos. 5648544, 5648545, 5648836) and crosses to generate tissue-specific lines are described in Supplementary Material and Supplementary Table T1. Male animals aged 8–30 weeks were used for metabolism studies. Knockout mice were backcrossed 3–7 generations to C57Bl6/J, at which point they were 94–99% C57Bl6/J based on SNP mapping (Charles River MaxBax Mouse 384 SNP panel). Animals were housed at 22 °C in individually ventilated cages with a 12 h light/dark cycle (9 AM–9PM) with free access to water and irradiated chow diet (LabDiet 5053). Animals were fasted in cages with synthetic bedding at 5 PM (overnight) or 9 AM (6 h). Ad lib blood samples were collected between 9 AM and 2 PM from conscious mice by tail or sub-mandibular bleed. Blood glucose was tested with a glucometer (OneTouch Ultra). Animals were fed 60% high fat diet (HFD, TestDiet 58G9) for up to 20 weeks. Glucose and pyruvate tolerance tests (GTT, PTT) were performed on overnight fasted mice, 1−1.5 g/kg glucose or 2 g/kg sodium pyruvate IP. For insulin tolerance tests (ITT), mice were fasted for 6 h followed by IP insulin (HumulinR®) injection (0.75–1 U/kg). For acute insulin signaling studies, mice were fasted for 4–6 h, injected IP with 0.75 (chow diet) or 1.5 (HFD) U/kg insulin and euthanized 15 min later. Recombinant AAV (AAV2/8-TBG-GFP or AAV2/8-TBG-Cre, 1.5 × 1011 vector genomes per animal, Penn Vector Core) was injected into the tail vein of 8–10 week old mice ≥7 days prior to testing. Body composition was determined by ECHO-MRI. Hyperinsulinemic-euglycemic clamps were performed in a similar manner to prior studies [21] on overnight fasted, unrestrained HFD-fed mice using a continuous infusion of regular insulin (Humulin R, 8 mU/min/kg body weight). Cold glucose (25%) was infused at a variable rate throughout the experiment to clamp blood glucose levels between 100 and 140 mg/dL (see also Supplementary Material). HPLC purified [3-3H]glucose was infused as previously described to determine glucose fluxes [21]. For glucose uptake during clamps, a 10 μCi bolus of [1-14C]2-deoxyglucose was injected 45 min before the end of the clamp, with blood samples collected at intervals over 45 min. Skeletal muscle glucose uptake during the clamp was calculated from the plasma [14C]2-deoxyglucose profile fitted with a double exponential curve and the tissue content of [14C]2-deoxyglucose-6-phosphate. Alternatively, 18F-labelled 2-deoxyglucose (Cyclotope, Houston TX) was injected (0.1 nmol, ∼0.4 mg/kg) into the tail vein of anesthetized mice fed HFD for 18 weeks followed immediately by an IP injection of insulin (0.75 mg/kg). 30 min later, animals were euthanized and tissues were collected for 18F quantification in a gamma counter, with data reported as % injected dose/g tissue weight.
2.2. Primary cells
Primary hepatocytes were prepared from anesthetized mice by hepatic perfusion with type IV collagenase (Sigma, C5138, 120 U/mL) as described [22]. Where indicated, hepatocytes were infected with 1–10 PFU recombinant adenovirus encoding unspecific or Sik1-selective shRNA [4] or transfected (Lipofectamine 2000, Life Technologies) with 5 nM control or Sik1-selective siRNA (Supplementary Table T1) 24 h prior to analysis. Cells were treated with glucagon (100 nM) or FSK/IBMX (forskolin 10 μM/isobutylmethylxanthine 18 μM, Sigma). For cycloheximide (CHX) chase assays, cells were washed in chase medium containing CHX (50 μg/mL) twice, then incubated in fresh medium containing CHX. MG-132 (10 μM, Cayman) was added 30 min prior to the first wash and maintained in the chase medium. Glucose output assays were performed on hepatocytes 24 h after isolation or 24–48 h after transfection as described [22]. Mature brown adipocytes were partially purified by digestion of fat pads from adult mice with type I collagenase (Worthington, 1500 U/mL) and collection of the floating adipocytes for immediate nucleic acid purification.
2.3. Nucleic acid analysis
Genomic DNA was purified by phenol: chloroform extraction and isopropanol precipitation. Total RNA was prepared using commercial kits with on-column DNase I digestion (5 Prime, Zymo). cDNA was prepared using MMLV reverse transcriptase (NEB ProtoScript II) with oligo-dT primers. Quantitative real-time PCR (qPCR) was performed on a Roche LC480 with Roche SYBR Master mix as described [22] and primers in Supplementary Table T1.
2.4. Protein analysis
Total protein extracts from cells were prepared in RIPA buffer [22] containing 0.1–2% SDS. Frozen tissues were pulverized in LN2 and homogenized in RIPA buffer using a rotor-stator homogenizer. Samples were sonicated and centrifuged (13,000 ×g, 15 min, 4 °C) prior to BCA assay and either immunoprecipitation (IP) or addition of 6X Laemmli sample buffer (LSB), boiling and SDS-PAGE resolution. SIK1 was detected on western blots with custom affinity purified anti-SIK1 antibodies: 7129 [GST-SIK1(343-776) [18]] or YZ5111, YZ51112 [custom rabbit polyclonal antibody (YenZym Antibodies, LLC) to GST-rSIK1(350-550), Supplementary Figure S1 A,B]. Commercial antibodies and IP methodology are described in Supplementary Material.
2.5. Metabolites
Plasma insulin levels were measured by immunoassay (Millipore Milliplex Mouse Metabolic Hormone panel MMHMAG-44 or Mercodia Mouse Insulin Elisa). For hepatic metabolites, tissue extracts were prepared from 30 mg of liver tissue in ice-cold RIPA buffer. For lipid extraction, 200 μL of homogenate was extracted in 800 μL of chloroform: methanol (2:1). The organic phase was collected, dried and re-suspended in 2% Triton X-100 (v/v) in chloroform. Glycogen was quantified as described [22]. Other analytes were quantified using commercial kits: Serum Triglyceride Determination Kit (Sigma), Free Fatty Acid Quantitation Kit (Sigma), Amplex® Red Cholesterol Assay Kit (Life Technologies), Adiponectin (mouse) total and high molecular weight ELISA (ALPCO).
2.6. Statistics
Statistical analysis was two-tailed Students t-test for all experiments comparing averages of 2 groups or ANOVA (2-way with repeated measures when applicable) with Tukey, Bonferroni or Sidak post-tests (with adjustment for multiple comparisons when applicable) using GraphPad Prism software. Chi-squared tests were used to analyze genotype frequency in mouse crosses.
