Abstract
Scaffolding proteins serve to assemble protein complexes in dynamic processes by means of specific protein-protein and protein-lipid binding domains. Many of these domains bind either proteins or lipids exclusively; however, it has become increasingly evident that certain domains are capable of binding both. Especially, many PDZ domains, which are highly abundant protein-protein binding domains, bind lipids and membranes. Here we provide an overview of recent large-scale studies trying to generalize and rationalize the binding patterns as well as specificity of PDZ domains towards membrane lipids. Moreover, we review how these PDZ-membrane interactions are regulated in the case of the synaptic scaffolding protein PICK1 and how this might affect cellular localization and function.
Keywords: PDZ domains, lipid-binding domains, PICK1, scaffolding proteins
1. Introduction
Scaffolding proteins are essential components required for spatial and temporal regulation of cellular processes, such as synaptic plasticity and cell-cell adhesion. The overall function of scaffolding proteins is characterized by a common ability to (I) bind its interaction partners via specific interactions using relatively short consensus motifs; and (II) bind to other cellular scaffolding proteins, mediators or specific intracellular membrane compartments. To this end, scaffolding proteins rely on both protein-protein interactions (PPIs) and protein-lipid interactions (PLIs), which can be attributed to various distinct domains or posttranslational modification (e.g., myristoylation or palmitoylation). Canonical examples of PPI domains include SRC homology 2/3 (SH2/3) domains [1,2,3,4], WW domains [5], phosphotyrosine-binding (PTB) domains [6,7], and homologous PSD-95/Disc-large (DLG1)/ZO-1 (PDZ) domains [8,9]. Prototypical membrane lipid binding domains include the phosphatidylinositide-binding Phox homology (PX) domains, Pleckstrin homology (PH) domains, C1/C2 domains, epsin N-terminal homology (ENTH) domains, and (Fab1/YotB/Vac1/EEA1) FYVE domains [10,11]. It is becoming evident, however, that many of these domains are functionally overlapping in terms of PPIs and PLIs.
The (PH) domain is one of the most prominent examples of a domain known to bind both lipid membranes and specific protein motifs [12]; however, recently several classical PPI domains have also been shown to directly interact with membrane lipids. Especially for PDZ domains, membrane lipid binding have been reported for several domains and there is accumulating evidence that the activity of PDZ domains might be tightly regulated by the spatiotemporal distribution of phosphatidylinositolphosphates (PIPs) in specific intracellular compartments [13,14].
2. PDZ Domain Structure and Canonical Ligand Binding
The PDZ domain is one of the most common PPI domains found in the mammalian proteome. They were first discovered in 1991 in imaginal discs of Drosophila [15] and in 1992 in rat brain homogenates [16], but was not conceptualized until 1995 [17], after a few other studies had reported the presence of “GLGF repeat” or “DHG” domains. Since then at least 266 unique PDZ domains in 150 different proteins have been identified in the human genome alone [18].
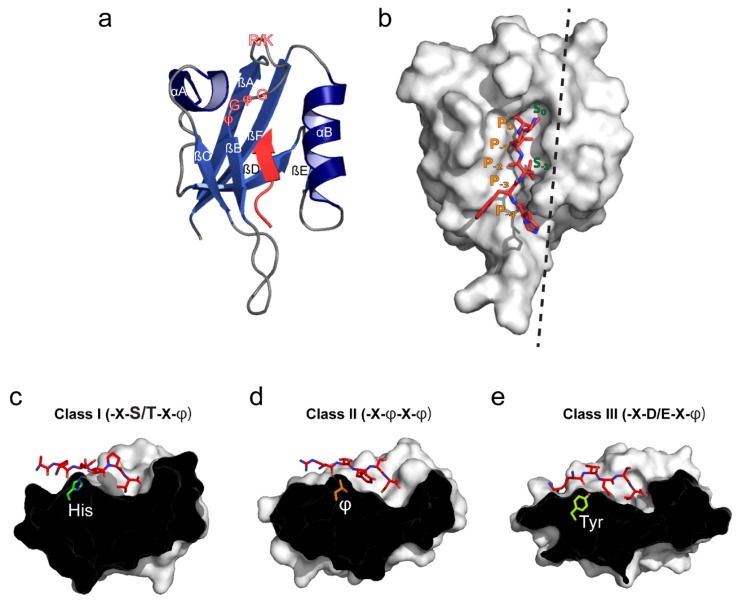
The first two studies describing the overall structural basis of PDZ domains were published in 1996. One study reported the crystal structure of the third PDZ domain (PDZ3) of PSD-95 in both a substrate-bound and a substrate-free state, whereas the other reported the substrate-free state of the homologous PDZ3 of SAP102 [9,19]. At present, 450 structures of PDZ domains are deposited in the PDB database; all sharing the same overall structural characteristics. The canonical PDZ domain is globular with an approximate diameter of 35 Å. The domain consists of ~90 residues forming a partially opened β-barrel including six β-strands (βA-βF) and two α-helices (αA and αB) capping the open sides of the β-barrel (Figure 1a). The key function of the PDZ domain is the ability to bind specific C-terminal motifs and is mediated by a confined, surface-exposed binding groove situated between the βB strand and the αB helix. The ligand is “stitched” into this binding groove, anti-parallel to the βB strand allowing for important backbone interactions. The bottom of the binding groove is constituted by a conserved R/K-X-X-X-G-φ-G-φ loop (X is any residue and φ is a hydrophobic residue) between the βA strand and the βB strand where the amides in the G-φ-G-φ motif, and possibly the basic side-chain of arginine/lysine tightly coordinate the carboxyl group of the most C-terminal residue of the ligand (denoted P0) [20,21] (Figure 1a,b). The actual binding groove is relatively small accommodating 3-5 C-terminal residues of the ligand, which is, in most cases, sufficient to obtain micromolar binding affinities [9,19].
Figure 1.
Structural overview of the canonical PDZ domain fold, ligand binding and peptide-based classification. (a) The canonical PDZ domain fold comprises two α-helices (αA and αB) and six β-strands (βA–βF) (PDB: 2LUI [22]). The ligand is shown in red and is stitched into the binding groove located between the αB helix and βB strand. The peptide/protein-binding groove is composed by the R/K-X-X-X-G-φ-G-φ (X—any residue; φ—hydrophobic residue) loop indicated in red outlined text; (b) Surface view of the canonical PDZ domain. Hydrophobic pockets S0 and S-2 (for class II domains) are indicated in green. Correspondingly, the five C-terminal ligand residues are denoted from P0 to P−4 in orange. The black dashed line indicates the cross section plane used in panels c-e; (c) Class I binding domains usually recognize ligands with either Ser or Thr at P−2. The αB1 residue is usually His (PDB: 1TP3 [23]); (d) Class II binding domains recognize ligands with hydrophobic residues at P−2. The αB1 residue is often hydrophobic and allows for formation of a second hydrophobic binding pocket (denoted S-2 in panel b) (PDB: 1N7F [24]); (e) Ligands binding class III domains typically have Asp or Glu at P−2 and Tyr at the αB1 position (PDB: 1B8Q [25]).
