Abstract
Cryptosporidiosis is a serious diarrheal disease in immunocompromised patients and malnourished children, and treatment is complicated by a lack of adequate drugs. Recent studies suggest that the natural occurrence of a small gatekeeper residue in serine threonine calcium-dependent protein kinase 1 (CDPK1) of Cryptosporidium parvum might be exploited to target this enzyme and block parasite growth. Here were explored the potency with which a series of pyrazolopyrimidine analogs, which are selective for small gatekeeper kinases, inhibit C. parvum CDPK1 and block C. parvum growth in tissue culture in vitro. Although these compounds potently inhibited kinase activity in vitro, most had no effect on parasite growth. Moreover, among those that were active against parasite growth, there was a very poor correlation with their 50% inhibitory concentrations against the enzyme. Active compounds also had no effect on cell invasion, unlike the situation in Toxoplasma gondii, where these compounds block CDPK1, prevent microneme secretion, and disrupt cell invasion. These findings suggest that CPDK1 is not essential for C. parvum host cell invasion or growth and therefore that it is not the optimal target for therapeutic intervention. Nonetheless, several inhibitors with low micromolar 50% effective concentrations were identified, and these may affect other essential targets in C. parvum that are worthy of further exploration.
INTRODUCTION
Cryptosporidiosis has recently gained increased recognition as a globally important cause of diarrheal disease in humans (1). Human cryptosporidiosis is caused primarily by two species, Cryptosporidium parvum and C. hominis. Although C. parvum also infects agricultural animals and is acquired as a zoonosis, C. hominis is transmitted from human to human. Infection occurs when oocysts shed in the feces contaminate food or water and are accidentally ingested. Oocysts are extremely environmentally resistant and highly infectious, making the life cycle difficult to interrupt (2). Following infection, most healthy individuals suffer only temporary discomfort and go on to self-cure; however, cryptosporidiosis is a serious, life-threatening infection in the immunocompromised (1). Recently, cryptosporidiosis has also been recognized as a serious cause of morbidity in young children in Africa and South Asia, being among the top three diarrhea-causing agents (3). Complicating this situation, there are few effective therapies for cryptosporidiosis, and the one FDA-approved drug, nitazoxanide, is not effective in immunocompromised patients or malnourished children (4).
Cryptosporidium is a deep-branching member of the phylum Apicomplexa, and it differs in many ways from distantly related parasites such as Toxoplasma gondii (5). Although Cryptosporidium is intracellular during its replicative phase, it exists in a membrane-bound compartment at the apical surface of intestinal epithelial cells, remaining separated from the cytosol by a feeding organelle (6). The genome of C. parvum is highly streamlined (7), lacking many metabolic pathways while containing a large number of transporters involved in nutrient uptake (8). As a result of its unique metabolism, and perhaps its unusual intracellular niche, few drugs that are effective against related parasites act on C. parvum or C. hominis (4). Therefore, there is a need to define new targets and to identify compounds that effectively inhibit parasite growth. An added challenge in working with Cryptosporidium is that it does not undergo continuous propagation in vitro, although short-term tissue culture assays have been used to screen for small-molecule inhibitors of C. parvum replication (9, 10).
Like other eukaryotes, Cryptosporidium contains a diverse complement of protein kinases (11), including members of the calcium-dependent protein kinase (CDPK) family that are expanded in apicomplexans (12). The fact that CDPKs are plantlike and absent from animal cells has made them attractive targets for consideration in developing therapeutic interventions (13). Cryptosporidium shares a unique property with T. gondii in having a small Gly gatekeeper residue in CDPK1 (14, 15), a feature that is unprecedented among kinases in animal cells (16). The importance of the gatekeeper residue has previously been used to engineer kinases to acquire sensitivity to pyrazolopyrimidine (PP) analogs, which are typically excluded by the bulky gatekeeper residue in the ATP-binding pocket of normal kinases (17, 18). Because of its naturally small gatekeeper, T. gondii CDPK1 (TgCDPK1) is highly sensitive to PP inhibitors (19, 20). Inhibition of TgCDPK1 blocks microneme secretion and prevents host cell invasion by T. gondii (19, 21). The gene for CDPK1 is essential in T. gondii (19), and a number of PP analogs have been developed to target this enzyme in vitro (22, 23). In addition to the chemical genetic evidence that large gatekeeper mutant forms of CDPK1 become refractory to inhibition (19), the tight correlation between enzyme in vitro 50% inhibitory concentrations (IC50s) and in vivo growth inhibition 50% effective concentrations (EC50s) (24) argues that CDPK is the primary target of PP inhibitors in T. gondii. Similarly, several studies have explored the sensitivity of C. parvum CDPK1 (CpCDPK1) in vitro by using series of PP inhibitors, showing that many were potent against the enzyme (25, 26). These findings were further extended by showing that one such inhibitor, called compound 1294, which was designed against TgCDPK1 (27), effectively blocks C. parvum growth in cell culture at submicromolar concentrations and reduces oocyst shedding in an immunocompromised-mouse model (28). These studies suggest that CpCDPK1 might be a good target for which to explore additional PP analogs.
Here we tested a broader array of PP-like compounds to establish the relative potencies with which they inhibit CpCDPK1 enzyme activity in vitro in comparison to their inhibition of parasite growth in an in vitro cell culture assay. We were surprised to find that although PP-like compounds are potent CpCDPK1 enzyme inhibitors, there was a poor correlation with their ability to inhibit parasite growth. This suggests that CDPK1 is either not essential or not the primary target of these compounds in vivo. Moreover, unlike the situation in T. gondii, the active analogs used here block only C. parvum growth and not parasite invasion. The findings indicate that although some members of the PP scaffold are effective against C. parvum, they may act on targets other than CpCDPK1, which may not be an appropriate target for the development of inhibitors to combat cryptosporidiosis.
MATERIALS AND METHODS
Compounds.
PP derivatives were synthesized as described previously (24, 29–31) and were >95% pure, as determined by liquid chromatography-mass spectrometry. Compound 22 is a trisubstituted pyrrole inhibitor of PKG (32), and it was provided by Merck under a material transfer agreement.
C. parvum strain propagation and purification of oocysts.
The AUCP-1 isolate of C. parvum was maintained by repeated passage in male Holstein calves (IACUC-approved protocol). Oocysts were purified from collected feces after sieve filtration, Sheather's sugar flotation (33), and discontinuous sucrose density gradient centrifugation (34). Oocysts were washed and stored at 4°C in 50 mM Tris–10 mM EDTA, pH 7.2. Oocysts were used within 1 to 2 weeks of initial isolation from stored feces, when viability remained above 80% as judged by excystation.
Excystation of oocysts and purification of sporozoites.
