Abstract
Clostridium difficile hyper-virulent ribotype 027 strain has become a significant concern globally, but has rarely been reported in Asian countries including China. Recently, a retrospective single-center study in Beijing, China, detected two ribotype 027 C. difficile isolates from two patients coming for outpatient visits in 2012 and 2013. We performed a systematic investigation of the two isolates (and patients). Both C. difficile isolates had the typical PCR ribotype 027 profile; were positive for tcdA, tcdB and binary toxin genes; belonged to multilocus sequence type 1 (ST1); had typical ribotype 027 deletions in the tcdC gene; and were highly-resistant to fluoroquinolones; but had a different MLVA profile and were not genetically related to any previously reported international ribotype 027 clones. A review of the patients’ medical records showed that neither received appropriate antimicrobial treatment and were lost to follow-up after outpatient visits. We propose that C. difficile infections caused by ribotype 027 are probably a neglected problem in China, and the subsequent impact of unawareness of this problem is worrying. Appropriate testing assays and multi-center or national level surveillance for C. difficile infections and specifically for ribotype 027 should be introduced to provide essential data and guide future clinical practice.
Clostridium difficile is a Gram-positive, spore-forming, anaerobic rod. C. difficile infection (CDI) is a major nosocomial disease which is associated with high morbidity and potential mortality1,2. Since the 2000s, there has been a significant increase in the incidence and severity of CDI in North America and Europe, placing a huge financial burden on health care systems worldwide1,3. The emergence of the hyper-virulent C. difficile strain BI/NAP1/027 (restriction endonuclease analysis group BI, North American pulse-field type 1, PCR ribotype 027) has substantially contributed to the rise in CDI incidence4. The hypervirulence of the epidemic strains is likely due to the polymorphisms in the tcdB receptor-binding domain, leading to a hypertoxic and antigenically variable form of TcdB5. In addition, this strain possesses an extra toxin known as binary toxin, which is associated with increased clinical severity. A further characteristic of C. difficile ribotype 027 is an increased in vitro sporulation rate in the absence or presence of non-chloride cleaning agents6. In contrast to historic strains of ribotype 027 C. difficile, the new epidemic strains are resistant to fluoroquinolones4,7. After the detection of the first case of ribotype 027 in North America, this epidemic strain has subsequently spread rapidly in Europe and North America8.
However, CDI is not widely recognized in Asia including China, possibly due to insufficient laboratory diagnostic capacity, low sample submission rate, and lack of high-quality surveillance systems8,9. Compared to North America and Europe, relatively few studies on C. difficile have been performed in Asia, with C. difficile ribotype 027 occasionally reported in the region to date. Thus the genuine extent of the disease and burden of C. difficile ribotype 027 in China and the majority of Asian countries remains largely unknown. Based on this background, and in order to gain some insights on CDI epidemiology in China, we retrospectively tested stool specimens (from suspected CDI cases), from a major hospital in Beijing (Peking Union Medical College Hospital, PUMCH), using culture and molecular tests.
In this retrospective study, two C. difficile ribotype 027 isolates were detected, which are the first reported cases in Beijing, China. Furthermore, we characterized isolates by molecular approaches and antimicrobial susceptibility testing and reviewed patients’ medical records, aiming to have a better understanding of the current situation for future clinical practice implications.
Results
Background of C. difficile isolates
Out of 336 stool specimens from suspected CDI cases, 94 (28.0%) were positive by culture, but only 36 (10.7%) were positive for toxin A and/or B, by enzyme immunoassay (EIA). Further toxin testing on the C. difficile isolates yielded four isolates positive for toxin A and B genes (tcdA, tcdB) and the binary toxin genes (cdtA and cdtB).
Confirmation of two C. difficile ribotype 027 isolates
By capillary sequencer-based PCR ribotyping and querying against WEBRIBO database, two of the four above-mentioned binary toxin gene positive C. difficile isolates, namely PUCD235 and PUCD301, were confirmed as ribotype 027. Both isolates produced eight peaks, which were of identical pattern to that produced by previous well-characterized PCR ribotype 027 strain CA210,11.