Upon canonical binding the side-chain of the P0 residue is guided into a hydrophobic cavity constituted by residues from βB and αB of the PDZ domain (S0) and, consequently, the vast majority of PDZ domain ligands have a hydrophobic residue at this position. The size and shape of S0 varies between the different PDZ domains, thereby controlling the preference of S0 towards different hydrophobic residues at P0. Most PDZ domains, however, prefer non-aromatic hydrophobic residues at P0, such as valine, isoleucine or leucine [26,27,28]. As a consequence of the extended anti-parallel β-strand ligand insertion, the side chains of residues at P−1 and P−3 will be solvent-exposed and, therefore, these residues are rarely directly involved in canonical PDZ domain binding (Figure 1b). Some studies have, however, suggested that amphipathic residues such as tryptophan or tyrosine might be slightly favored at these positions [29]. In contrast, the side chain of the P−2 residue points towards the PDZ domain surface, like the side chain of the P0 residue. Most importantly, it comes in direct contact with the side chain of the first residue of αB (αB1), but also several other residues (mostly hydrophobic) from αB and βB, which constitute a secondary binding pocket (S−2). The preference of S-2 towards specific residues at P−2 is much stricter than the residue preference of S0 and, consequently, the classification of PDZ domain ligands have been primarily based on the P−2 residue. Correspondingly, PDZ domains have been classified with respect to the first residue of the α-helix B (αB1) [20,30,31].
The majority of PDZ domains can be readily divided into three different classes. Class I PDZ domains, which is by far the largest class, typically recognizes a -X-S/T-X-φ motif using a histidine at the αB1 position to form a hydrogen bond with the P−2 serine/threonine (Figure 1c) [27]. PSD-95 (PDZ2 and 3) and syntrophin are classical examples of class I PDZ domains [9,32]. Class II domains recognize -X-φ-X-φ motifs using a hydrophobic residue at the αB1 position (Figure 1d). For this reason class II PDZ domains such as CASK, GRIP1 (PDZ 5 and 6) and syntenin, are described to have a second hydrophobic pocket formed by residues at positions αB1, αB4, αB5, and βB3 [24,33,34,35]. Class III PDZ domains recognize a -X-D/E-X-φ motif and generally have a tyrosine at αB1 position, allowing the hydroxyl group to interact with the carboxyl side-chain of aspartate or glutamate (Figure 1e). The PDZ domain of nNOS typically binds class III ligands [28].
3. Membrane Lipid Binding Is a General Property of PDZ Domains
The membrane lipid binding capacity of Syntenin PDZ1 and 2 was the first to be described, and the mechanism, as well as the functional relevance, has been characterized [36,37,38]. Subsequently, PDZ-lipid interactions of Par3-PDZ2 [39], PTP-BAS PDZ2 [40], ZO-1 [41], and PICK1 PDZ [42] have been reported. In the lack of direct structural insight, the PDZ-membrane lipid interactions have been investigated functionally using mutational analyses in combination with biochemical assays, such as liposome sedimentation, surface plasmon resonance (on liposomes) (SPR), lipid dot-blots, and NMR. Several recent papers have reviewed the detailed structural features underlying lipid binding for a selected subset of PDZ domains [13,43]. Here we review three studies aiming to provide a general framework for understanding PDZ-membrane lipid interactions by analyzing entire PDZ domain libraries.
To address the ability of PDZ domains to interact with membrane lipids in general terms, Wu et al. [39] measured the liposome binding capacity by sedimentation of 74 mammalian PDZ domains. From these 74 domains, 17 were found to have detectable membrane lipid binding capacities. Furthermore, six domains, originating from five different proteins (CASK PDZ, PICK1 PDZ, X11α PDZ, DLG5-PDZ2, and ZO-1 PDZ1 and 2), were found to have liposome affinities comparable to PAR3 PDZ2. Though this study did not further investigate how the composition of the liposomes could potentially influence the membrane binding, it has been reported that most PDZ domains bind anionic membranes [39,42].
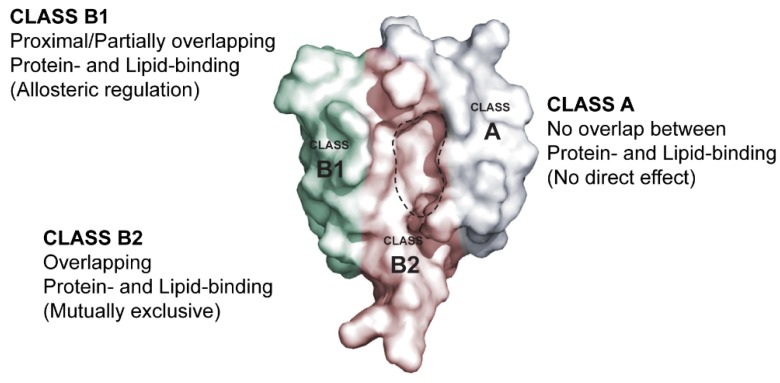
Another large-scale study by Chen et al. [44] tested membrane binding to such anionic lipids of 70 monomeric PDZ domains from 35 different mammalian proteins using SPR. Twenty-six of these were found to interact with affinities spanning three orders of magnitude (20 nM to 10 uM). Moreover, it was also stated that most of these PDZ domains did not show significant preference for specific PIPs. By using sequence information, Chen et al. built a prediction model using numerical vector representation as a function of primary and tertiary structure combined with residue specific information from which they predicted membrane binding of 2000 PDZ domains from 20 different species. They found that most membrane lipid binding PDZ domains have at least one cationic site. The position of the cationic site was subsequently used to divide PDZ domains into three distinct classes (Figure 2). Class A domains have a cationic site not overlapping with the peptide binding groove, whereas class B domains have a cationic site proximal to the peptide binding groove, typically clustered around the αA helix. This class was subdivided into classes B1 and B2. B1 domains typically have the cationic patch conserved at the C-terminal end of αA helix and are, therefore, able to simultaneously interact with peptides and membrane lipids. As an example, the presence of plasma membrane mimetic liposomes allosterically alters peptide binding for rhophillin2 PDZ. B2 domains, on the other hand, display overlap between the peptide binding groove and the membrane lipid binding cationic patch. As an example, binding to plasma membrane mimetic liposomes significantly reduced the binding to peptide ligands for tamalin PDZ. B2 domains seem to have the cationic patch conserved at the N-terminal end or scattered throughout the αA helix.
Figure 2.
Proposed cationic patch classification of membrane lipid binding PDZ domains. The classification is based on positioning of the cationic patch as proposed by Chen et al. [44]. Class A domains do not display overlap of the protein and membrane lipid binding sites. The cationic patch is likely to be positioned opposite to the binding groove and/or involving the βE strand. Class B1 domains display proximal or partially overlapping protein- and membrane lipid-binding sites that may allosterically regulate each other. Often the cationic patch of class B1 domains involves residues C-terminal to the αA helix. Finally, the class B2 domains have overlapping protein- and lipid-binding sites and therefore class B2 domains will almost exclusive bind one or the other. A dashed black line delineates the binding pocket.
Finally, a recent study sought to generalize the PDZ domain lipid binding and the nature of PDZ/peptide/lipid complexes using a fluorescence-based cell-localization assay [45]. Out of 246 investigated single domains, 53 displayed distinct cellular localization; either with discrete or strong plasma membrane localization or localization to cytosolic, or subnuclear organelles. Most PDZ domains displayed either discrete plasma membrane localization or localized to subnuclear organelles. Very few showed strong plasma membrane or cytosolic localization. Interestingly, localization was strongly cell-type dependent, which was hypothesized to be due to differences in ligand expression or lipid composition. It was further tested whether changes in the specific PIP composition of relevant cellular compartments would in fact also drive de- or relocalization of the PDZ domains. Neither strong and discrete plasma membrane localized PDZ domains showed significant PIP dependency. In fact, only the PDZ domain of CASK displayed sensitivity towards differences in the PIP plasma membrane composition. For the subnuclear localized domains; however, three out of six randomly-selected domains, including the deafness, autosomal recessive 31 PDZ1 (DFNB31_1), the sodium-hydrogen exchange regulatory cofactor NHERF2 PDZ1 (SLC9A3R2_1) and the Syntrophin, Gamma 1 PDZ domain (SNTG1) showed PI(4,5)P dependency, and displayed a more diffuse nuclear or cytoplasmic localization upon membrane lipid modifying treatments. The cytosolic PDZ domains, primarily localized to perioxisomes or appeared as cytosolic aggregates and showed no clear PIP dependency.