Sporozoites were excysted by a modified form of a previously described procedure (35). Briefly, purified oocysts (about 1 × 108) were washed in Hanks balanced salt solution (HBSS) or phosphate-buffered saline (PBS) and treated with an equal volume of 40% commercial laundry bleach for 10 min at 4°C. Oocysts were washed four times in PBS containing 1% (wt/vol) bovine serum albumin (BSA). Oocysts were suspended in HBSS, incubated for 60 min at 37°C, and then mixed with an equal volume of warm 1.5% sodium taurocholate in HBSS. After excystation for 60 min at 37°C, the sporozoite-oocyst-shell mixture was collected by centrifugation and suspended in RPMI 1640 medium containing 10% fetal bovine serum and sporozoites were separated from oocyst shells and unexcysted oocysts by filtration through 3-μm Whatman Nuclepore filters. Purified sporozoites were enumerated with a hemocytometer and used immediately for infection of cell monolayers.
C. parvum growth or invasion assays with cell monolayers.
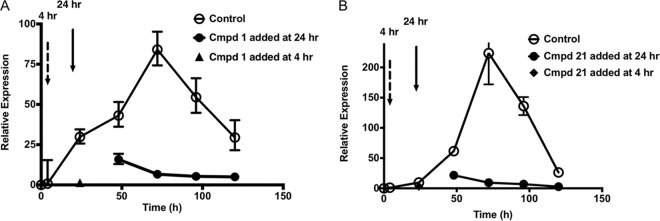
HCT-8 cells were obtained from the ATCC (CCL244) and cultured in RPMI 1640 medium (Sigma) supplemented with 2 g of sodium bicarbonate per liter, 2.5 g of glucose per liter, a final concentration of 10% fetal bovine serum (Bio-West), 1× antibiotic-antimycotic (Gibco), and 1× sodium pyruvate (Sigma). Cells were used to seed 48-well plates and grown to confluence within 2 to 3 days. For growth experiments, sporozoites (5 × 104 to 1 × 105/0.1 ml) in medium with serum were added to each well and incubated for 4 h at 37°C. Cells were rinsed three times with PBS, and then 0.3 ml of a compound at various concentrations or a dimethyl sulfoxide (DMSO) sham control was added to each well in triplicate. Incubation was continued for 48 h. For invasion experiments, sporozoites were premixed with various compounds or a DMSO sham control for 15 min and then 0.1-ml volumes were added to wells in triplicate. After 4 h at 37°C, wells were washed three times with PBS, followed by the addition of 0.3 ml of medium with serum and 24 or 48 h of continued growth at 37°C. At the end of each experiment, cells were chilled on ice and each sample consisting of three wells was extracted with 100 μl of cold TRIzol reagent (Invitrogen) per well, combined, frozen quickly in dry ice, and stored at minus 80°C until RNA harvest. For growth assays (see Fig. 5), sporozoites were added to host cells and cell layers were washed after 4 h at 37°C and then incubated for 24 h in medium without compounds. Following this establishment of infection, compound 21 (6.25 μM) or compound 1 (10 μM) was added at 24 h postinfection. At 24-h intervals, the medium was removed and fresh medium containing fresh drug at the original concentration was added to the remaining wells. As a positive control, one set of wells was incubated with either compound 21 (6.25 μM) or compound 1 (10 μM) immediately following the 4-h sporozoite invasion step and harvested at 24 h. At the end of each incubation step, cells were harvested with TRIzol reagent and frozen as described above until purification of RNA. Experiments were repeated two or more times with three technical replicates included in each experiment.
FIG 5.
Time dependence of the sensitivity of C. parvum growth to compounds. (A) Sensitivity of C. parvum growth to compound (Cmpd) 1. (B) Sensitivity of C. parvum growth to compound 21. Open circles, growth in the absence of inhibitors; black triangles, inhibitors added at 4 h postinfection (dashed arrow) representing a positive control demonstrating that compounds completely blocked C. parvum growth during the first 24 h; filled circles, inhibitors added at 24 h postinfection (solid arrow) and then replaced every 24 h thereafter (see Materials and Methods for additional details). Relative expression is defined as normalized expression (see Materials and Methods) at the time points indicated with and without the drug divided by the normalized expression at the initial 4-h time point (representing initial sporozoite invasion but no growth) as described in Materials and Methods. Values represent means ± standard deviations (n = 3).
C. parvum growth assays monitored by indirect fluorescent-antibody (IFA) assay.
Following growth experiments performed as described above, the medium was removed and cell layers were immediately fixed and stained with antibody to C. parvum (SporoGlo; Waterborn, Inc.) as previously described (36). Briefly, cells were fixed with methanol-acetic acid (9:1) for 2 min, rehydrated, permeabilized by two successive washes with a washing buffer containing 0.1% Triton X-100, blocked with 5% normal goat serum, and stained with antibody overnight at 4°C. The stained cells were washed twice with PBS, followed by water, and then observed with an inverted fluorescence microscope with a 20× objective. Infected cells containing replicating parasites appeared as fluorescent clusters of infectivity or foci. The numbers of fluorescent particles within each focus per 20× field were determined by automatic microscopic collection of nine fields per well of a 96-well plate. These nine fields were assembled into a single montaged image for each well, equating to approximately 75% of the surface area of each well. The number of fluorescent particles in each montage was determined with either Metamorph (Molecular Devices, Inc.) or Image J software and is expressed in fluorescent-focus units (FFU). Experiments were repeated two or more times with three technical replicates included in each experiment.
RNA isolation and RT-qPCR.
Total RNA from TRIzol samples was purified with Direct-zol RNA MiniPrep (Zymo Research) according to the manufacturer's directions, which included DNase1 digestion. Concentrations were determined and quality was assessed by measurement of absorbance at 230, 260, and 280 nm. First-strand cDNA was synthesized from total RNA with Superscript III (Invitrogen) according to the manufacturer's directions. Reverse transcription-quantitative PCR (RT-qPCR) was performed with Power SYBR green PCR master mix (Applied Biosystems Life Technologies) according to the manufacturer's directions. Expression of human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) employed primers 5′-CGACAGTCAGCCGCATCTT-3′ (forward) and 5′-ATCCGTTGACTCCGACCTTC-3′ (reverse). C. parvum 18S rRNA was detected with primers 5′-TAGAGATTGGAGGTTGTTCCT-3′ (forward) and 5′-CTCCACCAACTAAGAACGGCC-3′ (reverse). Samples were heated to 50°C for 2 min, followed by 95°C for 10 min, followed by 40 thermal cycles (95°C for 15 s and then 60°C for 1 min, followed by a 60°C-to-95°C melt curve) of PCR amplification with an Applied Biosystems 7500 real-time PCR detection system (Life Technologies). Triplicate reaction mixtures were prepared for each sample. Normalized C. parvum 18S rRNA gene expression relative to that of the reference gene for GAPDH was calculated with ABI 7500 software version 2.3. Normalized gene expression was linear from a dose of 103 sporozoites to a dose of 2.5 ×105 sporozoites.
Protein purification.