Genotypes
By MLST, the 94 C. difficile isolates were classified into 22 STs, and the majority (58.5%) belonged to clade 1. ST-3 was the most common ST (18.1%, 17/94), followed by ST-54 (16.0%, 15/94), ST-37 (10.6%, 10/94) and ST-81 (9.6%, 9/94). The two ribotype 027 isolates (PUCD235 and PUCD301) belonged to ST1. In addition, sequencing of the slpA gene of the two isolates revealed a single nucleotide polymorphism (SNP), C467T, which was different from most previous C. difficile ribotype 027 isolates world-wide (slpA sequence type gc-8, e.g. strains CD196, BI1, QCD-66c26, and CIP 107932, genome accession nos. FN538970, FN668941, CM000441 and CM000659, respectively)12,13, with an amino acid substitution from proline to leucine at residue 156, but was identical to the slpA gene sequence of strain 2007855 (genome accession no. FN665654) from USA12.
Multiple-locus variable-number tandem-repeat analysis (MLVA)
MLVA was performed to investigate the genetic relatedness among Chinese, European and North American C. difficile ribotype 027 isolates. In three previous studies, 69 ribotype 027 C. difficile isolates from the USA, Canada, United Kingdom, France and Netherlands, were investigated by MLVA10,14,15. Regarding the two ribotype 027 isolates, they differed by 3 markers (A6Cd, B7Cd and C6Cd), and had a similarity of about 57%. As shown in Fig. 1, the two ribotype 027 isolates were also not genetically related with other previously reported ribotype 027 strains (Fig. 1).
Figure 1. MLVA dendrogram based on profiles of seven markers for the two ribotype 027 isolates (n = 2) in this study and other previous reported strains (n = 69) 10,14,15.
Characterization of toxin genes
Of the 94 C. difficile isolates studied, 51 (54.3%) were tcdA and tcdB-positive, and cdtA/cdtB-negative (A+B+CDT−), whilst 20 (21.3%) were tcdA-negative, tcdB-negative and cdtA/cdtB-negative (A−B−CDT−), and 19 (20.2%) were tcdA-negative, tcdB-positive and cdtA/cdtB-negative (A−B+CDT−). The remaining four (4.2%) isolates were tcdA-positive, tcdB-positive and cdtA/cdtB-positive (A+B+CDT+). The tcdC gene sequences of the two ribotype 027 isolates revealed a single-nucleotide deletion at position 117 and an 18-bp deletion at position 330–347, resulting in an inactivating frame-shift mutation in the tcdC gene of these isolates when compared with the sequence of wild-type tcdC gene (C. difficile 630, genome accession no. NC_009089), which was identical to previous published ribotype 027 genome sequences13,16.
Antimicrobial susceptibilities
The two C. difficile PCR ribotype 027 isolates had similar antimicrobial susceptibilities, being resistant to clindamycin, erythromycin, fluoroquinolones (ciprofloxacin and levofloxacin), rifampicin, rifaximin, but susceptible to metronidazole, meropenem, piperacillin/tazobactam, tetracycline and vancomycin (Table 1).
Table 1. Antimicrobial susceptibility of the two C. difficile ribotype 027 isolates identified in this study.
| Antimicrobial agents | Resistant breakpoint (μg/mL) | MIC (μg/mL)/category |
|
|---|---|---|---|
| PUCD235 | PUCD301 | ||
| Erythromycin | ≥8b | ≥256/R | ≥256/R |
| Ciprofloxacin | ≥8b | 128/R | 64/R |
| Clindamycin | ≥8a | 256/R | 128/R |
| Levofloxacin | ≥8b | 256/R | 128/R |
| Meropenem | ≥16a | 2/S | 2/S |
| Metronidazole | ≥32a | 1/S | 1/S |
| Piperacillin/tazobactam | ≥128/4a | 4/4/S | 4/4/S |
| Rifampicin | ≥4b | ≥256/R | ≥256/R |
| Rifaximin | ≥16b | ≥256/R | ≥256/R |
| Tetracycline | ≥16a | 0.125/S | ≤0.064/S |
| Vancomycin | ≥32b | 2/S | 2/S |
Abbreviations: MIC, minimum inhibitory concentration; S: susceptible; R: resistant.
aBreakpoints per CLSI document M100-S25.
bBreakpoints per Huang et al.49.
