Abstract
The CLE (CLAVATA3/Endosperm surrounding region-related) peptide family is one of the best-studied secreted peptide families in plants. Accumulated data have revealed that CLE genes play vital roles on stem cell homeostasis in different types of meristems. Additionally, CLE genes have been found to perform various biological roles in plant growth and development, and in response to environmental stimuli. With recent advances on our understanding of CLE peptide function, it is showing that the existence of potential crosstalks of CLE peptides with phytohormones and external stimuli. Complex interactions exist in which CLE petides coordinate with hormones to regulate plant growth and development, and in response to external stimuli. In this article, we present recent advances in cell-cell communication that is mediated by CLE peptides combining with phytohormones and external stimuli, and suggest additional Arabidopsis CLE genes that are likely to be controlled by hormones and environmental cues.
Keywords: CLE peptide, phytohormone, crosstalk, Arabidopsis, receptor
Introduction
Phytohormones, as well as secreted peptides, are important in mediating intercellular communications to regulate numerous developmental and physiological activities, and respond to environmental cues (Betsuyaku et al., 2011; Murphy et al., 2012). The CLE (CLAVATA3/Endosperm surrounding region-related) peptide family is one of the best-studied secreted peptide families in plants. Over recent years, it is suggested that CLE peptide signaling is integrated with phytohormone signaling and is involved in responding to environmental cues to modulate a wide range of biological processes.
CLE genes are known to encode small, secreted peptides with a conserved C-terminal CLE motif (Cock and McCormick, 2001). The mature CLE peptides are cleaved from their precursor proteins after post-translational modification in the CLE motif such as hydroxylation and glycosylation (Matsubayashi, 2011). CLE genes have been identified in many plant species and some plant parasitic nematodes. In Arabidopsis, the CLE family comprised of 32 members, yet only a few CLE genes have been functionally characterized (Betsuyaku et al., 2011; Murphy et al., 2012). To date, CLE peptides have been implicated in the regulation of seed development, vascular formation, lateral root establishment, and the stem cell homeostasis in the shoot apical meristem (SAM), the root apical meristem (RAM) and (pro-)cambium (Czyzewicz et al., 2013; Ingram and Gutierrez-Marcos, 2015). Additionally, CLE peptides have been found to mediate responses to environmental stimuli including a notable role in sensing nitrate and controlling nodulation in legumes (Miyawaki et al., 2013). It is commonly recognized that CLE peptides are perceived by leucine-rich repeat receptor-like kinases (LRR-RLKs), forming the evolutionarily conserved CLE-RLK module to convey extracellular and intracellular signaling cascades (Betsuyaku et al., 2011; Murphy et al., 2012). Despite the large number of LRR-RLKs in plants, only a limited number of peptide-receptor pairs have been identified and assigned functionality. It is becoming increasingly apparent that CLE peptides are involved in various processes to establish, regulate and maintain plant development, and to respond to external stimuli.
In this perspective article, we focus on the characterization of CLE signaling pathways that integrating with phytohormone signaling and mediation of environmental stimuli to coordinate internal and external signals. We summarize studies that highlight the interactions of CLE peptides with hormones and external cues, and suggest additional Arabidopsis CLE genes that are likely to be regulated by phytohormones and/or environmental stimuli.
Orchestration of CLE Peptide Signaling and Phytohormone Signaling
It has revealed that CLE peptide signaling integrated with phytohormone signaling to control various biological processes in plants (Figure 1A; Table 1). CLE6 and CLE41/TDIF activated auxin transcriptional reporters and transporters including DR5pro:GUS, IAA2pro:GUS, PIN1pro:GUS, and PIN3pro:GUS, suggesting induction of an immediate auxin response upon CLE peptides (Whitford et al., 2008). In addition, CLE6 potentiated an effect of CLE41/TDIF on promoting procambium proliferation. Furthermore, the effect was synergistically strengthened in the presence of a synthetic auxin NAA, and weakened in the presence of an auxin polar transport inhibitor NPA (Whitford et al., 2008). Consistently, the CLE41/TDIF peptide-induced procambium proliferation was abolished by a mutation in the Monopteros/ARF5 gene, an auxin response factor required for mediating auxin stimuli (Whitford et al., 2008). Altogether, these results indicate that vascular patterning is regulated by different CLE peptides in conjunction with auxin signaling.
