Abstract
Objective
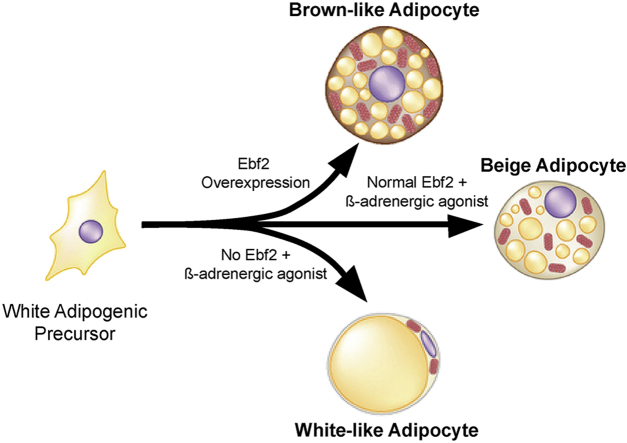
The induction of beige/brite adipose cells in white adipose tissue (WAT) is associated with protection against high fat diet-induced obesity and insulin resistance in animals. The helix-loop-helix transcription factor Early B-Cell Factor-2 (EBF2) regulates brown adipose tissue development. Here, we asked if EBF2 regulates beige fat cell biogenesis and protects animals against obesity.
Methods
In addition to primary cell culture studies, we used Ebf2 knockout mice and mice overexpressing EBF2 in the adipose tissue to study the necessity and sufficiency of EBF2 to induce beiging in vivo.
Results
We found that EBF2 is required for beige adipocyte development in mice. Subcutaneous WAT or primary adipose cell cultures from Ebf2 knockout mice did not induce Uncoupling Protein 1 (UCP1) or a thermogenic program following adrenergic stimulation. Conversely, over-expression of EBF2 in adipocyte cultures induced UCP1 expression and a brown-like/beige fat-selective differentiation program. Transgenic expression of Ebf2 in adipose tissues robustly stimulated beige adipocyte development in the WAT of mice, even while housed at thermoneutrality. EBF2 overexpression was sufficient to increase mitochondrial function in WAT and protect animals against high fat diet-induced weight gain.
Conclusions
Taken together, our results demonstrate that EBF2 controls the beiging process and suggest that activation of EBF2 in WAT could be used to reduce obesity.
Keywords: EBF2, Beige fat, Brown fat, Obesity, Thermogenesis
Abbreviations: BAT, brown adipose tissue; WAT, white adipose tissue; ingWAT, inguinal white adipose tissue; epiWAT, epididymal white adipose tissue; WT, wild type; TG, transgenic; HFD, high fat diet; TN, thermoneutrality; RT, room temperature
Graphical abstract
Highlights
-
•
Loss of Ebf2 prevents induction of beige adipocytes in inguinal WAT.
-
•
Ectopic expression of Ebf2 promotes beige fat induction in inguinal WAT.
-
•
Ectopic Ebf2 expression protects against high fat diet-induced obesity.
1. Introduction
Obesity is a major contributor to chronic illness and premature death in many countries. Unfortunately, there are limited therapeutic options to help people lose weight. A promising avenue to counteract weight gain is through increasing the activity of thermogenic brown and beige adipocytes [1]. Both of these cell types express high levels of Uncoupling Protein 1 (UCP1) in their mitochondria. When activated, UCP1 induces high rates of substrate oxidation with significant amounts of energy being expended in the form of heat [1]. Thermogenesis in brown and beige adipocytes is activated in response to cold-exposure and protects animals against hypothermia [2]. Additionally, UCP1 activity in brown and/or beige adipocytes regulates energy balance. Loss of UCP1 promotes weight gain in mice maintained at thermoneutrality and thus exempt from cold stress [3]. Conversely, animals with increased brown and beige fat activity are lean and have a healthy metabolic profile [4], [5], [6], [7], [8], [9]. Importantly, brown/beige fat activity levels in people are also correlated with leanness [10], [11], [12], [13].
Brown adipocytes develop in dedicated depots of brown adipose tissue (BAT) and express relatively high levels of UCP1 under a variety of environmental conditions. By contrast, beige adipocytes, which develop in WAT, are highly inducible [1]. In animals kept at normal vivarium conditions (∼22 °C), there are relatively few beige adipocytes. However, upon cold-exposure or chronic stimulation with β-adrenergic agonists such as CL 316, 243, there is a dramatic increase in beige fat cell recruitment and UCP1 levels. In mice, the cold-stimulated induction of beige adipocytes is particularly prominent in the inguinal WAT (ingWAT), a major subcutaneous fat depot. Importantly, recent studies have indicated that beige adipocyte activity affects systemic metabolism and contributes in a significant way to whole body insulin sensitivity [14], [15].
Brown adipocytes are distinct from the beige adipocytes found in ingWAT; they arise from separate lineages [16], but many studies have suggested that the differentiation and development of both cell types are controlled by a shared transcriptional program. Work by our lab and others has shown that the transcription factor PRDM16 is required for “beiging” of ingWAT and for the maintenance of BAT [6], [15], [17], [18]. PGC-1α, a transcriptional co-activator also plays important roles in both brown and beige fat cells [19], [20]. We recently identified Early B-cell factor-2 (EBF2) as a factor required for the BAT development [21]. Through an independent line of investigation, we also identified EBF2 as a selective marker of both brown and beige preadipose cells in embryonic and adult adipose depots [22]. This raised the question of whether EBF2 plays a critical role in beige fat development and function.
In this study, we report that EBF2 controls beige adipocyte development in WAT. We show that genetic loss of Ebf2 in mice blocks beige fat development and function without having any other obvious effects on WAT. EBF2 is required for the re-programming of primary ingWAT-derived preadipose cells into thermogenically competent beige/brown-like adipocytes in vitro. Finally, transgenic expression of Ebf2 in adipose tissue using the Fabp4 promoter/enhancer powerfully stimulates beige fat development and protects animals against high fat-diet-induced weight/fat gain. RNAseq analysis demonstrates that EBF2 robustly induces a mitochondrial gene program similar to that of cold-induced beige fat. Overall, our results demonstrate that EBF2 plays a central role in regulating beige fat function and could potentially be manipulated to improve overall systemic metabolism in mice.
