Abstract
West Nile virus (WNV) is a neurotropic flavivirus transmitted by the bite of mosquitoes that causes meningitis and encephalitis in humans, horses, and birds. Several studies have highlighted that flavivirus infection is highly dependent on cellular lipids for virus replication and infectious particle biogenesis. The first steps of lipid synthesis involve the carboxylation of acetyl coenzyme A (acetyl-CoA) to malonyl-CoA that is catalyzed by the acetyl-CoA carboxylase (ACC). This makes ACC a key enzyme of lipid synthesis that is currently being evaluated as a therapeutic target for different disorders, including cancers, obesity, diabetes, and viral infections. We have analyzed the effect of the ACC inhibitor 5-(tetradecyloxy)-2-furoic acid (TOFA) on infection by WNV. Lipidomic analysis of TOFA-treated cells confirmed that this drug reduced the cellular content of multiple lipids, including those directly implicated in the flavivirus life cycle (glycerophospholipids, sphingolipids, and cholesterol). Treatment with TOFA significantly inhibited the multiplication of WNV in a dose-dependent manner. Further analysis of the antiviral effect of this drug showed that the inhibitory effect was related to a reduction of viral replication. Furthermore, treatment with another ACC inhibitor, 3,3,14,14-tetramethylhexadecanedioic acid (MEDICA 16), also inhibited WNV infection. Interestingly, TOFA and MEDICA 16 also reduced the multiplication of Usutu virus (USUV), a WNV-related flavivirus. These results point to the ACC as a druggable cellular target suitable for antiviral development against WNV and other flaviviruses.
INTRODUCTION
West Nile virus (WNV) is a mosquito-borne neurotropic flavivirus responsible for recurrent outbreaks of meningitis and encephalitis affecting humans, horses, and birds in Africa, Europe, Asia, Oceania, and America (1). A great effort has been devoted in the past several years to decipher the molecular biology of WNV and its interaction with the host immune system (2, 3). Nevertheless, no licensed vaccine or therapy for human use against this pathogen is yet available.
The flavivirus life cycle (including that of WNV) is intimately associated with host cell lipids. The replication of the viral genomic RNA and flavivirus nascent virion assembly take place in modified membranes from the endoplasmic reticulum (4–7). To build an adequate microenvironment to support viral replication and particle biogenesis, flaviviruses rearrange host cell lipid metabolism by promoting the synthesis and accumulation of specific cellular lipids (i.e., fatty acids, glycerophospholipids [GPLs], sphingolipids [SLs], and cholesterol) (8–15). This makes the pharmacological manipulation of cellular lipids an attractive antiviral strategy against WNV and related flaviviruses (13, 14, 16, 17).
The first steps of lipid biogenesis involve the synthesis and elongation of fatty acids, which provide the building blocks for the synthesis of more-complex lipids. Hence, fatty acid synthesis and elongation have become key targets for antiviral therapy (13, 18, 19). Regarding the flaviviruses, the pharmacological blockage of the fatty acid synthase FASN (which catalyzes the synthesis of palmitate from acetyl coenzyme A [acetyl-CoA] and malonyl-CoA into long-chain saturated fatty acids) reduced the viral replication (11, 13). The enzyme preceding FASN in the fatty acid biosynthetic route is the acetyl-CoA carboxylase (ACC), which catalyzes the carboxylation of acetyl-CoA to malonyl-CoA. Due to its rate-limiting role in fatty acid synthesis, ACC is currently a target of increasing interest within the pharmacological industry (20, 21). However, to our knowledge, the involvement of ACC in the replication of WNV, or other related flaviviruses, has not yet been evaluated.
In this work, we have shown that ACC inhibitors alter the cellular lipid composition and reduce the levels of WNV infection in cultured cells. Furthermore, infection by Usutu virus (USUV; a related emerging flavivirus [22]) was also inhibited by the drugs used. Our results point to ACC as a potential druggable antiviral target against WNV and related flaviviruses.
MATERIALS AND METHODS
Cells, viruses, infections, and virus titrations.