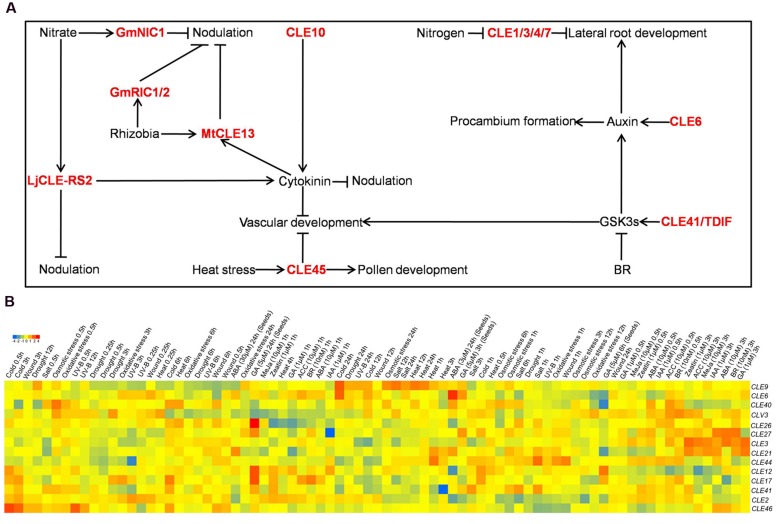
FIGURE 1.
The crosstalks of CLE peptides with diverse factors. (A) A schematic representation of the complex interactions of CLE peptides with external and internal factors. The arrow indicate positive, while barred line indicate negative effect. CLE peptides are indicated in red. (B) The expression pattern of AtCLE upon hormones and selected stresses. The microarray data were obtained from AtGenExpress initiative. A detail description of samples and experimental design would be found at https://www.arabidopsis.org/portals/expression/microarray/ATGenExpress.jsp. Gene expression is displayed as normalized log2-transformed values, which is visualized by the color scale.
Table 1.
A summary of crosstalks between CLE peptides with hormones and environmental stimuli.
| CLE peptide | Species | Factor(s) interacted | Function(s) | Reference(s) |
|---|---|---|---|---|
| CLE1/3/4/7 | Arabidopsis thaliana | Nitrogen | Inhibiting later root growth | Araya et al., 2014 |
| CLE6 | A. thaliana | Auxin/GA | Promoting procambium proliferation; compensation for GA deficiency | Whitford et al., 2008; Bidadi et al., 2014 |
| CLE10 | A. thaliana | Cytokinin | Inhibiting protoxylem vessel formation | Kondo et al., 2011 |
| CLE14/20 | A. thaliana | Cytokinin | Inhibiting root growth | Meng and Feldman, 2010 |
| CLE26 | A. thaliana | Auxin | Regulating root architecture and protophloem formation | Czyzewicz et al., 2015 |
| CLE40 | A. thaliana | ABA/auxin/cytokinin | Promoting ABA biosynthesis and signaling, repressing cytokinin signaling and differentially regulating auxin signaling | Pallakies and Simon, 2014 |
| CLE41/TDIF | A. thaliana | Auxin/BR | Later root development; xylem vessel formation | Cho et al., 2014; Kondo et al., 2014 |
| CLE45 | A. thaliana | Heat stress | Pollen growth and seed production | Endo et al., 2013 |
| OsCLE48 | Oryza sativa | Auxin | N/D | Guo et al., 2015 |
| LjCLE-RS1/2 | Lotus japonicus | Rhizobia/cytokinin/nitrate | Inhibiting nodule development | Okamoto et al., 2009; Okamoto et al., 2013; Sasaki et al., 2014 |
| LjCLE19/20 | L. japonicus | Phosphate | N/D | Funayama-Noguchi et al., 2011 |
| MtCLE12/13 | Medicago truncatula | Rhizobia | Inhibiting nodule development | Mortier et al., 2010; Mortier et al., 2012 |
| GmRIC1/2 | Glycine max | Rhizobia | Inhibiting nodule development | Reid et al., 2011 |
| GmNIC1 | G. max | Nitrate | Partially inhibiting nodulation | Reid et al., 2011 |
N/D, not determined.