3. Results
3.1. Global Sik1 knockout mice are normoglycemic on chow diet
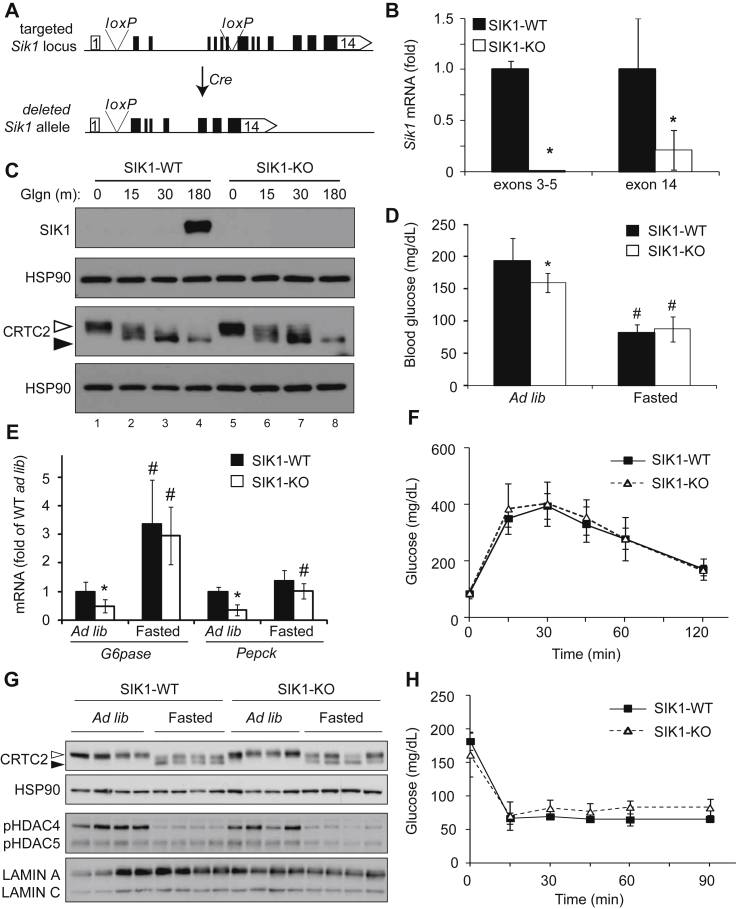
To characterize the functions of SIK1 in individual tissues in glucose metabolism, we created conditional SIK1 knockout mice by flanking the exons encoding the Sik1 kinase domain (exons 2–7) with loxP sites (Figure 1A). We deleted Sik1 in all tissues using a germline Cre driver (Gdf9) [23] and recovered fewer homozygous Sik1 knockout animals than expected, indicating a low rate of embryonic lethality that was more pronounced in males (Supplementary Table T2). Knockout animals were viable but displayed a defect in anabolic growth after weaning (8 week old males, WT 21.5 ± 1.35 gm; SIK1-KO 16.8 ± 1.72 gm, p < 0.001, n = 13–14 per genotype), which was also observed in an independent global SIK1 knockout line [16]. Full-length Sik1 mRNA was absent from liver tissue of global knockout mice when analyzed with PCR primers matching the deleted region of the gene (exons 3–5, Figure 1B). PCR primers recognizing the intact 3′UTR of the gene (exon 14) amplified a partial Sik1 mRNA (Figure 1B) that is a splice product from exon 1 to exon 8 (not shown). This truncated mRNA contains an in-frame ATG in exon 8 and is predicted to encode a truncated SIK1 protein (58 kDa) lacking the catalytic domain. Full-length SIK1 protein was absent in knockout hepatocytes (Figure 1C), and we did not detect a truncated protein in primary hepatocytes from global KO mice by direct western blotting (Supplementary Figure S1B). Interestingly, in primary hepatocytes, the phosphorylation state of CRTC2 was not affected by Sik1 deletion (Figure 1C). We did not detect an increase in SIK2 protein in SIK1-KO hepatocytes, nor did we observe increased Sik2 or Sik3 mRNA in SIK1-KO liver tissue (Supplementary Figure S1C,D), although this does not rule out functional compensation.
Figure 1.
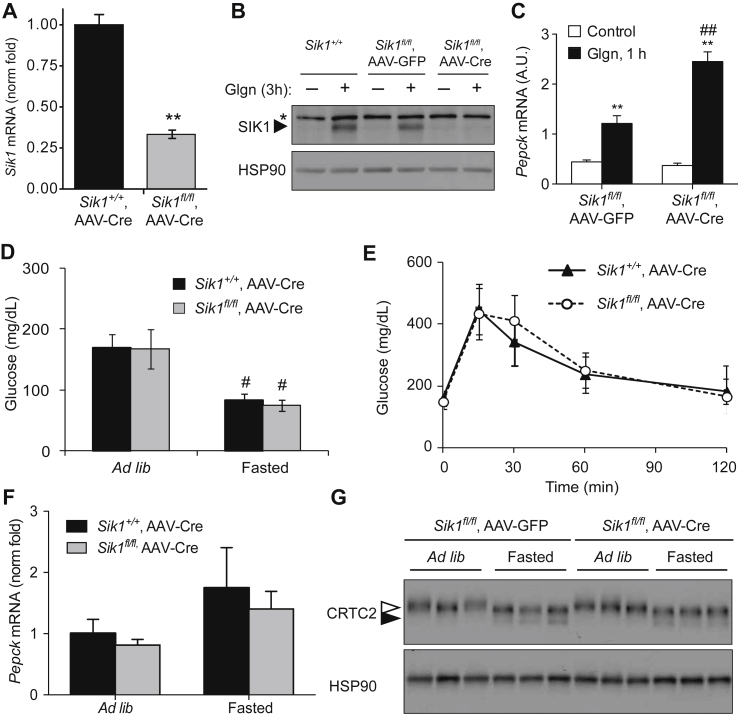
Global SIK1 knockout mice are normoglycemic. A) Schematic of mouse Sik1 locus with loxP sites shown. Cre-mediated recombination removes exons 2–7 (kinase domain). B) qPCR of Sik1 mRNA in the deleted region (exons 3–5) or intact genomic region (exon 14), normalized to Rpl32, fold of WT (mean ± stdev; n = 5 per genotype; *, p < 0.05). C) SIK1, CRTC2 and HSP90 proteins in glgn-treated primary WT and SIK1-KO hepatocytes. Open arrowhead, phospho-CRTC2; filled arrowhead, dephosphorylated CRTC2. D) Ad libitum and fasting blood glucose in global SIK1-KO mice (mean ± stdev; n = 10 per genotype; *, p < 0.05 KO vs WT (ad lib); #, p < 0.05 fasted vs ad lib within genotype). E) Gluconeogenic gene expression in SIK1 knockout liver, ad libitum fed or after 16 h fasting, (mean ± stdev; n = 4–6 per genotype; *, p < 0.05 KO vs WT (ad lib); #, p < 0.05 fasted vs ad lib within genotype). F) GTT of wild type and SIK1-KO mice (mean ± stdev; n = 5–6 per genotype). G) CRTC2 and class II HDAC phosphorylation in liver tissue of SIK1-WT and SIK1-KO mice ad libitum fed or 16 h fasted. H) ITT of SIK1-WT and SIK1-KO mice (mean ± stdev, n = 5–6 per genotype).
We tested the hypothesis that in SIK1 knockout mice de-repression of CRTC2/CREB activity would lead to hyperglycemia and glucose intolerance. Surprisingly, global SIK1 knockout animals were normoglycemic, with slightly lower blood glucose than WT control animals in the ad libitum fed state and unaltered blood glucose in the fasted state (Figure 1D). Hepatic expression of rate-limiting gluconeogenic genes Pepck and G6pase paralleled the blood glucose levels, with significantly reduced expression in ad libitum fed SIK1-KO animals and comparable amounts of Pepck and G6pase mRNA after fasting (Figure 1E). Glucose tolerance on chow diet was also unaffected by global Sik1 deletion (Figure 1F). We hypothesized that SIK1 limits fasting gluconeogenic gene expression in liver via inhibitory phosphorylation of CRTC2 [4], [11] or the class II histone deacetylase HDAC4 [17], [24], [25], which co-activate CREB and FOXO1, respectively. Both CRTC2 and HDAC4 were normally phosphorylated in ad libitum fed SIK1-KO liver and appropriately dephosphorylated after overnight fasting (Figure 1G). Together these data indicate that compensation by other AMPK-related kinases is sufficient to maintain normal CRTC and HDAC phosphorylation in liver, in agreement with recent reports of other SIK family knockout mice [26], [27], [16], [28].
To determine if lower ad libitum glucose levels in SIK1-KO mice were a result of enhanced insulin whole body sensitivity, we performed insulin tolerance tests. SIK1-WT and global SIK1-KO animals had comparable insulin sensitivity (Figure 1H) and hepatic AKT activation following a bolus insulin injection, with slightly reduced phosphorylation on AKT(T308) in the knockout liver (Supplementary Figure S1E,F). Mild hypoglycemia in chow fed SIK1-KO animals might also be due to enhanced insulin secretion [16]. In contrast to the findings of Kim et al., plasma insulin levels were unaltered in our global SIK1-knockout mouse line on chow diet (Supplementary Figure S2A). Similarly, hepatic cholesterol, glycogen and triglycerides as well as plasma free fatty acids were unaffected in SIK1-KO mice (Supplementary Figure S2B–E). We conclude that global deletion of the Sik1 catalytic domain does not affect glucose homeostasis on normal chow diet.