2. Materials and methods
2.1. Animals
All animal experiments were performed in accordance with an approved University of Pennsylvania Institutional Animal Care and Use Committee protocol. Mice were fed standard chow unless otherwise specified and kept on a 12 h light/dark cycle. Ebf2 whole body knockout animals were obtained from R. Reed (Johns Hopkins, Baltimore, MD, USA) and have been described previously [23]. Fabp4-Ebf2 transgenic mice were generated by cloning full-length Ebf2 cDNA downstream of the 5.4 kb Fabp4 promoter/enhancer. A bovine growth hormone polyadenylation site was inserted 3′ to the Ebf2 cDNA. The construct was injected into FVB mouse oocytes by the University of Pennsylvania Transgenic and Chimeric Mouse Facility. For CL 316,243 treatment, Ebf2 knockout mice and wild type (WT) littermate controls were raised at thermoneutrality (30 °C) and injected with 1 mg/kg CL 316,243 diluted in PBS every day for 6 days. Mice were sacrificed for analysis on day 7. For weight gain analysis, Fabp4-Ebf2 mice were housed at thermoneutrality and fed a 45% high-fat diet (Research Diets).
2.2. Cell culture
Primary adipogenic precursor cells were isolated from mouse ingWAT as described previously [24]. For adipocyte differentiation assays, cells were grown to confluence and then treated with differentiation induction medium (DMEM/F12 with 10% FBS, 1 μM dexamethasone, 0.5 mM isobutylmethylxanthine, 125 nM indomethacin, 20 nM insulin, and 1 nM T3). Induction medium was removed after 48 h and cells were then grown in DMEM/F12 medium containing 10% FBS, 1 nM T3 and 20 nM insulin until they were harvested. To stimulate a thermogenic program, cells were incubated either with 10 μM isoproterenol for 3 h or with 1 μM rosiglitazone for the entire differentiation time course. For Oil Red O staining, cell cultures were washed with PBS, fixed with 4% paraformaldehyde for 15 min and stained with Oil Red O solution for 30 min. All primary cell experiments were done using three technical replicates.
2.3. Western blot analysis
Cells or tissues were lysed in RIPA buffer containing 0.5% NP-40, 0.1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-Cl, pH 7.5, protease inhibitor cocktail (Complete; Roche) and 1 mM phenylmethylsulfonyl fluoride (PMSF). Protein concentrations were quantified using detergent-compatible (DC) protein assay kit (Bio-Rad). Lysates or nuclear fractions were run on Bis-Tris NuPAGE gels (Invitrogen), transferred to PVDF membrane (Millipore), and probed with primary antibody. Antibodies used were sheep anti-Ebf2 (R&D systems, AF7006), mouse anti-Ucp1 (R&D systems, MAB6158), rabbit anti-VDAC (Cell Signaling, 4866), mouse anti-actin (clone C4, Millipore, MAB1501) and mouse anti-tubulin (Sigma, T6199).
2.4. RT-qPCR analysis
Total RNA was isolated from cultured cells or whole tissue using TRIzol extraction (Invitrogen). RNA was then purified using PureLink RNA columns (Life Technologies). RNA was reverse transcribed to generate cDNA using a High-Capacity cDNA Synthesis kit (Applied Biosystems). Real-time PCR was used to quantify transcript levels using SYBR Green master mix (Applied Biosystems) on a 7900 Fast HT real time PCR machine (Applied Biosystems). For all real-time PCR, Tata-binding protein (Tbp) was used as an internal normalization control. P-values were determined in Excel using a two-tailed T-test. Primer sequences are in Table S1.
2.5. Histology
For immunohistochemistry, whole inguinal fat pads were fixed in 4% PFA overnight, dehydrated, and embedded in paraffin for sectioning. Sections were stained with hematoxylin and eosin using standard methods. For whole mount immunofluorescence staining of fat pads, mice were transcardially perfused with 2% paraformaldehyde in PBS. Inguinal fat pads were removed and 6 mm punch biopsies were taken from the fat pad region immediately adjacent to the lymph node. Tissues were incubated in 4% paraformaldehyde overnight, washed with PBS and blocked in PBS with 5% Normal Donkey Serum, 0.5% Triton X at 4 °C overnight. Tissues were incubated in primary antibody diluted in blocking solution overnight and secondary antibody diluted in block for 2 h at room temperature. Tissues were mounted on concavity slides in Prolong Gold antifade mounting medium. Slides were imaged on a Leica TCS Sp8 confocal microscope. Primary antibodies used were rabbit anti-UCP1 (1:1000, AZ) and sheep anti-EBF2 (1:200, R&D Systems). Secondary Alexa fluor-conjugated antibodies were from Life Technologies.
2.6. Tissue O2 consumption
A 6 mm punch biopsy was taken from each ingWAT sample directly below the lymph node. For BAT, one lobe was used. Tissues were weighed and minced in a respiration buffer (2% BSA, 1.1 mM sodium pyruvate, and 25 mM glucose in PBS). Oxygen consumption was measured for each sample in an MT200A Respirometer Cell (Strathkelvin) until oxygen consumption rate was stable (for approximately 5 min). Oxygen consumption rates were normalized to tissue weight. Two measurements were taken from each mouse.
2.7. RNAseq
An RNAseq library was prepared using isolated RNA from whole ingWAT pads with lymph nodes removed using the Illumina TrueSeq Library preparation kit. High-throughput sequencing was performed using a HiSeq 2000. All the high throughput sequencing was performed by the Functional Genomics Core at the University of Pennsylvania using HiSeq 2000 or 2500. RNA-seq reads were aligned to the UCSC mm9 using RUM [25]. Differential gene expression analysis was performed using edgeR [26], where differentially regulated genes with FDR < 0.05 were selected for downstream analysis. Hierarchical clustering was performed based on log2-scaled gene expression value using 1− (correlation coefficient) as a distance measure and following Ward's criterion. Gene ontology analysis was done for up or down regulated genes separately using DAVID [27], [28] and significantly overrepresented terms from GOTERM_CC_FAT category were selected for presentation. Sequence data has been deposited into the Gene Expression Omnibus (GEO) database under the accession number GSE74366.
