Abstract
Epidemiological and individual risk factors for colonization by enterobacteria producing extended-spectrum beta-lactamases (E-ESBL) have been studied extensively, but whether such colonization is associated with significant changes in the composition of the rest of the microbiota is still unknown. To address this issue, we assessed in an isolated Amerindian Guianese community whether intestinal carriage of E-ESBL was associated with specificities in gut microbiota using metagenomic and metatranscriptomic approaches. While the richness of taxa of the active microbiota of carriers was similar to that of noncarriers, the taxa were less homogeneous. In addition, species of four genera, Desulfovibrio, Oscillospira, Parabacteroides, and Coprococcus, were significantly more abundant in the active microbiota of noncarriers than in the active microbiota of carriers, whereas such was the case only for species of Desulfovibrio and Oscillospira in the total microbiota. Differential genera in noncarrier microbiota could either be associated with resistance to colonization or be the consequence of the colonization by E-ESBL.
INTRODUCTION
Antibiotics have revolutionized the treatment of bacterial infections since the first use of penicillin. However, antibiotic overuse during recent decades has led to an increase in bacterial resistance (1). For a long time, this increased resistance has been masked by the continuous discovery and use of new antibiotics. Innovation in the field has, however, stopped, and we now find patients who are infected by bacteria which are resistant to most, if not all, available antibiotics. For many years, such multiresistant bacteria were observed only in hospitalized patients, but they are currently increasingly isolated from members of the community at large. Enterobacteria that produce extended-spectrum beta-lactamases (E-ESBLs), which are not only resistant to all beta-lactams (except carbapenems) but are also very often coresistant to the most commonly used antibiotics and can cause very common infections, such as urinary tract infections and pyelonephritis, are particularly feared (2). Besides causing infections, such E-ESBLs can very often colonize the intestinal microbiota of healthy individuals (3, 4). Intestinal colonization is recognized as the cornerstone of E-ESBL dissemination. Epidemiological and individual risk factors for intestinal colonization by E-ESBLs have been studied extensively (4, 5), but whether colonization is associated with significant changes in the composition of the rest of the microbiota, as a cause or as a consequence, is still unknown.
Currently, culture-independent methods based on extensive sequencing of total DNA are preferred to describe intestinal microbiota because large numbers of species are noncultivable. In addition, analysis based on RNA sequencing allows a functional approach to study the intestinal flora. An important function of the intestinal microbiota is to prevent colonization by exogenous bacteria, including resistant potentially pathogenic microorganisms. This “colonization resistance” is associated with the anaerobic intestinal microbiota (6, 7), but the precise mechanism involved is still largely unknown. Recently, the presence of the Barnesiella genus in the intestinal microbiota was shown to prevent vancomycin-resistant Enterococcus domination (8).
Here, we explored and compared the total gut microbiota and active gut microbiota in relation to E-ESBL carriage using DNA and RNA high-throughput sequencing methods and found some features that might be of high interest for infection control.
MATERIALS AND METHODS
Study subjects and sampling.
The volunteers enrolled in the study were healthy adult Wayampi Amerindians from the village of Trois-Sauts, in French Guyana, where huts with no modern facilities are disseminated over ∼3 km along the Oyapok river. Their living conditions and microbiological follow-up since 2001 has been extensively described elsewhere (9–13). This is a genetically homogenous population of ∼500 inhabitants, still living mostly in a traditional manner, including alimentation, which is provided by hunting, fishing, and raising crops. However, a resident paramedic ensures fully free access to allopathic medicine and records all health care deliveries, including antibiotics. During the last sampling campaign, which took place in October 2010, we tested 151 individuals for E-ESBL intestinal carriage, as described previously (9, 11, 13). In addition, a fecal sample of about 5 g that had recently passed (less than 2 h earlier) was diluted 1:10 in RNAlater (Applied Biosystems, Villebon-sur-Yvette, France), thoroughly mixed, and immediately frozen at −20°C before being taken to the laboratory, without defrosting, and stored at −80°C until harvesting.
Eight (5.3%) of the 151 individuals were found to be E-ESBL carriers. Among them, 4 were carrying a CTX-M-1 ESBL, 2 were carrying a CTX-M-8 ESBL, and 1 was carrying a CTX-M-2 ESBL. The last carrier was carrying both CTX-M-2 and CTX-M-8 ESBLs (13). In order to identify possible specificities in the microbiota of the carriers, we also analyzed the microbiota of noncarriers, used as controls. Each of the 8 ESBL carriers was matched with 3 noncarrier controls chosen among the 143 possible ones (the 151 sampled individuals less the 8 carriers). Controls were chosen for being as close as possible to the corresponding case subject in terms of gender, age, and antibiotic exposure within the year preceding sampling. The first control was part of the carrier's family, the second was living in the same household, and the last was nonrelated to the carrier for the two criteria. Controls were available in all instances except for one carrier for whom only two adequate controls could be matched (a properly matched family noncarrier was not available).