In addition to the interaction between CLE41/TDIF and auxin, CLE41/TDIF, along with brassinosteroids (BR), was found to determine the xylem vessel formation by regulating the GSK3s activity (Kondo et al., 2014). CLE41/TDIF prevents the differentiation of procambial cells into xylem cells, whereas BR positively regulates xylem differentiation (Hirakawa et al., 2008; Clouse, 2011). It has been shown that TDR/PXY, the CLE41/TDIF receptor, interacts with GSK3s, and activates GSK3s in a CLE41/TDIF-dependent manner. BES1, as a downstream target of GSK3s, mediates CLE41/TDIF-TDR-GSK3s signaling to suppress xylem differentiation (Kondo et al., 2014). Additionally, CLE41/TDIF-initiated TDR signaling activates BIN2, which further interacts with auxin signaling by phosphorylating ARF7 and ARF19 to inhibit their interactions with AUX/IAAs and positively regulates their target genes LBD16 and LBD29 to modulate the lateral root development (Cho et al., 2014). However, as BR exerts no effect on regulation of BIN2 activity, it suggests an immediate CLE41/TDIF-induced regulation of BIN2 in lateral root formation (Cho et al., 2014). This provides an example of complex interaction among CLE41/TDIF, BR and auxin signaling to regulate root development.
The CLE10 peptide, similar to cytokinin, suppressed protoxylem formation in Arabidopsis roots, implying a possible crosstalk between CLE peptide and cytokinin. Further investigation revealed the expression of ARR5 and ARR6, two negative regulators of cytokinin signaling, was repressed by CLE10 (Kondo et al., 2011). Consistently, protoxylem formation of the lateral root, but not the primary root, was inhibited in arr5 arr6 double mutants despite no alteration is observed in either single mutant. Intriguingly, the clv2 mutant exhibited insensitivity to CLE10 peptides in the suppression of protoxylem formation, suggesting that CLE10 acts through CLV2 to regulate the protoxylem formation (Kondo et al., 2011). Additionally, ARR10 and ARR12, two positive regulators of cytokinin signaling, were shown to be necessary for CLE10 induced protoxylem inhibition as the arr10 arr12 double mutant was unresponsive to the CLE10 peptide (Kondo et al., 2011). It is therefore suggested that CLE10 suppresses the expression of ARR5 and ARR6, by which results in promoting cytokinin signaling to inhibit protoxylem formation.
In vitro application of either CLE14 or CLE20 peptides, or overexpression of CLE14 and CLE20 resulted in short-root phenotype by reducing cell division rates in the RAM (Meng and Feldman, 2010). The short-root phenotype caused by the exogenous application of the CLE14 or CLE20 peptide cannot be overcome by auxin or cytokinin treatment, while cytokinin partially rescued the short-root phenotype induced by overexpression of CLE14 or CLE20 in planta (Meng and Feldman, 2010). This result implies that cytokinin, but not auxin, may influence CLE14/CLE20 functions by affecting the post-translational regulation of CLE peptides in vivo, probably resulting in an alteration in the availabilities and/or abundances of CLE14/CLE20 peptides. Alternatively, overexpression of CLE14 or CLE20 may down-regulate cytokinin biosynthesis gene(s) or promote cytokinin metabolic gene(s) which can be compensated by exogenous cytokinin. However, the molecular mode of action of crosstalk between CLE and cytokinin in this process awaits elucidation.
As reported recently, in additional to CLE45, CLE26 affected primary root protophloem (Czyzewicz et al., 2015). CLE26 is expressed in the stele at the phloem pole. Expression of CLE26 is increased significantly upon auxin treatment, indicating a possible interaction between CLE26 and auxin (Czyzewicz et al., 2015). Further studies revealed that exogenously applied CLE26 peptide resulted in altered auxin responses as evidenced by reduced auxin response marker DR5pro:GUS and elevated auxin sensor DII:VENUS. In addition, pPIN1::PIN1:GFP is reduced in the presence of CLE26 peptide, although no effect on the PIN1 gene expression. This indicates that CLE26 influences auxin signaling through modulating the activity of the polar auxin transporter (Czyzewicz et al., 2015). Collectively, the CLE26 peptide represses the distribution and/or abundance of auxin in the RAM, possibly by decreasing the abundance of PIN1 through post-translational regulation. However, the biological significance of auxin alteration induced by CLE26 is unclear. Nevertheless, it is also intriguingly to unravel the mechanism by which auxin transcriptionally regulates the CLE26 and the biological consequences by elevating its transcripts.