3. Results
3.1. EBF2 is required for beige adipocyte development
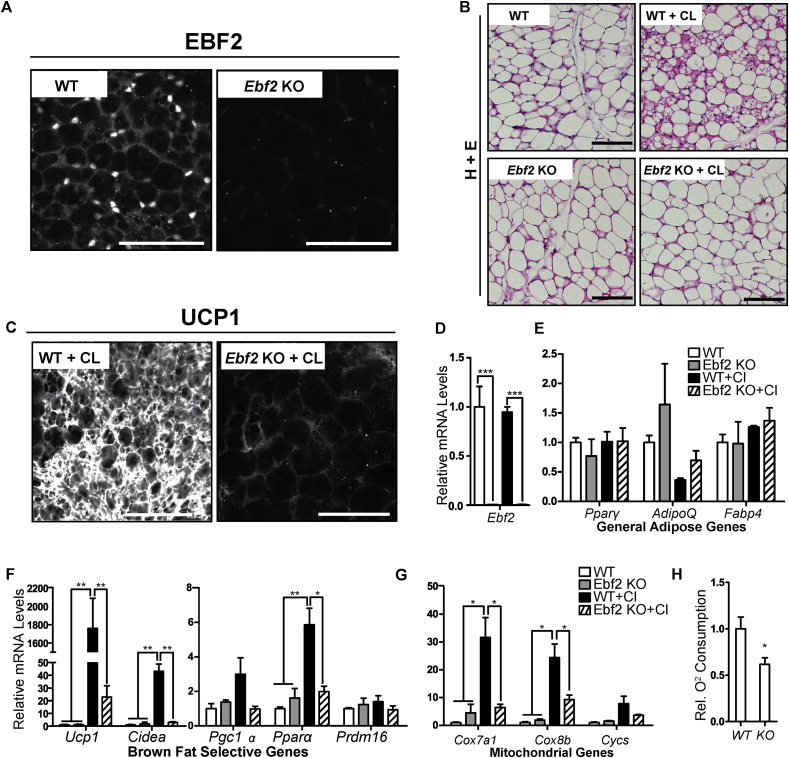
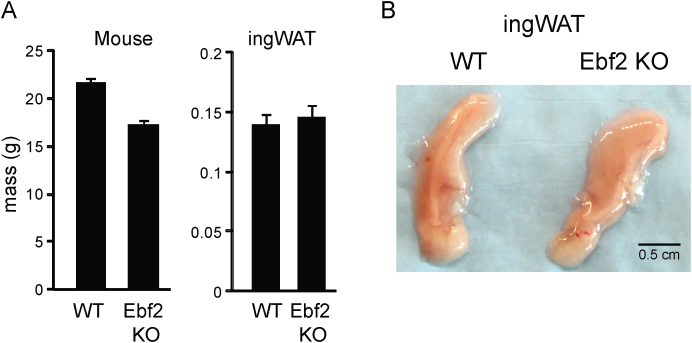
To determine if EBF2 regulates beige adipocyte development, we analyzed the beiging response of ingWAT in WT and Ebf2 knockout (KO) animals. After outcrossing of Ebf2 KO mice into the C57Black6 background, we found that, in contrast to previous studies [21], [23], the KO mice survive into adulthood at approximately 40% of expected Mendelian ratios when they are bred at thermoneutrality. The KO mice used in our experiments had slightly though not significantly lower body weights (Figure S1A). ingWAT pads from Ebf2 KO mice were comparable in both appearance and mass to WT (Figure S1B).
To stimulate the beiging of WAT, we treated WT and KO animals with CL 316,243 (CL), a β3-selective adrenergic agonist [29]. We confirmed by immunofluorescent staining a complete loss of EBF2 protein in the KO ingWAT (Figure 1A). Hematoxylin and eosin (H&E) staining prior to CL-treatment showed that WT and Ebf2 KO ingWAT had a similar appearance, consisting mostly of large adipocytes with unilocular lipid droplets (Figure 1B). Following CL treatment, WT ingWAT showed regions of dense, multilocular adipocytes that were absent in CL-treated Ebf2 KO ingWAT and non-treated controls (Figure 1B). Moreover, CL-treatment dramatically increased UCP1 protein expression in WT ingWAT with minimal induction of UCP1 in Ebf2 KO ingWAT (Figure 1C).
Figure 1.
Ebf2 is required for in vivo beiging of ingWAT. (A) Confocal immunofluorescence imaging of whole-mount ingWAT tissue. EBF2 protein is present in wild type (WT) but not Ebf2 KO ingWAT. (B) H&E staining of ingWAT from WT and Ebf2 KO mice before (left panels) or after (right panels) injection with CL-316,243 to induce beiging. (C) Confocal immunofluorescence imaging of whole-mount ingWAT tissue. UCP1 protein is highly expressed in WT but not Ebf2 KO IngWAT following CL 316,243 injection. (D–G) Relative mRNA levels from in WT or Ebf2 knockout ingWAT before and after CL 316,243 injection as measured by RT-qPCR. Levels of (D)Ebf2, (E) the general adipose genes Pparϒ, AdipoQ and Fabp4, (F) the brown selective genes Ucp1, Cidea, Pgc1α, Pparα and Prdm16, and (G) the mitochondrial genes Cox7a1, Cox8b, Cycs. (n = 3). (H) Relative oxygen consumption of ingWAT from WT and Ebf2 KO mice. (n = 4). P-values are represented with asterisks (*, p-value ≤ .05; **, p-value ≤ .01; ***, p-value ≤ .001) and error bars represent SEM. Scale bars = 10 microns.
To better characterize the beiging response, we used RT-qPCR to measure the expression levels of key genes involved in adipose function in WT and KO mice before and after CL injection. CL-treatment of WT mice did not affect Ebf2 mRNA levels (Figure 1D). The levels of general adipogenic genes such as Pparγ, AdipoQ and Fabp4 were not significantly affected by CL treatment, and levels of these factors were similar between WT and Ebf2 KO tissue (Figure 1E), suggesting that EBF2 does not play a major role in general adipose development and maintenance. However, the CL-mediated induction of brown fat-selective genes like Ucp1, Cidea, Pgc1α and Pparα was severely blunted or nonexistent in Ebf2 KO animals (Figure 1F). For example, CL increased Ucp1 mRNA levels ∼1,700-fold in WT ingWAT and only ∼20-fold in KO tissue. The CL-mediated induction of Cidea, Pparα and Pgc1α expression was completely dependent on Ebf2 (Figure 1F). Prdm16 mRNA levels were not affected by Ebf2 deletion or CL-treatment (Figure 1F).
Because the beiging process is also characterized by increased mitochondrial biogenesis and function, we assessed the levels of the mitochondrial genes and mitochondrial function in WT and Ebf2 KO ingWAT. We found that the CL-mediated induction of mitochondrial genes (e.g. Cox7a1, Cox8b and Cytochrome C (Cycs)) was severely blunted or completely blocked by Ebf2-deficiency (Figure 1G). Consistent with the reduced expression levels of mitochondrial genes in Ebf2 KO tissue, we found that oxygen consumption from Ebf2 KO ingWAT was ∼40 percent less than oxygen consumption in WT ingWAT in mice that were housed at room temperature (Figure 1H). Taken together, these data show that Ebf2 is required for induction of the beige fat-selective program in ingWAT.