Signed informed consent was obtained from all subjects, and the study protocol was approved by the ad hoc Ethical Committee (Comité de Protection des Personnes Sud-Ouest et Outre Mer III; 2010-A00682-37).
Purification of nucleic acids.
The fecal samples were defrosted and homogenized, and 5 ml of each sample was diluted with 5 ml of phosphate-buffered saline (PBS) (containing, per liter, 8 g of NaCl, 0.2 g of KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 [pH 7.2]). Then, they were centrifuged at 1,250 × g at 4°C for 2 min to remove fecal debris. The supernatant was centrifuged at 13,000 rpm for 5 min to pellet the cells. Total DNA and RNA were extracted from pelleted cells using an AllPrep DNA/RNA/Protein minikit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The DNA and RNA extracts were checked by running a standard agarose gel electrophoresis procedure and were quantified using a Nanodrop-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
Amplification of the 16S rDNA gene.
For each sample, a region of the 16S ribosomal DNA (rDNA) gene was amplified by PCR with the universal primers E8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 357R (5′-TGCTGCCTCCCGTAGGAGT-3′) using a sample-specific Multiplex Identifier (MID) for pyrosequencing. The amplified region comprises hypervariable regions V1 and V2. For each sample, a 50-μl PCR mix was prepared that contained 5 μl of buffer Taq (10×) with 20 mM MgCl2, 2 μl of deoxynucleoside triphosphates (dNTPs) (10 mM), 1 μl of each primer (10 mM), 0.4 μl of Taq Fast start polymerase (5 U/μl), 39.6 μl of nuclease-free water, and 1 μl of DNA template. PCR was run under the following conditions: 95° for 2 min followed by 25 cycles of 95° for 30 s, 52° for 1 min, and 72° for 1 min and a final extension step at 72° for 10 min. The amplification process was checked by electrophoresis using an agarose gel (1.4%). PCR products were purified using a NucleoFast 96 PCR Clean-Up kit (Macherey-Nagel) and quantified using a Nanodrop-1000 spectrophotometer (Thermo Scientific) and a QuantiT PicoGreen double-stranded DNA (dsDNA) assay kit (Invitrogen). The pooled PCR products were directly pyrosequenced.
cDNA synthesis.
Total RNA was first treated with DNase (Ambion), and then it was retrotranscribed into single-stranded cDNA using a High-Capacity cDNA reverse transcription kit (Ambion, Carlsbad, CA, USA) as recommended by the manufacturer, and double-stranded cDNA (ds-cDNA) was then synthesized following standard procedures. The products were quantified using a QuantiT PicoGreen dsDNA assay kit (Invitrogen, Carlsbad, CA, USA). A standard agarose gel electrophoresis procedure was run to check the integrity of the double-stranded cDNA. Total ds-cDNAs obtained from each fecal sample were directly sequenced.
Pyrosequencing and sequence analysis.
Pyrosequencing was performed using a Roche GS FLX sequencer and Titanium chemistry in the Centre for Public Health Research (FISABIO-Salud Pública, Valencia, Spain). All sequences were deposited in the European Bioinformatics Institute (http://www.ebi.ac.uk/) database under accession number ERP001842.
16S rDNA reads with a low quality score (<20) and short read lengths (<100 nucleotides) were removed. Potential chimeras were also removed from the remaining sequences using the Chimera-Slayer package of mothur software (14).
To select the 16S rRNA reads among the total RNA sequences, we compared the reads against the Small Subunit rRNA Reference Database (SSUrdb) as described previously (15), using BLASTN with an E value of 10−16 (16).
Taxonomic information concerning the sequences of 16S rRNA (active bacteria) and 16S rDNA (total bacteria) was obtained by comparisons using Ribosomal Database Project II (RDP) (17) and the pick_otus_through_otu_table.py pipeline available in QIIME v1.4.0 software (18). Operational taxonomic units (OTUs) were created by the use of Uclust, applying cluster criteria of 97% similarity. The most representative sequences for each OTU were then compared against the Qiime cluster version of the Greengenes database (database gg_97_otus_4feb2011_aligned.fasta). We accepted the annotation if the bootstrap confidence estimation value was over 0.8, stopping the assignation at the last well-identified phylogenetic level. Representative sequences were aligned using Pynast against the clustered version of the Greengenes database (database gg_97_otus_4feb2011_aligned.fasta) (19). The aligned sequences were used to reconstruct the phylogenetic tree using FastTree software (20).
We computed the Chao1 richness estimator (21) to assess the expected number of bacterial taxa in the bacterial communities and the Shannon diversity index (22) and the Shannon equitability (evenness) index (23) to measure the homogeneity in the microbiota composition. Shannon equitability indicates the distribution of individuals over genera, while the Shannon diversity index takes into account two features, richness and evenness. The value of the diversity index increases both when richness increases and when evenness increases. For a given number of genera, the value of a diversity index is maximized when all genera are equally abundant.
Epidemiological data and statistical methods.
The data on antibiotic exposure during the year preceding sampling, demography, and living habits were collected as described previously (9, 10, 11, 13). Epidemiological characteristics of carriers and noncarrier controls were compared using the permutation Student t test for quantitative variables and the binomial exact test for paired data in cases of qualitative variables (24).