It has been shown that CLE40 integrated with phytohormone pathways by regulating hormone synthesis, signaling, and their target genes (Pallakies and Simon, 2014). Genes involving in abscisic acid biosynthesis and signaling were down-regulated in cle40 mutants, whereas auxin-related genes were differentially expressed. CLE40 represses cytokinin signaling by supressing the expression of key genes in cytokinin signaling and biosynthesis (Pallakies and Simon, 2014). Altogether, it is suggested that CLE40 modulates phytohormone signaling in distinct modes through multi-pronged targets that comprise phytohormone synthesis, signaling, and downstream genes.
The application of gibberellin (GA) promoted CLE6 expression, suggesting a direct and long-distant regulation of CLE6 by GA hormone. Conversely, application of CLE6 peptide exerted no effect on the growth and development of GA-deficient mutant plants (Bidadi et al., 2014). However, over-expression of CLE6 in a GA-deficient mutant partially rescued the mutant phenotype, suggesting that CLE6 could compensate for the GA deficiency. Grafting of GA-deficient mutant plants to 35S::CLE6 transgenic plants complemented the shoot phenotype associated with GA deficiency, suggesting CLE6 can exert its action over a long distance (Bidadi et al., 2014). However, whether the CLE6 peptide itself moves through the vascular system is yet to be demonstrated.
Forty-seven CLE genes have been identified from rice (Kinoshita et al., 2007). OsCLE48, a rice CLE peptide, is significantly induced by auxin (Guo et al., 2015). OsCLE48 rescued the clv3-2 mutant phenotype when driven by the native CLV3 promoter. However, overexpression of OsCLE48 in rice failed to alter the shoot development (Guo et al., 2015), implying the functional divergence of CLE genes in Arabidopsis and rice. It is still unclear that function of OsCLE48 and the biological consequence of transcriptionally regulating of OsCLE48 by auxin.
CLE Peptide Signaling Upon Environmental Stimuli
Plants are continuously exposed to a wide range of environmental stimuli. Considerable advances have been made in our understanding of interactions between CLE peptides and environmental stimuli (Figure 1A; Table 1). A study on CLE45 peptide provided an example of a CLE peptide mediating the signal of an environmental cue. In addition to its role on inhibiting protophloem differentiation via BAM3 in roots, CLE45 is implicated in pollen-pistil interaction upon heat stress (Depuydt et al., 2013; Endo et al., 2013). CLE45 is preferentially expressed in the stigma at normal temperature, whereas its expression domain expand into the transmitting tract at elevated temperature, suggesting a temperature-dependent function of CLE45 (Endo et al., 2013). Two RLKs, SKM1 and SKM2, are expressed preferentially in pollen and pollen tubes. Additionally, pollen tube growth of the skm1 skm2 double mutant displayed complete insensitivity to CLE45 peptide, suggesting that SKM1 and SKM2 may function as receptors of CLE45 in this process (Endo et al., 2013). This was confirmed by a genetic study showing that skm1 skm2 double mutants phenocopied the CLE45-RNAi plants. Furthermore, a direct and specific binding of CLE45 peptide with SKM1 protein was demonstrated (Endo et al., 2013). In conclusion, CLE45 mitigates heat stress by binding with SKM1/SKM2 to sustain pollen growth under higher temperatures and maintain successful seed production.
It is known that lateral roots stop growing under severe deficiency of nitrogen (N), while the expression of CLE1/3/4/7 were induced under N-deficient conditions (Araya et al., 2014). The clv1 mutant exhibited progressive growth of lateral roots under N-deficient conditions. Conversely, overexpression of CLE1/3/4/7 repressed the emergence and growth of lateral roots. However, this inhibitory action of CLE3 was abolished in the clv1 mutant (Araya et al., 2014). CLE1/3/4/7 are predominantly expressed in the root pericycle, while the location of CLV1 is restricted in phloem companion cells (Araya et al., 2014). Altogether, these results indicate that CLV1 mediates a N-responsive CLE peptide signaling pathway that negatively regulates lateral root development under N-deficient conditions.