Plasmids expressing residues T71 to E538 of CpCDPK1 (15) or full-length TgCDPK1 (24) were transformed into Escherichia coli BL21(DE3)-V2R-pACYC-LIC+LamP phosphatase-expressing cells, which were described previously (15). A single colony was inoculated into 5 ml of Terrific broth (TB) with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) and cultured overnight at 37°C. The next day, the overnight culture was diluted into 250 ml of fresh TB with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml). After growth for 3 to 3.5 h at 37°C, cultures were cooled to 15°C and induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for CpCDPK1 or 1 mM IPTG for TgCDPK1 during growth overnight. For CpCDPK1, the cell pellet was then sonicated and purified with HIS-Select nickel affinity gel (Sigma). For TgCDPK1, the cell pellet was lysed in CelLytic B 2× (Sigma) with added lysozyme, Benzonase, and a protease inhibitor cocktail. The proteins were purified with HIS-Select nickel affinity gel (Sigma) and dialyzed in a buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.125% Chelex 100. Glycerol was added to 25%, dithiothreitol (DTT) was added to a final concentration of 0.5 mM, and the proteins were stored at −80°C. Protein concentrations were determined by SDS-PAGE separation and SYPRO-Ruby Protein Gel staining (Invitrogen) in comparison with a BSA standard.
CpCDPK1 enzyme assay.
A coupled enzyme ATPase assay was used to measure CpCDPK1 activity as described previously (15). The coupled assay was carried out with 150 μM reduced β-NAD (NADH), 300 μM phosphoenolpyruvic acid, 50 mM KCl, and a 3-U/ml mixture of pyruvate kinase and lactate dehydrogenase (Sigma). The following conditions were used for IC50 determination: 13 nM CpCDPK1, 50 μM ATP, 50 μM Syntide-2 peptide (Calbiochem), and serial dilutions of inhibitors (10 pM to 20 μM) in a buffer containing 20 mM HEPES (pH 7.5), 30 mM NaCl, 10 mM MgCl2, 1 mM CaCl2, 2 μg/ml BSA, 10 mM DTT, and 0.01% (vol/vol) Tween 20. Compound dilutions were incubated with CpCDPK1 for 15 min at room temperature, and then ATP was added to initiate the kinase reaction. After incubation for 40 min at 30°C, reactions were read by measuring absorbance at 340 nm on a Cytation3 plate reader (BioTek). Assays were repeated two or more times, and average values were used to determine IC50s by nonlinear curve fitting of normalized data with Prism (GraphPad).
TgCDPK1 expression and enzyme assay.
An enzyme-linked immunosorbent assay (ELISA)-based method was used to measure TgCDPK1 activity as described previously (24). Syntide-2 peptide (Calbiochem) was used to coat 96-well plates (Immulon 4HBX; Thermo Scientific) at 10 μg/ml (100 μl/well) in carbonate coating buffer, pH 9.6, at 4°C overnight. After being washed with 50 mM Tris-HCl (pH 7.5)–0.2% Tween 20, the plates were blocked with 250 μl of 3% BSA for 2 h at room temperature. Kinase reactions were carried out at 30°C for 30 min in a kinase reaction buffer consisting of 20 mM HEPES/KOH (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.1 mM EGTA, 2.5 mM CaCl2, and 0.005% Tween 20. Purified TgCDPK1 (15 nM) was added to serial dilutions of inhibitors (10 pM to 10 μM) in kinase reaction buffer and incubated for 15 min at room temperature. Reactions were initiated by the addition of ATP to a final concentration of 50 μM. After 30 min, the solution was removed and phosphorylated Syntide2 was detected with monoclonal antibody MS-6E6 (Cyclex Co.), followed by peroxidase-conjugated secondary goat anti-mouse IgG. Reaction products were developed with the substrate 3,3′,5,5′-tetramethylbenzidine and detected by measuring absorbance at 450 nm. Assays were repeated two or more times, and average values were used to determine IC50s by nonlinear curve fitting of normalized data with Prism (GraphPad).
RESULTS
Choice of PP analogs.
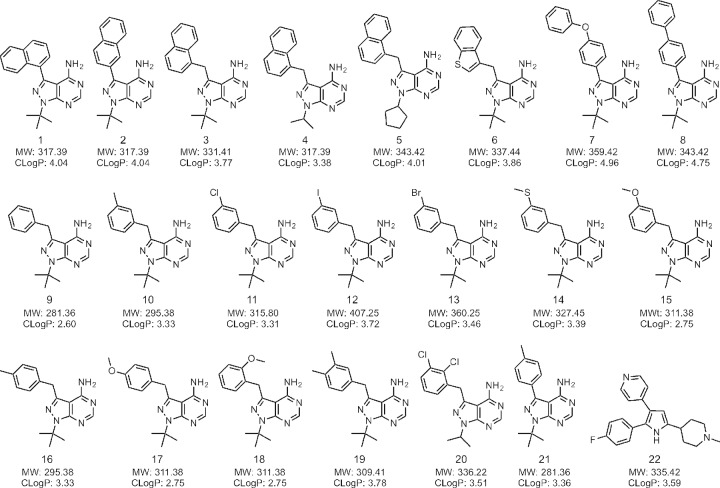
We selected a series of previously described PP analogs designed to probe the expanded hydrophobic cavity of CDPK1 that is created by the small gatekeeper residue. Included were commonly used analogs such as 1-naphthyl-PP (1NA, compound 1), 2-naphthyl-PP (2NA, compound 2), and 1-naphthyl-methyl-PP (2NM, compound 3), as well as derivatives containing isopropyl (compound 4) or cyclopentyl (compound 5) at the N1 position (Fig. 1) or modified at the C-3 position to contain a benzothiaphene (compound 6). We also included several compounds with bulkier phenyl-oxy-phenol or biphenyl groups at the C-3 position (compounds 7, 8). Benzyl groups modified at the C-3 position (meta) have previously proven to be potent inhibitors of TgCDPK1, so we included a series of these analogs here (compounds 9 to 15), as well as similar compounds modified at C-4 (para) (compounds 16 to 17) or C-2 (ortho) (compound 18) and several with dual substitutions on the benzyl ring (compounds 19 and 20). For comparison, we included the original PP1 compound, 4-methyl-phenyl PP (compound 21) (Fig. 1). Compound 21 was originally described as an inhibitor of Src, which contains Thr at the gatekeeper, although like most of these compounds, it is more potent inhibitor of kinases with small (i.e., Ala, Gly) residues at the gatekeeper position (37). We also included a previously described trisubstituted pyrrole inhibitor of PKG in apicomplexans (compound 22 here, often referred to as compound 1 in the Toxoplasma literature) (32).
FIG 1.
Structures of the compounds used in this study. Compound 21 is the original PP inhibitor, previously referred to as PP1 (49). Compound 22 is an unrelated trisubstituted pyrrole that inhibits PKG (44). MW, molecular weight.
PP analogs inhibit CpCDPK1 and TgCDPK1 with similar potencies.