Medical records review
The two ribotype 027 isolates were from two individual patients who visited an outpatient clinic at Peking Union Medical College Hospital. The first isolate, PUCD235, was from a 58-year-old male patient who came to the outpatient clinic on December 11, 2012 post rectal cancer surgery at another hospital. The second isolate, strain PUCD301, was from a 59-year-old female patient who visited the hospital for cholecystitis on March 17, 2013. Both patients had symptoms of diarrhea upon visiting, and increased white blood cell counts and neutrophils were noticed. However, laboratory routine testing for C. difficile toxins A and B by the commercial EIA methods for both patients were negative, and no additional tests (e.g. C. difficile culture) were ordered by the clinicians. Consequently, CDI was not diagnosed, and thus the patients did not receive appropriate antimicrobial treatment and were subsequently lost to follow-up.
C. difficile ribotype 027 reported in Asian countries
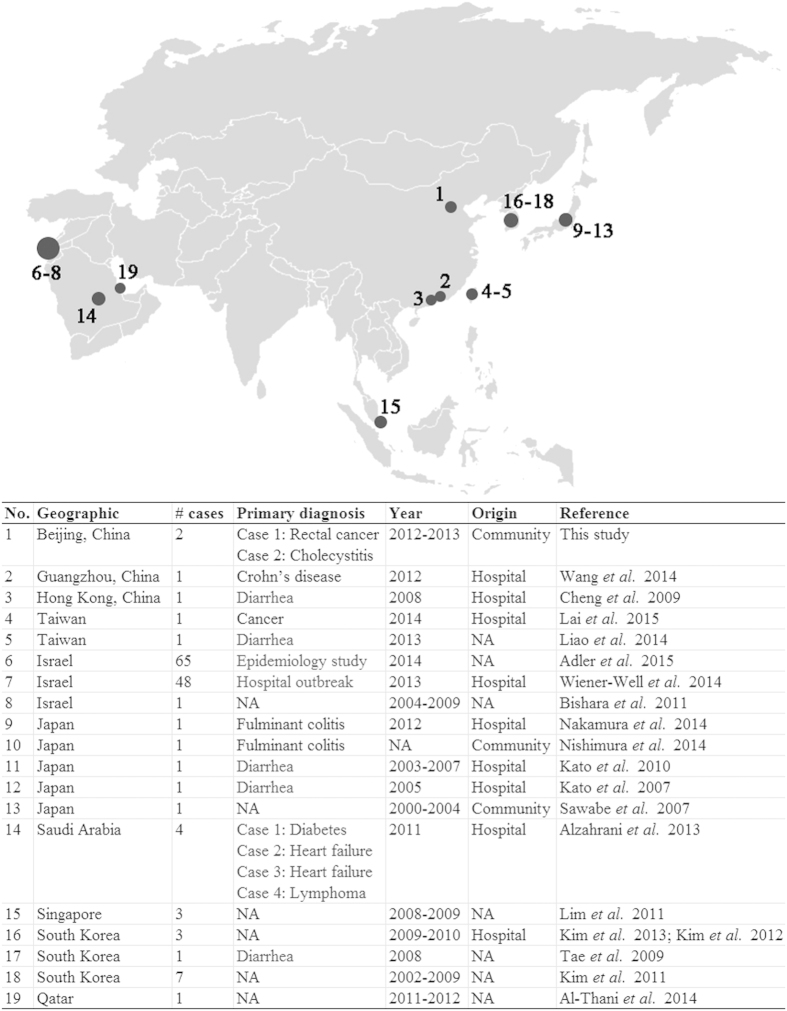
By searching on PubMed (http://www.ncbi.nlm.nih.gov/pubmed) using “Clostridium difficile” and “Asia” as the keywords, a total of 206 articles were found as of November 6, 2015. Among these, 19 full text articles reporting on the detection of C. difficile ribotype 027 isolates were found, with 142 cases identified in eight regions (Fig. 2)17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35. In 2000, the first C. difficile ribotype 027 case was reported in Japan by Sawabe et al.29. To date, most of the C. difficile ribotype 027 isolates identified in Asia (114/142, 80.3%) were reported in Israel17,20,31, including 65 isolates from a local national epidemiology study17, and 48 isolates from an outbreak31. In addition, 11 (7.7%) isolates were reported in South Korea23,24,30,34. The remaining 17 C. difficile ribotype 027 isolates were sporadically reported case-by-case from Japan (n = 5)22,28,29,32,33, Saudi Arabia (n = 4)19, Singapore (n = 3)27, mainland China (n = 1)35, Taiwan (n = 2)25,26, Hong Kong (n = 1)21, and Qatar (n = 1)18 (Fig. 2).