Funayama-Noguchi et al. (2011) identified two LjCLE genes, LjCLE19 and LjCLE20, which respond specifically in the presence of phosphate (Pi). LjCLE19 and LjCLE20 were up-regulated specifically and significantly upon excess Pi. Along with the increase in Pi level, expressions of LjCLE19 and LjCLE20 increased prior to the increment of Pi content in plants. However, the Pi content in plants decreased prior to the reduction of LjCLE19 and LjCLE20 expression with external Pi decreased (Funayama-Noguchi et al., 2011). Nevertheless, it remains largely unknown how exactly LjCLE19 and LjCLE20 mediated the phosphate signaling to modulate plant growth and development.
CLE Peptide Signaling in Nodulation
It is well known that many CLE peptides play vital roles in legume-rhizobium symbioses (Miyawaki et al., 2013). A search of the Lotus japonicus genome database has identified 39 LjCLE genes, among which LjCLE-RS1 and LjCLE-RS2 are significantly up-regulated in rhizobial inoculated roots (Okamoto et al., 2009). HAR1 encodes a LRR-RLK which is highly similar to CLV1. The har1 mutant exhibits a hypernodulation phenotype (Krusell et al., 2002; Nishimura et al., 2002). Overexpression of LjCLE-RS1 and LjCLE-RS2 suppress nodule development. However, this inhibitory effect is abolished in the har1 mutant, implying that LjCLE-RS1 and LjCLE-RS2 inhibit nodulation in HAR1-dependent manner (Okamoto et al., 2009). Further studies revealed that LjCLE-RS1/2, as long-distance signals, were root-derived signals that were recognized by HAR1 in the shoot. Once perceived, the CLE-RS1/2-HAR1 signaling activated the shoot-derived cytokinins which systemically inhibited nodulation (Okamoto et al., 2013; Sasaki et al., 2014). Constitutive activation of LjCLE genes incurred disappearance of auxin responses (Suzaki et al., 2012, 2013). In addition to HAR1, KLV, and LjCLV2 encode a RPK2-like RLK and a CLV2-like protein, respectively (Miyazawa et al., 2010; Krusell et al., 2011). Genetic and biochemical studies revealed that HAR1 and KLV function in the same pathway via forming a receptor complex by which LjCLE-RS1 and LjCLE-RS2 signaling are transmitted (Krusell et al., 2011). Similarly, mutations in LjCLV2 result in increased nodulation (Miyazawa et al., 2010). However, it is not known by what mechanism LjCLV2 controls the nodulation. Furthermore, LjCLE-RS2 is strongly up-regulated upon excess nitrate application in roots, suggesting that LjCLE-RS2 plays key roles in nitrate sensing (Okamoto et al., 2009). It is known that accumulation of nitrate suppresses nodulation. Thus, LjCLE-RS2 may sense the nitrate to negatively regulate nodulation.
MtCLE12 and MtCLE13, two MtCLEs identified from Medicago truncatula, were also implicated during nodulation (Mortier et al., 2010). MtCLE12 and MtCLE13 were up-regulated in nodulated roots, whereas the expression of MtCLE13 increased much earlier than that of MtCLE12. Moreover, MtCLE13 expression was induced by cytokinin, while that of MtCLE12 was unaffected (Mortier et al., 2010, 2012). Nevertheless, the CLE domain sequences of MtCLE12 and MtCLE13 are highly similar to the CLE domains of LjCLE-RS1 and LjCLE-RS2, suggesting that MtCLE12/MtCLE13 and LjCLE-RS1/LjCLE-RS2 may exert a comparable function during nodulation. Indeed, the ectopic expression of MtCLE12 and MtCLE13 inhibited nodulation, which was mediated by a LRR-RLK, SUNN (Schnabel et al., 2005; Mortier et al., 2010).
Reid et al. (2011) identified three CLE peptides, namely GmRIC1, GmRIC2, and GmNIC1, in soybean. The expression of GmRIC1 and GmRIC2 were induced by rhizobial inoculation, while the expression of GmNIC1 was up-regulated by nitrate (Reid et al., 2011). Overexpression of GmRIC1 and GmRIC2 inhibited soybean nodulation systemically and required the presence of GmNARK which encodes a LRR-RLK (Searle et al., 2003; Reid et al., 2011; Lim et al., 2011). In contrast, overexpression of GmNIC1 partially reduced nodulation locally, which also required GmNARK (Reid et al., 2011). These results suggest the requirement of GmNARK, possibly as the receptor for GmRIC1/2 and GmNIC1, in both inoculation- and nitrate-induced regulation of nodulation in soybean.