Concerning the microbiota, the values were expressed as means ± standard deviations (SD). Differences in Shannon diversity index, Shannon equitability index, and Chao1 estimator values were analyzed by performing permutations using the Wilcoxon signed-rank test with a significance level of <0.05 (25). Heat maps of taxon abundance and composition and clustering based on the Bray-Curtis distance measurements were analyzed using the free statistical package R. Biodiversity between groups of samples was evaluated using the pipeline jackknifed_beta_diversity.py available in Qiime v1.4.0. The rooting tree method was performed by adding two archaeal 16S sequences (AB294259 and AB294260) to the phylogenetic tree, the rest of the settings being left at default values. Weighted Unifraq was chosen to estimate beta diversity because its components better explain the genetic variability between samples.
We used the linear discriminant analysis (LDA) effect size (LEfSe) algorithm from the Galaxy software package (26) to identify specific taxa as biomarkers for carriers and noncarriers. LEfSe first identifies the significant differences in taxon composition between groups (carriers and noncarriers) by applying the Kruskall-Wallis test (27). Then, the Wilcoxon test (28) was used to check all pairwise comparisons by considering that for each group there were individuals treated or not treated with antibiotics. We fixed a cutoff α value of <0.05 for both type of tests. The bacterial taxa with significant differences between samples were used to build the LDA model and to estimate its effect as a discriminative feature among them. The threshold used to consider a discriminative feature for the logarithmic LDA score was set to >2.0.
RESULTS
Comparison of general features between E-ESBL carriers and noncarrier controls.
The gut microbiota of carriers and noncarrier controls did not differ significantly with respect to any of the epidemiological characteristics, including antibiotic exposure (Table 1). The three exposed carriers received metronidazole, co-trimoxazole, and amoxicillin, respectively, whereas the three exposed noncarrier controls received co-amoxiclav (one subject) or metronidazole (two subjects). It is worth noticing that administration of all courses of antibiotic stopped at least 6 weeks before sampling.
TABLE 1.
Epidemiological characteristics of ESBL carriers and noncarrier controls among the Wayampis Amerindians of Trois-Sautsa
| Epidemiological parameter | Value(s) |
|||
|---|---|---|---|---|
| Carriers (n = 8) | Nonrelated controls (n = 8) (P value) | Household controls (n = 8) (P value) | Family controls (n = 7) (P value) | |
| Sociodemographic data | ||||
| Avg age, yrs [range]b | 46.1 [33–65] | 45.6 [28–67] | 42.4 [25–55] | 35.7 [24–54] |
| Sex ratio (female/male)b | 6:2 | 5:3 | 2:6 | 5:2 |
| No. of subjects in a marriage or stable relationship | 7 | 5 (0.63) | 8 (1.00) | 7 (1.00) |
| Median no. of children | 6 | 5 (0.36) | 7 (0.52) | 6 (0.36) |
| Median no. of close relativesc | 6 | 6 (0.99) | 6 (0.99) | 7 (0.46) |
| Household | ||||
| Median no. of inhabitants | 7 | 7 (0.93) | 7 (1.00) | 8 (0.35) |
| No. of animals in the household | 6 | 6 (1.00) | 6 (1.00) | 5 (1.00) |
| No. of subjects with indicated lifestyle | ||||