Figure 2. Summary of C. difficile PCR ribotype 027 reported in Asia.
Size of circles indicated the number of cases identified in each region. NA, data not available. The map was generated by GNU Image Manipulation Program (version 2.8.14, the GIMP Team, USA).
Discussion
CDI is a major medical and infection control threat to health care facilities, including hospitals, long-term care facilities, and nursing homes around the world. The majority of CDI cases reported recently are largely associated with the emergence of epidemic ribotype 027 strain, with most of it reported from North America and Europe36,37,38. However, according to our literature review, only 142 ribotype 027 cases were reported in Asia from 19 publications in eight regions, with 80.3% of these cases reported from Israel, where ribotype 027 has become the major clone disseminated in that country17,20,31. For most Asian countries, C. difficile ribotype 027 cases have only been reported sporadically.
The few reported cases of C. difficile ribotype 027 in Asia may be a tip of the iceberg as many Asian countries have inadequate laboratory diagnostic capacity, low submission rate of samples, and lack high-quality and multiple-center surveillance systems for CDI. According to our previous survey data, as of Dec 2014, only 70 amongst over 1700 tertiary hospitals in China, routinely carried out EIA for detection of C. difficile toxins A and B (unpublished data), which was the only commercial C. difficile testing method approved by China Food and Drug Administration then. Furthermore, few laboratories perform culture or molecular-based detection of C. difficile. Therefore, the magnitude of the CDI problem may be much under-estimated8,9.
To date, only one CDI case due to C. difficile ribotype 027 has been reported in mainland China, so the two cases identified in the present study are the second and third reported cases in China. Of note, the two patients involved did not get accurate diagnosis of CDI upon outpatient visit, mainly because the EIA testing for C. difficile toxins A and B were negative, although both patients had symptoms of diarrhea and abnormal routine blood test results. It has been reported that EIA-based methods are comparably less sensitive and have lower positive predictive value than toxigenic culture or molecular tests, in detecting C. difficile toxins. Moreover, the VIDAS EIA assay also appears to have lower sensitivity than other EIA-based methods for the detection of CDI39, prompting many centers to implement a two-pronged strategy that combines glutamate dehydrogenase (GDH) assay and toxin detection results to diagnose CDI. The two patients did not receive any appropriate treatment and were lost to follow-up which is a concern as these un-recognized C. difficile cases may become important transmission sources and a potential threat to public health40. Our findings suggest that C. difficile ribotype 027 has existed in northern China, therefore, it is important to improve laboratory diagnostic capacity and carry out active surveillance for the emergence of C. difficile hyper-virulent clones to avoid potential epidemic spread.