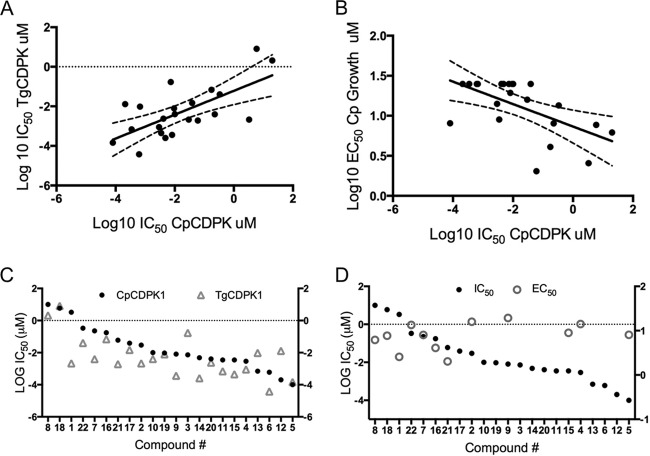
We compared the potencies with which PP compounds inhibit the activities of the CpCDPK1 and TgCDPK1 enzymes, which were recombinantly expressed in E. coli and purified to homogeneity as described previously (15, 19). A previously described ELISA was used to monitor the activity of TgCDPK1 (24), as it provided consistent results between replicates. However, this assay proved more variable when monitoring the activity of CpCDPK1, and therefore, a more sensitive coupled enzymatic assay was used to monitor the activity of this enzyme, as described previously (25). PP analogs potently inhibited both enzymes, with IC50s ranging from low nanomolar to low micromolar (Table 1). Because of different assay conditions were used, it is not possible to directly compare the potencies with which a given compound inhibits the two enzymes. Nevertheless, the relative rank of potencies should remain the same if they have the same structure-activity relationship. Comparison of the IC50s for TgCDPK1 and those for CpCDPK1 shows that there is a significant correlation between the potencies of the compounds (Pearson correlation coefficient [r], 0.6734; P ≤ 0.001) (Fig. 2A). The exceptions to this pattern were the much more potent activities of compounds 1, 7, 9, and 21 against TgCDPK1 versus CpCDPK1; in contrast, compounds 12, 3, and 13 were more potent against CpCDPK1 (Fig. 2B). Compound 22, an inhibitor of PKG, was a reasonably potent inhibitor of both TgCDPK1 and CpCDPK1 in vitro, an effect consistent with previous reports that this compound also targets small gatekeeper kinases in vitro (38).
TABLE 1.
Comparison of enzyme inhbition (IC50) and growth inhibition (EC50)
| Compound | Compound no. | IC50 (μM) for T. gondiia |
C. parvum |
|
|---|---|---|---|---|
| IC50 (μM)a | EC50 (μM)b | |||
| 1NA | 1 | 0.00213 | 3.31050 | 2.56 |
| 2NA | 2 | 0.00213 | 0.02970 | 15.90 |
| 1NM | 3 | 0.16950 | 0.00730 | NIc |
| CZ33 | 4 | 0.00089 | 0.00290 | 14.11 |
| CZ34 | 5 | 0.00015 | 0.00010 | 8.06 |
| CZ43 | 6 | 0.00004 | 0.00060 | NI |
| CZ29 | 7 | 0.00397 | 0.22560 | 8.02 |
| CZ30 | 8 | 2.08600 | 10.00000 | 6.18 |
| Bn | 9 | 0.00036 | 0.00810 | 19.34 |
| 3MB | 10 | 0.00399 | 0.01000 | NI |
| 3ClB | 11 | 0.00069 | 0.00350 | NI |
| 3IB | 12 | 0.01295 | 0.00020 | NI |
| 3BrB | 13 | 0.00964 | 0.00670 | NI |
| CZ42 | 14 | 0.00026 | 0.00480 | NI |
| CZ31 | 15 | 0.00044 | 0.00350 | 8.96 |
| CZ15 | 16 | 0.06840 | 0.17530 | 4.08 |
| CZ20 | 17 | 8.15800 | 5.89800 | 7.68 |
| CZ19 | 18 | 0.01471 | 0.03870 | NI |
| CZ23 | 19 | 0.00787 | 0.00950 | NI |
| CZ72 | 20 | 0.00238 | 0.00410 | NI |
| PP1 | 21 | 0.00192 | 0.06020 | 2.03 |
| Compound 1 | 22 | 0.04041 | 0.33360 | 13.47 |
IC50s are enzyme assay results.
EC50s reflect growth as determined by PCR assay.
NI, no inhibition at 25 μM.
FIG 2.
Comparison of the activities of TgCDPK1 and CpCDPK1 enzyme inhibitors and inhibition of C. parvum growth. (A) Comparison of the sensitivities of the CpCDPK1 and TgCDPK1 enzymes to inhibitors in vitro. Linear regression is shown; dashed lines indicate 95% confidence intervals. (B) Plot of IC50s of individual compounds for the CpCDPK1 and TgCDPK1 enzymes. Linear regression is shown; dashed lines indicate 95% confidence intervals. (C) Sensitivity of CpCDPK1 enzyme activity (IC50) versus growth (EC50) of C. parvum in culture to inhibitors. (D) Plot of IC50s of individual compounds for CpCPKD1 enzyme activity versus EC50s against C. parvum growth. IC50s and EC50s are from Table 1.
In comparing the relative potencies with which the various PP analogs inhibit CpCDPK1 enzyme activity in vitro, several patterns were evident (Table 1; Fig. 1). Among the naphthyl derivatives modified at the C-3 position, compound 3 (1-naphthyl-methylene) was the most potent, and its activity increased with the replacement of the tertiary butyl group at N1 with either isopropyl (compound 4) or cyclopentyl (compound 5). Biaryl C-3 substituents such as compounds 7 and 8 lost potency, which likely reflects the different angle imposed by both the direct aryl linkage and the bulkier diphenyl groups. Compound 6 showed improved potency, suggesting that the benzothiophene makes a favorable interaction in the pocket. In terms of the smaller benzyl substituents, modifications to the meta position (C-3) of the benzyl group were all quite potent against CpCDPK1 (compounds 9 to 15). Modification at the para position (C-4) with a methyl group (compound 16) lost potency relative to that of the C-3-substituted form (compound 10), and this was further diminished by the substitution of a bulkier methoxy group at C-4 (compound 17). In comparison, substitution of a similar group at C-2 showed reasonable potency against both CpCDPK1 and TgCDPK1 (compound 18). Double-methyl substitutions at C-3 and C-4 (compound 19) improved potency over that of the single para substitution (compound 16), while Cl substitutions at C-2 and C-3 (compound 20) were similar to the single C-3 substitution (compound 11) (Table 1; Fig. 1). Overall, most of the PP inhibitors tested reasonably potently inhibited both CpCDPK1 and TgCDPK1.
Lack of correlation between enzyme inhibition and parasite growth inhibition.