To investigate if the subcellular localization could be explained directly by specificity towards different PIPs, the affinities for liposomes with different PIP contents were quantified using SPR. Out of 19 randomly selected domains, 10 domains displayed micromolar affinities towards tri- or biphosphorylated PIPs. Only one PDZ domain displayed micromolar affinity for monophosphorylated PIPs (DFNB31_1). Importantly, all of the four domains found to respond to cellular lipid modification also displayed PIP specificity. The authors further tested the affinity towards several other membrane lipids (e.g., cardiolipin, phosphatidylserine (PS), phospatidylcholine (PC), and cholesterol, etc.) using a lipid dot blot assay. This assay revealed that a few domains also have the ability to bind non-phosphorylated lipids. Interestingly, the PIP affinities determined from the SPR experiments was different from affinities obtained using dot blots, suggesting that some of these interactions relies on the presence of intact membranes. In agreement with findings by Chen et al. and adding to the general understanding of PDZ-membrane lipid interactions, it was suggested that high pI (>9) seems to be a common feature of the lipid binding PDZ domains, and that conserved cationic patches are important for PIP specificity and cellular localization [45].
Though it has become well established that lipid/membrane binding is a common feature shared by several PDZ domains, the molecular mechanisms underlying the binding are still poorly understood. As presented above, attempts to classify the domains according to either their cellular localization, or their cationic properties, have provided a valuable framework for understanding how these interactions might be electrostatically favored. To emphasize how these general properties may relate to regulation by e.g., redox state, ion binding, and posttranslational modifications we review the case of the lipid and membrane binding by the PICK1 PDZ domain below.
4. PICK1 Is a Flexible Membrane-Associated Scaffolding Protein
Protein Interacting with C kinase 1 (PICK1) is ~47 kDa membrane associated scaffolding protein which was first discovered in a yeast two hybrid experiment investigating interaction partners of activated Protein Kinase C (PKC) [46]. The overall size varies from 400 to 500 aa across different species with the largest difference being found in the length of the C-termini [47]. In vertebrates, PICK1 is highly expressed in the brain (detected in high levels in Cerebellar Purkinje neurons, and hippocampal neurons), in the pancreas, and in the testis. Additionally, heart, lung, spleen, liver, and muscle tissue were found to contain modest levels of PICK1 [48,49].
Functionally, PICK1 plays a pivotal role in regulation of synaptic plasticity involving trafficking of the AMPA-type ionotropic glutamate receptors (AMPARs), which is thought to be the neuronal basis for learning and memory [50]. PICK1 interacts directly with the GluA2 subunit of the AMPAR via its PDZ domain, and has been shown to promote activity-stimulated AMPAR internalization [51]. Moreover, PICK1 reduces the recycling rate of internalized AMPARs giving rise to an overall intracellular accumulation [52,53]. As a result of these separate functions PICK1 is necessary for long-term depression [49], as well as some types of long term potentiation of neuronal synapses [54]. Through the functional interaction with the AMPARs PICK1 is, moreover, implicated in the pathogenesis of several psychiatric and neurological disorders [55] and is, consequently, considered a promising novel drug target. More recently, PICK1 has been shown to associate with the Golgi compartment and regulate the biogenesis of dense core vesicles in the pancreas, pituitary, and adrenal glands, thereby affecting metabolic homeostasis in flies and mice [56,57,58].
At the structural level, PICK1 contains two highly conserved domains; the ~90 aa N-terminal PDZ domain and by a ~210 aa Bin/amphipysin/Rvs (BAR) domain which mediates strong homo-dimerization. PICK1 is the only protein featuring these two domains and they are separated by a ~40 aa linker region capable of forming a four-turn membrane binding amphipatic helix (AH). The domains and are flanked by a ~20 aa acidic N-terminal region (NAR) and a ~50 aa acidic C-terminal region (CAR). The PICK1 homo-dimers adopt, overall, an elongated crescent shape defined by the BAR domain [59,60,61] which form a coiled-coil six helix bundle upon dimerization as seen for i.e., amphiphysin, arfaptin, and endophilin [62,63,64]. Two recent studies utilized small angle X-ray scattering (SAXS) to determine the entire quaternary structure of the PICK1 homo-dimer [60,61]. The studies largely agree on the overall structure of the BAR domain and the dimerization interface, but differ considerably with respect to the positioning of the PDZ domains relative to the BAR domain. Madasu et al. suggest that the PDZ domains are associated with the BAR domain and the linker between the adjacent domain forms a permanent helix [61], whereas we suggest the PDZ domain to be flexibly attached through an unstructured linker that only folds into an amphipathic helix in the presence of membrane binding [60]. This controversy is central to the understanding of PICK1 function, since the PDZ domain has been suggested to auto-inhibit the membrane binding capacity of the BAR domain through steric hindrance [53,65,66,67].
5. PICK1 Contains a Non-Canonical Promiscuous PDZ Domain
Each monomer of PICK1 contains a single PDZ domain, which is highly conserved between species from mollusks to vertebrates. The PDZ domain of PICK1 interacts with more than 40 different ligands, including GluA2, the dopamine transporter (DAT), mGluR7, Glt1b, and ASIC1a through their respective C-termini [22,47,68,69,70]. Remarkably, these ligands belong both to class I and II and several ligands fall entirely outside the classification. Only a few other PDZ domains, e.g., PAR3-PDZ3[71], have been reported to bind ligands of distinct classes. Though the PICK1 PDZ domain is indeed highly promiscuous, class II ligands are preferred over class I and non-classified ligands. The binding affinity of the PICK1-PDZ domain with this class II distal C-terminus of DAT (0.8 μM) is almost 20-fold better than the interaction between PICK1-PDZ and the Protein Kinase C α (PKCα) class I C-terminus (14 μM) [22].