The development of molecular typing methods is of great significance for better understanding of C. difficile epidemiology. Our study identified the similarities and differences between the present C. difficile ribotype 027 isolates and globally described epidemic strains. Specifically, if only looking at MLST data, both strains belonged to ST1, suggesting possible similar origin source. However, a number of the 027 isolates previously reported in Asia were from a “historic” strain, which was fluoroquinolone susceptible, which is in contrast to the strains described in this study that were clearly fluoroquinolone resistant. In addition, sequencing of the slpA gene revealed that the two strains shared a SNP C467T, which differed from most global ribotype 027 strains, but was similar to one USA strain (2007855) recovered from animal rather than human source12. Further MLVA typing results also supported that the two ribotype 027 isolates were genetically different as they did not cluster together with strains from previously reported cases in North America and European countries (Fig. 1). The high discriminatory power of MLVA typing, if used properly (with suggested reasonable cut-off value) can be a potential valuable tool for investigating future ribotype 027 outbreaks41. In addition, if an association can be established between specific ribotype 027 MLVA subtypes, and virulence and clinical outcome of CDI patients42, this would be valuable for infection control and may help explain why our two strains appeared to have low virulence.
In conclusion, our study characterized the first two C. difficile PCR ribotype 027 isolates identified in Beijing, China. Our findings highlight the importance of raising public awareness, improving the laboratory diagnostic capacity, as well as implementing active surveillance systems for CDI in China, so as to capture this neglected potential public health threat and for providing useful indications for future clinical and infection control practices.
Methods
Ethics
The study was approved by the Human Research Ethics Committee of Peking Union Medical College Hospital (No. S-263), and the study was carried out in accordance with the approved guidelines. Informed consents were obtained from all the patients.
Background for local laboratory diagnosis of C. difficile infections
Stool specimens from suspected CDI cases are routinely sent to the PUMCH laboratory in Beijing for C. difficile detection. The available tests for C. difficile detection at this hospital are toxin A and B detection using a commercial enzyme immunoassay (EIA) (VIDAS C. difficile Toxin A&B, bioMerieux, Marcy l’Etiole, France). However, C. difficile culture and molecular testing are rarely performed in most Chinese hospitals.
Retrospective analysis of faecal specimens for C. difficile, including culture and molecular assays
A total of 336 consecutive, non-repetitive faecal specimens from 336 patients (stored at −80 °C before use) were collected between August 2012 and July 2014. All the specimens were tested by culture and molecular assays. Generally, the specimens were cultured on cycloserine-cefoxitin fructose agar (CCFA) in anaerobic condition at 35 °C for 48 h, and suspected colonies were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Presence of toxin-encoding genes was then detected by a multiplex PCR assay to all confirmed C. difficile isolates as described below.
DNA extraction and toxin gene detection
One to five typical colonies were picked up from pure cultures of C. difficile isolates, and bacterial suspensions equivalent to 1 McFarland turbidity standard in 200μl double-distilled water, were made. Genomic DNA was extracted with QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. A 5-plex PCR was used to detect tcdA, tcdB, cdtA and cdtB genes and the 16S rDNA, as previously described by Persson et al.43. We also sequenced the tcdC gene, a negative regulator of tcdA and tcdB43.
Capillary sequencing-based PCR ribotyping
Capillary sequencer-based PCR ribotyping was performed as described by Indra et al.44. Generally, primers 16S-F (5′-GTGCGGCTGGATCACCTCCT-3′) and 23S-R (5′-CCCTGCACCCTTAATAACTTGACC-3′) were used for amplification, with 16S primer labeled at the 5′ end with carboxyfluorescein (FAM). The PCR product was diluted 1:10 in water before loading. Fragment separation was performed on an ABI 3730 sequencer (Applied Biosystems, Carlsbad, USA) with a 50 cm POP 7 gel. Sample injection was at 1.6 kV over 15 seconds, with a total running time of 6200 seconds. A GeneScan™ 1200 LIZ (Applied Biosystems) size standard was used as internal marker. The results were analyzed by GeneMarker software (Version2.2.0, SoftGenetics, State College, PA, USA), and queried against WEBRIBO database (https://webribo.ages.at/) for ribotypes. A previously well-characterized PCR ribotype 027 isolate, strain CA2, was used as internal control for PCR ribotyping10.
Multilocus sequence typing (MLST)
MLST was performed by sequencing seven gene loci (adk, atpA, dxr, glyA, recA, sodA and tpi) as previously described by Griffiths et al.45. DNA sequences were submitted to the PubMLST sequence query page (http://pubmlst.org/cdifficile/) to obtain the sequence type (ST) and clade.