In summary, CLE peptides play critical roles in nodulation formation in legume. A recurrent theme in CLE-mediated nodulation formation is the requirement of a LRR-RLK to perceive the CLE peptide signaling, suggesting evolutionary conservation and commonality in the regulation of nodulation. In addition, LjCLE-RS2 and GmNIC1 are nitrate-responsive, while MtCLE13 expression is induced by cytokinin, implying a greater level of complexity in the interactions between CLE peptides, hormones and external stimuli on controlling nodulation.
The Expression of Many Arabidopsis CLE Genes is Perturbed by Phytohormones and Environmental Stimuli
Microarray datasets allowed us to identify Arabidopsis CLE genes that are regulated by phytohormones and environmental stimuli. In silico expression data for 14 out of 32 Arabidopsis CLE genes are available for further analysis (Kilian et al., 2007; Goda et al., 2008). Notably, CLE3 is dramatically up-regulated by all tested hormones correlating with the time course, while its expression was repressed by cold, salt and UV-B (Figure 1B). CLE2 is generally inhibited by hormones and external stresses under sustained treatment, and induced under short-term treatment. The expression of CLE9 is not significantly induced by hormones, but was dramatically induced by almost all selected stresses under sustained treatment (Figure 1B). CLV3, CLE2, CLE3, CLE21, and CLE46 exhibit diverse responses upon hormones and abiotic stresses. Much to our surprise, CLV3, known as a key player in regulation of stem cell homoeostasis, is shown not only respond to hormones, but also display variable responses to selected stresses (Figure 1B). In contrast, CLE12, CLE17, CLE26, CLE27, CLE40, CLE41, and CLE44 remain unchanged, with the exception of CLE27 which is prominently repressed by auxin (Figure 1B). Collectively, these data suggest that Arabidopsis CLE genes are distinctly perturbed by phytohormones and environmental stimuli depending on time of treatment and concentration. The challenge will be to develop ways to attribute specific functions to these CLE genes that are transcriptionally regulated by hormones and stresses, and to understand how exactly they are controlled and their roles in adaptive plant growth.
Conclusion and Perspectives
Over recent years, crosstalks between CLE peptides with phytohormones and environmental cues have been shown (Figure 1A; Table 1). Clearly, concluding from the examples presented, one could speculate that there are complicated and diverse regulatory networks involved in these crosstalks, but most await further elucidation. Regarding the functional dissection of CLE genes, antagonistic peptide technology and CRISPR gene editing technology may be helpful to overcome functional redundancies and difficulties in obtaining loss-of-function mutants (Song et al., 2013; Ma et al., 2015). CLE peptides are perceived by the extracellular domains of RLKs, but information on peptide–receptor pairs with assigned functions is scarce. The identification and characterization of peptide-receptor pairs will continue to yield insights into how they are integrated with diverse factors. To this end, it is essential to understand the spatial and temporal control of CLE gene expression in various plant species, as has been shown for many CLE genes in Arabidopsis (Jun et al., 2010). It is also important to identify internal and external factors that regulate the expression of CLE genes. As a first step, it is shown that many Arabidopsis CLE genes are perturbed by phytohormones and environmental stimuli (Figure 1B), which open avenues to gain insights into the crosstalk of Arabidopsis CLE peptides with diverse factors. The future challenge will be to develop ways to attribute specific functions to these responsive CLE genes, and ultimately match their functions to the supposed complex regulatory networks that integrating with phytohormones and/or environmental stimuli.
Author Contributions
GW conceived and wrote the manuscript; GZ and MW critically reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Research in our group was supported by the National Natural Science Foundation of China (31271575; 31200902), the Fundamental Research Funds for the Central Universities (GK201103005), the Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education of China (20120202120009), and the Natural Science Basic Research Plan in Shaanxi Province of China (2014JM3064).