| Drinking water | ||||
| River | 3 | 2 (1.00) | 4 (1.00) | 4 (1.00) |
| Cove | 2 | 3 (1.00) | 3 (1.00) | 3 (1.00) |
| Tap | 6 | 5 (1.00) | 6 (1.00) | 3 (0.25) |
| Duty | ||||
| Hunter | 2 | 4 (0.63) | 5 (0.38) | 2 (1.00) |
| Fisher | 7 | 6 (1.00) | 7 (1.00) | 6 (1.00) |
| Manioc culture worker | 7 | 7 (1.00) | 6 (1.00) | 7 (1.00) |
| Cook | 7 | 7 (1.00) | 6 (1.00) | 5 (0.62) |
| Cachiri preparerd | 6 | 5 (1.00) | 3 (0.38) | 5 (1.00) |
| Pirogue driver | 4 | 4 (1.00) | 7 (0.38) | 3 (1.00) |
| Health worker | 1 | 0 (1.00) | 1 (1.00) | 0 (1.00) |
| Babysitter for children ≤5 yrs of age | 8 | 4 (0.13) | 6 (0.50) | 6 (0.50) |
| Travel outside Trois-Sauts during past yr | 6 | 7 (1.00) | 6 (1.00) | 7 (1.00) |
| Having a child at school outside Trois-Sauts | 3 | 2 (1.00) | 2 (1.00) | 2 (1.00) |
| No. of subjects with indicated medical history during past yr | ||||
| Hospitalization | 0 | 0 (1.00) | 1 (1.00) | 1 (1.00) |
| Surgery | 0 | 0 (1.00) | 0 (1.00) | 0 (1.00) |
| Serious medical event | 1 | 0 (1.00) | 1 (1.00) | 0 (1.00) |
| H1N1 vaccination | 2 | 2 (1.00) | 1 (1.00) | 2 (1.00) |
| Antibiotic usee | 3 | 2 (1.00) | 1 (0.50) | 0 (0.25) |
| Antibiotic use among relatives | 6 | 8 (1.00) | 7 (1.00) | 6 (1.00) |
| Cocolonization | ||||
| Persistent Staphylococcus aureus nasal carriage | 1 | 3 (0.63) | 4 (0.38) | 1 (1.00) |
| Intestinal colonization with Candida albicansf | 2 | 1 (1.00) | 0 (0.50) | 2 (1.00) |
| Intestinal colonization with Candida kruseif | 7 | 5 (0.63) | 6 (1.00) | 6 (1.00) |
| Intestinal colonization with Saccharomyces cerevisiaef | 8 | 6 (0.50) | 7 (1.00) | 5 (0.25) |
| Biodiversity of total microbiota | ||||
| Shannon diversity index (mean ± SD) | 1.94 ± 0.50 | 2.10 ± 0.48 (0.43) | 2.04 ± 0.48 (0.51) | 2.36 ± 0.50 (0.06) |
| Shannon equitability index (mean ± SD) | 0.53 ± 0.10 | 0.56 ± 0.10 (0.34) | 0.54 ± 0.10 (0.89) | 0.63 ± 0.10 (0.06) |
| Chao1 estimator (mean ± SD) | 43.91 ± 10.47 | 46.62 ± 9.94 (0.90) | 53.15 ± 9.94 (0.13) | 52.52 ± 10.47 (0.19) |
| Biodiversity of active microbiota | ||||
| Shanon diversity index (mean ± SD) | 1.90 ± 0.70 | 2.46 ± 0.65 (0.07) | 2.30 ± 0.65 (0.50) | 2.79 ± 0.70 (0.01) |
| Shanon equitability index (mean ± SD) | 0.45 ± 0.15 | 0.57 ± 0.14 (0.04) | 0.55 ± 0.14 (0.35) | 0.64 ± 0.15 (0.01) |
| Chao1 estimator (mean ± SD) | 85.99 ± 27.41 | 86.26 ± 25.39 (0.69) | 74.80 ± 25.39 (0.28) | 89.34 ± 27.41 (0.94) |
Univariate analysis was performed using a permutation Student t test and a binomial exact test for paired data. Differences in Shannon index values and Chao1 richness estimator values were analyzed using the Wilcoxon signed-rank test. These values were expressed as means ± standard deviations (SD). P data represent probability at α = 0.05.
Statistical testing was not performed on matching variables.
Close relatives were defined as members of the same family living in the same hut. In cases of multiple life partners, second or third wives and their children were included as family members even if they lived in a different hut. The number of relatives ranged from 1 to 11.
Cachiri is a locally prepared fermented beverage.