3.2. The requirement for EBF2 in beige fat programming is adipose cell-autonomous
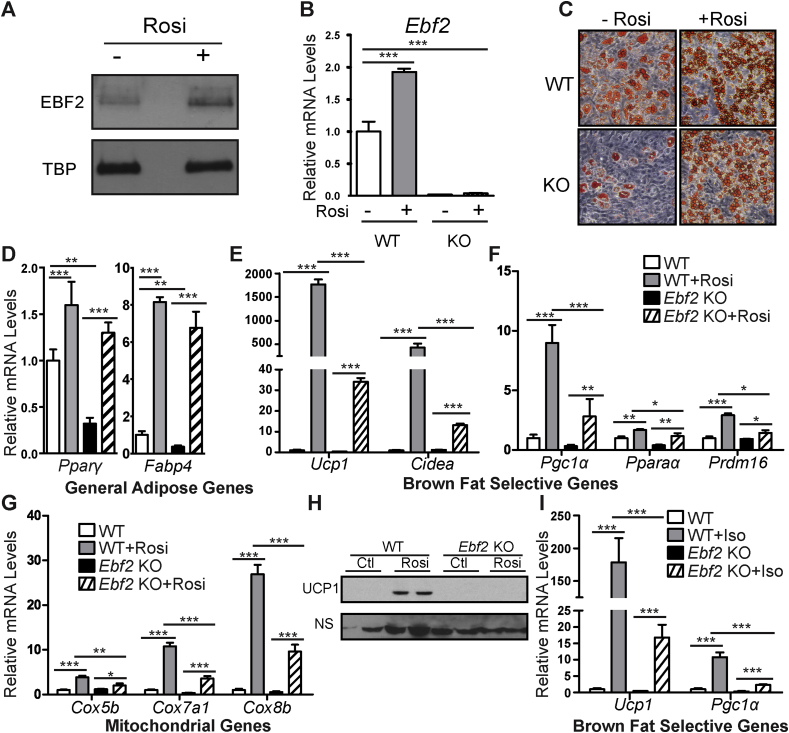
We next examined whether EBF2 was required for the differentiation of beige adipocytes in a cell culture model. Primary adipogenic precursor cells from ingWAT can be induced to activate the brown/beige fat program in response to chronic treatment with the thiazolidinedione, rosiglitazone (rosi) [30], [31]. Notably, rosi treatment increased EBF2 mRNA and protein levels by ∼2-fold during the adipogenic differentiation of ingWAT-derived precursor cells (Figure 2A–B). To determine the requirement for Ebf2 in the beiging process, we isolated and cultured WT and Ebf2 KO ingWAT preadipose cells and confirmed that the KO cells did not express Ebf2 (Figure 2B) These cultures were induced to undergo adipocyte differentiation and treated with rosi to induce beiging. Under basal conditions (without rosi), Ebf2 KO cells differentiated into adipocytes less efficiently than WT cells as revealed by Oil Red O staining of lipid accumulation (Figure 2C). Consistent with reduced adipocyte differentiation, Ebf2 KO cultures expressed lower levels the pan-adipocyte genes Pparγ and Fabp4 (Figure 2D). This morphological and molecular differentiation defect was rescued, almost completely, by rosi treatment (Figure 2C–D). As anticipated, rosi treatment dramatically increased the expression of brown fat-selective genes (Ucp1, Cidea, Pgc1α, Pparα, and Prdm16) in WT adipocytes (Figure 2E–F). However, there was a severely blunted induction of these genes by rosi in Ebf2 KO cells (Figure 2E–F). For example, rosi induced Ucp1 expression by > 1500-fold in WT cells and by only 30-fold in KO cells (Figure 2E). The rosi-mediated increases in the expression levels of mitochondrial genes (Cox5b, Cox7a1, Cox8b) were also drastically diminished in KO relative to WT cells (Figure 2G). Western blot analysis showed that rosi treatment induced UCP1 protein expression in WT but not in Ebf2 KO adipocyte cultures (Figure 2H).
Figure 2.
Ebf2 is required for beiging. (A) Relative EBF2 protein levels from WT primary ingWAT stromal vascular cells plus or minus rosi treatment as assayed by western blot. (B) Relative Ebf2 mRNA levels from WT or Ebf2 KO primary ingWAT stromal vascular cells before and after rosi treatment as measured by RT-qPCR. (C) Oil Red O staining of differentiated and rosi treated WT or Ebf2 KO primary ingWAT stromal vascular cells. (D–G) Relative mRNA levels from WT or Ebf2 KO primary ingWAT stromal vascular cells plus or minus rosi treatment as measured by RT-qPCR. Levels of (D) the general adipose genes Pparϒ and Fabp4, (E) the brown selective genes Ucp1 and Cidea, (F) the brown selective genes Pgc1α, Pparα and Prdm16, and (G) the mitochondrial genes Cox5b, Cox7a1, Cox8b. (H) Western blot to measure protein levels of UCP1 in differentiated and rosi treated WT or Ebf2 knockout primary ingWAT stromal vascular cells. NS = non-specific band loading control. (I) Relative Ucp1 and Pgc1α mRNA levels from WT or Ebf2 knockout primary ingWAT stromal vascular cells before and after Iso treatment as measured by RT-qPCR. P-values are represented with asterisks (*, p-value ≤ .05; **, p-value ≤ .01; ***, p-value ≤ .001) and error bars represent standard deviation.
Thermogenic genes such as Ucp1 are also acutely increased in mature brown/beige adipocytes in response to β-adrenergic agonists. To test if this requires Ebf2, we treated differentiated WT and KO primary adipocytes with the synthetic pan β-adrenergic agonist Isoproterenol (iso) for 3 h. Iso-stimulation robustly increased the expression levels of Ucp1 and Pgc1α in WT adipocytes but only led to small increases in Ebf2 KO adipocytes (Figure 2I). Altogether, these results reveal a critical requirement for Ebf2 in the rosi- or β-agonist-mediated browning response of cultured adipocytes and strongly suggest that EBF2 acts in an adipose cell-autonomous manner to direct beige adipocyte development.