References
- Araya T., Miyamoto M., Wibowo J., Suzuki A., Kojima S., Tsuchiya Y. N., et al. (2014). CLE-CLV1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111 2029–2034. 10.1073/pnas.1319953111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S., Sawa S., Yamada M. (2011). The function of the CLE peptides in plant development and plant-microbe interactions. Arabidopsis Book 9 e0149 10.1199/tab.0149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidadi H., Matsuoka K., Sage-Ono K., Fukushima J., Pitaksaringkarn W., Asahina M., et al. (2014). CLE6 expression recovers gibberellins deficiency to promote shoot growth in Arabidopsis. Plant J. 78 241–252. 10.1111/tpj.12475 [DOI] [PubMed] [Google Scholar]
- Cho H., Ryu H., Rho S., Hill K., Smith S., Audenaert D., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16 66–76. 10.1038/ncb2893 [DOI] [PubMed] [Google Scholar]
- Clouse S. D. (2011). Brassinosteroids. Arabidopsis Book 9 e0151 10.1199/tab.0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock J. M., McCormick S. (2001). A large family of genes that share homology with CLAVATA3. Plant Physiol. 126 939–942. 10.1104/pp.126.3.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N., Shi C. L., Vu L. D., Van De Cotte B., Hodgman C., Butenko M. A., et al. (2015). Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. J. Exp. Bot. 66 5229–5243. 10.1093/jxb/erv360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzewicz N., Yue K., Beeckman T., De Smet I. (2013). Message in a bottle: small signalling peptide outputs during growth and development. J. Exp. Bot. 64 5281–5296. 10.1093/jxb/ert283 [DOI] [PubMed] [Google Scholar]
- Depuydt S., Rodriguez-Villalon A., Santuari L., Wyser-Rmili C., Ragni L., Hardtke C. S. (2013). Suppression of Arabidopsis protophloem differentiation and root meristem growth by CLE45 requires the receptor-like kinase BAM3. Proc. Natl. Acad. Sci. U.S.A. 110 7074–7079. 10.1073/pnas.1222314110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S., Shinohara H., Matsubayashi Y., Fukuda H. (2013). A novel pollen-pistil interaction conferring high-temperature tolerance during reproduction via CLE45 signaling. Curr. Biol. 23 1670–1676. 10.1016/j.cub.2013.06.060 [DOI] [PubMed] [Google Scholar]
- Funayama-Noguchi S., Noguchi K., Yoshida C., Kawaguchi M. (2011). Two CLE genes are induced by phosphate in roots of Lotus japonicus. J. Plant Res. 124 155–163. 10.1007/s10265-010-0342-5 [DOI] [PubMed] [Google Scholar]
- Goda H., Sasaki E., Akiyama K., Maruyama-Nakashita A., Nakabayashi K., Li W., et al. (2008). The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J. 55 526–542. 10.1111/j.0960-7412.2008.03510.x [DOI] [PubMed] [Google Scholar]
- Guo H., Zhang W., Tian H., Zheng K., Dai X., Liu S., et al. (2015). An auxin responsive CLE gene regulates shoot apical meristem development in Arabidopsis. Front. Plant Sci. 6:295 10.3389/fpls.2015.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., et al. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. U.S.A. 105 15208-15213. 10.1073/pnas.0808444105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram G., Gutierrez-Marcos J. (2015). Peptide signalling during angiosperm seed development. J. Exp. Bot. 66 5151–5159. 10.1093/jxb/erv336 [DOI] [PubMed] [Google Scholar]
- Jun J., Fiume E., Roeder A. H., Meng L., Sharma V. K., Osmont K. S., et al. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154 1721–1736. 10.1104/pp.110.163683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50 347–363. 10.1111/j.1365-313X.2007.03052.x [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48 1821–1825. 10.1093/pcp/pcm154 [DOI] [PubMed] [Google Scholar]
- Kondo Y., Hirakawa Y., Kieber J. J., Fukuda H. (2011). CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52 37–48. 10.1093/pcp/pcq129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y., Ito T., Nakagami H., Hirakawa Y., Saito M., Tamaki T., et al. (2014). Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5 3504 10.1038/ncomms4504 [DOI] [PubMed] [Google Scholar]
- Krusell L., Madsen L. H., Sato S., Aubert G., Genua A., Szczyglowski K., et al. (2002). Shoot control of root development and nodulation is mediated by a receptor like kinase. Nature 420 422–426. 10.1038/nature01207 [DOI] [PubMed] [Google Scholar]