We were interested in comparing the in vitro inhibition of CpCDPK1 enzyme activity to inhibition of parasite growth in cell culture. The expectation is that if inhibition of CpCDPK1 is important for growth, there should be a correlation between activity against the enzyme and the ability to block parasite growth. For these comparisons, we monitored the growth of C. parvum in monolayers of HCT-8 cells by either a PCR-based method or an immunofluorescence-based assay as described previously (36). Under these conditions, C. parvum expands for several days before becoming growth arrested. Inhibition is expressed as the growth level relative to that of an untreated control. We utilized freshly isolated and purified sporozoites for infection in order to maximize the chance to measure inhibition of invasion, although inhibitors were left in for the duration of the assay used for EC50 determination.
Unlike the enzyme activity assay, where nearly all of the compounds were potent inhibitors, many of them failed to inhibit parasite growth at the highest concentration tested (25 μM) (Table 1). Approximately half of the compounds showed inhibition in this range, allowing us to determine EC50s that ranged from ∼2 to 20 μM (Table 1). Although some loss of potency might be expected when treating whole-cell cultures, these values were 100- to 1,000-fold higher than those necessary for effective inhibition of the enzyme (Table 1). Moreover, when we compared the relative enzyme inhibitory potencies of the compounds in vitro with their abilities to block parasite growth, there was a significant and negative correlation between the EC50s and IC50s (r = −0.58; P ≤ 0.005) (Fig. 2B). When the paired IC50s-EC50s of each compound were plotted together, it was apparent that (i) many PP analogs lack activity against the parasite, (ii) those that are active are only modestly so, and (iii) there is little correlation between the inhibition of enzyme activity and parasite growth (Fig. 2D).
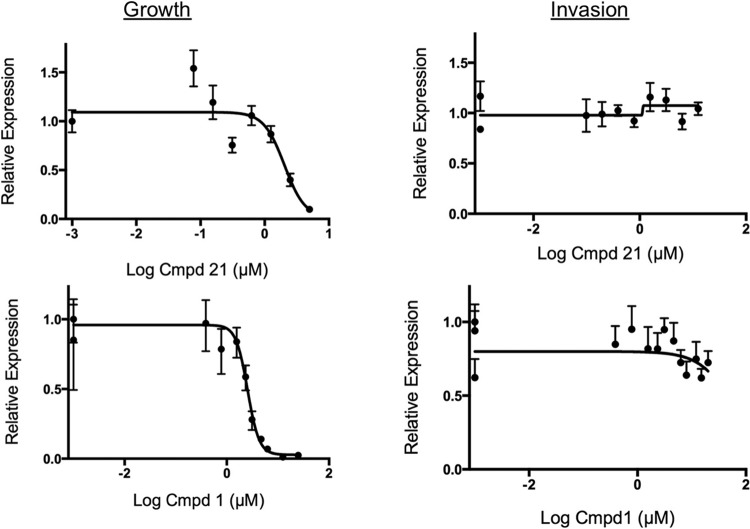
PP analogs act on growth, not invasion, of C. parvum.
Previous studies have shown that PP analogs inhibit TgCDPK1, block microneme secretion, and therefore prevent cell invasion (19). To determine if the active PP analogs studied here have a similar mechanism, we compared the effects of potent inhibitors of invasion with C. parvum growth by using specific assays to separate these phases of infection. For invasion, we treated sporozoites briefly (15 min) with compounds, added them to cell monolayers, and incubated them for 4 h before washing out the compound and culturing the cells in fresh medium. For growth, the compounds were added 4 h after infection with purified sporozoites and then left in for the duration of the experiment. Under these conditions, compounds 1 and 21 effectively inhibited growth only when added at 4 h postinfection and left in the culture continuously (Fig. 3). The same result, that the compounds were specific for inhibition of growth, was obtained with a PCR-based assay (Fig. 3) and an IFA assay that measured parasite foci by semiautomated microscopy (Fig. 4). To extend these findings further, we used concentrations that were approximately three times the EC90s of compounds 1 and 21 and added the compounds at 24 h after infection. Even when added at this late time point, the compounds effectively prevented growth over an extended culture period. These same doses of compounds provided sterilizing protection when added at 4 h postinfection (Fig. 5). In composite, these finding clearly show that effective PP analogs, at least compounds 1 and 21, work only by blocking growth and have no effect on C. parvum sporozoite invasion.
FIG 3.
Comparison of the effects of compounds on invasion and growth of C. parvum in HCT-8 cells in vitro. Compounds were tested in C. parvum growth or invasion assays by using RT-qPCR assays to quantify parasite numbers as described in Materials and Methods. Invasion was monitored by pretreatment (15 min) of isolated sporozoites, followed by challenge of monolayers and removal of the compound (Cmpd) at 4 h. Growth was monitored by inoculation of isolated sporozoites, followed by the addition of compounds at 4 h postinfection and continual culture in the compounds. Relative expression values represent the values of experimental samples (plus drug) normalized to those of control samples (no drug) (ratio of gene expression at each drug concentration/control). Triplicate PCR assays were performed, and relative expression values are presented as means ± standard deviations (n = 3). Curve fitting was performed by nonlinear regression with Prism (GraphPad). EC50s are provided in Table 1.
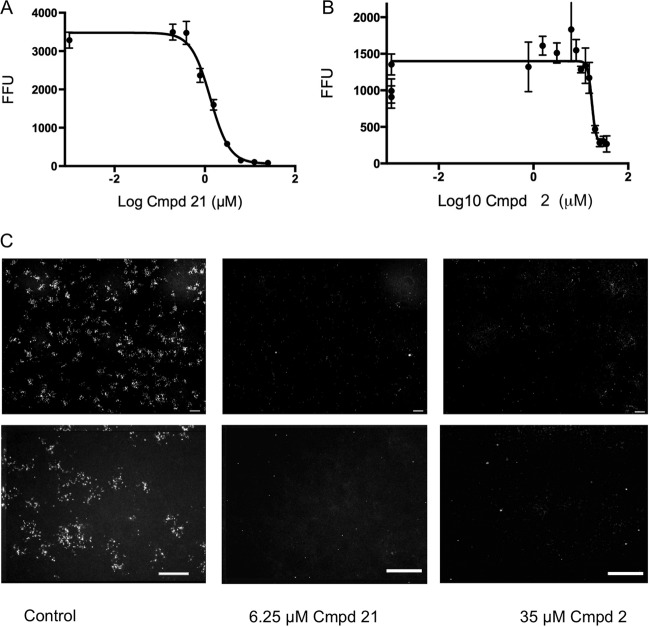
FIG 4.
Effects of selected compounds on C. parvum growth as determined by IFA assays. (A) Dose response of inhibition of growth was monitored by IFA assay detection of intracellular C. parvum forms as described in Materials and Methods. FFU values represent means ± standard deviations (n = 3). Curve fitting was performed by nonlinear regression with Prism (GraphPad). (B) Representative images of IFA staining of control and compound (Cmpd) 1- and 21-treated cultures. The upper set of images depicts representative single-well montage photomicrographs (comprising nine fields) of a 96-well plate as described in Materials and Methods. The lower images are each enlargements representing one of the nine image frames shown in the corresponding upper images. EC50s are provided in Table 1. Scale bars, 200 μm.