3.2. Global SIK1-KO mice have heightened insulin sensitivity on high-fat diet
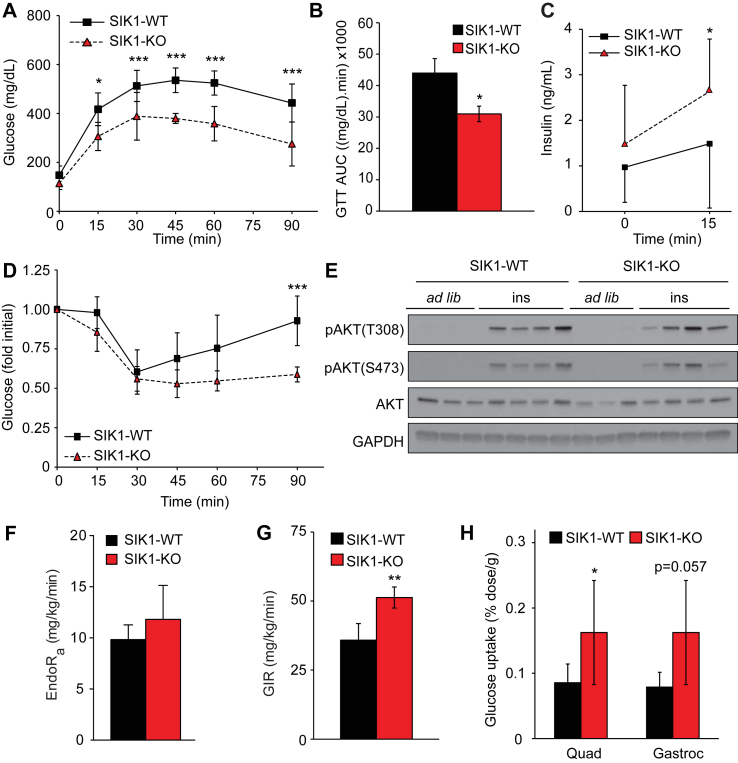
CREB/CRTC2 activity in liver becomes unrestrained in obese db/db mice [5], and Sik1 mRNA consequently increases [15]. Despite accumulation, SIK1 does not inhibit CREB/CRTC activity in the livers of genetically obese mice, either because SIK1 is not required for CRTC2 regulation in states of obesity or because O-glycosylation of CRTC2 on SIK consensus phosphorylation sites in obese mice blocks phosphorylation by SIKs [8]. To genetically interrogate the impact of Sik1 on glucose metabolism in obesity, we challenged SIK1-KO mice with 60% high fat diet (HFD) for up to 20 weeks. SIK1-KO mice maintained lower body weight that WT controls throughout the experiment, but weight gain as percent of initial weight was similar (Supplementary Figure S3A,B). Moreover, SIK1-KO and WT mice had comparable fat and lean mass as well as proportions of organs, including fat pads (Supplementary Figure S3C,D). We anticipated that SIK1-KO animals would have exacerbated glucose intolerance on HFD. Remarkably, SIK1-KO mice were protected from HFD-induced glucose intolerance, with reduced total glucose excursion during glucose tolerance tests (Figure 2A,B). This is partly explained by increased circulating insulin in response to glucose bolus in SIK-KO mice under HFD conditions (Figure 2C), consistent with recent findings that Sik1-deficient beta cells have enhanced glucose-stimulated insulin secretion [16]. However, we reasoned that enhanced beta cell function is not sufficient to explain the improved glucose tolerance, as SIK1-KO mice also have heightened insulin sensitivity judged by insulin tolerance test (Figure 2D, Supplementary Figure S3E). Surprisingly, 15 min after insulin injection, AKT phosphorylation on both Thr308 and Ser473 was comparable in skeletal muscle (Figure 2E, Supplementary Figure S3F) and liver (Supplementary Figure S3G,H) of WT and SIK1-KO mice. Neither of the key endogenous insulin-sensitizing hormones, adiponectin or FGF-21, was induced in SIK1-KO animals to a pharmacologically significant level (Supplementary Table T3, Supplementary Figure S3I), although plasma FGF21 was elevated in fasted SIK1-KO mice fed high fat diet.
Figure 2.
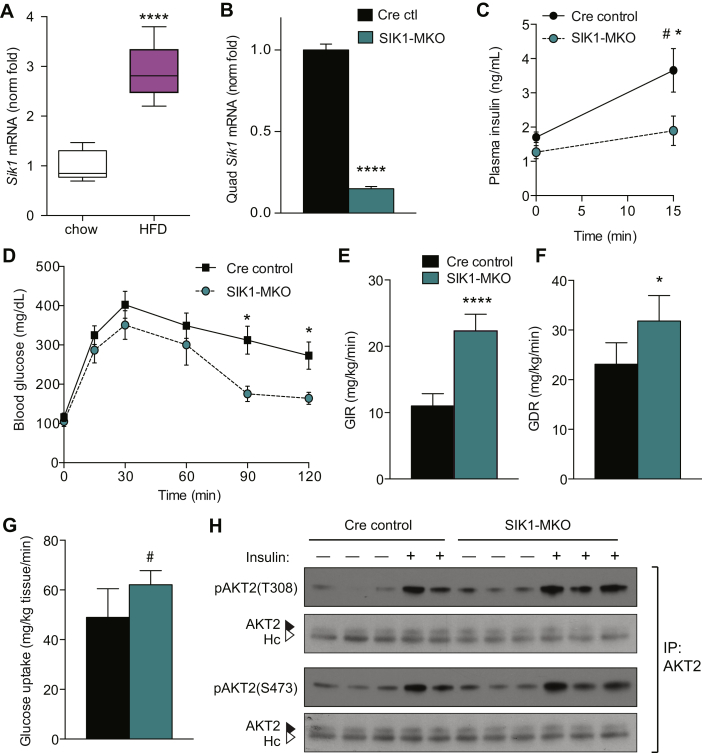
SIK1-knockout mice are protected from insulin resistance in obesity. A) GTT of male global SIK1-KO mice after 16–18 weeks of high fat diet feeding (mean ± stdev; n = 6–8 per genotype among 2 cohorts; 2-way repeated measures ANOVA p < 0.0001; Bonferroni post-tests: *, p < 0.05; ***, p < 0.001 vs WT at indicated time points). B) Area under curve analysis of GTT in panel A (mean ± stdev; n = 6–8 per genotype; *, p < 0.05). C) Plasma insulin in HFD fed male SIK1-KO mice after overnight fast and 15 min after glucose injection (mean ± stdev; n = 11–12 per genotype among 2 cohorts; *, p < 0.05). D) ITT of male HFD SIK1-KO mice (mean ± stdev; n = 4–5 per genotype; 2-way repeated measures ANOVA p = 0.06; Bonferroni post-test: ***, p < 0.001 vs WT at indicated time points). E) Western blots of phosphorylated and total AKT in quadriceps muscle from male HFD-fed SIK1-WT and KO animals euthanized ad libitum or after 6 h fast and IP insulin (15 min). Each lane represents one animal (n = 4 per condition). See Supplementary Figure S3F for densitometry. F) Rate of endogenous glucose appearance (EndoRa) in fasted HFD male SIK1-WT and SIK1-KO mice 60 min after [3-3H]-glucose tracer infusion (mean ± stdev; n = 4–5 per genotype). G) Glucose infusion rate (GIR) at the final time point of hyperinsulinemic euglycemic clamp in dynamic steady state (mean ± stdev; n = 4–5 per genotype; **, p < 0.01). See Supplementary Figure S3M,N for time courses. H) Insulin-stimulated biodistribution of 18F-fluorodeoxyglucose in male HFD fed mice (mean ± stdev; n = 4–5 per genotype; *, p < 0.05 vs WT).