Among the carriers, one received a course of metronidazole in September 2010; one received a course of co-trimoxazole in December 2009 and one a course of amoxicillin in October 2009. Among the noncarriers, one received a course of amoxicillin and clavulanic acid in July 2010; one received a course of metrodinazole and spiramycine in June 2010 and one a course of metronidazole in September 2009.
See reference 9.
Biodiversity and composition of the total microbiota.
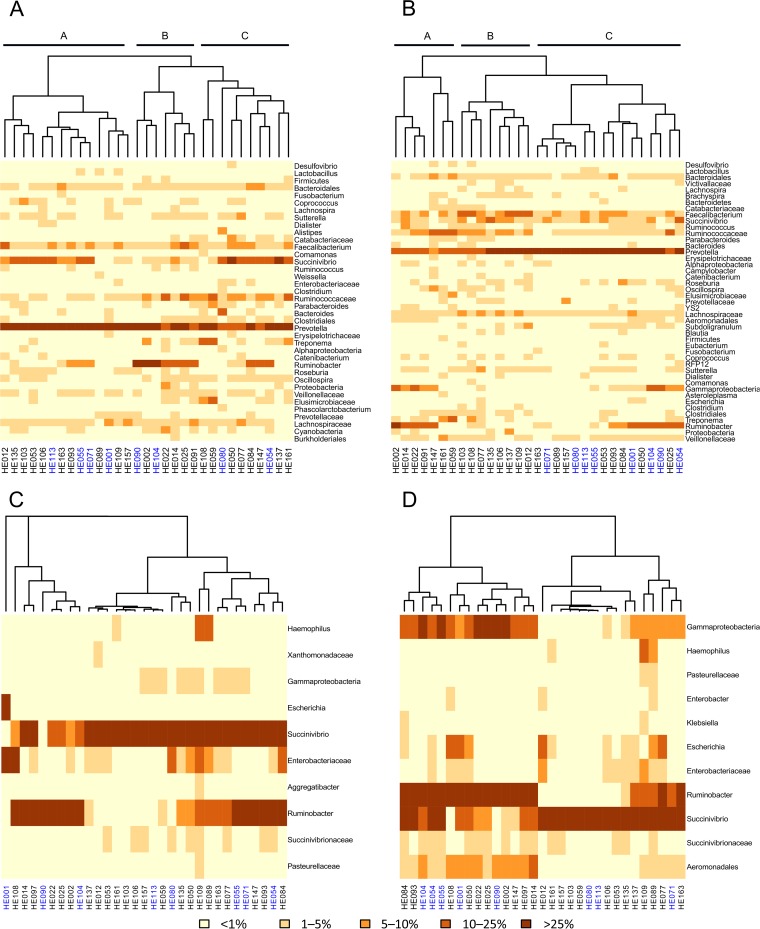
An average of 2,500 16S rDNA sequences per sample were obtained, with the phyla Bacteroidetes (45%) and Firmicutes (40%) being the most abundant, followed by Proteobacteria (12%), Spirochaetes (1.8%), and Tenericutes (0.27%). Actinobacteria were present in only four individuals and with few sequences. Figure 1A shows the heat map based on taxon abundance and sample composition. Three major clusters (named A, B, and C) were obtained, and the 8 E-ESBL carriers were scattered among them (4, 2, and 2 in clusters A, B, and C, respectively). In almost all the samples, the Prevotella genus (Bacteroidetes phylum) was the most abundant taxon. Most of the Firmicutes sequences belonged to the families Lachnospiraceae, Ruminococcaceae, and Clostridiaceae. Focusing only on Gammaproteobacteria, we found that the Succinivibrio and Ruminobacter genera, both members of the family Succinivibrionaceae, were the main taxa, whereas the members of the Enterobacteriaceae family (to which Escherichia coli belongs) were less abundant, even in the 8 E-ESBL carriers (Fig. 1C).
FIG 1.
Gut microbiota composition. (A and B) Heat maps and clustering based on taxon composition and abundance (Bray-Curtis distance) for the total microbiota (16S rDNA) (A) and the active microbiota (16S rRNA) (B). (C and D) We also focused the analysis on gammaproteobacteria from total microbiota (C) and active microbiota (D). Colors in the figures depict the percentage range of sequences assigned to main taxa (abundance of >1% in at least one sample). The ESBL carriers are indicated in blue and noncarriers in black.
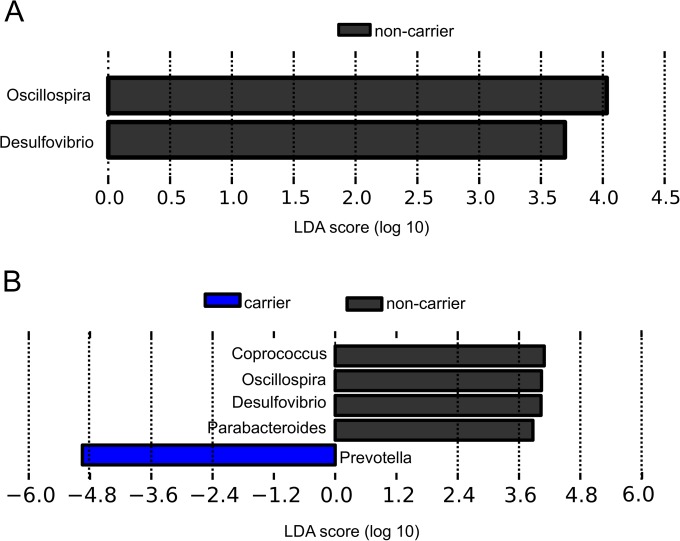
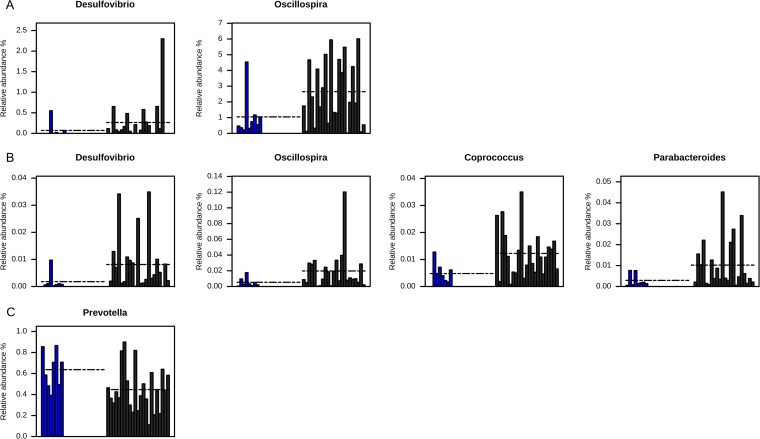
To quantify the diversity of each bacterial community, we used Chao1 richness estimator, Shannon equitability index, and Shannon diversity index values. The values determined for the Shannon diversity index, the Shannon equitability index, and the Chao1 richness estimator did not differ significantly between carriers and the three noncarrier controls (Table 1). The weighted UniFrac analysis was not able to separate carriers and noncarrier controls either (see Fig. S1A in the supplemental material). In addition, no cluster was obtained on the basis of the type of CTX-M genes carried (see Fig. S2A). The results of LEfSe analysis revealed, however, that two biomarkers, Desulfovibrio and Oscillospira, were significantly more abundant in the noncarriers than in the carriers (0.264 ± 0.492 versus 0.072 ± 0.181 [P = 0.05] and 2.652 ± 2.025 versus 1.048 ± 1.349 [P = 0.04], respectively) (Fig. 2A and 3A). Both genera are low-abundance taxa, but, unlike Desulfovibrio, detected only in two carriers, Oscillospira were present in all the samples and were, on average, 2.5 times more abundant in noncarriers than in carriers (Fig. 3A). No statistically significantly abundant taxa were found in total microbiota for noncarriers (Fig. 2A).