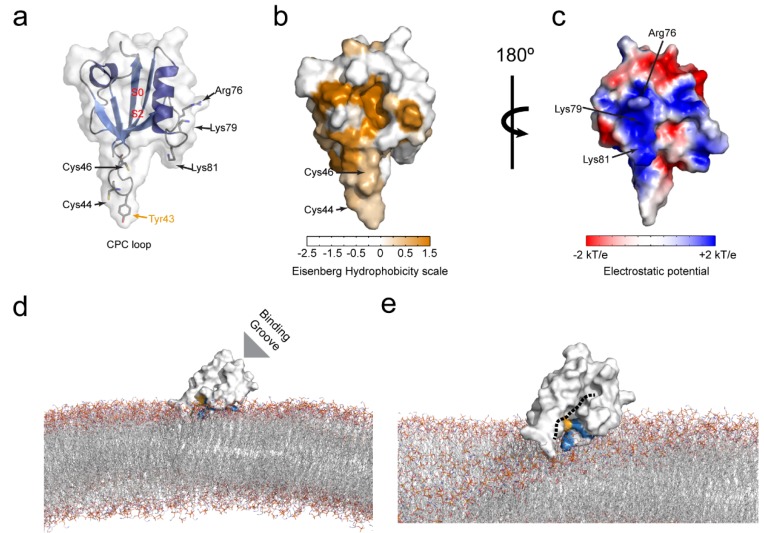
The isolated PICK1 PDZ domain has been fully structurally characterized using both X-ray crystallography and NMR spectroscopy, in complex with canonical class II ligands; DAT, GluA2, and EphrinB1. The truncated PDZ domain is only stable upon direct fusion to ligands which can effectively occupy the PDZ binding groove, however, the canonical overall globular fold consisting of six β-strands and two α-helices is conserved [22,42,72,73]. For these class II ligands the interaction is primarily facilitated via two hydrophobic pockets, S0 and S2 (see Figure 1b and Figure 3a). Hydrophobic pocket I (S0) consists of the G-φ-G-φ motif involving Ile33, Gly34, Ile35 but also a conserved Ile90 at position αB8. A preference for P0 residues of Val > Ile > Leu was previously found for S0 [68]. Hydrophobic pocket II (S2) consists of residues at positions αB1, αB4, αB5 and βB3; Lys83, Val86, Ala87, and Ile37. Notably, the αB1 residue is a highly conserved Lysine (Lys83), which is unique for the PICK1 PDZ domain. The amine side-chain of Lys83 can interact with the peptide backbone in P−4 of the ligand, and the aliphatic chain can complement the hydrophobic pocket II to make a favorable environment for a hydrophobic residue at P-2 in class II ligands [72]. The bottom of the PICK1-PDZ binding groove contains a conserved lysine (Lys27) that in the crystal structures has been shown to interact through a highly ordered water molecule, with the carboxyl C-terminal of the ligand [72].
Figure 3.
Molecular determinants of the PICK1 PDZ domain lipid membrane binding. (a) Cartoon and surface view of the PICK1 PDZ (PDB: 2LUI) [22] domain. Hydrophobic pocket I and II are depicted by S0 and S2, respectively. Cys44 and Cys46 in the CPC loop are indicated by arrows and shown as sticks. Arg76, Lys79, and Lys81, comprising the cationic patch, juxtapose to the canonical binding groove are also indicated by arrows and shown as sticks. Tyr43 involved in the upstream peptide-binding mode in indicated in orange; (b) The PICK1 PDZ domain colored by hydrophobicity (Eisenberg scale). Evidently, the binding pocket is highly hydrophobic, but also the CPC loop is predominantly hydrophobic; (c) The cationic patch is clearly visible when calculating and displaying the electrostatic potential of the domain; (d) Proposed orientation of the PICK PDZ domain in context of a negatively charged membrane. When the hydrophobic CPC loop is partly inserted into the membrane (involving one or both Cys residues, as well as Tyr43) and the cationic patch (shown in blue) facing the negatively charged membrane surface, the binding groove will still be completely accessible; (e) A rotated view of the proposed PICK1 PDZ membrane interaction suggests that the binding pocket (indicated by dashed black line) is optimally positioned to bind the C-termini of membrane embedded ligands.
PICK1 PDZ promiscuity relies on a combination of increased tolerance in the canonical binding pocket and additional non-canonical binding motifs [22]. Thus, the interaction with Class I ligand, PKCα, depends on residues upstream from the canonical binding sequence, which are likely to interact with a flexible loop between βB and βC (Tyr43 in particular, see Figure 3a) of the PDZ domain. Several additional ligands (e.g., GluA2, HER2, mGluR7b, UNC5H1, Glt1b) have bulky hydrophobic residues in the P-7 position, which may favor interaction with this part of the PICK1 PDZ domain. Furthermore, it was demonstrated that the unconventional ligand ASIC1a has a dual binding mode involving a canonical insertion and a non-canonical internal insertion with the two most C-terminal residues (P0 and P-1) forming interactions outside the groove.
6. Membrane Lipid Binding of the PICK1 PDZ Domain
6.1. Molecular Determinants of the PICK1 PDZ Domain Membrane Lipid Binding
The first studies of the membrane lipid binding capacities of PICK1 were focused exclusively on the BAR domain and suggested that the involvement of the PDZ domains was primarily to regulate the accessibility of the BAR domain to the membranes [49,53,65,66,67]. Nevertheless, truncation studies suggested that the PDZ domain rather reinforces the membrane binding capability of the BAR domain [66].
Direct binding studies with the truncated PDZ domain stabilized with the self-binding GluA2 peptide confirmed membrane lipid binding both on dot blots and in context of membranes [42]. Using lipid dot-blots it was found that PICK1 PDZ binds weakly to PI3P, PI4P, PI5P, PI(3,5)P, PI(4,5)P, and PI(3,4,5)P compared to the stronger binding observed for the PAR3 PDZ2 domain. These findings were supported by liposome sedimentation studies where PICK1 PDZ displayed dose-dependent interaction with bovine brain liposomes with an apparent Kd of 3.6 ± 0.5 ug/mg. To specifically probe the preference towards different PIPs, PC/PS (80/20) liposomes containing 10% PI3P, PI(4,5)P or PI(3,4,5)P were also tested. Consistent with the findings in the lipid dot-blot assay, PICK1 PDZ displayed preference towards the various PIPs in the following order: PI(3,4,5)P > PI(4,5)P ≥ PI3P. Interestingly it was shown that PICK1 seems to also weakly bind to PC/PS liposomes. By strategic mutations it was demonstrated that the lipid binding capacity of PICK PDZ towards bovine brain liposomes was mediated by two distinct binding motifs: a cationic patch on the opposite site of the PICK1 PDZ binding groove involving three positively charged residues, Arg76, Lys79 and Lys81 (Figure 3a,c), and a flexible loop between βB and βC involving a conserved CPC (Cys44-Pro45-Cys46) motif (Figure 3a,b). Mutation of Lys79 and Lys81 residues in the cationic patch into either Ala or Glu (K79/81A,E) significantly compromised liposome binding, without fully eliminating it. Mutation of both or individual Cys residues (C44G, C46G) in the CPC motif, on the other hand, completely abolished the PICK1 PDZ liposome binding capacity. The importance of the CPC loop was further substantiated by chemical modification of the cysteine residues into S-carboxymethyl-cysteine using iodoacetic acid, which similarly disrupted liposome binding. Functional studies furthermore demonstrated that mutation of the CPC loop compromised synaptic localization of PICK1 in hippocampal neurons, as well as the role of PICK1 in regulation of AMPAR trafficking. More recently, we further demonstrated that mutation of the CPC loop likewise compromised the PICK1 function in biogenesis of dense core vesicles in chromafin cells from the adrenal gland [74].
It was previously shown for the lipid binding of the PAR3-PDZ2 domain how a hydrophobic motif was inserted into the cytosolic bilayer of the membrane, thereby stabilizing the interaction [39]. To test whether the CPC loop of PICK1 PDZ could rely on the same insertion-based mechanism, the accessibility of the Cys residue was probed using Ellmans reagent (DTNB). The accessibility of the Cys residues was slightly reduced in presence of PC/PS liposomes but significantly reduced in presence of PI(4,5)P liposomes for the wild-type protein. In contrast, both the C44G and the C46G mutation introduced separately allowed full accessibility of the remaining Cys residue. Interestingly, the K79/81A,E mutations caused an increase in the Cys accessibility in presence of PI(4,5)P liposomes, but not in presence of PC/PS liposomes. Together these findings suggested that the cationic patch might serve to recruit the PICK1 PDZ domain to PIP–enriched liposomes via non-specific electrostatic interactions but that the PICK1 PDZ domain liposome binding is facilitated via an insertion-based mechanism, dependent on the two aliphatic cysteine side chains in the CPC loop. Furthermore, in all of the above experiments, the PICK1 PDZ domain was fused to the GluA2 C-terminus in order to stabilize it. This suggests that peptide binding and lipid binding are not mutually exclusive which concurs with the position of the cationic patch (Figure 3c), making the domain a typical class A membrane lipid-binding PDZ domain. We note that Tyr43, which is important for binding of the PKCα C-terminal peptide and likely several other ligands, is probably embedded in the lipid bilayer together with the CPC loop upon membrane binding of the PDZ domain (Figure 3a). However, the interplay between PDZ membrane and peptide binding has not yet been directly addressed experimentally (Figure 3d,e).