To obtain a more accurate measure of glucose turnover and insulin sensitivity in these animals, we used tracer techniques and hyperinsulinemic-euglycemic clamps. We found that basal glucose production (EndoRa) was not significantly increased in fasted SIK1-KO animals versus controls (Figure 2F), consistent with pyruvate tolerance tests and unchanged gluconeogenic gene expression (Supplementary Figure S3J–L). In response to hyperinsulinemia, we found that the glucose infusion rate required to achieve and maintain euglycemia in SIK1-KO animals was higher than in controls (Figure 2G, Supplementary Figure S3M,N), indicating improved whole-body insulin sensitivity in agreement with ITT results (see Figure 2D). Utilizing a glucose analog tracer, we observed elevated insulin-stimulated glucose uptake in the skeletal muscles of HFD-fed SIK1-KO animals (Figure 2H). These data show that global SIK1 knockout does not exacerbate HFD-induced hyperglycemia. On the contrary, SIK1-KO mice have improved glucose tolerance due in part to enhanced peripheral insulin sensitivity and skeletal muscle glucose disposal.
3.3. SIK1 synthesis and degradation are highly regulated in hepatocytes
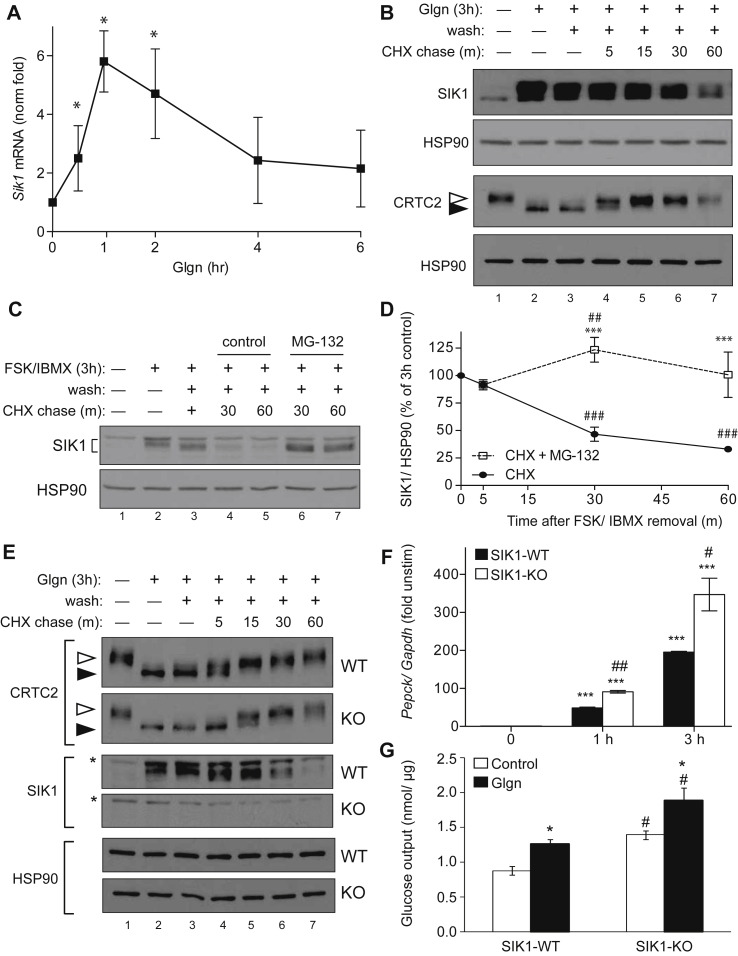
Our results in global SIK1 knockout animals prompted two major questions: 1) why is gluconeogenesis unaffected and 2) how and where does SIK1 promote insulin resistance in obesity? We reasoned that lack of increased gluconeogenesis in global SIK1 knockout mice could result from A) functional compensation by other AMPK-related kinases, B) the impact of disrupted growth pathways on systemic metabolism, or C) an effect of Sik1 deletion in extra-hepatic tissues that could mask liver effects. To explore these hypotheses, we first investigated SIK1 regulation and function in isolated mouse hepatocytes. SIK1 was highly induced in hepatocytes in response to glucagon [4], peaking from 1 h (mRNA) to 3 h (protein) (Figure 3A,B; Supplementary Figure S4A). Notably, neither Sik2 nor Sik3 mRNA was induced by glucagon in a similar fashion (Supplementary Figure S4B). Newly translated SIK1 is thought to contribute to CRTC2 inhibition after feeding, although it is striking that at peak SIK1 protein expression, CRTC2 is maximally de-phosphorylated (Figure 3B, lane 2). This is most likely because sustained cAMP signaling causes direct PKA phosphorylation and nuclear export of SIKs, blocking phosphorylation of nuclear SIK substrates [29], [18]. When the priming glucagon stimulus was washed away and hepatocytes were chased in control medium, SIK1 protein was dephosphorylated (Figure 3B, lanes 3–7) and presumably entered the nucleus where it phosphorylates CRTCs. Indeed, CRTC2 was re-phosphorylated over the same time course (Figure 3B). We noticed that SIK1 is very unstable in hepatocytes, with an estimated half-life of 30 min, and that the proteasome inhibitor MG-132 stabilized SIK1 protein (Figure 3C,D). Thus, SIK1 is induced in hepatocytes but only has approximately 30–60 min to act on CRTCs or other substrates before the protein is degraded. Although SIK2 does not accumulate in response to glucagon, SIK2 protein was also somewhat destabilized after glucagon removal (Supplementary Figure S4C, D), indicating that these two kinases may be degraded by a common molecular mechanism in hepatocytes.
Figure 3.
SIK1 is dynamically regulated and regulates CRTC2 in primary hepatocytes. A) qPCR analysis of Sik1 mRNA in primary mouse hepatocytes treated with glucagon (Glgn, 100 nM). Fold change of Sik1 from untreated cells, normalized to Gapdh (mean ± stdev; n = 3; *, p < 0.05 vs untreated. B) Primary mouse hepatocytes were left untreated (lane 1) or stimulated with Glgn (100 nM) for 3 h (lanes 2–7) followed by wash and chase in normal growth medium containing cycloheximide (CHX, 50 μg/mL) for the times indicated (lanes 3–7). Open arrowhead, phospho-CRTC2; filled arrowhead, dephosphorylated CRTC2. C) SIK1 protein in primary hepatocytes treated as in panel B. In lanes 6–7, MG-132 (10 μM) was added for the final 30 min of FSK/IBMX stimulation and during the CHX chase. SIK1 and HSP90 proteins shown. D) Percent SIK1 protein normalized to HSP90 during CHX chase assays (mean % remaining ± stdev, n = 4; ***, p < 0.001 vs CHX; ##, p < 0.01 or ###, p < 0.001 vs no chase. E) Glgn priming and CHX chase assay on WT and SIK1-KO hepatocytes as in Figure 1B. Western blots show CRTC2 (open arrowhead, phosphorylated; filled arrowhead, dephosphorylated), SIK1 and HSP90 proteins. *, non-specific band. F) Pepck mRNA in SIK1-WT and SIK1-KO primary hepatocytes treated with glgn for up to 3 h. (Avg of n = 2 biological replicates ± stdev, fold of unstimulated of same genotype; ***, p < 0.001 vs control within genotype; #, p < 0.05 or ##, p < 0.01 KO vs WT each time point). Representative assay of 4 experiments. G) Glucose output assay of SIK1-WT and SIK1-KO primary hepatocytes incubated with pyruvate, lactate, and either glgn or vehicle (control) (mean ± stdev; n = 3; *, p < 0.05 glgn vs control within genotype; #, p < 0.01 KO vs WT). All panels represent n ≥ 3 individual experiments on hepatocytes isolated on at least two separate days.
To test the contribution of newly synthesized SIK1 to CRTC2 re-phosphorylation, we performed similar glucagon priming and chase assays in hepatocytes from WT and SIK1-KO mice. As expected, CRTC2 was dephosphorylated in both WT and SIK1-KO hepatocytes after glucagon treatment (Figure 3E). During the chase period, CRTC2 re-phosphorylation was delayed in cells genetically deficient for Sik1, although CRTC2 phosphorylation was eventually fully restored in cells of both genotypes (Figure 3E). In keeping with the proposed model of SIK1 action in hepatocytes, SIK1-KO hepatocytes also had stronger transcriptional responses to glucagon (Figure 3F) and synthesized more glucose ex vivo than control hepatocytes (Figure 3G). Therefore, newly synthesized SIK1 contributes to inactivation of CRTC2 and control of glucose synthesis ex vivo. However, other CRTC2 kinases such as SIK2, SIK3 or AMPK isoforms are sufficient to maintain CRTC2 phosphorylation in resting hepatocytes or at longer intervals after cessation of glucagon signaling. There is little SIK1 expressed in resting hepatocytes (Figure 3) or ad libitum fed liver [4], and it is yet unknown what specific role SIK1 may have in the fed state.