FIG 2.
Linear discriminant analysis between noncarriers and carriers. (A) Total microbiota. (B) active microbiota. The LDA (log 10) scores for the statistically significant taxa in the noncarrier samples are represented by the black bars in the positive scale, whereas the differential taxa in carriers correspond to the blue bar with a LDA-negative score.
FIG 3.
Relative abundances of the bacterial biomarkers. (A) Noncarriers' biomarkers in total microbiota. (B) Noncarriers' biomarkers in active microbiota. (C) Carriers' biomarkers in active microbiota. Dashed lines represent mean values.
Biodiversity and composition of the active microbiota.
Direct sequencing of the cDNA products gave a mean of 17,972 16S rRNA sequences per sample with an average length of 313 bp. The heat map (Fig. 1B) showed that samples also grouped into three major clusters but that they did so in a way different from that observed in the total microbiota. In fact, all eight carriers were grouped in cluster C whereas the noncarrier controls were evenly distributed among clusters. Similarly to the total microbiota, the active microbiota was characterized by a high abundance of members of the genus Prevotella, ranging from 25% to 80% of the total bacterial taxa detected. There was also a low abundance of members of the Enterobacteriaceae family, with the genus Escherichia, ranging from 0% to 2% of total sequences (Fig. 1D). When we compared the diversities of the active microbiotas of the carriers and noncarrier controls, we found significant differences with the Shannon diversity index (family controls) and the Shannon equitability index (nonrelated and family controls) results but not with the Chao1 estimator results (Table 1). Since the active bacteria of ESBL carriers were as taxon rich as those of noncarriers and since the individuals in the community were distributed less equitably among the taxon categories (Shannon equitability index), the active microbiota in carriers was less diverse than in controls. In analyzing the composition of the active microbiota using the UniFrac metric to determine beta diversity, we found that the ESBL carriers clustered as a group (see Fig. S1B in the supplemental material), indicating that they showed bacterial compositions that were more similar. However, all clusters according to the CTX-M allele type were found in active microbiota (see Fig. S2B).
Using LEfSe biomarker discovery software, we identified several genera discriminating between carriers and noncarrier controls, as shown by the LDA score data in Fig. 2B. Noncarriers had 4.5 times more Desulfovibrio (0.0081 ± 0.0100 versus 0.0017 ± 0.0030, P = 0.01), 3.7 times more Oscillospira (0.0197 ± 0.025 versus 0.0053 ± 0.0050, P = 0.03), 3.5 times more Parabacteroides (0.0102 ± 0.0120 versus 0.0029 ± 0.0030, P = 0.04), and 2.5 times more Coprococcus (0.0122 ± 0.008 versus 0.0048 ± 0.0040, P = 0.01) bacteria than carriers (Fig. 3B). Carriers, on the other hand, had 1.4 times more abundant Prevotella bacteria than noncarriers (0.63 ± 0.17 versus 0.44 ± 0.20, P = 0.02) (Fig. 2B and 3C).
DISCUSSION
The main result of this study was that we observed tentative evidence for differences between the compositions of the microbiota of E-ESBL carriers and noncarrier controls, in the abundance and/or in the expression of some specific taxa, which could be considered biomarkers. As far as we know, this observation has not been made previously. Members of two anaerobic taxa, belonging to the genera Desulfovibrio (Desulfovibrionaceae) and Oscillospira (Ruminococcaceae), appeared significantly more abundant in the total microbiota of noncarrier controls than in carriers. When the active microbiota was considered, members of two additional genera (Parabacteroides and Coprococcus) were also significantly more abundant in noncarriers. In contrast, there was only one genus (Prevotella) whose species were more abundant in the active microbiota of carriers than in noncarriers. It is tempting to suggest there is a relationship between the more abundant genera in the microbiota of noncarriers and resistance to E-ESBL colonization. However, the cross-sectional case-control nature of the study prevents our identifying a conclusive causal relationship. Indeed, the differences observed might as well be the consequences of E-ESBL carriage as the causes. It is important to point out, however, that members of the Enterobacteriaceae family were present in very low abundance in the total microbiota as well as in the active microbiota of carriers. This low abundance suggests that enterobacteria, whether E-ESBL or not, are not numerous enough, and are thus insufficiently metabolically active, to underlie the microbiota composition. However, low abundance does not always exclude high gene expression levels (29); thus, we cannot exclude a possible role of E-ESBL in shaping the microbiota. One limitation of the study was that the sample size was small. Therefore, matching between ESBL carriers and noncarriers was not fully successful. Moreover, the differential genera that were discriminated by LEfSe were based on a small number of reads. Thus, further studies should be done to give support to such findings with respect to ESBL biomarkers.