Sequencing of the slpA gene
As described previously by Kato et al.46, the partial slpA gene was amplified by the primers slpAcom19 (5′-GTTGGGAGGAATTTAAGRAATG-3′) and slpAcom22 (5′-GCWGTYTCTATTCTATCDTYWCC-3′). Both strands of the amplified products were sequenced and comparisons were made with the amino acid sequences deduced from the DNA sequences.
Multilocus variable-number tandem repeat analysis (MLVA)
The genetic relatedness of the toxigenic isolates was investigated by MLVA, using the set of 7 loci (A6Cd, B7Cd, C6Cd, E7Cd, F3Cd, G8Cd, and H9Cd) as previously described by van den Berg et al.15. Repeat numbers were analyzed using BioNumerics software v6.5 (Applied Maths, Texas, USA) for cluster analysis. A dendrogram was constructed using the unweighted-pair group method with arithmetic mean clustering (UPGMA) with the multistate categorical similarity coefficient (MCSC).
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by the agar dilution method according to Clinical and Laboratory Standards Institute (CLSI) guidelines (document M11-A8)47. The following 11 antimicrobial agents were chosen: ciprofloxacin, clindamycin, erythromycin, levofloxacin, meropenem, metronidazole, piperacillin/tazobactam, rifampicin, rifaximin, tetracycline and vancomycin. Interpretation of testing results was based on CLSI M100-S2548, or according to the criteria suggested by Huang et al.49 for the drugs whose breakpoints were not available in CLSI documents, as summarized in Table 1.
Review of C. difficile ribotype 027 reported in Asian countries
To have a more comprehensive understanding of current epidemiology of C. difficile ribotype 027 in Asia, we reviewed and summarized all related published literature in PubMed (http://www.ncbi.nlm.nih.gov/pubmed) database as of November 6, 2015.
Additional Information
How to cite this article: Cheng, J.-W. et al. The First Two Clostridium difficile Ribotype 027/ST1 Isolates Identified in Beijing, China–an Emerging Problem or a Neglected Threat? Sci. Rep. 6, 18834; doi: 10.1038/srep18834 (2016).
Acknowledgments
This study was financially supported by a Natural Science Foundation of China (grant number 81501807), Beijing outstanding young talents project (grant number 2015000020124G071) and a National Research Special Fund for Public Welfare Industry of Health of China (grant number 201402001).
Footnotes
Author Contributions J.W.C., M.X., T.K. and F.K. wrote the manuscript; Z.P.X., X.H., L.Z. and X.F. collaborated in molecular investigations of the strains; L.Y. Sun summarized the patient’s medical records; Y.C.X. designed and supervised the study.
References
- Jones A. M., Kuijper E. J. & Wilcox M. H. Clostridium difficile: a European perspective. J Infect 66, 115–128 (2013). [DOI] [PubMed] [Google Scholar]
- Kelly C. P. & LaMont J. T. Clostridium difficile–more difficult than ever. N Engl J Med 359, 1932–1940 (2008). [DOI] [PubMed] [Google Scholar]
- Lessa F. C. et al. Burden of Clostridium difficile infection in the United States. N Engl J Med 372, 825–834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45, 109–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanis J. M., Heinlen L. D., James J. A. & Ballard J. D. Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog 9, e1003523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerlund T. et al. Increased sporulation rate of epidemic Clostridium difficile Type 027/NAP1. J Clin Microbiol 46, 1530–1533 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L. C. et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 353, 2433–2441 (2005). [DOI] [PubMed] [Google Scholar]