SIK1-KO animals have a small body size that could influence metabolism of the isolated hepatocytes. These animals also express a partial Sik1 mRNA (Figure 1B) predicted to encode a catalytically inactive truncated protein. We therefore acutely inhibited Sik1 expression in hepatocytes by two additional methods. Transfection of a Sik1-selective siRNA that targets the remaining 3′ end of the mRNA resulted in higher glucose output after 6 h glucagon treatment, but not in un-stimulated cells (Supplementary Figure S5A,B). We also isolated hepatocytes from Sik1fl/fl animals and treated the cells successively with adenovirus encoding GFP or Cre recombinase and control siRNA or the same si-SIK1. Cre-mediated Sik1 deletion ex vivo also potentiated glucagon-stimulated glucose output, which was not further enhanced by transfection with si-SIK1 (Supplementary Figure S5C,D). Together these data show that eliminating SIK1 expression in primary hepatocytes promotes glucagon-stimulated glucose production ex vivo and confirm that the remaining truncated Sik1 mRNA does not contribute to glucose regulation in hepatocytes. Interestingly, acute inhibition of Sik1 expression in hepatocytes does not result in increased basal glucose production as we observed in hepatocytes from global SIK1-KO mice (Figure 3G), indicating that either longer-term deletion of Sik1 stimulates autophagy [30] or amino acid utilization for gluconeogenesis [31] or that the endocrine environment in the global SIK1-KO animals might influence hepatocyte metabolism even after isolation.
3.4. Liver-specific Sik1 knockout does not affect glucose metabolism
Having observed increased glucose output and gluconeogenesis in isolated hepatocytes with Sik1 knockout or knock-down, we reasoned that Sik1 deletion in an extra-hepatic tissue might mask these effects in vivo in global SIK1-KO mice. We therefore acutely deleted Sik1 in hepatocytes of adult Sik1fl/fl mice by injecting adeno-associated virus (AAV) that expresses Cre from the hepatocyte-specific thyroxine-binding globulin (TBG) promoter [32]. Compared with controls, Sik1fl/fl mice infected with AAV-TBG-Cre (SIK1-AAV-LKO mice) had 75% Sik1 deletion in whole liver tissue (Figure 4A, Supplementary Figure S6A) that did not occur in other tissues (Supplementary Figure S6B). In hepatocytes isolated from AAV infected animals, neither full-length SIK1 protein nor Sik1 mRNA was detectable (Figure 4B, Supplementary Figure S6C). Similar to hepatocytes isolated from SIK1-KO mice, hepatocytes from SIK1-AAV-LKO mice exhibited a stronger gluconeogenic transcriptional response to glucagon ex vivo (Figure 4C, Supplementary Figure S6D). Homozygous Sik1 floxed animals show a mild (7%) reduction in body weight (Supplementary Figure S6E), in contrast to the ∼25% reduction in body weight of homozygous global SIK1-KO animals. AAV encoding Cre or Gfp had no further effect, indicating that the floxed allele has reduced function to promote anabolic growth in a different tissue. Unlike the mild hypoglycemia observed in global SIK1-KO mice, chow-fed SIK1-AAV-LKO animals had identical blood glucose under both ad libitum fed and fasted conditions (Figure 4D) as well as normal glucose tolerance (Figure 4E) compared with Cre control animals. Consistently, we observed no difference in hepatic Pepck mRNA (Figure 4F) or CRTC2 phosphorylation in liver of mice lacking Sik1 specifically in hepatocytes (Figure 4G). We performed the same experiments in Sik1 floxed mice crossed to Alb-Cre transgenic mice [33] to induce chronic postnatal hepatocyte specific Sik1 deletion. As expected, Sik1 was only knocked out in liver, with 90% mRNA reduction in hepatocytes isolated from SIK1-LKO mice (Supplementary Figure S7A–C). Just as in AAV-LKO mice, SIK1-LKO animals showed no alteration in fasting or fed blood glucose levels or glucose tolerance on chow diet (Supplementary Figure S7D,E). We conclude that hepatocyte SIK1 is not required in vivo for regulation of CREB/CRTC2 activity or glucose homeostasis on chow diet.
Figure 4.
Hepatocyte-specific SIK1 knockout mice are normoglycemic on chow diet. A) Sik1 mRNA (exon 14) in liver of SIK1-AAV-LKO mice or controls, normalized to Rpl32 (mean fold of control ± SEM; n = 2 per genotype; **, p = 0.01). B) Western blot of SIK1 protein in glgn-stimulated hepatocytes isolated from wild type (Sik1+/+) or floxed (Sik1fl/fl) animals injected with AAV-TBG-GFP or AAV-TBG-Cre. *, non-specific band. C) Pepck mRNA in primary hepatocytes from an AAV infected animal (mean ± stdev; n = 3 biological replicates; **, p < 0.01 control vs glgn; ##, p < 0.01 glgn GFP vs glgn Cre). D) Blood glucose in male control (Sik1+/+,AAV-Cre) and SIK1-AAV-LKO (Sik1fl/fl,AAV-Cre) (mean ± stdev; n = 12–14 per genotype; #p < 0.05 to ad lib of same genotype). E) GTT in male control and SIK1-AAV-LKO animals (mean ± stdev; n = 8–14 per genotype). F) qPCR of Pepck (mean ± stdev; n = 5 per genotype per condition) and G) western blot of CRTC2 in control and SIK1-AAV-LKO liver, ad libitum fed and overnight fasted (n = 3 animals per condition).
Hepatic CRTC2/CREB has been shown to contribute to excessive gluconeogenesis in obese rodents [8], [9], [6]. Although we found that basal glucose production in HFD-fed global SIK1-knockout mice was comparable to control mice, we tested glucose metabolism in HFD-fed SIK1-LKO animals to rule out impact of another tissue on the liver in the global SIK1-KO model. Liver-specific SIK1-LKO mice (Alb-Cre) fed HFD had comparable body weight and food intake to littermate control mice (Supplementary Figure S8A,B). Interestingly, obese SIK1-LKO animals showed neither worsened nor improved glucose or insulin tolerance compared to littermate Cre controls (Supplementary Figure S8C–E). These data provide the first rigorous genetic evidence that hepatic SIK1 is dispensable for regulation of glucose metabolism on chow or HFD and that the observed increase of Sik1 in db/db livers, likely a consequence of activated CREB activity, does not serve an important modulatory role on glucose metabolism. Moreover, the data clearly indicate that loss of Sik1 in hepatocytes does not contribute to the improved insulin sensitivity on high fat diet that is observed in global KO mice.
3.5. Skeletal muscle SIK1 promotes insulin resistance in obesity
We have shown that despite the fact that primary hepatocytes lacking SIK1 produce more glucose, mice lacking SIK1 are not hyperglycemic. Instead, global SIK1-KO mice fed high fat diet have markedly enhanced peripheral insulin sensitivity and increased insulin-stimulated skeletal muscle glucose disposal (see Figure 2, Supplementary Figure S3). Sik1 mRNA has been shown to increase in skeletal muscle, liver and adipose tissue of obese db/db mice [15]. Similarly, we found that Sik1 mRNA increases three-fold in skeletal muscle after 12 weeks of 60% HFD feeding in mice (Figure 5A), suggesting that SIK1 might contribute to insulin resistance in obesity by a muscle autonomous mechanism. To test this hypothesis, we crossed Myf5Cre/+ knock-in animals [34] with Sik1 floxed mice to efficiently delete the kinase domain of Sik1 in the developing myotome. Myf5-Cre deleted exons encoding the Sik1 catalytic domain in skeletal muscle and brown adipose tissue but not other adult tissues we tested, including heart and pancreas (Supplementary Figure S9A). SIK1-MKO mice had grossly normal muscle structure and did not show evidence of degeneration in histological sections (not shown). Sik1 mRNA was almost undetectable in both skeletal muscle and BAT of SIK1-MKO mice (Figure 5B, Supplementary Figure S9B). Deletion of Sik1 in BAT could confound analysis of skeletal muscle Sik1 action in this mouse line. To circumvent this, we crossed Sik1 floxed animals with Adiponectin-Cre BAC transgenic mice [35] to generate fat-specific SIK1 knockout mice (SIK1-FKO). Similar to SIK1-LKO mice, SIK1-FKO mice did not display any glucose homeostasis phenotypes on either chow or HFD (Supplementary Figure S10), indicating that adipose tissue Sik1 does not make a major contribution to glucose metabolism in obesity. We therefore term the Sik1fl/fl;Myf5Cre/+ mice “muscle-specific” KO (SIK1-MKO) and interpret phenotypes as driven primarily by skeletal muscle Sik1 deletion.