3.3. Ectopic expression of EBF2 is sufficient to drive conversion of white adipocytes to brown-like adipocytes
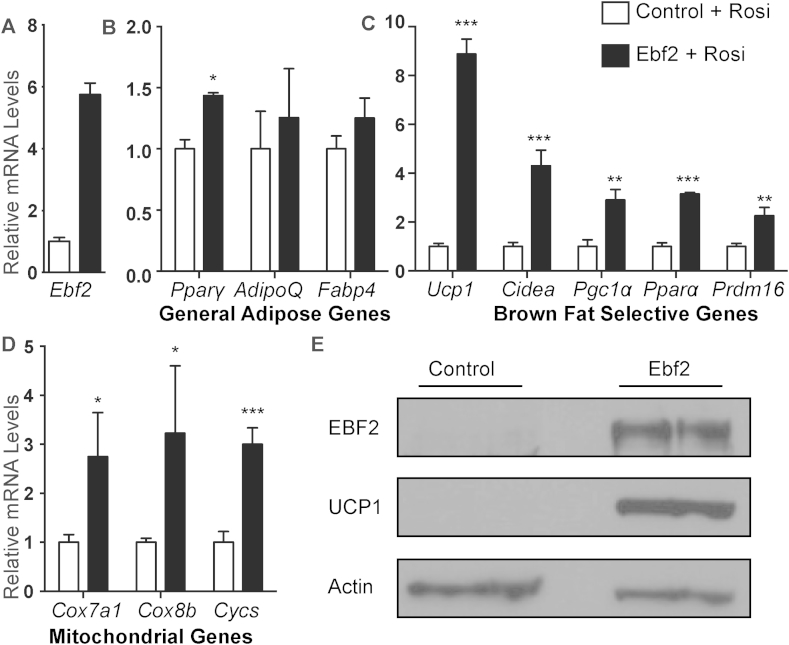
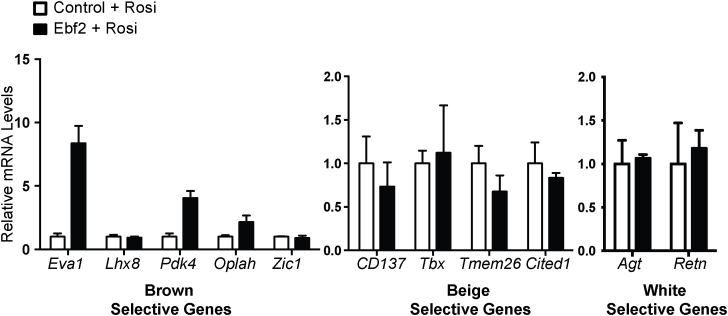
To determine if EBF2 is able to drive beige fat differentiation, we used retrovirus to overexpress EBF2 in primary ingWAT preadipocytes isolated from adult mice and treated with rosi (Figure 3A). EBF2-expression resulted in a subtle increase in the levels of the general adipogenic marker Pparγ although levels of Adipoq and Fabp4 were unchanged (Figure 3B), suggesting that EBF2 overexpression has very little impact on adipocyte differentiation. By contrast, EBF2 dramatically increased the levels of brown fat-selective and mitochondrial genes, including: Ucp1, Cidea, Pgc1α, Pparα, Prdm16, Cox7a1, Cox8b and Cycs as compared to control cells (Figure 3C–D). While rosi treatment normally induced a large increase in Ucp1 mRNA expression in primary ingWAT, EBF2-overexpression increased UCP1 even further at both the RNA (Figure 3C) and the protein levels (Figure 3E). Finally, we measured the levels of brown and beige adipocyte-selective marker genes [16], [32] in control and EBF2-expressing adipocytes. We found that while expression of EBF2 induced some brown-specific marker genes (Eva1, Pdk4, Oplah), it did not significantly increase in beige-specific genes (Figure S2). Further, EBF2-expression did not reduce classical markers of white fat including Agt and Retn in these cultures (Figure S2). Overall, we conclude that EBF2 drives a brown fat-like differentiation program when expressed in inguinal white preadipose cells.
Figure 3.
EBF2-expression drives a brown fat/thermogenic profile in primary adipocytes. (A–C) Relative mRNA levels from control or retroviral EBF2-expressing primary ingWAT stromal vascular cells with or without rosi treatment as measured by RT-qPCR. Levels of (A) Ebf2, (B) the general adipose genes Pparϒ, AdipoQ and Fabp4, and (C) the brown selective genes Ucp1, Cidea, Pgc1α, Pparα and Prdm16, and (D) the mitochondrial genes Cox7a1, Cox8b, Cycs. (E) Western blot to measure protein levels of EBF2 and UCP1 levels in control or retroviral EBF2-expressing primary ingWAT stromal vascular cells following rosi treatment. P-values are represented with asterisks (*, p-value ≤ .05; **, p-value ≤ .01; ***, p-value ≤ .001) and error bars represent standard deviation.
3.4. Increased EBF2 activity in adipose tissue drives beiging
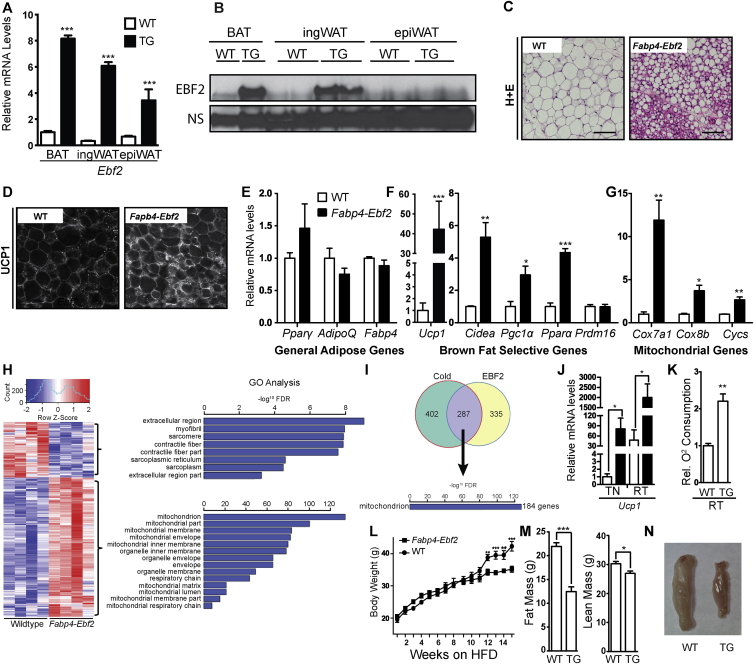
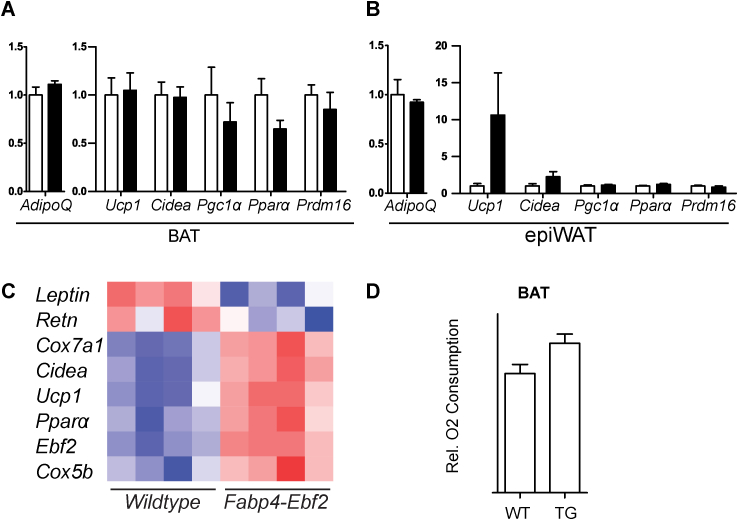
To determine if increasing EBF2 expression in WAT promotes beige adipocyte biogenesis, we analyzed transgenic mice that ectopically express Ebf2 under control of the Fabp4 enhancer/promoter, which drives expression in an adipose-selective manner. We first studied Fabp4-Ebf2 and WT littermate adult male mice at room temperature. RT-qPCR analysis showed that Ebf2 mRNA was elevated by between 4 and 8 fold in BAT, ingWAT and epididymal WAT (epiWAT) (Figure 4A). Western blot analysis indicated that EBF2 protein was overexpressed in BAT and ingWAT but not in epiWAT (Figure 4B). We first focused on the characterization of EBF2 overexpression in ingWAT. H&E staining revealed that the ingWAT of Fabp4-Ebf2 mice accumulated large numbers of multilocular adipocytes, while the ingWAT of WT littermates had mostly large adipocytes with unilocular lipid inclusions (Figure 4C). Immunofluorescence staining showed that ingWAT from Fabp4-Ebf2 mice displayed much higher UCP1 protein expression than ingWAT from WT littermates (Figure 4D). RT-qPCR analyses showed that transgenic expression of EBF2 did not alter the expression levels of general adipose markers (Ppary, AdipoQ or Fabp4) in ingWAT, indicating that the increased EBF2 expression does not affect overall development or maintenance of adipocytes (Figure 4E). However, in ingWAT, EBF2-overexpression led to a marked increase in the expression of many brown fat-selective genes including a ∼40-fold increase in Ucp1 and 3- to 15-fold elevated levels of Cidea, Pgc1α, Ppara and mitochondrial genes (Cox7a, Cox8b, Cycs) relative to their levels in WT ingWAT (Figure 4F–G). Analysis of BAT and epiWAT tissues overexpressing EBF2 showed no significant change in mRNA levels for either general adipose genes or levels of brown fat-selective genes (Figure S3A–B). This indicates that overexpressing EBF2 in adipose tissue primarily affects the ingWAT depot, which is the depot that is most permissive to beiging.