Two characteristics of the population that we studied make these results particularly meaningful. First, the study was performed on subjects from an isolated community, in which the antibiotic exposure of each individual in the year preceding sampling was known exactly. The exposures were infrequent and were not significantly different in carriers and noncarriers. Thus, differences in antibiotic exposure would are not likely the reason for the differences observed. Second, the subjects from this community have a very traditional lifestyle and eat exclusively what they cultivate, fish, or hunt. Their diet is not very varied, and they all have approximately the same one. Thus, differences in diet, which are known to influence the composition of the intestinal microbiota (30–34), were minimized in our study. Last, the population is ethnically very homogeneous (10) and it is thus unlikely that genetic differences are the cause of the differences observed between carriers and noncarriers.
The microbiota of the Amerindians was found to be characterized by a high abundance of members of the Prevotella genus, as described in other rural populations (31–33), where it has been related to the vegetarian diet. This is possibly because these taxa ferment the dietary fiber contained in whole grain, producing short-chain fatty acids (SCFAs), important metabolites in host physiology. Species of Faecalibacterium (Firmicutes), another SCFA producer, and of the Lachnospiraceae and Ruminococcaceae families, known as important pectin and cellulose degraders in colonic fermentation of dietary fibers (35–37), were also highly represented in the microbiota of the Amerindians. In addition, species of Ruminobacter and Succinivibrio (Succinivibrionaceae family), also present in the Amerindians, are known inhabitants of the rumen of cattle and sheep, playing an important role in the digestion of alpha-linked glucose molecules (maltose, maltodextrins, and starch). Thus, the overall intestinal microbiota seemed to be shaped to obtain the largest amount of energy from dietary fiber, which may be a reflection of the diet of this population.
In conclusion, our results showed that the active microbiota of the Wayampi was less diverse in E-ESBL carriers than in noncarrier controls. Noncarriers had differential anaerobic bacteria in both total microbiota and active microbiota that could play a role in colonization resistance, while Prevotella appeared as a biomarker for the active microbiota of carriers. Indeed, the actual relationships between these specificities and E-ESBL colonization still have to be explored prospectively and studied in animal models; however, this will not be possible for those of these species which, such as Oscillospira species, are not cultivable (38). Clearly, because the results were obtained using a small sample pool and matching was imperfect, the results will have to be confirmed in a larger, more diversified, and prospective cohort. Nonetheless, our results open up interesting avenues for modulating the intestinal microbiota and reducing the burden associated with E-ESBL colonization.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Spanish Ministry of Economy and Competitiveness (BFU2009-04501-E, SAF2009-13032-C02-01, SAF2012-31187, and SAF2013-49788-EXP) and by “Generalitat Valenciana” (PrometeoII/2014/065). J.F.V.-C. was supported by a fellowship, “Ayudas Predoctorales de Formación en Investigación en Salud,” from the Instituto de Salud Carlos III, Spain. The work was supported in France by ANR (05/9114), ANSES (ES-05-01 and EST-09-21), and, partially, by EU FP7 EvoTAR and R-Gnosis consortia.
Pyrosequencing was carried out by Nuria Jiménez in the Sequencing Service of FISABIO-Salud Pública (Valencia, Spain). We thank Alejandro Artacho for bioinformatics support.
We declare that we have no conflicts of interests.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01528-15.