Figure 5.
Skeletal muscle deletion of Sik1 enhances insulin sensitivity in obesity. A) Skeletal muscle Sik1 mRNA in male mice fed chow diet or 12 weeks 60% HFD (n = 5–6 per genotype; ****, p < 0.0001). B) Sik1 mRNA in quadriceps skeletal muscle of female Cre control (Myf5Cre/+) and SIK1-MKO (mean ± stdev, n = 5 per genotype, ****, p < 0.0001). C) Plasma insulin in male HFD fed SIK1-MKO mice, fasted (0) and 15 min after glucose injection IP (mean ± SEM, n = 4–5 per genotype, non-significant by 2-way RM ANOVA; *, p < 0.05 Cre:MKO by Sidak's multiple comparison test; #, p < 0.05 0:15 min glucose in Cre control by t-test). D) Glucose tolerance tests of male Cre control and SIK1-MKO fed 60% HFD for 12 weeks (mean ± SEM, n = 5–9 per genotype, p < 0.05 by 2-way repeated measures ANOVA, effect of genotype p = 0.1, *p < 0.05 at indicated time points by individual t-tests). E) Glucose infusion rate (GIR) and F) glucose disposal rate (GDR) at final time point of dynamic steady state in hyperinsulinemic euglycemic clamps of male Cre control and SIK1-MKO mice fed HFD for 19 weeks (mean ± stdev, n = 5 per genotype, ****, p < 0.0001, *, p < 0.05 by t-test). See Supplementary Figure S9K,L for time courses. G) Skeletal muscle 2-deoxyglucose uptake rate during last 45 min of hyperinsulinemic euglycemic clamps (mean ± stdev, n = 5 per genotype, #, p = 0.05 by t-test). H) Western blots of AKT2 immunoprecipitates from male 12-week HFD-fed Cre control and MKO animals fasted for 6 h and/or administered insulin IP 15 min prior to euthanization. See Supplementary Figure S9N for densitometry.
On chow diet, SIK1-MKO mice had normal glucose tolerance and muscle AKT2 activation in response to insulin (Supplementary Figure S9C–E). Unlike global SIK1-KO animals, SIK1-MKO mice did not exhibit smaller body weight when compared to control animals (Supplementary Figure S9F). In response to 60% HFD feeding, SIK1-MKO and Cre controls gained a comparable amount of weight and had similar percentages of lean and fat mass (Supplementary Figure S9F–H). Importantly, HFD fed SIK1-MKO animals exhibited a slightly weaker insulin response to glucose than Cre controls (Figure 5C), indicating that the improvement in beta cells observed in global KO mice (Figure 2C and cit [16]) does not occur in the SIK1-MKO line. In spite of this, glucose tolerance tests of HFD-fed SIK1-MKO mice exhibited a trend toward improved glucose tolerance (p = 0.1 for effect of genotype by ANOVA), which was significant at the late time points by individual t-tests (Figure 5D, Supplementary Figure S9I). To precisely test insulin sensitivity in these animals, we performed hyperinsulinemic euglycemic clamps. Basal endogenous glucose production (EndoRa) was slightly reduced in SIK1-MKO animals, and insulin reduced endogenous glucose production in both genotypes (Supplementary Figure S9J). Similar to results in global SIK1-KO mice, substantially more exogenous glucose was required to achieve and maintain euglycemia in HFD-fed SIK1-MKO mice than in control animals (Figure 5E, Supplementary Figure S9K,L), indicating improved whole body insulin sensitivity. The glucose disposal and skeletal muscle 2-deoxyglucose uptake rates at dynamic steady state were also significantly elevated in SIK1-MKO mice (Figure 5F,G). Enhanced glucose disposal was not due to increased Glut4, Glut1 or Glut12 mRNA expression in SIK1-MKO muscle (Supplementary Figure S9M). Surprisingly, basal and insulin-stimulated AKT2 phosphorylation in skeletal muscle was also not significantly induced in obese SIK1-MKO versus control animals (Figure 5H and Supplementary Figure S9N). Altogether these data demonstrate that Sik1 is upregulated in skeletal muscle of mice with diet-induced obesity, where it contributes to impaired insulin-stimulated glucose uptake.
4. Discussion
During fasting CREB and its co-activator CRTC2 stimulate gluconeogenic gene transcription in hepatocytes and thus form a crucial regulatory node in the hepatic response to fasting [3], [4], [5], [6], [7]. CRTC2 is a dynamic component within this transcriptional complex due to its regulated subcellular localization controlled by inhibitory phosphorylation by several kinases including SIK1-3, MARK2 and AMPK isoforms. Of particular interest is SIK1, which is a CREB transcriptional target poised to act as a feedback inhibitor of CREB activity and limit the response to fasting [4]. In addition to its proposed role as a CREB/CRTC2 inhibitor in liver, SIK1 couples the CREB and MEF2 pathways in muscle, promoting survival in adult muscle [17] and differentiation of muscle progenitor cells [18]. Sik1 expression has also been shown to increase in skeletal muscle of obese db/db [15], HFD fed (Figure 5A), injured [20], and exercise trained mice [19]. SIK1 is also expressed in other tissues, such as vascular smooth muscle where it regulates Na/K-ATPase activity and blood pressure [36]. To test the hypothesis that SIK1 represses hepatic glucose output and to potentially identify novel functions of SIK1 in other tissues, we created conditional Sik1 knockout mice and tested glucose metabolism phenotypes in global SIK1-KO as well as selective liver-, adipose- or skeletal muscle SIK1-knockout mice.
Mice lacking the catalytic domain of SIK1 in all tissues are viable, but display a reduction in post-weaning anabolic growth, which could be a manifestation of a complex endocrine phenotype in these animals that also impacts hepatocyte glucose production ex vivo (see Figure 3). In future studies, it would be useful to employ an inducible Cre for additional analysis of phenotypes resulting from global Sik1 deletion after development. Nonetheless, similar to a recent report in an independent SIK1 knockout line [16], global KO animals are largely normoglycemic on chow diet but exhibit strikingly improved glucose tolerance on high fat diet, with increased circulating insulin after glucose challenge. In addition to this mechanism, we show for the first time that SIK1-knockout animals have profoundly increased peripheral insulin sensitivity judged by insulin tolerance test, 2-deoxyglucose uptake and hyperinsulinemic-euglycemic clamps. It is possible that this phenotype was overlooked in the prior report because clamps were not performed.
Using conditional knockout mice, we conclusively demonstrate that hepatocyte SIK1 is not required to regulate gluconeogenesis in vivo. These data contrast the effects of acute Sik1 knock-down in hepatocytes (see Figure 3 and Supplementary Figure S5) and livers [4], which is sufficient to de-repress CREB/CRTC2 activity and glucose production. The discrepancies between these studies could reflect the contribution of inflammation to the former studies or the method of deletion. Indeed, the Sik1-selective shRNA employed causes destruction of the Sik1 transcript [17], while a partial Sik1 mRNA remains in our knockout mouse liver. However, an independent global SIK1-KO mouse also showed no obvious hyperglycemia or hyperlipidemia [16], and we observed no further potentiation of glucose output when we transfected primary hepatocytes with an siRNA that would inhibit the partial mRNA expressed in our KO line. Therefore, although glucagon signaling dynamically regulates SIK1 mRNA and protein in hepatocytes by both synthesis and regulated protein degradation, and newly synthesized SIK1 contributes to CRTC2 regulation in primary hepatocytes, other CRTC kinases can functionally compensate for Sik1 knockout in vivo. Our results, along with findings from global SIK2 knockout mice, SIK3 knockout hepatocytes, and combination AMPK-alpha1/alpha2 knockout mice, indicate that there is significant functional redundancy among AMPK-related kinases in liver to maintain CRTC2 and HDAC4 phosphorylation and prevent excessive glucose output that is observed in Lkb1-deficient hepatocytes [37], [38], [26], [27], [16], [28]. As LKB1 is the requisite upstream activating kinase for all AMPK-related kinases [39], it is not surprising that CRTC2 and HDAC4 phosphorylation are more severely affected by Lkb1 deletion.