Figure 4.
Ectopic ingWAT expression of EBF2 induces beiging in vivo. (A) qPCR of relative Ebf2 mRNA levels from WT (white bar) and Fabp4-Ebf2 (black bar) mice in BAT, ingWAT and epiWAT (mice housed at room temperature). (n = 3). (B) Western blot to measure EBF2 protein levels in WT or Fabp4-Ebf2 (TG) mice in ingWAT, BAT and epiWAT (mice housed at room temperature). (C) H&E staining of ingWAT from WT and Fabp4-Ebf2 mice. (D) Confocal immunofluorescence imaging of UCP1 protein levels in ingWAT from WT or Fabp4-Ebf2 mice. (E–G)) Relative mRNA levels from WT and Fabp4-Ebf2 ingWAT as measured by RT-qPCR. Levels of (E) the general adipose genes Pparϒ, AdipoQ and Fabp4, (F) the brown selective genes Cidea, Pgc1α, Pparα and Prdm16 and (G) mitochondrial genes Cox7a1, Cox8b, Cycs. (H) Heat map of the top up-regulated and down-regulated genes in Fabp4-Ebf2 ingWAT compared to WT, with corresponding GO analysis. (I) Cross comparison between RNAseq data comparing WT and Fabp4-Ebf2 ingWAT gene expression and microarray comparing thermoneutral and cold exposed ingWAT. The majority of overlapping upregulated genes are related to mitochondrial function. (J) Relative UCP1 mRNA levels of WT (white bars) and Fabp4-Ebf2 (black bars) ingWAT from mice housed at thermoneutrality (TN, 30 °C) or room temperature (RT) as measured by RT-qPCR. (K) Relative oxygen consumption of ingWAT from WT and TG Fabp4-Ebf2 mice housed at room temperature, n = 4. (L) Body weight of wild type (square) and TG Fabp4-Ebf2 (circle) for mice housed at thermoneutrality on HFD beginning at 5 weeks of age. (n = 5). (M) Fat mass and lean weight mass of WT or TG Fabp4-Ebf2 (n = 5) mice after 25 weeks on HFD at TN. (N) Gross histology of ingWAT fat pads from WT or TG Fabp4-Ebf2 mice after 25 weeks on HFD at TN. P-values are represented with asterisks (*, p-value ≤ .05; **, p-value ≤ .01; ***, p-value ≤ .001) and error bars denote SEM. Scale bars = 10 microns.
We then further examined global transcriptional effect of EBF2-overexpression using RNA-sequencing (RNAseq) to compare the gene expression profiles of ingWAT from WT and Fabp4-Ebf2 mice (Figure 4H–I). RNAseq analysis showed that EBF2 activated a brown fat gene program, matching what we observed by RT-qPCR (Figure 4F–G, Figure S3C). For example, the levels of white fat-specific genes, including Retn and Leptin were decreased by EBF2-expression while levels of brown fat-specific and mitochondrial genes such as Ucp1, Cidea, Cox7a1, Cox8b, and Pparα were increased. Gene Ontology analysis showed a high enrichment of metabolism and mitochondrial genes among the genes upregulated in Fabp4-Ebf2 tissue (Figure 4H). Many of the genes induced by EBF2-overexpression in ingWAT were also previously shown to be induced in WAT by environmental cold-exposure (Figure 4I) [33].
All the above analyses were performed in animals that were housed at room temperature. Under these conditions, the expression of Ucp1 and other thermogenic/mitochondrial genes in ingWAT are stimulated, to some degree, by sympathetic nerve activity. To determine if the EBF2-mediated induction of thermogenic programming in ingWAT depends on sympathetic nerve activity, we studied WT and Fabp4-Ebf2 mice that were housed at thermoneutrality to exempt the animals from cold stress. As anticipated, Ucp1 expression in the ingWAT of WT mice was much higher at room temperature (∼30-fold) as compared to thermoneutrality (Figure 4J). Interestingly, EBF2-expression led to a larger fold increase in Ucp1 levels under thermoneutral conditions than under room temperature condition (∼80 fold vs. ∼40 fold). This result shows that EBF2 can induce thermogenic gene expression in the absence of thermal stress (Figure 4J).
3.5. EBF2-expression increases mitochondrial activity in ingWAT and counteracts high fat diet-induced weight gain
To determine if elevated EBF2 levels increased mitochondrial activity in ingWAT, we measured the oxygen consumption of isolated ingWAT from WT and Fabp4-Ebf2 mice housed at room temperature using a Clark electrode. We found that the transgenic tissue had a 2.5-fold higher rate of oxygen consumption than WT tissue (Figure 4K). Notably, oxygen consumption of BAT was not increased in the transgenic animals (Figure S3D). These results further suggest that increased EBF2 expression selectively increases the mitochondrial activity of ingWAT.