6.2. The CPC Loop of PICK1 PDZ Is a Proposed Zinc Binding Site
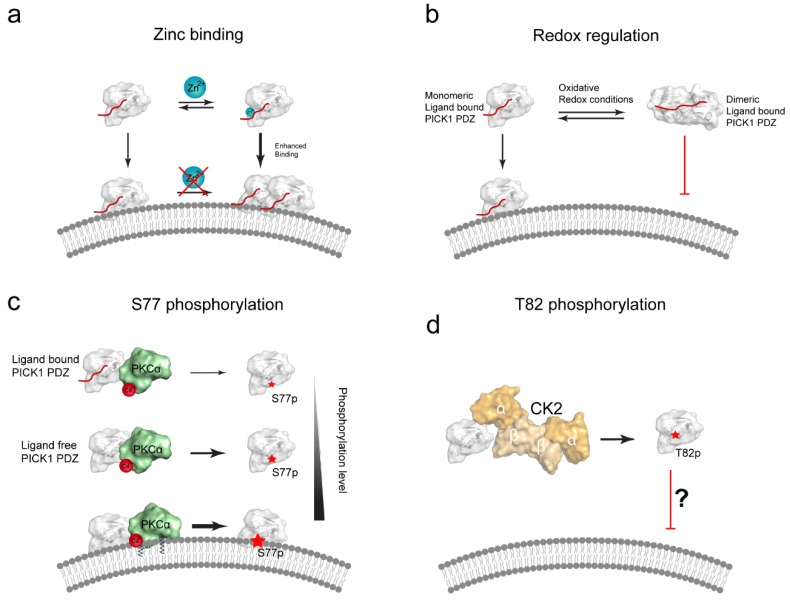
Interestingly, CPC motifs are conserved in P1B ATPases, where they localize to the center of the sixth transmembrane helix (TM6) and are directly involved in coordinating both mono- and bivalent cations (e.g., Cu+, Cu2+, Cd2+ and Zn2+) during transport [75]. Using fluorescence spectroscopy Shi et al. showed that Zn2+ can also bind specifically to the CPC loop of the PICK1 PDZ domain covalently conjugated to the GluA2 C-terminus [76]. The stoichiometry was determined to be 1:1 and an association constant to be 8.17 ± 0.23 × 105/M. The Kd was estimated to be in the lower micromolar range. Although zinc was not found to be necessary for retaining PICK1 PDZ liposome binding, it was found that Zn2+ significantly enhanced the liposome binding capacity of PICK1 PDZ, but that the interaction between liposomes and PICK1 PDZ decreased the ability of PICK1 PDZ to bind zinc. It was hypothesized that the Zn2+-binding might increase liposome binding by stabilization of the interaction itself or by stimulation of a PDZ/PDZ dimer (Figure 4a). The PDZ dimer, however, was later shown to have compromised liposome binding (see next paragraph) [73].
Figure 4.
Proposed regulatory mechanisms for the PICK1 PDZ membrane binding. (a) Zn2+-binding has been suggested to positively regulate the membrane binding of the isolated, ligand bound PICK1 PDZ domain. This interaction was shown to involve the Cys44 and Cys46 in the CPC loop. Remarkably, Zn2+-binding could be completely abolished when the PDZ domain was already bound to liposomes, suggesting that this mechanism does not require/allow Zn2+ to be inserted into the membrane [78]; (b) Proposed mechanism for redox regulation of the PICK1 PDZ domain. PICK1 forms reversible dimeric complexes under oxidative conditions, via interdomain cystines involving Cys44 and Cys46. In contrast to the monomeric ligand bound (ligand shown in red) PDZ domain, the dimeric ligand bound PDZ domain complex is not capable of binding liposomes (Dimeric PICK1-PDZ PDB:3HPK) [73]; (c) Ser77, positioned in the cationic patch, is a PKCα (shown in green, PDB: 3GPE [79]) substrate [77]. Phosphorylation of Ser77 in full length PICK1 was decreased in presence of a PDZ domain ligand, compared to a ligand free PDZ domain. Conversely, the phosphorylation level was significantly enhanced in the presence of liposomes. A decrease in clustering in COS7 cells suggested that the S77D phospho-mimetic could weaken the PDZ:lipid interaction in cells; (d) T82, positioned at the edge of the binding groove, is a putative phosphorylation site for CK2 (shown in shades of orange, PDB: 1JWH [80]). T82E likewise displays decrease cellular PICK1:GluA2 co-clusters, in a mechanism possible alleviating subcellular localization by reduced PDZ domain lipid binding capacity.
6.3. PICK1 Membrane Binding May by Redox-Regulated
A study from Shi et al. [73] showed that the CPC loop Cys residues form inter-domain disulphide bonds under mild oxidizing conditions both in vitro and in cells under oxidative stress and thereby facilitate stable covalently linked PICK1 PDZ domain dimer complexes. This dimeric PDZ arrangement allowed Shi et al. to solve a dimeric PICK1-PDZ crystal structure. Interestingly the authors also showed that the domain dimerization significantly reduced the GluA2 peptide binding affinity and, furthermore, completely obliterated the liposome binding capacity of the PICK1 PDZ domain. On this basis it was hypothesized that mild oxidation of one or both of the Cys residues could be directly involved in regulating the ability of the PICK1 PDZ domain to bind lipid membranes (Figure 4b). However, it remains to be determined to which extent such oxidative conditions and consequently the modifications are relevant in vivo.
6.4. Posttranslational Modifications of PICK1 PDZ Domain Can Regulate Membrane Binding
Posttranslational modifications (PTMs) might also play an important role in regulating the membrane lipid binding of the PICK1 PDZ domain. Ser77, which is part of the proposed lipid-binding motif opposite the binding groove, is particularly interesting. Recently, Ammendrup-Johnsen et al. [77], showed that Ser77 is phosphorylated by PKCα. Though the C-terminus of PKCα interacts with the PICK1 PDZ domain ligand [46] the phosphorylation level did not depend on a direct PDZ-PKCα interaction, nor did the phospho-mimetic mutant, S77D, alter PDZ affinities in solution. On the contrary, it was found that PDZ domain ligand binding slightly decreased the phosphorylation level of Ser77.
In presence of liposomes the phosphorylation level of Ser77 by PKCα increased a staggering 10-fold. As PKCα is also capable of binding to lipid membranes [81], it was rationalized that the membrane could act as a scaffold to facilitate phosphorylation by bringing both PKCα and PICK1 in proximity to the membrane (Figure 4c). Since Ser77 is positioned in the actual cationic patch of the PICK1 PDZ domain it is interesting that membrane binding does not sterically hinder the phosphorylation but rather promotes it. This could simply be due to the fact that dynamics/flexibility is still sufficient to allow Ser77 to access the catalytic site of PKCα even on the membrane, or it could perhaps suggest a hitherto unseen alternative insertion mechanism. Although S77D did not affect PICK1 membrane binding, as determined by the liposome sedimentation assay, it can be speculated that the functional role of this phosphorylation might be to facilitate dissociation of PICK1 from the membrane and subsequently facilitate its cellular redistribution. This hypothesis remains to be tested.