REFERENCES
- 1.Appelbaum PC. 2012. 2012 and beyond: potential for the start of a second pre-antibiotic era? J Antimicrob Chemother 67:2062–2068. doi: 10.1093/jac/dks213. [DOI] [PubMed] [Google Scholar]
- 2.Savard P, Perl TM. 2012. A call for action: managing the emergence of multidrug-resistant Enterobacteriaceae in the acute care settings. Curr Opin Infect Dis 25:371–377. doi: 10.1097/QCO.0b013e3283558c17. [DOI] [PubMed] [Google Scholar]
- 3.Carlet J. 2012. The gut is the epicentre of antibiotic resistance. Antimicrob Resist Infect Control 1:39. doi: 10.1186/2047-2994-1-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woerther P-L, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum β-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruppé E, Pitsch A, Tubach F, de Lastours V, Chau F, Pasquet B, Lucet J-C, Andremont A, Fantin B. 2012. Clinical predictive values of extended-spectrum beta-lactamase carriage in patients admitted to medical wards. Eur J Clin Microbiol Infect Dis 31:319–325. doi: 10.1007/s10096-011-1313-z. [DOI] [PubMed] [Google Scholar]
- 6.Lawley TD, Walker AW. 2013. Intestinal colonization resistance. Immunology 138:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donskey CJ. 2006. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis 43(Suppl 2):S62–S69. doi: 10.1086/504481. [DOI] [PubMed] [Google Scholar]
- 8.Ubeda C, Bucci V, Caballero S, Djukovic A, Toussaint NC, Equinda M, Lipuma L, Ling L, Gobourne A, No D, Taur Y, Jenq RR, van den Brink MRM, Xavier JB, Pamer EG. 2013. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun 81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angebault C, Djossou F, Abélanet S, Permal E, Soltana MB, Diancourt L, Bouchier C, Woerther P-L, Catzeflis F, Andremont A, D'Enfert C, Bougnoux M-E. 2013. Candida albicans is not always the preferential yeast colonising humans: a study in Wayampi Amerindians. J Infect Dis 208:1705–1716. doi: 10.1093/infdis/jit389. [DOI] [PubMed] [Google Scholar]
- 10.Grenet K, Guillemot D, Jarlier V, Moreau B, Dubourdieu S, Ruimy R, Armand-Lefevre L, Bau P, Andremont A. 2004. Antibacterial resistance, Wayampis Amerindians, French Guyana. Emerg Infect Dis 10:1150–1153. doi: 10.3201/eid1006.031015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruimy R, Angebault C, Djossou F, Dupont C, Epelboin L, Jarraud S, Lefevre LA, Bes M, Lixandru BE, Bertine M, Miniai AE, Renard M, Bettinger RM, Lescat M, Clermont O, Peroz G, Lina G, Tavakol M, Vandenesch F, van Belkum A, Rousset F, Andremont A. 2010. Are host genetics the predominant determinant of persistent nasal Staphylococcus aureus carriage in humans? J Infect Dis 202:924–934. doi: 10.1086/655901. [DOI] [PubMed] [Google Scholar]
- 12.Woerther P-L, Angebault C, Lescat M, Ruppé E, Skurnik D, Mniai AE, Clermont O, Jacquier H, Costa AD, Renard M, Bettinger RM, Epelboin L, Dupont C, Guillemot D, Rousset F, Arlet G, Denamur E, Djossou F, Andremont A. 2010. Emergence and dissemination of extended-spectrum beta-lactamase-producing Escherichia coli in the community: lessons from the study of a remote and controlled population. J Infect Dis 202:515–523. doi: 10.1086/654883. [DOI] [PubMed] [Google Scholar]
- 13.Woerther P-L, Angebault C, Jacquier H, Clermont O, El Mniai A, Moreau B, Djossou F, Peroz G, Catzeflis F, Denamur E, Andremont A. 2013. Characterization of fecal extended-spectrum-β-lactamase-producing Escherichia coli in a remote community during a long time period. Antimicrob Agents Chemother 57:5060–5066. doi: 10.1128/AAC.00848-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Mcgarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(Database issue):D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caporaso JG, Kuczynski JS. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nchembio.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chao A, Hwang W-H, Chen Y-C, Kuo C-Y. 2000. Estimating the number of shared species in two communities. Stat Sin 10:227–246. [Google Scholar]
- 22.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423, 623–656.23. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 23.Help CHR, Herman PMJ, Soetaert K. 1998. Indices of diversity and evenness. Oceanis 24:61–87. [Google Scholar]
- 24.Fisher RA. 1936. The use of multiple measurements in taxonomic problems. Ann Eugen 7:179–188. doi: 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- 25.Mann HB, Whitney DR. 1947. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat 18:50–60. doi: 10.1214/aoms/1177730491. [DOI] [Google Scholar]
- 26.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 28.Wilcoxon F. 1945. Individual comparisons by ranking methods. Biometrics 1:80–83. doi: 10.2307/3001968. [DOI] [Google Scholar]
- 29.Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, Neulinger SC, Däumer C, Heinsen F-A, Latorre A, Barbas C, Seifert J, Dos Santos VM, Ott SJ, Ferrer M, Moya A. 2013. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nam Y-D, Jung M-J, Roh SW, Kim M-S, Bae J-W. 2011. Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One 6:e22109. doi: 10.1371/journal.pone.0022109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 35.Dusková D, Marounek M. 2001. Fermentation of pectin and glucose, and activity of pectin-degrading enzymes in the rumen bacterium Lachnospira multiparus. Lett Appl Microbiol 33:159–163. [DOI] [PubMed] [Google Scholar]
- 36.Marounek M, Duskova D. 1999. Metabolism of pectin in rumen bacteria Butyrivibrio fibrisolvens and Prevotella ruminicola. Lett Appl Microbiol 29:429–433. doi: 10.1046/j.1472-765X.1999.00671.x. [DOI] [Google Scholar]
- 37.Rode LM, Genthner BR, Bryant MP. 1981. Syntrophic association by cocultures of the ethanol- and C02-H2-utilizing species Eubacterium limosum and pectin-fermenting Lachnospira multiparus during growth in a pectin medium. Appl Environ Microbiol 42:20–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mackie RI, Aminov RI, Hu W, Athol V, Ouwerkerk D, Sundset MA. 2003. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol 69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.