We next examined if Fabp4-Ebf2 animals were protected against high fat diet (HFD) -induced weight gain. We placed WT and Fabp4-Ebf2 mice on a HFD at thermoneutrality (30 °C) beginning at 5-weeks of age. After 12-weeks, Fabp4-Ebf2 mice showed significantly reduced weight gain compared to WT mice (Figure 4L). By 15-weeks on a HFD, WT mice showed a percent weight gain of 116% while Fabp4-Ebf2 had a percent weight gain of only 71%. NMR studies of body composition showed that Fabp4-Ebf2 mice had ∼50% reduced fat mass with only a slight reduction in lean mass as compared to WT controls, and ingWAT pads were visibly reduced in size (Figure 4M,N). These results demonstrate that overexpression of EBF2 in ingWAT can suppress HFD-induced weight gain.
4. Discussion
The storage of excess energy in WAT is associated with many health problems including heart disease, diabetes and cancer. The thermogenic capacity of brown fat has attracted interest as a potential target for anti-obesity therapy [1]. While many mammals, including human infants, have interscapular brown fat, adult humans also have additional depots that contain beige adipocytes [16], [32], [34], [35]. Our lack of understanding of the factors that control the induction of beige fat limits our ability to target beige adipocytes for anti-obesity therapy. In this study, we show that EBF2 is a critical regulator of beige fat development and that its overexpression in adipose tissues activates a thermogenic and anti-obesity program.
EBF2 is selectively expressed in brown and beige adipogenic precursor cells relative to muscle or white fat precursors [22]. Moreover, EBF2 regulates the expression of many preadipose-selective genes at the precursor stage, suggesting that it has an important role in preadipocyte commitment. In mature adipocytes, EBF2 directly regulates the expression of Ucp1 and other brown fat-specific genes [21]. An interesting question is whether EBF2 drives the beiging process mainly through functions in adipogenic precursors and/or adipocytes. The Fabp4 promoter is active in mature adipocytes and in a subset of adipogenic precursors [36]. Therefore, beige induction in the WAT depots of Fabp4-Ebf2 mice may depend on EBF2 activation before or during differentiation as well as in mature adipocytes. Future studies will determine if EBF2-activation in a fully mature adipocyte can stimulate thermogenic programming in vivo.
EBF2 cooperates with the master adipocyte regulator PPARγ at enhancers of brown fat-selective genes to activate transcription [21]. Synthetic PPARγ activators, particularly in the TZD class, are among the most effective inducers of the beige fat differentiation program in mouse and human adipocytes [30], [31], [37], [38], [39], [40]. Interestingly, we found that rosi treatment and EBF2-expression synergized to drive beiging in cultured adipocytes. A recent study suggests that rosi drives the beige program through activation of SIRT1, a de-acetylase [8]. Mechanistically, SIRT1 de-acetylates PPARγ, which can then recruit PRDM16 and activate brown fat genes. In light of our findings, it would be interesting to examine whether de-acetylated PPARγ has higher affinity for EBF2-bound enhancer regions in brown/beige-fat selective genes.
Elevated EBF2 activity stimulates beiging in ingWAT but does not increase the levels of UCP1 and other brown fat genes in the BAT. This suggests that increases in the thermogenic capacity of ingWAT and possibly other WAT depots are responsible for reducing weight gain in Fabp4-Ebf2 animals. In our study, Fabp4-Ebf2 mice gained less weight than WT animals while housed at thermoneutrality and fed a HFD. HFD-induced thermogenesis, which depends on UCP1-function, is known to make an important contribution to energy expenditure in mice that are housed at thermoneutrality [3]. We assume that HFD activates UCP1 and thermogenesis in EBF2-induced beige fat, presumably via increases in sympathetic outflow to adipose. Future studies will determine if EBF2-activation in adipose produces additional systemic metabolic benefits beyond its effects on body weight.
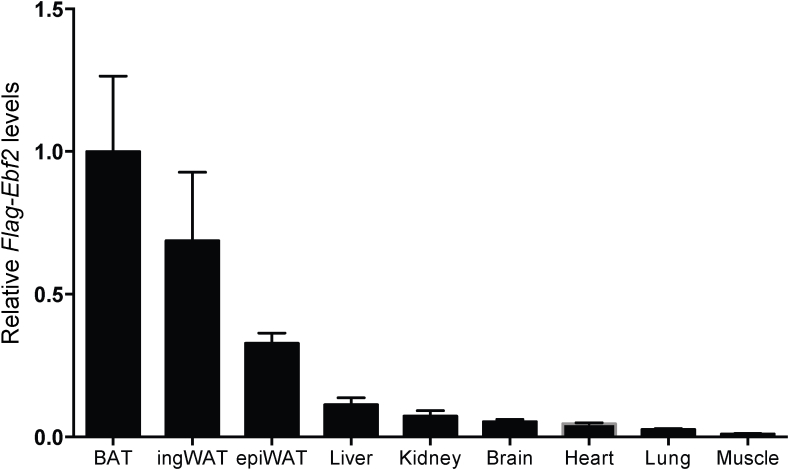
The epididymal WAT of Fabp4-Ebf2 mice had increased Ebf2 mRNA but not protein expression. This is reminiscent of analogous results obtained using a similar Fabp4 cassette to express Prdm16 and Zfp516 in adipose tissues [6], [7]. In our work on Fabp4-Prdm16 mice [6], we speculated that PRDM16 might be translated less efficiently or de-stabilized in the epididymal depot for interesting biological reasons. But, given that at least three cDNAs expressed from a related transgene construct display the same behavior, it is more likely that the 3′ UTR (e.g. 3′ UTR from Bovine growth hormone) may somehow prevent efficient protein translation in epididymal WAT. This means that the effect of overexpressing beiging factors like PRDM16, ZFP516 or EBF2 in epididymal WAT remains unknown but of substantial interest. It is also important to point out that while the Fabp4 promoter/enhancer cassette is an extremely useful and convenient tool to drive transgene expression in adipose tissue, Fabp4 expression is not restricted to adipose tissue [41], [42]. In line with this, we found very low but detectable levels of transgene expression in non-adipose tissues (Figure S4). Thus, while the reduced weight gain in Fabp4-Ebf2 mice is very likely due to the adipose-autonomous action of EBF2, we cannot rule out the possibility that ectopic expression of EBF2 in other tissues affects the metabolism of these mice.
In summary, we have established that EBF2 is a powerful regulator of brown and beige fat biogenesis and that activation of EBF2 in adipose tissue has anti-obesity effects. Although the role of EBF2 in human body weight regulation remains to be explored, genome-wide association studies have identified a SNP (rs10503776) in exon 11 of EBF2 that is highly associated with an increase in mean BMI [43], indicating that the role of EBF2 in energy balance might be conserved in humans. Increased brown/beige fat activity in humans correlates with decreased BMI [12], so promising targets to increase the activity of these tissues in people could be of great therapeutic value.