Finally, Thr82, which is situated at the edge of the binding pocket, has been predicted to be a potential Casein Kinase (CK2) phosphorylation site. The effect of this particular phosphorylation, was recently investigated by using the PICK1 T82E mutation phospho-mimetic [78]. In this study it was found that PICK1 T82E decreased the cellular co-clustering (not the co-localization, as such) with GluA2 and also failed to regulate its trafficking. Interestingly, the T82E mutation does not seem to directly affect the binding of the GluA2 C-terminus in vitro (Erlendsson, Madsen, unpublished) suggesting that the redistribution from punctate PICK1-GluA2 co-clusters into more diffuse cellular co-localization might be controlled by an altered ability of PICK1 T82E to bind specific subcellular compartments (Figure 4d). It is still unclear under which conditions and by which kinase T82 is actually phosphorylated.
7. Future Directions
The work reviewed here suggests that a general consensus regarding the mechanisms underlying lipid/membrane binding by PDZ domains is developing—yet direct structural evidence is still lacking and general conclusions might be compromised by the methodological gap between SPR based approaches, dot-blots and steady state cellular studies using confocal microscopy. The exact binding mechanism is likely to differ between domains; however, we review two prevailing mechanisms that have been shown to facilitate PDZ-membrane lipid interactions. Interestingly, and possibly important for cellular localization, several PDZ domains have different specificity towards negatively charged lipids (i.e., PIPs), which are prominent in the cytosolic leaflets of cellular membranes. This feature seems to rely on a surface-exposed cationic patch. The positioning of the cationic patch with respect to the peptide-binding groove differs between domains and, therefore, membrane binding also affects PDZ peptide/protein binding very differently. Another mechanism that has not been yet been described in general terms, is the insertion of hydrophobic appendices into the leaflet of the membrane bilayer as seen for the PDZ domain of PICK. Recent findings suggest that such insertion based membrane binding share several common features, including high functional affinity, slow kinetics, and membrane curvature sensing [82]. It will be of great interest to see whether this also applies to the membrane binding of PDZ domains and how these features relate to the function of full-length scaffolding proteins.
Author Contributions
Simon Erlendsson and Kenneth L. Madsen wrote the manuscript and prepared all figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tong L., Warren T.C., King J., Betageri R., Rose J., Jakes S. Crystal structures of the human p56lck SH2 domain in complex with two short phosphotyrosyl peptides at 1.0 A and 1.8 A resolution. J. Mol. Biol. 1996;256:601–610. doi: 10.1006/jmbi.1996.0112. [DOI] [PubMed] [Google Scholar]
- 2.Russell R.B., Breed J., Barton G.J. Conservation analysis and structure prediction of the SH2 family of phosphotyrosine binding domains. FEBS Lett. 1992;304:15–20. doi: 10.1016/0014-5793(92)80579-6. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T., Schlessingert J. SH2 and SH3 domains. Curr. Biol. 1993;3:434–442. doi: 10.1016/0960-9822(93)90350-W. [DOI] [PubMed] [Google Scholar]
- 4.Mayer B.J. SH3 domains: Complexity in moderation. J. Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 5.Macias M.J., Hyvönen M., Baraldi E., Schultz J., Sudol M., Saraste M., Oschkinat H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature. 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 6.Eck M.J., Dhe-Paganon S., Trüb T., Nolte R.T., Shoelson S.E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/S0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M.M., Huang B., Olejniczak E.T., Meadows R.P., Shuker S.B., Miyazaki M., Trüb T., Shoelson S.E., Fesik S.W. Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain. Nat. Struct Biol. 1996;3:388–393. doi: 10.1038/nsb0496-388. [DOI] [PubMed] [Google Scholar]
- 8.Feng W., Zhang M. Organization and dynamics of PDZ-domain-related supramodules in the postsynaptic density. Nat. Rev. Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 9.Doyle D.A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: Molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/S0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 10.Stahelin R.V. Lipid binding domains: More than simple lipid effectors. J. Lipid Res. 2009;50:S299–S304. doi: 10.1194/jlr.R800078-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stahelin R.V., Scott J.L., Frick C.T. Cellular and molecular interactions of phosphoinositides and peripheral proteins. Chem. Phys. Lipids. 2014;182:3–18. doi: 10.1016/j.chemphyslip.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemmon M.A. Pleckstrin homology (PH) domains and phosphoinositides. Biochem. Soc. Symp. 2007;74:81–93. doi: 10.1042/BSS2007c08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wawrzyniak A.M., Kashyap R., Zimmermann P. Phosphoinositides and PDZ domain scaffolds. Adv. Exp. Med. Biol. 2013;991:41–57. doi: 10.1007/978-94-007-6331-9_4. [DOI] [PubMed] [Google Scholar]
- 14.Van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woods D.F., Bryant P.J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-X. [DOI] [PubMed] [Google Scholar]
- 16.Cho K.O., Hunt C.A., Kennedy M.B. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy M.B. Origin of PDZ (DHR, GLGF) domains. Trends Biochem. Sci. 1995;20:350. doi: 10.1016/S0968-0004(00)89074-X. [DOI] [PubMed] [Google Scholar]
- 18.Luck K., Charbonnier S., Travé G. The emerging contribution of sequence context to the specificity of protein interactions mediated by PDZ domains. FEBS Lett. 2012;586:2648–2661. doi: 10.1016/j.febslet.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 19.Morais Cabral J.H., Petosa C., Sutcliffe M.J., Raza S., Byron O., Poy F., Marfatia S.M., Chishti A.H., Liddington R.C. Crystal structure of a PDZ domain. Nature. 1996;382:649–652. doi: 10.1038/382649a0. [DOI] [PubMed] [Google Scholar]
- 20.Van Ham M., Hendriks W. PDZ domains-glue and guide. Mol. Biol. Rep. 2003;30:69–82. doi: 10.1023/A:1023941703493. [DOI] [PubMed] [Google Scholar]
- 21.Harris B.Z., Lim W.A. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 22.Erlendsson S., Rathje M., Heidarsson P.O., Poulsen F.M., Madsen K.L., Teilum K., Gether U. Protein interacting with C-kinase 1 (PICK1) binding promiscuity relies on unconventional PSD-95/discs-large/ZO-1 homology (PDZ) binding modes for nonclass II PDZ ligands. J. Biol. Chem. 2014;289:25327–25340. doi: 10.1074/jbc.M114.548743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saro D., Martin P., Vickrey J.F., Griffin A., Kovari L.C., Spaller M.R. Structure of the Third PDZ Domain of PSD-95 Protein Complexed with KKETPV Peptide Ligand. [(accessed on 20 September 2005)]. Available online: http://www.rcsb.org/pdb/explore.do?structureId=1tp3.
