Abstract
Colistin-resistant mutants were obtained from 17 colistin-susceptible strains of Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli. The stability of colistin resistance in these mutants was investigated. Three of four colistin-resistant P. aeruginosa mutants recovered colistin susceptibility in colistin-free medium; however, colistin-susceptible revertants were obtained from only one strain each of A. baumannii and E. coli. No susceptible revertants were obtained from K. pneumoniae mutants.
TEXT
Colistin resistance has been observed in Gram-negative pathogens (1–3). Colistin resistance is mediated by mutations in the PmrAB or PhoPQ two-component regulatory systems, the loss of lipopolysaccharide, or MgrB inactivation (4). Colistin resistance is described as a type of adaptive resistance with the rapid development of resistance in the presence of antibiotics and reversal to susceptibility in the absence of the same (5). This suggests that resistance to colistin may diminish in the absence of colistin or by limiting the extracellular concentration of divalent cations. In this study, we developed colistin resistance in vitro in four Gram-negative bacteria—Acinetobacter baumannii, Pseudomonas aeruginosa, Klebsiella pneumoniae, and Escherichia coli. We also examined the stability of the resistant strains.
Seventeen strains, which were randomly isolated from patients suffering from bacteremia or urinary tract infections in South Korea, were used in this study (Table 1). The patients had not received intravenous or inhaled colistimethate. For all isolates, multilocus sequence typing (MLST) was performed as described previously (6–9). MICs were determined by a broth microdilution method using cation-adjusted Mueller-Hinton broth and interpreted according to CLSI breakpoints (10) for A. baumannii and P. aeruginosa and EUCAST breakpoints (11) for E. coli and K. pneumoniae.
TABLE 1.
Gram-negative rod-shaped bacterial strains used in this study, their MICs for colistin, heteroresistance, mutation frequency, and amino acid alterations in colistin-resistant mutants and colistin-susceptible revertants
| Species | Strain | ST | Colistin MIC (μg/ml) for: |
Heteroresistancea | Mutation frequencyb | Amino acid alteration in: |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parent | Resistant mutant | Susceptible revertant | Colistin-resistant mutants |
Colistin-susceptible revertantsc | ||||||||
| PmrA | PmrB | PhoP | PhoQ | |||||||||
| A. baumannii | H07-988 | 220 | 1 | >64 | HR | 8.69 × 10−7 | H263R, V444A | NA | ||||
| H05-513 | 20 | 1 | >64 | 1 | HR | 5.58 × 10−6 | I235T, G390V | T235I, V390G in PmrB | ||||
| H09-673 | 92 | 1 | >64 | 6.64 × 10−7 | H263R | NA | ||||||
| H09-968 | 138 | 1 | >64 | 1.84 × 10−7 | M12R | NA | ||||||
| C095 | 110 | 0.5 | >64 | 2.72 × 10−7 | NA | |||||||
| P. aeruginosa | P5 | 235 | 1 | >64 | 1 | 5.74 × 10−6 | K123Q V260G | R117L in PhoP, Q123K in PhoQ | ||||
| P6 | 1340 | 2 | 64 | 0.5 | HR | 8.29 × 10−7 | A67T | T67A in PmrB | ||||
| P33 | 641 | 1 | >64 | 4.81 × 10−7 | V15I | NA | ||||||
| P155 | 17 | 0.5 | >64 | 0.5 | HR | 3.41 × 10−6 | L167P | P167L in PmrB, A110V in PhoP, Q411* in PhoQ | ||||
| K. pneumoniae | B0608-134 | 730 | 1 | >64 | HR | 2.94 × 10−6 | NA | |||||
| B0704-039 | 11 | 0.5 | >64 | HR | 4.28 × 10−6 | NA | ||||||
| 08-B063 | 23 | 0.5 | >64 | HR | 2.94 × 10−6 | Y268S, del14–18 | NA | |||||
| B0701-068 | 152 | 0.5 | >64 | HR | 2.16 × 10−6 | NA | ||||||
| E. coli | E015 | 405 | 0.25 | 64 | 0.25 | HR | 3.23 × 10−7 | del133–136 | V24E and del162–165 in PmrB | |||
| E139 | 131 | 0.25 | >64 | 4.23 × 10−7 | P94L | NA | ||||||
| E154 | 38 | 0.25 | 64 | 8.06 × 10−8 | A159V | NA | ||||||
| E188 | 410 | 0.125 | 64 | 1.03 × 10−7 | V125E | NA | ||||||
Heteroresistance (HR) was defined as the presence of colonies more than the LOQ on the agar plate containing 10 μg/ml colistin.