Acknowledgments
We would like to thank the UPenn Transgenic and Chimeric Mouse Facility, the UPenn Next Generation Sequencing Core and the UPenn Histology and Gene Expression Core. We thank R. Reed for providing Ebf2 KO mice. Our funding was provided by NIDDK grants 5R01DK10300802 (P. Seale) and 1F32DK10574301 (R. Stine).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2015.11.001.
Conflict of interest
All authors declare no conflict of interest.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Harms M., Seale P. Brown and beige fat: development, function and therapeutic potential. Nature Medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 2.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 3.Feldmann H.M., Golozoubova V., Cannon B., Nedergaard J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metabolism. 2009;9:203–209. doi: 10.1016/j.cmet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Cederberg A., Gronning L.M., Ahren B., Tasken K., Carlsson P., Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 5.Kopecky J., Clarke G., Enerback S., Spiegelman B., Kozak L.P. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. Journal of Clinical Investigation. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seale P., Conroe H.M., Estall J., Kajimura S., Frontini A., Ishibashi J. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. Journal of Clinical Investigation. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dempersmier J., Sambeat A., Gulyaeva O., Paul S.M., Hudak C.S., Raposo H.F. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Molecular Cell. 2015;57:235–246. doi: 10.1016/j.molcel.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiang L., Wang L., Kon N., Zhao W., Lee S., Zhang Y. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDonald M.E., Li C., Bian H., Smith B.D., Layne M.D., Farmer S.R. Myocardin-related transcription factor a regulates conversion of progenitors to beige adipocytes. Cell. 2015;160:105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushita M., Yoneshiro T., Aita S., Kameya T., Sugie H., Saito M. Impact of brown adipose tissue on body fatness and glucose metabolism in healthy humans. International Journal of Obesity. 2014;38:812–817. doi: 10.1038/ijo.2013.206. [DOI] [PubMed] [Google Scholar]
- 11.Saito M., Okamatsu-Ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-Kobayashi J. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. The New England Journal of Medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. The New England Journal of Medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 14.Qiu Y., Nguyen K.D., Odegaard J.I., Cui X., Tian X., Locksley R.M. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen P., Levy J.D., Zhang Y., Frontini A., Kolodin D.P., Svensson K.J. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J., Bostrom P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harms M.J., Ishibashi J., Wang W., Lim H.W., Goyama S., Sato T. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metabolism. 2014;19:593–604. doi: 10.1016/j.cmet.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harms M.J., Lim H.W., Ho Y., Shapira S.N., Ishibashi J., Rajakumari S. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Development. 2015;29:298–307. doi: 10.1101/gad.252734.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner S., Mepani R.J., Laznik D., Ye L., Jurczak M.J., Jornayvaz F.R. Development of insulin resistance in mice lacking PGC-1alpha in adipose tissues. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9635–9640. doi: 10.1073/pnas.1207287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajakumari S., Wu J., Ishibashi J., Lim H.W., Giang A.H., Won K.J. EBF2 determines and maintains brown adipocyte identity. Cell Metabolism. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W., Kissig M., Rajakumari S., Huang L., Lim H.W., Won K.J. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang S.S., Lewcock J.W., Feinstein P., Mombaerts P., Reed R.R. Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development. 2004;131:1377–1388. doi: 10.1242/dev.01009. [DOI] [PubMed] [Google Scholar]
- 24.Seale P., Kajimura S., Yang W., Chin S., Rohas L.M., Uldry M. Transcriptional control of brown fat determination by PRDM16. Cell Metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant G.R., Farkas M.H., Pizarro A.D., Lahens N.F., Schug J., Brunk B.P. Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM) Bioinformatics. 2011;27:2518–2528. doi: 10.1093/bioinformatics/btr427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Tan Q., Kir J., Liu D., Bryant D. DAVID bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Research. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himms-Hagen J., Melnyk A., Zingaretti M.C., Ceresi E., Barbatelli G., Cinti S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. American Journal of Physiology. Cell Physiology. 2000;279:C670–C681. doi: 10.1152/ajpcell.2000.279.3.C670. [DOI] [PubMed] [Google Scholar]
- 30.Ohno H., Shinoda K., Spiegelman B.M., Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metabolism. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. The Journal of Biological Chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp L.Z., Shinoda K., Ohno H., Scheel D.W., Tomoda E., Ruiz L. Human BAT possesses molecular signatures that resemble beige/brite cells. PloS One. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue Y., Petrovic N., Cao R., Larsson O., Lim S., Chen S. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metabolism. 2009;9:99–109. doi: 10.1016/j.cmet.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Jespersen N.Z., Larsen T.J., Peijs L., Daugaard S., Homoe P., Loft A. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metabolism. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Lidell M.E., Betz M.J., Dahlqvist Leinhard O., Heglind M., Elander L., Slawik M. Evidence for two types of brown adipose tissue in humans. Nature Medicine. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 36.Shan T., Liu W., Kuang S. Fatty acid binding protein 4 expression marks a population of adipocyte progenitors in white and brown adipose tissues. FASEB Journal. 2013;27:277–287. doi: 10.1096/fj.12-211516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson-Fritch L., Nicoloro S., Chouinard M., Lazar M.A., Chui P.C., Leszyk J. Mitochondrial remodeling in adipose tissue associated with obesity and treatment with rosiglitazone. Journal of Clinical Investigation. 2004;114:1281–1289. doi: 10.1172/JCI21752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elabd C., Chiellini C., Carmona M., Galitzky J., Cochet O., Petersen R. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells. 2009;27:2753–2760. doi: 10.1002/stem.200. [DOI] [PubMed] [Google Scholar]
- 39.Bartesaghi S., Hallen S., Huang L., Svensson P.A., Momo R.A., Wallin S. Thermogenic activity of UCP1 in human white fat-derived beige adipocytes. Molecular Endocrinology. 2015;29:130–139. doi: 10.1210/me.2014-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loft A., Forss I., Siersbaek M.S., Schmidt S.F., Larsen A.S., Madsen J.G. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes Development. 2015;29:7–22. doi: 10.1101/gad.250829.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee K.Y., Russell S.J., Ussar S., Boucher J., Vernochet C., Mori M.A. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hynes M.A., Gitt M., Barondes S.H., Jessell T.M., Buck L.B. Selective expression of an endogenous lactose-binding lectin gene in subsets of central and peripheral neurons. Journal of Neuroscience. 1990;10:1004–1013. doi: 10.1523/JNEUROSCI.10-03-01004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fox C.S., Heard-Costa N., Cupples L.A., Dupuis J., Vasan R.S., Atwood L.D. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genetics. 2007;8(Suppl. 1):S18. doi: 10.1186/1471-2350-8-S1-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.