- Collins D. A., Hawkey P. M. & Riley T. V. Epidemiology of Clostridium difficile infection in Asia. Antimicrob Resist Infect Control 2, 21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey P. M. et al. Molecular epidemiology of Clostridium difficile infection in a major chinese hospital: an underrecognized problem in Asia? J Clin Microbiol 51, 3308–3313 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killgore G. et al. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J Clin Microbiol 46, 431–437 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M. et al. Comparison of two capillary gel electrophoresis systems for Clostridium difficile ribotyping, using a panel of ribotype 027 isolates and whole-genome sequences as a reference standard. J Clin Microbiol 50, 2755–2760 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. et al. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci USA 107, 7527–7532 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabler R. A. et al. In-depth genetic analysis of Clostridium difficile PCR-ribotype 027 strains reveals high genome fluidity including point mutations and inversions. Gut Microbes 1, 269–276 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C., Vromman F., Halkovich A. & Barbut F. Multilocus variable-number tandem repeat analysis: a helpful tool for subtyping French Clostridium difficile PCR ribotype 027 isolates. J Med Microbiol 60, 1088–1094 (2011). [DOI] [PubMed] [Google Scholar]
- van den Berg R. J., Schaap I., Templeton K. E., Klaassen C. H. & Kuijper E. J. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J Clin Microbiol 45, 1024–1028 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton T. et al. Complete Genome Sequence of the Hypervirulent Bacterium Clostridium difficile Strain G46, Ribotype 027. Genome Announc 3, 10.1128/genomeA.00073-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler A. et al. A national survey of the molecular epidemiology of Clostridium difficile in Israel: the dissemination of the ribotype 027 strain with reduced susceptibility to vancomycin and metronidazole. Diagn Microbiol Infect Dis 83, 21–24 (2015). [DOI] [PubMed] [Google Scholar]
- Al-Thani A. A., Hamdi W. S., Al-Ansari N. A., Doiphode S. H. & Wilson G. J. Polymerase chain reaction ribotyping of Clostridium difficile isolates in Qatar: a hospital-based study. BMC Infect Dis 14, 502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzahrani N. & Johani S. A. Emergence of a highly resistant Clostridium difficile strain (NAP/BI/027) in a tertiary care center in Saudi Arabia. Ann Saudi Med 33, 198–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishara J., Goldberg E., Madar-Shapiro L., Behor J. & Samra Z. Molecular epidemiology of clostridium difficile in a tertiary medical center in Israel: emergence of the polymerase chain reaction ribotype 027. Isr Med Assoc J 13, 338–341 (2011). [PubMed] [Google Scholar]
- Cheng V. C. et al. Clostridium difficile ribotype 027 arrives in Hong Kong. Int J Antimicrob Agents 34, 492–493 (2009). [DOI] [PubMed] [Google Scholar]
- Kato H. et al. Typing of Clostridium difficile isolates endemic in Japan by sequencing of slpA and its application to direct typing. J Med Microbiol 59, 556–562 (2010). [DOI] [PubMed] [Google Scholar]
- Kim H. et al. Emergence of Clostridium difficile ribotype 027 in Korea. Korean J Lab Med 31, 191–196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. et al. Epidemiology of Clostridium difficile infections in a tertiary-care hospital in Korea. Clin Microbiol Infect 19, 521–527 (2013). [DOI] [PubMed] [Google Scholar]
- Lai M. J., Chiueh T. S., Huang Z. Y. & Lin J. C. The first Clostridium difficile ribotype 027 strain isolated in Taiwan. J Formos Med Assoc, 10.1016/j.jfma.2015.02.006 (2015). [DOI] [PubMed] [Google Scholar]
- Liao T. L., Lin C. F., Chiou C. S., Shen G. H. & Wang J. Clostridium difficile PCR Ribotype 027 Emerges in Taiwan. Jpn J Infect Dis 68, 338–340 (2015). [DOI] [PubMed] [Google Scholar]
- Lim P. L. et al. Isolation of the first three cases of Clostridium difficile polymerase chain reaction ribotype 027 in Singapore. Singapore Med J 52, 361–364 (2011). [PubMed] [Google Scholar]
- Nakamura I. et al. Fulminant colitis from Clostridium difficile infection, the epidemic strain ribotype 027, in Japan. J Infect Chemother 20, 380–383 (2014). [DOI] [PubMed] [Google Scholar]