The ratio of the CFU on a plate containing 4 μg/ml colistin to that on an antibiotic-free plate.
NA, not available; *, premature termination.
Colistin-resistant mutants were developed from the colistin-susceptible wild-type strains. Starting with a single colony of each wild-type strain, colistin-resistant mutants were chosen by serial passage, using progressively increasing concentrations of colistin (12). At the end of the induction period, the spontaneous mutants growing in Luria-Bertani (LB) medium containing 16 μg/ml colistin were reinoculated on LB agar plates containing 32 μg/ml colistin in order to obtain single resistant populations.
To investigate the stability of the colistin resistance developed, the mutants were repeatedly subcultured in the absence of colistin. Overnight cultures of all induced colistin-resistant mutants were diluted 1:1,000 in fresh LB medium without colistin and incubated with vigorous shaking (220 rpm) at 37°C for 24 h. Colistin MICs for the pooled populations diluted in saline were estimated for all serially transferred cultures. For E. coli and P. aeruginosa, the maximum number of passages was 32 days, and A. baumannii and K. pneumoniae cells were transferred serially for 62 and 42 days, respectively.
Heteroresistance to colistin was identified by population analysis profiling by spreading a 0.1-ml aliquot from a 24-h culture of parental susceptible strains (13). Heteroresistance was defined as the presence of colonies more than the limit of quantification (LOQ) (400 CFU/ml) on the agar plate containing 10 μg/ml colistin (13, 14). Mutation frequency was investigated using cultures that were subjected to several serial passages in antibiotic-free LB broth medium. Mutation frequency was defined as the ratio of the CFU on a plate containing 4 μg/ml colistin to that on an antibiotic-free plate for each strain.
Amino acid substitutions were identified in pmrAB for A. baumannii, P. aeruginosa, K. pneumoniae, and E. coli, phoPQ for P. aeruginosa, K. pneumoniae, and E. coli, and mgrB for K. pneumoniae using primers described previously (12, 15, 16).
In this study, colistin-resistant mutants were obtained from all susceptible parental strains (Table 1). Colistin-resistant mutants were selected in vitro from all cultures grown in medium containing 0.5 to 16 μg/ml colistin, which indicates that colistin resistance can be readily developed under antibiotic pressure. The colistin-resistant mutants had a colistin MIC of ≥64 μg/ml. Rapid development of colistin resistance in some bacterial species has previously been reported (12, 17, 18). A previous mutant prevention concentration study also indicated that colistin resistance can be readily induced during drug therapy by single-step mutation in A. baumannii, P. aeruginosa, and K. pneumoniae (19). While MgrB mutations were readily found in other colistin-resistant K. pneumoniae strains or mutants (20–23), no mutations of MgrB were identified in this study.
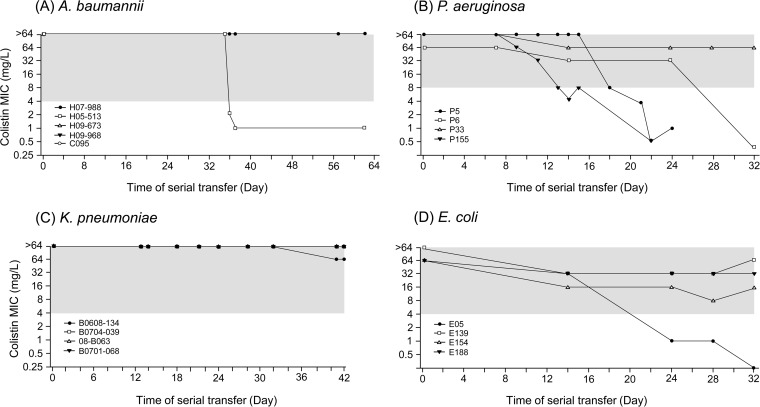
Contrary to the nature of development of colistin resistance, the stability of colistin resistance differed between strains. Colistin-susceptible revertants were obtained from only 5 of the 17 colistin-resistant mutants: one A. baumannii and three P. aeruginosa strains and one E. coli strain (Table 1 and Fig. 1). None of the K. pneumoniae mutants produced any colistin-susceptible revertants.