- 24.Im Y.J., Im Y.J., Park S.H., Rho S.-H., Lee J.H., Kang G.B., Sheng M., Kim E., Eom S.H. Crystal Structure of GRIP1 PDZ6-Peptide Complex Reveals the Structural Basis for Class II PDZ Target Recognition and PDZ Domain-mediated Multimerization. J. Biol. Chem. 2002;278:8501–8507. doi: 10.1074/jbc.M212263200. [DOI] [PubMed] [Google Scholar]
- 25.Tochio H., Zhang Q., Mandal P., Li M., Zhang M. Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide. Nat. Struct. Biol. 1999;6:417–421. doi: 10.1038/8216. [DOI] [PubMed] [Google Scholar]
- 26.Hung A.Y., Sheng M. PDZ domains: Structural modules for protein complex assembly. J. Biol. Chem. 2002;277:5699–5702. doi: 10.1074/jbc.R100065200. [DOI] [PubMed] [Google Scholar]
- 27.Songyang Z. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- 28.Stricker N.L., Christopherson K.S., Yi B.A., Schatz P.J., Raab R.W., Dawes G., Bassett D.E., Bredt D.S., Li M. PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol. 1997;15:336–342. doi: 10.1038/nbt0497-336. [DOI] [PubMed] [Google Scholar]
- 29.Tonikian R., Zhang Y., Sazinsky S.L., Currell B., Yeh J.-H., Reva B., Held H.A., Appleton B.A., Evangelista M., Wu Y., et al. A specificity map for the PDZ domain family. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M., Wang W. Organization of signaling complexes by PDZ-domain scaffold proteins. Acc. Chem. Res. 2003;36:530–538. doi: 10.1021/ar020210b. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M., Sheng M., Sala C., Sala C. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Schultz J., Hoffmüller U., Krause G., Ashurst J., Macias M.J., Schmieder P., Schneider-Mergener J., Oschkinat H. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nat. Struct. Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 33.Grootjans J.J., Reekmans G., Ceulemans H., David G. Syntenin-syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J. Biol. Chem. 2000;275:19933–19941. doi: 10.1074/jbc.M002459200. [DOI] [PubMed] [Google Scholar]
- 34.Daniels D.L., Cohen A.R., Anderson J.M., Brünger A.T. Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat. Struct. Biol. 1998;5:317–325. doi: 10.1038/nsb0498-317. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann P., Tomatis D., Rosas M., Grootjans J., Leenaerts I., Degeest G., Reekmans G., Coomans C., David G. Characterization of syntenin, a syndecan-binding PDZ protein, as a component of cell adhesion sites and microfilaments. Mol. Biol. Cell. 2001;12:339–350. doi: 10.1091/mbc.12.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J.V., Vandekerckhove J., David G., Gettemans J. PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell. 2002;9:1215–1225. doi: 10.1016/S1097-2765(02)00549-X. [DOI] [PubMed] [Google Scholar]
- 37.Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M.Y., Degeest G., David G., Zimmermann P. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 2005;24:2556–2565. doi: 10.1038/sj.emboj.7600722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugi T., Oyama T., Morikawa K., Jingami H. Structural insights into the PIP2 recognition by syntenin-1 PDZ domain. Biochem. Biophys. Res. Commun. 2008;366:373–378. doi: 10.1016/j.bbrc.2007.11.138. [DOI] [PubMed] [Google Scholar]
- 39.Wu H., Feng W., Chen J., Chan L.-N., Huang S., Zhang M. PDZ domains of Par-3 as potential phosphoinositide signaling integrators. Mol. Cell. 2007;28:886–898. doi: 10.1016/j.molcel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 40.Kachel N., Erdmann K.S., Kremer W., Wolff P., Gronwald W., Heumann R., Kalbitzer H.R. Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): Splicing-induced modulation of ligand specificity. J. Mol. Biol. 2003;334:143–155. doi: 10.1016/j.jmb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 41.Meerschaert K., Tun M.P., Remue E., de Ganck A., Boucherie C., Vanloo B., Degeest G., Vandekerckhove J., Zimmermann P., Bhardwaj N., et al. The PDZ2 domain of zonula occludens-1 and -2 is a phosphoinositide binding domain. Cell. Mol. Life Sci. 2009;66:3951–3966. doi: 10.1007/s00018-009-0156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan L., Wu H., Shen C., Shi Y., Jin W., Xia J., Zhang M. Clustering and synaptic targeting of PICK1 requires direct interaction between the PDZ domain and lipid membranes. EMBO J. 2007;26:4576–4587. doi: 10.1038/sj.emboj.7601860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallardo R., Ivarsson Y., Schymkowitz J., Rousseau F., Zimmermann P. Structural diversity of PDZ-lipid interactions. ChemBioChem. 2010;11:456–467. doi: 10.1002/cbic.200900616. [DOI] [PubMed] [Google Scholar]
- 44.Chen Y., Sheng R., Källberg M., Silkov A., Tun M.P., Bhardwaj N., Kurilova S., Hall R.A., Honig B., Lu H., Cho W. Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell. 2012;46:226–237. doi: 10.1016/j.molcel.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivarsson Y., Wawrzyniak A.M., Kashyap R., Polanowska J., Betzi S., Lembo F., Vermeiren E., Chiheb D., Lenfant N., Morelli X., et al. Prevalence, specificity and determinants of lipid-interacting PDZ domains from an in-cell screen and in vitro binding experiments. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0054581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staudinger J., Zhou J., Burgess R., Elledge S.J., Olson E.N. PICK1: A perinuclear binding protein and substrate for protein kinase C isolated by the yeast two-hybrid system. J. Cell Biol. 1995;128:263–271. doi: 10.1083/jcb.128.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xia J., Xu J. Structure and Function of PICK1. Neuro Signals. 2006;15:190–201. doi: 10.1159/000098482. [DOI] [PubMed] [Google Scholar]
- 48.Cao M., Kam C., Xu J., Shen C., Huganir R.L., Xia J., Xu J., Shen C., Kam C., Huganir R.L., et al. PICK1 ICA69 Heteromeric BAR Domain Complex Regulates Synaptic Targeting and Surface Expression of AMPA Receptors. J. Neurosci. 2007;27:12945–12956. doi: 10.1523/JNEUROSCI.2040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steinberg J.P., Shen Y., Rubio M.E., Linden D.J., Huganir R.L., Jin W., Thomas G.M., Xia J., Yu S., Takamiya K. Targeted In Vivo Mutations of the AMPA Receptor Subunit GluR2 and Its Interacting Protein PICK1 Eliminate Cerebellar Long-Term Depression. Neuron. 2006;49:845–860. doi: 10.1016/j.neuron.2006.02.025. [DOI] [PubMed] [Google Scholar]
- 50.Anggono V., Huganir R.L. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 2012;22:461–469. doi: 10.1016/j.conb.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hanley J.G. Molecular mechanisms for regulation of AMPAR trafficking by PICK1. Biochem. Soc. Trans. 2006;34:931–935. doi: 10.1042/BST0340931. [DOI] [PubMed] [Google Scholar]