- Krusell L., Sato N., Fukuhara I., Koch B. E., Grossmann C., Okamoto S., et al. (2011). The Clavata2 genes of pea and Lotus japonicas affect autoregulation of nodulation. Plant J. 65 861–871. 10.1111/j.1365-313X.2010.04474.x [DOI] [PubMed] [Google Scholar]
- Lim C. W., Lee Y. W., Hwang C. H. (2011). Soybean nodule-enhanced CLE peptides in roots act as signals in GmNARK-mediated nodulation suppression. Plant Cell Physiol. 52 1613–1627. 10.1093/pcp/pcr091 [DOI] [PubMed] [Google Scholar]
- Ma X., Zhang Q., Zhu Q., Liu W., Chen Y., Qiu R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8 1274–1284. 10.1016/j.molp.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. (2011). Small post-translationally modified Peptide signals in Arabidopsis. Arabidopsis Book 9 e0150 10.1199/tab.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L., Feldman L. J. (2010). CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinase to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232 1061–1074. 10.1007/s00425-010-1236-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki K., Tabata R., Sawa S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis and parasitism. Curr. Opin. Plant Biol. 16 598–606. 10.1016/j.pbi.2013.08.008 [DOI] [PubMed] [Google Scholar]
- Miyazawa H., Oka-Kira E., Sato N., Takahashi H., Wu G. J., Sato S., et al. (2010). The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicas. Development 137 4317–4325. 10.1242/dev.058891 [DOI] [PubMed] [Google Scholar]
- Mortier V., De Wever E., Vuylsteke M., Holsters M., Goormachtig S. (2012). Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J. 70 367–376. 10.1111/j.1365-313X.2011.04881.x [DOI] [PubMed] [Google Scholar]
- Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D’Haeseleer K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153 222–237. 10.1104/pp.110.153718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24 3198–3217. 10.1105/tpc.112.099010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura R., Hayashi M., Wu G. J., Kouchi H., Imaizumi-Anraku H., Murakami Y., et al. (2002). HAR1 mediates systemic regulation of symbiotic organ development. Nature 420 426–429. 10.1038/nature01231 [DOI] [PubMed] [Google Scholar]
- Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., et al. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50 67–77. 10.1093/pcp/pcn194 [DOI] [PubMed] [Google Scholar]
- Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4 2191 10.1038/ncomms3191 [DOI] [PubMed] [Google Scholar]
- Pallakies H., Simon R. (2014). The CLE40 and CRN/CLV2 signaling pathways antagonistically control root meristem growth in Arabidopsis. Mol. Plant 7 1619–1636. 10.1093/mp/ssu094 [DOI] [PubMed] [Google Scholar]
- Reid D. E., Ferguson B. J., Gresshoff P. M. (2011). Inoculation– and nitrate– Induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 24 606–618. 10.1094/MPMI-09-10-0207 [DOI] [PubMed] [Google Scholar]
- Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. (2014). Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 5 4983 10.1038/ncomms5983 [DOI] [PubMed] [Google Scholar]
- Schnabel E., Journet E. P., de Carvalho-Niebel F., Duc G., Frugoli J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58 809–822. 10.1007/s11103-005-8102-y [DOI] [PubMed] [Google Scholar]
- Searle I. R., Men A. E., Laniya T. S., Buzas D. M., Iturbe-Ormaetxe I., Carroll B. J., et al. (2003). Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109–112. 10.1126/science.1077937 [DOI] [PubMed] [Google Scholar]
- Song X. F., Guo P., Ren S. C., Xu T. T., Liu C. M. (2013). Antagonistic peptide technology for functional dissection of CLV3/ESR genes in Arabidopsis. Plant Physiol. 161 1076–1085. 10.1104/pp.112.211029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Ito M., Kawaguchi M. (2013). Genetic basis of cytokinin and auxin functions during root nodule development. Front. Plant Sci. 4:42 10.3389/fpls.2013.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T., Yano K., Ito M., Umehara Y., Suganuma N., Kawaguchi M. (2012). Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139 3997–4006. 10.1242/dev.084079 [DOI] [PubMed] [Google Scholar]
- Whitford R., Fernandez A., De Groodt R., Ortega E., Hilson P. (2008). Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. U.S.A. 105 18625–18630. 10.1073/pnas.0809395105 [DOI] [PMC free article] [PubMed] [Google Scholar]