DISCUSSION
We have used a series of well-studied PP analogs that target small gatekeeper kinases to explore the relationship between inhibition of CDPK1 activity in vitro and blocking of C. parvum growth in cell culture. As expected, PP analogs were potent inhibitors of the CpCDPK1 enzyme and their activities were largely similar to that against TgCDPK1, although there were notable differences in some compounds. In contrast to previous studies with T. gondii, there was little correlation between the activities of PP inhibitors against the CpCDPK1 enzyme in vitro and their abilities to inhibit C. parvum growth in cell culture. Moreover, the activity of PP analogs that did inhibit growth was not dependent on blocking of cell invasion, unlike the situation for T. gondii. These findings suggest that although C. parvum has a conserved small gatekeeper residue in CPKD1, this enzyme may not be a good target for the development of inhibitors to prevent growth in vivo.
The majority of the PP inhibitors tested were similarly potent against TgCDPK1 and CpCDPK1 enzymes in vitro; however, differences in the potencies of some analogs were seen. These differences likely result from differences in the structure of the ATP-binding pocket, despite the closely similar crystal structures of these two proteins (14, 15, 20). Previous cocrystal structures with selected PP analogs (14, 20) could be extended to further refine these differences and design compounds with improved potency; however, in the case of CpCDPK1, this effort may not be warranted, given the discordant in vivo sensitivity of C. parvum growth to these inhibitor, as discussed below.
Consistent with previous reports (25, 26), we observed that a number of PP analogs were potent inhibitors of CpCDPK1 activity in vitro. Among the naphthyl derivatives modified at the C-3 position, linkage through a methylene group, as in compound 3 (1-naphthyl-methylene), was more potent that the direct linkage in compounds 1 and 2. Modification of the N1 position from tertiary butyl (compound 3) to isopropyl (compound 4) or cyclopentyl (compound 5) further improved the potency of compound 3 against the CpCDPK1 enzyme. However, even among this small set, it was already evident that there was a lack of correlation between inhibition of enzyme activity and inhibition of parasite growth, as the two most potent 1-naphthyl-methylene derivatives (compounds 5 and 6) had no activity against the parasite. Indeed, four of the six most potent inhibitors of enzyme activity (compounds 3, 4, 5, 6, 12, and 13) had no activity against C. parvum growth, and this pattern continued when the entire series was compared. One possible explanation for these findings is that the PP analogs tested here were not able to gain access to the target in vivo. However, this seems unlikely, given that they all conform to Lipinski's rule of five, which characterizes bioactive drugs on the basis of their small size (molecular mass of <500 Da) and low hydrophobicity (calculated log octanol/water partition coefficient [CLogP], <5) (39), as listed in Fig. 1. Moreover, previous studies have shown that these compounds easily enter mammalian cells (18, 30) and are active against the related parasite T. gondii even when it is intracellular (24). Instead, the lack of correlation between IC50s and EC50s suggests that CDPK1 is likely not an essential enzyme in C. parvum and that CDPK1 is not the target of those PP analogs that are effective against parasite growth. Recent advances in the development of molecular genetics for C. parvum (40) might be used to test this hypothesis; however, the current system used to generate transgenic parasites still requires propagation in mice and thus is not amenable to essentiality testing in vitro.
Our findings differ from observations with T. gondii showing that PP analogs block the function of CDPK1 in controlling microneme secretion and hence prevent host cell invasion (24). In contrast, those PP analogs that were potent in the C. parvum growth inhibition assay had no activity when added only during invasion. In T. gondii, microneme secretion is controlled by calcium fluxes that are thought to activate CDPK1 (41) and facilitate other steps in microneme exocytosis (42). Recent evidence also implicates CDPK3 and PKG in microneme secretion control in T. gondii (43). This pathway is likely conserved in the coccidian parasite Eimeria tenella, where treatment with compound 22, an inhibitor of PKG, blocked microneme secretion and host cell invasion (44). Hence, it was surprising to find that neither PP inhibitors that were active against CpCDPK1 in vitro nor compound 22, a PKG inhibitor that also acts on CDPK1 in vitro, were able to block C. parvum invasion. Cryptosporidium also contains apical organelles called micronemes, which are enriched in adhesive domains involved in cell attachment (45). Although less well studied than in T. gondii, elevated calcium also appears to be important in the release of microneme proteins in sporozoites of C. parvum (46). The failure of PP inhibitors to block cell invasion by C. parvum suggests that the process of microneme secretion differs from that described in T. gondii. It is possible that sporozoites have already upregulated micronemal protein on their surface during excystation, such that inhibition does not block invasion. CpCPDK1 has been shown to be consistently expressed relatively late in infection, peaking after 24 h and continuing beyond 48 h (47), suggesting that it may be involved in a later stage of intracellular growth. Hence, CDPK1 might become important in controlling microneme secretion during the transition from type I to type II meronts, between which merozoites exit from one cell and invade new cells. However, we failed to observe a consistent relationship between PP inhibitors that block growth and the potency with which they inhibit CpCDPK1 enzyme activity. These results suggest that CpCDPK1 may not be involved in the release of micronemes or in cell invasion by C. parvum. In this regard, it is instructive that the ortholog of CDPK1 in P. berghei (called CDPK4 for historic reasons) is not required for microneme secretion but rather functions in male gametocyte exflagellation (41). Given the deep-branching position of Cryptosporidium (5), it is not surprising that CDPK1 orthologues have diverged in function since the common origin of the Apicomplexa.
Our findings differ from a previous report demonstrating that a modified PP analog called 1294 was potent in blocking C. parvum invasion when the drug was added for only 2 h during the initial infection. Compound 1294 contains a 6-ethoxy-naphthyl in direct aryl linkage with C-3 of the PP scaffold and a methyl-piperidine at the N1 position (27). Compound 1294 is a potent inhibitor of C. parvum CDPK1 enzyme activity in vitro (27). However, the above evidence that the potency with which PP analogs inhibit CpCDPK1 enzyme activity does not correlate with growth inhibition suggests that the potency of 1294 may be due to its action on an unrelated yet essential target. Others have shown that mitogen-activated protein kinase 1 (MAPK1) is a secondary target of some PP analogs in T. gondii, notably, 1NM (compound 3) (21, 48). TgMAPK contains a Ser gatekeeper residue, and mutations in the nucleotide-binding pocket render it resistant to PP analogs, including 1NM (compound 3). Therefore, it may be instructive to explore other kinases to define the target(s) of PP analogs that are active against C. parvum. Cryptosporidium contains more than 70 protein kinases, including seven CDPKs, although of these, only CDPK1 contains a glycine gatekeeper (13, 25). It is possible that additional protein kinases with smallish residues such as A/T/S are alternative targets for the PP inhibitors examined here. In this regard, the compounds that blocked C. parvum growth the most potently were 1, 16, and 21. Although genetic approaches have been very powerful in validating the target of PP analogs in T. gondii (19), it has not been possible to use such techniques with C. parvum previously. With the advent of stable transformation (40), such tools that can be used to both validate and discover the true targets of inhibitors in C. parvum may be available soon.