FIG 1.
Change in colistin MIC of resistant mutants obtained by serial passage in colistin-free medium. (A) A. baumannii, (B) P. aeruginosa, (C) K. pneumoniae, and (D) E. coli colistin-resistant mutant strains. The y axis represents the colistin MIC in the log2 scale. Colistin-susceptible revertants were obtained from three P. aeruginosa strains and one strain each of A. baumannii and E. coli resistant mutants. Dashed lines indicate the breakpoint of colistin resistance for each species.
Heteroresistance to colistin was identified in all four K. pneumoniae strains, and two P. aeruginosa and two A. baumannii strains and one E. coli strain were heteroresistant to colistin (Table 1). The correlation between colistin heteroresistance and stability of colistin resistance may not be supported because the heteroresistant K. pneumoniae strains did not lose colistin resistance in antibiotic-free medium. In addition, A. baumannii H07-988 showed heteroresistance to colistin, but it did not develop a colistin-susceptible revertant, and P. aeruginosa P5 showed a completely opposite nature. Furthermore, mutation frequency might not be associated with the heteroresistance and stability of colistin resistance (Table 1).
We identified several mutations in PhoPQ and PmrAB in colistin-resistant mutants. However, it was not proven that the mutations are associated with colistin resistance. In colistin-susceptible revertants of P. aeruginosa P5 and P155 and E. coli E015, additional mutations were found compared to their colistin-resistant progenitors (Table 1). However, such compensatory mutations were not observed in colistin-susceptible revertants of A. baumannii H05-513 and P. aeruginosa P6, in which only genetic reversions were identified. Such genetic reversion was also identified in P. aeruginosa P5 and P155.
The induced colistin resistance was eliminated in most P. aeruginosa strains in a colistin-free medium, but it remained stable in the other species tested (A. baumannii, K. pneumoniae, and E. coli). Therefore, the principle of adaptive resistance can be applied to P. aeruginosa but not to the others. The stability of colistin resistance has already been observed in A. baumannii (18). However, this stability is a major concern in the other three Gram-negative species, as newly emerged resistance in these species can be preserved and disseminated even in the absence of antibiotic pressure. Many studies have discussed the factors affecting the fitness cost of colistin resistance, such as increased susceptibility to other antibiotics, growth retardation, and reduced virulence (15, 24, 25), which may prevent an increase in the cases of colistin resistance in hospitals. However, compensatory mutations can change this situation, making it more difficult to treat the infections caused by Gram-negative pathogens.
The colistin resistance developed in patients treated with colistin for Gram-negative pathogenic infections may be preserved is a valid concern in the public health domain, with respect to preventing further development of resistance to the antibiotic. In addition, the mechanisms underlying the stability of colistin resistance, which has marked implications for the therapeutic options, need to be investigated.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been submitted to the GenBank database under accession no. KT716084 to KT716131, KT716132 to KT716179, KT719393, and KT719394.
ACKNOWLEDGMENTS
The colistin-susceptible strains used in this study were obtained from the Asian Bacterial Bank (ABB) of the Asia Pacific Foundation for Infectious Diseases (APFID, Seoul, South Korea).
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT and Future Planning (NRF-2013R1A2A2A0101413). This research was supported partly by the Korea Medical Institute (KMI, Seoul, South Korea).