- 52.Madsen K.L., Thorsen T.S., Rahbek-Clemmensen T., Eriksen J., Gether U. Protein interacting with C kinase 1 (PICK1) reduces reinsertion rates of interaction partners sorted to Rab11-dependent slow recycling pathway. J. Biol. Chem. 2012;287:12293–12308. doi: 10.1074/jbc.M111.294702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Citri A., Bhattacharyya S., Ma C., Morishita W., Fang S., Rizo J., Malenka R.C., Citri A., Bhattacharyya S., Ma C., et al. Calcium Binding to PICK1 Is Essential for the Intracellular Retention of AMPA Receptors Underlying Long-Term Depression. J. Neurosci. 2010;30:16437–16452. doi: 10.1523/JNEUROSCI.4478-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volk L., Kim C.-H., Takamiya K., Yu Y., Huganir R.L. Developmental regulation of protein interacting with C kinase 1 (PICK1) function in hippocampal synaptic plasticity and learning. Proc. Natl. Acad. Sci. USA. 2010;107:21784–21789. doi: 10.1073/pnas.1016103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Focant M.C., Hermans E. Protein interacting with C kinase and neurological disorders. Synapse. 2013;67:532–540. doi: 10.1002/syn.21657. [DOI] [PubMed] [Google Scholar]
- 56.Holst B., Madsen K.L., Jansen A.M., Jin C., Rickhag M., Lund V.K., Jensen M., Bhatia V., Sørensen G., Madsen A.N., et al. PICK1 Deficiency Impairs Secretory Vesicle Biogenesis and Leads to Growth Retardation and Decreased Glucose Tolerance. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen A.M., Nässel D.R., Madsen K.L., Jung A.G., Gether U., Kjaerulff O. PICK1 expression in the Drosophila central nervous system primarily occurs in the neuroendocrine system. J. Comp. Neurol. 2009;517:313–332. doi: 10.1002/cne.22155. [DOI] [PubMed] [Google Scholar]
- 58.Cao M., Mao Z., Kam C., Xiao N., Cao X., Shen C., Cheng K.K.Y., Xu A., Lee K.-M., Jiang L., et al. PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He Y., Weinstein H., Liwo A., Scheraga H.A., He Y. PDZ Binding to the BAR Domain of PICK1 is Elucidated by Coarse-grained Molecular Dynamics. J. Mol. Biol. 2011;405:298–314. doi: 10.1016/j.jmb.2010.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karlsen M.L., Thorsen T.S., Johner N., Ammendrup-Johnsen I., Erlendsson S., Tian X., Simonsen J.B., Høiberg-Nielsen R., Christensen N.M., Khelashvili G., et al. Structure of Dimeric and Tetrameric Complexes of the BAR Domain Protein PICK1 Determined by Small-Angle X-Ray Scattering. Structure. 2015;23:1258–1270. doi: 10.1016/j.str.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Madasu Y., Yang C., Boczkowska M., Bethoney K.A., Zwolak A., Rebowski G., Svitkina T., Dominguez R. PICK1 is implicated in organelle motility in an Arp2/3 complex-independent manner. Mol. Biol. Cell. 2015;26:1308–1322. doi: 10.1091/mbc.E14-10-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peter B.J., Kent H.M., Mills I.G., Vallis Y., Butler P.J. G., Evans P.R., McMahon H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 63.Gallop J.L., Jao C.C., Kent H.M., Butler P.J. G., Evans P.R., Langen R., McMahon H.T. Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tarricone C., Xiao B., Justin N., Walker P.A., Rittinger K., Gamblin S.J., Smerdon S.J. The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature. 2001;411:215–219. doi: 10.1038/35075620. [DOI] [PubMed] [Google Scholar]
- 65.Lu W., Ziff E.B. PICK1 interacts with ABP/GRIP to regulate AMPA receptor trafficking. Neuron. 2005;47:407–421. doi: 10.1016/j.neuron.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 66.Jin W., Ge W.-P., Xu J., Cao M., Peng L., Yung W., Liao D., Duan S., Zhang M., Xia J. Lipid binding regulates synaptic targeting of PICK1, AMPA receptor trafficking, and synaptic plasticity. J. Neurosci. 2006;26:2380–2390. doi: 10.1523/JNEUROSCI.3503-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanley J.G. PICK1: A multi-talented modulator of AMPA receptor trafficking. Pharmacol. Ther. 2008;118:152–160. doi: 10.1016/j.pharmthera.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 68.Madsen K.L., Beuming T., Niv M.Y., Chang C.-W., Dev K.K., Weinstein H., Gether U. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem. 2005;280:20539–20548. doi: 10.1074/jbc.M500577200. [DOI] [PubMed] [Google Scholar]
- 69.Dev K.K. PDZ domain protein-protein interactions: A case study with PICK1. Curr. Top. Med. Chem. 2007;7:3–20. doi: 10.2174/156802607779318343. [DOI] [PubMed] [Google Scholar]
- 70.Keskin O., Dev K.K., Bolia A., Gerek Z.N., Banu Ozkan S., Ozkan S.B. The binding affinities of proteins interacting with the PDZ domain of PICK1. Proteins. 2012;80:1393–1408. doi: 10.1002/prot.24034. [DOI] [PubMed] [Google Scholar]
- 71.Tyler R.C., Peterson F.C., Volkman B.F. Distal interactions within the par3-VE-cadherin complex. Biochemistry. 2010;49:951–957. doi: 10.1021/bi9017335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elkins J.M., Papagrigoriou E., Berridge G., Yang X., Phillips C., Gileadi C., Savitsky P., Doyle D.A. Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions. Protein Sci. 2007;16:683–694. doi: 10.1110/ps.062657507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi Y., Yu J., Jia Y., Pan L., Shen C., Xia J., Zhang M. Redox-regulated lipid membrane binding of the PICK1 PDZ domain. Biochemistry. 2010;49:4432–4439. doi: 10.1021/bi100269t. [DOI] [PubMed] [Google Scholar]
- 74.Pinheiro P.S., Jansen A.M., de Wit H., Tawfik B., Madsen K.L., Verhage M., Gether U., Sørensen J.B. The BAR domain protein PICK1 controls vesicle number and size in adrenal chromaffin cells. J. Neurosci. 2014;34:10688–10700. doi: 10.1523/JNEUROSCI.5132-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Argüello J.M., Eren E., González-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 76.Shi Y., Zhang L., Yuan J., Xiao H., Yang X., Niu L. Zinc binding site in PICK1 is dominantly located at the CPC motif of its PDZ domain. J. Neurochem. 2008;106:1027–1034. doi: 10.1111/j.1471-4159.2008.05434.x. [DOI] [PubMed] [Google Scholar]
- 77.Ammendrup-Johnsen I., Thorsen T.S., Gether U., Madsen K.L. Serine 77 in the PDZ domain of PICK1 is a protein kinase Cα phosphorylation site regulated by lipid membrane binding. Biochemistry. 2012;51:586–596. doi: 10.1021/bi2014689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shao X., Zhu L., Wang Y., Lu Y., Wang W., Zhu J., Shen Y., Xia J., Luo J. Threonine 82 at the PDZ domain of PICK1 is critical for AMPA receptor interaction and localization. Neurochem. Int. 2010;56:962–970. doi: 10.1016/j.neuint.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 79.Guerrero-Valero M., Ferrer-Orta C., Querol-Audí J., Marin-Vicente C., Fita I., Gómez-Fernández J.C., Verdaguer N., Corbalán-García S. Structural and mechanistic insights into the association of PKCalpha-C2 domain to PtdIns(4,5)P2. Proc. Natl. Acad. Sci. USA. 2009;106:6603–6607. doi: 10.1073/pnas.0813099106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niefind K., Guerra B., Ermakowa I., Issinger O.G. Crystal structure of human protein kinase CK2: Insights into basic properties of the CK2 holoenzyme. EMBO J. 2001;20:5320–5331. doi: 10.1093/emboj/20.19.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kohout S.C., Corbalán-García S., Gómez-Fernández J.C., Falke J.J. C2 domain of protein kinase C alpha: Elucidation of the membrane docking surface by site-directed fluorescence and spin labeling. Biochemistry. 2003;42:1254–1265. doi: 10.1021/bi026596f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Madsen K.L., Bhatia V.K., Gether U., Stamou D. BAR domains, amphipathic helices and membrane-anchored proteins use the same mechanism to sense membrane curvature. FEBS Lett. 2010;584:1848–1855. doi: 10.1016/j.febslet.2010.01.053. [DOI] [PubMed] [Google Scholar]