ACKNOWLEDGMENTS
We are grateful to Martin John Rogers for his early support of this project, Ray Hui for helpful advice, Bahaa A. Fadl-Alla for his expert assistance with RT-qPCR assays, and Jennifer Barks for technical assistance with cell culture.
REFERENCES
- 1.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzipori S, Widmer G. 2008. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol 24:184–189. doi: 10.1016/j.pt.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acacio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 4.Pantenburg B, Cabada MM, White AC Jr. 2009. Treatment of cryptosporidiosis. Expert Rev Anti Infect Ther 7:385–391. doi: 10.1586/eri.09.24. [DOI] [PubMed] [Google Scholar]
- 5.Barta JR, Thompson RC. 2006. What is Cryptosporidium? Reappraising its biology and phylogenetic affinities. Trends Parasitol 22:463–468. [DOI] [PubMed] [Google Scholar]
- 6.Bouzid M, Hunter PR, Chalmers RM, Tyler KM. 2013. Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev 26:115–134. doi: 10.1128/CMR.00076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Lakshminarayan I, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 8.Rider SD Jr, Zhu G. 2010. Cryptosporidium: genomic and biochemical features. Exp Parasitol 124:2–9. doi: 10.1016/j.exppara.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bessoff K, Sateriale A, Lee KK, Huston CD. 2013. Drug repurposing screen reveals FDA-approved inhibitors of human HMG-CoA reductase and isoprenoid synthesis that block Cryptosporidium parvum growth. Antimicrob Agents Chemother 57:1804–1814. doi: 10.1128/AAC.02460-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessoff K, Spangenberg T, Foderaro J E, Jumani RS, Ward GE, Huston CD. 2014. Identification of Cryptosporidium parvum active chemical series by repurposing the open access malaria box. Antimicrob Agents Chemother 58:2731–2739. doi: 10.1128/AAC.02641-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda-Saavedra D, Gabaldon T, Barton GJ, Langsley G, Doerig C. 2012. The kinomes of apicomplexan parasites. Microbes Infect 14:796–810. doi: 10.1016/j.micinf.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Billker O, Lourido S, Sibley LD. 2009. Calcium-dependent signaling and kinases in apicomplexan parasites. Cell Host Microbe 5:612–622. doi: 10.1016/j.chom.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hui R, El Bakkouri M, Sibley LD. 2015. Designing selective inhibitors for calcium-dependent protein kinases in apicomplexans. Trends Pharmacol Sci 36:452–460. doi: 10.1016/j.tips.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wernimont AK, Amani M, Qiu W, Pizarro JC, Artz JD, Lin YH, Lew J, Hutchinson A, Hui R. 2011. Structures of parasitic CDPK domains point to a common mechanism of activation. Proteins 79:803–820. doi: 10.1002/prot.22919. [DOI] [PubMed] [Google Scholar]
- 15.Wernimont AK, Artz JD, Finerty P, Lin Y, Amani M, Allali-Hassani A, senisterra G, Vedadi M, Tempel W, Mackenzie F, Chau I, Lourido S, Sibley LD, Hui R. 2010. Structures of apicomplexan calcium-dependent protein kinases reveal mechanism of activation by calcium. Nat Struct Mol Biol 17:596–601. doi: 10.1038/nsmb.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 17.Bishop AC, Shokat KM. 1999. Acquisition of inhibitor-sensitive protein kinases through protein design. Pharmacol Ther 82:337–346. doi: 10.1016/S0163-7258(98)00060-6. [DOI] [PubMed] [Google Scholar]
- 18.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. 2000. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 19.Lourido S, Shuman J, Zhang C, Shokat KM, Hui R, Sibley LD. 2010. Calcium-dependent protein kinase 1 is an essential regulator of exocytosis in Toxoplasma. Nature 465:359–362. doi: 10.1038/nature09022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ojo KK, Larson ET, Keyloun KR, Castaneda LJ, DeRoucher AE, KInampudi KK, Kim J E, Arakaki TL, Murphy RC, Zhang L, Napuli AJ, Maly DJ, Verlinde CLMJ, Buckner FS, Parsons M, Hol WGJ, Meritt EA, Van Voorhis C. 2010. Toxoplasma gondii calcium-dependent protein kinase 1 is a target for selective kinase inhibitors. Nat Struct Mol Biol 17:602–607. doi: 10.1038/nsmb.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugi T, Kato K, Kobayashi K, Watanabe S, Kurokawa H, Gong H, Pandey K, Takemae H, Akashi H. 2010. Use of the kinase inhibitor analog 1NM-PP1 reveals a role for Toxoplasma gondii CDPK1 in the invasion step. Eukaryot Cell 9:667–1670. doi: 10.1128/EC.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson SM, Murphy RC, Geiger JA, DeRocher AE, Zhang Z, Ojo KK, Larson ET, Perera BG, Dale EJ, He P, Reid MC, Fox AM, Mueller NR, Merritt EA, Fan E, Parsons M, Van Voorhis WC, Maly DJ. 2012. Development of Toxoplasma gondii calcium-dependent protein kinase 1 (TgCDPK1) inhibitors with potent anti-toxoplasma activity. J Med Chem 55:2416–2426. doi: 10.1021/jm201713h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larson ET, Ojo KK, Murphy RC, Johnson SM, Zhang Z, Kim JE, Leibly DJ, Fox AM, Reid MC, Dale EJ, Perera BG, Kim J, Hewitt SN, Hol WG, Verlinde CL, Fan E, Van Voorhis WC, Maly DJ, Merritt EA. 2012. Multiple determinants for selective inhibition of apicomplexan calcium-dependent protein kinase CDPK1. J Med Chem 55:2803–2810. doi: 10.1021/jm201725v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lourido S, Zhang C, Lopez MS, Tang K, Barks J, Wang Q, Wildman SA, Shokat KM, Sibley LD. 2013. Optimizing small molecule inhibitors of calcium-dependent protein kinase 1 to prevent infection by Toxoplasma gondii. J Med Chem 56:3068–3077. doi: 10.1021/jm4001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Artz JD, Wernimont AK, Allali-Hassani A, Zhao Y, Amani M, Lin YH, Senisterra G, Wasney GA, Fedorov O, King O, Roos A, Lunin VV, Qiu W, Finerty P Jr, Hutchinson A, Chau I, von Delft F, MacKenzie F, Lew J, Kozieradzki I, Vedadi M, Schapira M, Zhang C, Shokat K, Heightman T, Hui R. 2011. The Cryptosporidium parvum kinome. BMC Genomics 12:478. doi: 10.1186/1471-2164-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy RC, Ojo KK, Larson ET, Castellanos-Gonzalez A, Perera BG, Keyloun KR, Kim J E, Bhandari JG, Muller NR, Verlinde CL, White AC, Merritt