- Sawabe E. et al. Molecular analysis of Clostridium difficile at a university teaching hospital in Japan: a shift in the predominant type over a five-year period. Eur J Clin Microbiol Infect Dis 26, 695–703 (2007). [DOI] [PubMed] [Google Scholar]
- Tae C. H. et al. The first case of antibiotic-associated colitis by Clostridium difficile PCR ribotype 027 in Korea. J Korean Med Sci 24, 520–524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener-Well Y. et al. Clinical and molecular characteristics of an outbreak caused by the pandemic (BI/NAP1/027) Clostridium difficile clone in a single center in Israel. Infect Control Hosp Epidemiol 35, 1306–1308 (2014). [DOI] [PubMed] [Google Scholar]
- Kato H., Ito Y., van den Berg R. J., Kuijper E. J. & Arakawa Y. First isolation of Clostridium difficile 027 in Japan. Euro Surveill 12, E070111 070113 (2007). [DOI] [PubMed] [Google Scholar]
- Nishimura S. et al. Fulminant pseudomembranous colitis caused by Clostridium difficile PCR ribotype 027 in a healthy young woman in Japan. J Infect Chemother 20, 729–731 (2014). [DOI] [PubMed] [Google Scholar]
- Kim J., Kang J. O., Pai H. & Choi T. Y. Association between PCR ribotypes and antimicrobial susceptibility among Clostridium difficile isolates from healthcare-associated infections in South Korea. Int J Antimicrob Agents 40, 24–29 (2012). [DOI] [PubMed] [Google Scholar]
- Wang P. et al. Identification of Clostridium difficile ribotype 027 for the first time in Mainland China. Infect Control Hosp Epidemiol 35, 95–98 (2014). [DOI] [PubMed] [Google Scholar]
- Murabata M. et al. Intestinal colonization and nosocomial spread of Clostridium difficile in pediatric cancer patients under long-term hospitalization. Kansenshogaku Zasshi 82, 419–426 (2008). [DOI] [PubMed] [Google Scholar]
- Bartlett J. G. & Perl T. M. The new Clostridium difficile–what does it mean? N Engl J Med 353, 2503–2505 (2005). [DOI] [PubMed] [Google Scholar]
- Warny M. et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366, 1079–1084 (2005). [DOI] [PubMed] [Google Scholar]
- Planche T. et al. Diagnosis of Clostridium difficile infection by toxin detection kits: a systematic review. Lancet Infect Dis 8, 777–784 (2008). [DOI] [PubMed] [Google Scholar]
- Riggs M. M. et al. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis 45, 992–998 (2007). [DOI] [PubMed] [Google Scholar]
- Eyre D. W. et al. Comparison of multilocus variable-number tandem-repeat analysis and whole-genome sequencing for investigation of Clostridium difficile transmission. J Clin Microbiol 51, 4141–4149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawley W. N. et al. Use of highly discriminatory fingerprinting to analyze clusters of Clostridium difficile infection cases due to epidemic ribotype 027 strains. J Clin Microbiol 46, 954–960 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Torpdahl M. & Olsen K. E. New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection. Clin Microbiol Infect 14, 1057–1064 (2008). [DOI] [PubMed] [Google Scholar]
- Indra A. et al. Characterization of Clostridium difficile isolates using capillary gel electrophoresis-based PCR ribotyping. J Med Microbiol 57, 1377–1382 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D. et al. Multilocus sequence typing of Clostridium difficile. J Clin Microbiol 48, 770–778 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Yokoyama T. & Arakawa Y. Typing by sequencing the slpA gene of Clostridium difficile strains causing multiple outbreaks in Japan. J Med Microbiol 54, 167–171 (2005). [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institude. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard-eighth edition. Document M11-A8. Clinical and Laboratory Standards Institude, Wayne, PA (2012).
- Clinical and Laboratory Standards Institude. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. Document M100-S25. Clinical and Laboratory Standards Institude, Wayne, PA (2015).
- Huang H. et al. Clostridium difficile infections in a Shanghai hospital: antimicrobial resistance, toxin profiles and ribotypes. Int J Antimicrob Agents 33, 339–342 (2009). [DOI] [PubMed] [Google Scholar]