Our results that skeletal muscle knockout of SIK1 specifically improves insulin sensitivity on high fat diet (compared with liver or adipose tissue SIK1-KO) have begun to reveal new and unexpected functions of this kinase that are distinct from its closely related family members SIK2 and SIK3, despite the fact that each of these kinases is capable of phosphorylating and inhibiting CRTCs and class II HDACs. Sik3 knockout results in highly penetrant embryonic lethality due to defects in chondrocyte hypertrophy, which occurs through a SIK3-class II HDAC pathway [40]. Surviving animals have complex metabolic phenotypes [41], and hepatocytes from these animals exhibit the expected increase in CREB-mediated gluconeogenesis [28]. Global SIK2 knockout mice have enhanced CRTC2/CREB activity in white adipocytes, which contributes to insulin resistance, hyperglycemia, and hypertriglyceridemia in that model [27]. In adipocytes, SIK2 stimulates Glut4 mRNA expression and glucose uptake [42], which is apparently opposite to the overall action of SIK1 in skeletal muscle. SIK1, on the other hand promotes myogenic differentiation in vitro [18], myofiber survival [17] and blood pressure regulation [36], and we now show that SIK1 promotes insulin resistance in skeletal muscle in the context of over-nutrition. In contrast to Sik2 and Sik3, deletion of Sik1 in mice improves insulin sensitivity and glucose disposal in skeletal muscle.
The molecular mechanism responsible for improved physiological insulin sensitivity in Sik1-deficient muscle remains elusive. It is surprising that insulin-stimulated AKT2 phosphorylation is not elevated in HFD SIK1-MKO muscles compared to controls. Although PI(3)-kinase stimulates glucose uptake in myocytes by AKT2-dependent [43] and AKT2-independent mechanisms [44], it seems unlikely that SIK1 alters insulin signaling to PI(3)-kinase without also affecting AKT2. The best-characterized SIK1 substrates in skeletal muscle are transcriptional regulators CRTC and class II HDACs. Loss of SIK1 would cause hyper-activation of both CRTC2 and class II HDACs, the latter resulting in MEF2 inhibition. Notably, an AMPK-HDAC-MEF2 pathway has been shown to stimulate Glut4 mRNA expression after exercise in skeletal muscle [45]. However, the phenotype of increased glucose transport in SIK1-KO muscle is opposite to the expected effect of reduced MEF2 activity, and Glut4 mRNA is not altered in SIK1-KO muscles. Our data do not, however, rule out altered GLUT transporter trafficking in the SIK1-KO skeletal muscle. Gain of CRTC function is another intriguing possible mechanism. Indeed, transgenic overexpression of constitutively activated CRTC2 in skeletal muscle resulted in hypertrophy and improved glycogen storage [19]. It is possible that increased glucose utilization could establish a steeper glucose concentration gradient across the sarcolemma, which could account for the increased glucose uptake we observe in SIK1-deficient skeletal muscle. Of note, global CRTC2 knockout mice were also shown have enhanced insulin sensitivity [6], suggesting that CRTC2 gain of function may not account for improved insulin sensitivity in SIK1 knockout mice. Whether SIK1 could regulate muscle glucose uptake and utilization by direct phosphorylation of enzymes involved in glucose metabolism or by a transcriptional mechanism remains to be determined. Finally, acute knock-down of full-length Sik1 mRNA in undifferentiated skeletal myoblasts impairs myogenic differentiation [18] but skeletal muscle development appears grossly unaffected in mice lacking the catalytic domain of SIK1 either globally or in Myf5-expressing muscle progenitor cells. This could be due to either compensation by other proteins in vivo for loss of Sik1 or expression of a partial SIK1 protein in skeletal myocytes that supports differentiation. We therefore do not believe that a defect in muscle development accounts for the phenotype of improved insulin sensitivity on high fat diet. In future studies, it will be important to rule this out using an inducible myofiber-selective Cre.
Our findings are significant in establishing a new role for Sik1 in extra-hepatic tissues that is of potential clinical importance. Sik1 transcription increases in skeletal muscle of obese animals, and genetic deletion of Sik1 ameliorates insulin resistance on high fat diet. These findings coupled with the finding that deletion of Sik1 in all tissues or specifically in hepatocytes has no detectable impact on hepatic gluconeogenesis, renders SIK1 a promising therapeutic target to improve peripheral insulin sensitivity in overweight or obese patients without deleterious consequences on hepatic gluconeogenesis. It will be important in future studies to identify SIK1-selective catalytic inhibitors and test their efficacy in preclinical models of type 2 diabetes as well as to delineate the molecular mechanism underlying SIK1-mediated insulin resistance in skeletal muscle.
Author contributions
MN, RSF, JF, DA and RB prepared figures. MN, RSF, JF, KR, DA, MGMR, YARM, MG, NJJ and RB performed experiments and analyzed data. EB analyzed and interpreted data. MN and RB wrote the manuscript, which all authors edited and approved. RB and NJJ designed the mouse strains. RB designed and oversaw the study.
Acknowledgments
This study was supported by grants from the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01-DK092590 and DK092590-03S1 to RB, P30-DK079638 supporting the Baylor College of Medicine Diabetes Research Center, P30-DK056338 supporting the Texas Medical Center Digestive Disease Center, and T35-DK007676-21 supporting MG; National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) R01-AR059847 to RB; National Institute on Aging K01-AG036738 to NJJ, as well as startup funds from the University of Texas Health Science Center at Houston. The authors thank Isabel Lorenzo in the Baylor College of Medicine Mouse Embryonic Stem Cell Core and Dr. Eva Zsigmond in the UT Health Transgenic and Stem Cell Service Unit for assistance with creating knock-in mice. We are grateful to Dr. Pradip Saha in the Mouse Metabolism Core (Baylor College of Medicine Diabetes Research Center) for assistance with clamp studies and the Texas Medical Center Digestive Diseases Center (DDC) Cores for provision of primary hepatocytes for pilot studies and assistance with histology. We thank Drs. Eva M. Sevick-Muraca and Ali Azhdarinia and the staff of the Institute for Molecular Medicine Center for Molecular Imaging (supported by U54-CA136404 and a Texas Star Award to EMS), University of Texas Health Science Center at Houston, for help with glucose biodistribution studies.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.10.004.