EA, Van Voorhis WC, Maly DJ. 2010. Discovery of potent and selective inhibitors of calcium-dependent protein kinase 1 (CDPK1) from C. parvum and T. gondii. ACS Med Chem Lett 1:331–335. doi: 10.1021/ml100096t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doggett JS, Ojo KK, Fan E, Maly DJ, Van Voorhis WC. 2014. Bumped kinase inhibitor 1294 treats established Toxoplasma gondii infection. Antimicrob Agents Chemother 58:3547–3549. doi: 10.1128/AAC.01823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castellanos-Gonzalez A, White AC Jr, Ojo KK, Vidadala RS, Zhang Z, Reid MC, Fox AM, Keyloun KR, Rivas K, Irani A, Dann SM, Fan E, Maly DJ, Van Voorhis WC. 2013. A novel calcium-dependent protein kinase inhibitor as a lead compound for treating cryptosporidiosis. J Infect Dis 208:1342–1348. doi: 10.1093/infdis/jit327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, Hoffman R, Williams RL, Shokat KM, Knight ZA. 2008. Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat Chem Biol 4:691–699. doi: 10.1038/nchembio.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bishop A, Kung C, Shah K, Witucki L, Shokat KM, Liu Y. 1999. Generation of monospecific nanomolar tyrosine kinase inhibitors via chemical genetic approaches. J Am Chem Soc 121:627–631. doi: 10.1021/ja983267v. [DOI] [Google Scholar]
- 31.Zhang C, Lopez MS, Dar AC, Ladow E, Finkbeiner S, Yun CH, Eck MJ, Shokat KM. 2013. Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology. ACS Chem Biol 8:1931–1938. doi: 10.1021/cb400376p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurnett AM, Liberator PA, Dulski PM, Salowe SP, Donald RG, Anderson JW, Wiltsie J, Diaz CA, Harris G, Chang B, Darkin-Rattray SJ, Nare B, Crumley T, Blum PS, Misura AS, Tamas T, Sardana MK, Yuan J, Biftu T, Schmatz DM. 2002. Purification and molecular characterization of cGMP-dependent protein kinase from apicomplexan parasites. A novel chemotherapeutic target. J Biol Chem 277:15913–15922. doi: 10.1074/jbc.M108393200. [DOI] [PubMed] [Google Scholar]
- 33.Current WL. 1990. Techniques and laboratory maintenance of Cryptosporidium, p 44–77. In Dubey JP, Speer CA, Fayer R (ed), Cryptosporidiosis of man and animals. CRC Press, Boca Raton, FL. [Google Scholar]
- 34.Arrowood MJ, Sterling CR. 1987. Isolation of Cryptosporidium oocysts and sporozoites using discontinuous sucrose and isopycnic Percoll gradients. J Parasitol 73:314–319. doi: 10.2307/3282084. [DOI] [PubMed] [Google Scholar]
- 35.Johnson JK, Schmidt J, Gelberg HB, Kuhlenschmidt MS. 2004. Microbial adhesion of Cryptosporidium parvum sporozoites: purification of an inhibitory lipid from bovine mucosa. J Parasitol 90:980–990. doi: 10.1645/GE-231R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu C, Dong S, Kuhlenschmidt MS, Kuhlenschmidt TB, Drnevich J, Nguyen TH. 2015. Inactivation mechanisms of Cryptosporidium parvum oocysts by solar ultraviolet irradiation. Environ Sci Water Res Technol 1:188–198. doi: 10.1039/C4EW00079J. [DOI] [Google Scholar]
- 37.Liu Y, Bishop A, Witucki L, Kraybill B, Shimizu E, Tsien J, Ubersax J, Blethrow J, Morgan DO, Shokat KM. 1999. Structural basis for selective inhibition of Src family kinases by PP1. Chem Biol 6:671–678. doi: 10.1016/S1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- 38.Donald RG, Zhong T, Wiersma H, Nare B, Yao D, Lee A, Allocco J, Liberator PA. 2006. Anticoccidial kinase inhibitors: identification of protein kinase targets secondary to cGMP-dependent protein kinase. Mol Biochem Parasitol 149:86–98. doi: 10.1016/j.molbiopara.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 39.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 40.Vinayak S, Pawlowic MC, Sateriale A, Brooks CF, Studstill CJ, Bar-Peled Y, Cipriano MJ, Striepen B. 2015. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523:477–480. doi: 10.1038/nature14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Billker O, Dechamps S, Tewari R, Wenig G, Franke-Fayard B, Brinkmann V. 2004. Calcium and a calcium-dependent protein kinase regulate gamete formation and mosquito transmission in a malaria parasite. Cell 117:503–514. doi: 10.1016/S0092-8674(04)00449-0. [DOI] [PubMed] [Google Scholar]
- 42.Lourido S, Moreno SN. 2015. The calcium signaling toolkit of the apicomplexan parasites Toxoplasma gondii and Plasmodium spp. Cell Calcium 57:186–193. doi: 10.1016/j.ceca.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lourido S, Tang K, Sibley LD. 2012. Distinct signalling pathways control Toxoplasma egress and host-cell invasion. EMBO J 31:4524–4534. doi: 10.1038/emboj.2012.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiersma HI, Galuska SE, Tomley FM, Sibley LD, Liberator PA, Donald RGK. 2004. A role for coccidian cGMP-dependent protein kinase in motility and invasion. Int J Parasitol 34:369–380. doi: 10.1016/j.ijpara.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 45.Carruthers VB, Tomley FM. 2008. Microneme proteins in apicomplexans. Subcell Biochem 47:33–45. doi: 10.1007/978-0-387-78267-6_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen XM, O'Hara SP, Huang BQ, Nelson JB, Lin JJ, Zhu G, Ward HD, LaRusso NF. 2004. Apical organelle discharge by Cryptosporidium parvum is temperature, cytoskeleton, and intracellular calcium dependent and required for host cell invasion. Infect Immun 72:6806–6816. doi: 10.1128/IAI.72.12.6806-6816.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Etzold M, Lendner M, Daugschies A, Dyachenko V. 2014. CDPKs of Cryptosporidium parvum—stage-specific expression in vitro. Parasitol Res 113:2525–2533. doi: 10.1007/s00436-014-3902-0. [DOI] [PubMed] [Google Scholar]
- 48.Sugi T, Kobayashi K, Takemae H, Gong H, Ishiwa A, Murakoshi F, Recuenco FC, Iwanaga T, Horimoto T, Akashi H, Kato K. 2013. Identification of mutations in TgMAPK1 of Toxoplasma gondii conferring resistance to 1NM-PP1. Int J Parasitol Drugs Drug Resist 3:93–101. doi: 10.1016/j.ijpddr.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bishop AC, Shah K, Liu Y, Witucki L, Kung C, Shokat KM. 1998. Design of allele-specific inhibitors to probe protein kinase signaling. Curr Biol 8:257–266. doi: 10.1016/S0960-9822(98)70198-8. [DOI] [PubMed] [Google Scholar]