REFERENCES
- 1.Ko KS, Suh JY, Kwan KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. 2007. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother 60:1163–1167. doi: 10.1093/jac/dkm305. [DOI] [PubMed] [Google Scholar]
- 2.Lee JY, Song JH, Ko KS. 2011. Identification of nonclonal Pseudomonas aeruginosa isolates with reduced colistin susceptibility in Korea. Microb Drug Resist 17:299–304. doi: 10.1089/mdr.2010.0145. [DOI] [PubMed] [Google Scholar]
- 3.Suh JY, Son JS, Chung DR, Peck KR, Ko KS, Song JH. 2010. Nonclonal emergence of colistin-resistant Klebsiella pneumoniae isolates from blood samples in South Korea. Antimicrob Agents Chemother 54:560–562. doi: 10.1128/AAC.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skiada A, Markogiannakis A, Plachouras D, Daikos GL. 2011. Adaptive resistance to cationic compounds in Pseudomonas aeruginosa. Int J Antimicrob Agents 37:187–193. doi: 10.1016/j.ijantimicag.2010.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Bartual SG, Seifert H, Hippler C, Luzon MA, Whisplinghoff H, Rodriguez-Valera F. 2005. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 43:4382–4390. doi: 10.1128/JCM.43.9.4382-4390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. 2004. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol 42:5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, Fox AJ, Upton M. 2008. Major uropathogenic Escherichia coli strain isolated in the northwest of England by multilocus sequence typing. J Clin Microbiol 46:1076–1080. doi: 10.1128/JCM.02065-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial susceptibility testing: twenty-first informational supplement M100-S23. CLSI, Wayne, PA. [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing. 2015. Breakpoint tables for interpretation of MICs and zone diameters. Version 5.0. http://www.eucast.org/.
- 12.Park YK, Choi JY, Shin D, Ko KS. 2011. Correlation between overexpression and amino acid substitution of the PmrAB locus and colistin resistance in Acinetobacter baumannii. Int J Antimicrob Agents 37:525–530. doi: 10.1016/j.ijantimicag.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, Nation RL, Li J. 2009. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J Infect 58:138–144. doi: 10.1016/j.jinf.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 14.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choi MJ, Ko KS. 17 August 2015. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JY, Ko KS. 2014. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn Microbiol Infect Dis 78:271–276. doi: 10.1016/j.diagmicrobio.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 17.Lee JY, Lim MH, Heo ST, Ko KS. 2012. Repeated isolation of Pseudomonas aeruginosa isolates resistant to both polymyxins and carbapenems from 1 patient. Diagn Microbiol Infect Dis 72:267–271. doi: 10.1016/j.diagmicrobio.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Snitkin ES, Zelazny AM, Gupta J, NISC Comparative Sequencing Program, Palmore TN, Murray PR, Segre JA. 2013. Genomic insights into the fate of colistin resistance and Acinetobacter baumannii during patient treatment. Genome Res 23:1155–1162. doi: 10.1101/gr.154328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choi MJ, Ko KS. 2014. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother 69:275–277. doi: 10.1093/jac/dkt315. [DOI] [PubMed] [Google Scholar]
- 20.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olaitan AO, Diene SM, Kempf M, Berrazeg M, Bakour S, Gupta SK, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Chaisiri K, Komalamisra C, Adelowo OO, Fagade OE, Banjo OA, Oke AJ, Adler A, Assous MV, Morand S, Raoult D, Rolain JM. 2014. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 44:500–507. doi: 10.1016/j.ijantimicag.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Poirel L, Jayol A, Bontron S, Villegas MV, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 23.Wright MS, Jones MB, Marshall SH, Rudin SD, van Duln D, Kaye K, Jacobs MR, Bonomo RA, Adams MD. 2015. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother 59:536–543. doi: 10.1128/AAC.04037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Rojas R, Jimenez-Mejias ME, Lepe JA, Pachon J. 2011. Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J Infect Dis 204:1147–1148. doi: 10.1093/infdis/jir476. [DOI] [PubMed] [Google Scholar]
- 25.Pournaras S, Poulou A, Dafopoulou K, Chabane YN, Kristo I, Makris D, Hardouin J, Cosette P, Tsakris A, Dé E. 2014. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 58:828–832. doi: 10.1128/AAC.01439-13. [DOI] [PMC free article] [PubMed] [Google Scholar]