Conflict of interest
The authors declare that no conflict of interest exists.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Ramnanan C.J., Edgerton D.S., Kraft G., Cherrington A.D. Physiologic action of glucagon on liver glucose metabolism. Diabetes, Obesity & Metabolism. 2011;13(Suppl 1):118–125. doi: 10.1111/j.1463-1326.2011.01454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saltiel A.R., Kahn C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 3.Herzig S., Long F., Jhala U.S., Hedrick S., Quinn R., Bauer A. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 4.Koo S.H., Flechner L., Qi L., Zhang X., Screaton R.A., Jeffries S. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 5.Dentin R., Liu Y., Koo S.H., Hedrick S., Vargas T., Heredia J. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Inoue H., Ravnskjaer K., Viste K., Miller N., Liu Y. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Lay J., Tuteja G., White P., Dhir R., Ahima R., Kaestner K.H. CRTC2 (TORC2) contributes to the transcriptional response to fasting in the liver but is not required for the maintenance of glucose homeostasis. Cell Metabolism. 2009;10:55–62. doi: 10.1016/j.cmet.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dentin R., Hedrick S., Xie J., Yates J., 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 9.Saberi M., Bjelica D., Schenk S., Imamura T., Bandyopadhyay G., Li P. Novel liver-specific TORC2 siRNA corrects hyperglycemia in rodent models of type 2 diabetes. American Journal of Physiology - Endocrinology and Metabolism. 2009 doi: 10.1152/ajpendo.00158.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erion D.M., Ignatova I.D., Yonemitsu S., Nagai Y., Chatterjee P., Weismann D. Prevention of hepatic steatosis and hepatic insulin resistance by knockdown of cAMP response element-binding protein. Cell Metabolism. 2009;10:499–506. doi: 10.1016/j.cmet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Screaton R.A., Conkright M.D., Katoh Y., Best J.L., Canettieri G., Jeffries S. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Katoh Y., Takemori H., Lin X.Z., Tamura M., Muraoka M., Satoh T. Silencing the constitutive active transcription factor CREB by the LKB1-SIK signaling cascade. FEBS Journal. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 13.Jansson D., Ng A.C., Fu A., Depatie C., Al Azzabi M., Screaton R.A. Glucose controls CREB activity in islet cells via regulated phosphorylation of TORC2. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10161–10166. doi: 10.1073/pnas.0800796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berdeaux R. Metabolic regulation by salt inducible kinases. Frontiers in Biology. 2011;6:231–241. [Google Scholar]
- 15.Horike N., Takemori H., Katoh Y., Doi J., Min L., Asano T. Adipose-specific expression, phosphorylation of Ser794 in insulin receptor substrate-1, and activation in diabetic animals of salt-inducible kinase-2. Journal of Biological Chemistry. 2003;278:18440–18447. doi: 10.1074/jbc.M211770200. [DOI] [PubMed] [Google Scholar]
- 16.Kim M.J., Park S.K., Lee J.H., Jung C.Y., Sung D.J., Park J.H. Salt inducible kinase 1 terminates cAMP signaling by an evolutionarily conserved negative feedback loop in beta cells. Diabetes. 2015 doi: 10.2337/db14-1240. [DOI] [PubMed] [Google Scholar]
- 17.Berdeaux R., Goebel N., Banaszynski L., Takemori H., Wandless T., Shelton G.D. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nature Medicine. 2007;13:597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 18.Stewart R., Akhmedov D., Robb C., Leiter C., Berdeaux R. Regulation of SIK1 abundance and stability is critical for myogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:117–122. doi: 10.1073/pnas.1212676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bruno N.E., Kelly K.A., Hawkins R., Bramah-Lawani M., Amelio A.L., Nwachukwu J.C. Creb coactivators direct anabolic responses and enhance performance of skeletal muscle. EMBO Journal. 2014;33:1027–1043. doi: 10.1002/embj.201386145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart R., Flechner L., Montminy M., Berdeaux R. CREB is activated by muscle injury and promotes muscle regeneration. PLoS One. 2011;6:e24714. doi: 10.1371/journal.pone.0024714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma K., Saha P.K., Chan L., Moore D.D. Farnesoid X receptor is essential for normal glucose homeostasis. Journal of Clinical Investigation. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu J., Akhmedov D., Berdeaux R. The short isoform of the ubiquitin ligase NEDD4L is a CREB target gene in hepatocytes. PLoS One. 2013;8:e78522. doi: 10.1371/journal.pone.0078522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan Z.J., Xu X., Cooney A.J. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biology of Reproduction. 2004;71:1469–1474. doi: 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- 24.Wang B., Moya N., Niessen S., Hoover H., Mihaylova M.M., Shaw R.J. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihaylova M.M., Vasquez D.S., Ravnskjaer K., Denechaud P.D., Yu R.T., Alvarez J.G. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel K., Foretz M., Marion A., Campbell D.G., Gourlay R., Boudaba N. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nature Communications. 2014;5:4535. doi: 10.1038/ncomms5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park J., Yoon Y.S., Han H.S., Kim Y.H., Ogawa Y., Park K.G. SIK2 is critical in the regulation of lipid homeostasis and adipogenesis in vivo. Diabetes. 2014;63:3659–3673. doi: 10.2337/db13-1423. [DOI] [PubMed] [Google Scholar]
- 28.Itoh Y., Sanosaka M., Fuchino H., Yahara Y., Kumagai A., Takemoto D. Salt inducible kinase 3 signaling is important for the gluconeogenic programs in mouse hepatocytes. Journal of Biological Chemistry. 2015 doi: 10.1074/jbc.M115.640821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh Y., Takemori H., Doi J., Okamoto M. Identification of the nuclear localization domain of salt-inducible kinase. Endocrine Research. 2002;28:315–318. doi: 10.1081/erc-120016802. [DOI] [PubMed] [Google Scholar]
- 30.Seok S., Fu T., Choi S.E., Li Y., Zhu R., Kumar S. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014;516:108–111. doi: 10.1038/nature13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Erion D.M., Kotas M.E., McGlashon J., Yonemitsu S., Hsiao J.J., Nagai Y. cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 2 (CRTC2) promotes glucagon clearance and hepatic amino acid catabolism to regulate glucose homeostasis. Journal of Biological Chemistry. 2013;288:16167–16176. doi: 10.1074/jbc.M113.460246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Z., Miller R.A., Patel R.T., Chen J., Dhir R., Wang H. Hepatic Hdac3 promotes gluconeogenesis by repressing lipid synthesis and sequestration. Nature Medicine. 2012;18:934–942. doi: 10.1038/nm.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. Journal of Biological Chemistry. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 34.Tallquist M.D., Weismann K.E., Hellstrom M., Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development. 2000;127:5059–5070. doi: 10.1242/dev.127.23.5059. [DOI] [PubMed] [Google Scholar]
- 35.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bertorello A.M., Pires N., Igreja B., Pinho M.J., Vorkapic E., Wagsater D. Increased arterial blood pressure and vascular remodeling in mice lacking salt-inducible kinase 1 (SIK1) Circulation Research. 2015;116:642–652. doi: 10.1161/CIRCRESAHA.116.304529. [DOI] [PubMed] [Google Scholar]
- 37.Shaw R.J., Lamia K.A., Vasquez D., Koo S.H., Bardeesy N., Depinho R.A. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foretz M., Hebrard S., Leclerc J., Zarrinpashneh E., Soty M., Mithieux G. Metformin inhibits hepatic gluconeogenesis in mice independently of the LKB1/AMPK pathway via a decrease in hepatic energy state. Journal of Clinical Investigation. 2010;120:2355–2369. doi: 10.1172/JCI40671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lizcano J.M., Goransson O., Toth R., Deak M., Morrice N.A., Boudeau J. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO Journal. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasagawa S., Takemori H., Uebi T., Ikegami D., Hiramatsu K., Ikegawa S. SIK3 is essential for chondrocyte hypertrophy during skeletal development in mice. Development. 2012;139:1153–1163. doi: 10.1242/dev.072652. [DOI] [PubMed] [Google Scholar]
- 41.Uebi T., Itoh Y., Hatano O., Kumagai A., Sanosaka M., Sasaki T. Involvement of SIK3 in glucose and lipid homeostasis in mice. PLoS One. 2012;7:e37803. doi: 10.1371/journal.pone.0037803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henriksson E., Sall J., Gormand A., Wasserstrom S., Morrice N.A., Fritzen A.M. SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. Journal of Cell Science. 2015;128:472–486. doi: 10.1242/jcs.153932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho H., Mu J., Kim J.K., Thorvaldsen J.L., Chu Q., Crenshaw E.B., 3rd Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 44.JeBailey L., Wanono O., Niu W., Roessler J., Rudich A., Klip A. Ceramide- and oxidant-induced insulin resistance involve loss of insulin-dependent Rac-activation and actin remodeling in muscle cells. Diabetes. 2007;56:394–403. doi: 10.2337/db06-0823. [DOI] [PubMed] [Google Scholar]
- 45.Richter E.A., Hargreaves M. Exercise, GLUT4, and skeletal muscle glucose uptake. Physiological Reviews. 2013;93:993–1017. doi: 10.1152/physrev.00038.2012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.