All infectious virus manipulations were performed in biosafety level 3 (BSL-3) facilities. Vero, HeLa, and Neuro2a (N2a) cell lines were cultured as described previously (10, 23). Cells were incubated with the corresponding virus, WNV strain NY99 or USUV strain SAAR-1776 (24), for 1 h at 37°C; viral inoculum was removed; and infected cells were incubated in culture medium containing 1% fetal bovine serum (time that was considered 1 h postinfection [p.i.]). Viral titer was determined 24 h p.i. by plaque assay in semisolid agarose medium using Vero cells (25). The multiplicity of infection (MOI) used in each experiment was expressed as PFU per cell and is indicated in the corresponding figure legend.
Drug treatments.
5-(Tetradecyloxy)-2-furoic acid (TOFA) and 3,3,14,14-tetramethylhexadecanedioic acid (MEDICA 16) were from Sigma (St. Louis, MO). Control cells were treated in parallel with the same amount of drug vehicle (dimethyl sulfoxide [DMSO]). Unless otherwise specified, drugs were added after the first hour of infection, when viral inoculum was replaced by medium containing 1% fetal bovine serum. Drug toxicity was examined by measuring the cellular ATP content with the CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI).
Microscopy.
Antibodies and procedures used for immunofluorescence and confocal microscopy, as well as sample processing for transmission electron microscopy, have been described previously (11). The analysis of fluorescence intensity was performed using ImageJ software (http://imagej.nih.gov/ij/).
Quantitative RT-PCR.
Viral RNA was extracted from the supernatant of infected cultures with the Speedtools RNA virus extraction kit (Biotools, Madrid, Spain). For quantification of cell-associated viral RNA, supernatants from infected cells were removed, cell monolayers were subjected to three freeze-thaw cycles, and RNA was extracted as described above. The amount of viral RNA was determined by real-time fluorogenic reverse transcriptase PCR (RT-PCR) as reported previously (26). The forward primer 5′-CAGACCACGCTACGGCG-3′, the reverse primer 5′-CTAGGGCCGCGTGGG-3′, and the probe 5′-FAM (6-carboxyfluorescein)-TCTGCGGAGAGTGCAGTCTGCGAT-3′-BHQ-1 (black hole quencher 1) were used. Quantification was performed using the High Scriptools-Quantimix Easy Probes kit (Biotools) and Rotor-Gene RG-3000 equipment (Corbett Research). Genomic equivalents to PFU per milliliter were calculated by comparison with 10-fold serial dilutions of viral RNA extracted from previously titrated samples (27).
Lipid analysis.
Vero cells were treated with 10 μg/ml TOFA or with DMSO and infected or not with WNV at a high MOI (50 PFU/cell) to ensure that all cells were initially infected. Cells were detached from the flasks and resuspended in phosphate-buffered saline (PBS) at 24 h p.i. (10). Infections were performed in serum-free medium to avoid possible biases introduced by lipids present in the serum. The number of cells in each sample was determined, and aliquots containing 106 cells were subjected to lipid extractions. Procedures for lipid extractions, identification, and quantification by mass spectrometry have been previously described (10, 28, 29). Annotation of lipid species followed the recommendations indicated in reference 30. Glycerophospholipids, diacylglycerol (DAG), triacylglycerol (TAG), and cholesteryl esters (CHOL) were annotated as “lipid subclass” followed by “total fatty acyl chain length:total number of unsaturated bonds” (e.g., PC 36:2). Plasmalogens were annotated as described above, except that “p” was added. Lysophospholipids were annotated as described above except that “lyso” was added. Sphingolipids were annotated as “lipid subclass” followed by “total fatty acyl chain length:total number of unsaturated bonds” (e.g., CER 24:1). If the sphingoid base residue was dihydrosphingosine, the lipid class contained a “dh” prefix. For ceramide derivatives containing sugar moieties, the lipid class contained a “hex” prefix to denote monohexosylceramides (glucosylceramides and galactosylceramides) or a “lac” prefix for lactosylceramides.
Data analysis.
Data are presented as means ± standard deviations (SDs). Analysis of variance (ANOVA) and Student's t test were performed with SPSS 15 (SPSS Inc., Chicago, IL). Bonferroni's correction was applied for multiple comparisons. Statistically significant differences were considered at a P value of <0.05.
RESULTS
Effect of inhibition of ACC on cellular lipid content.
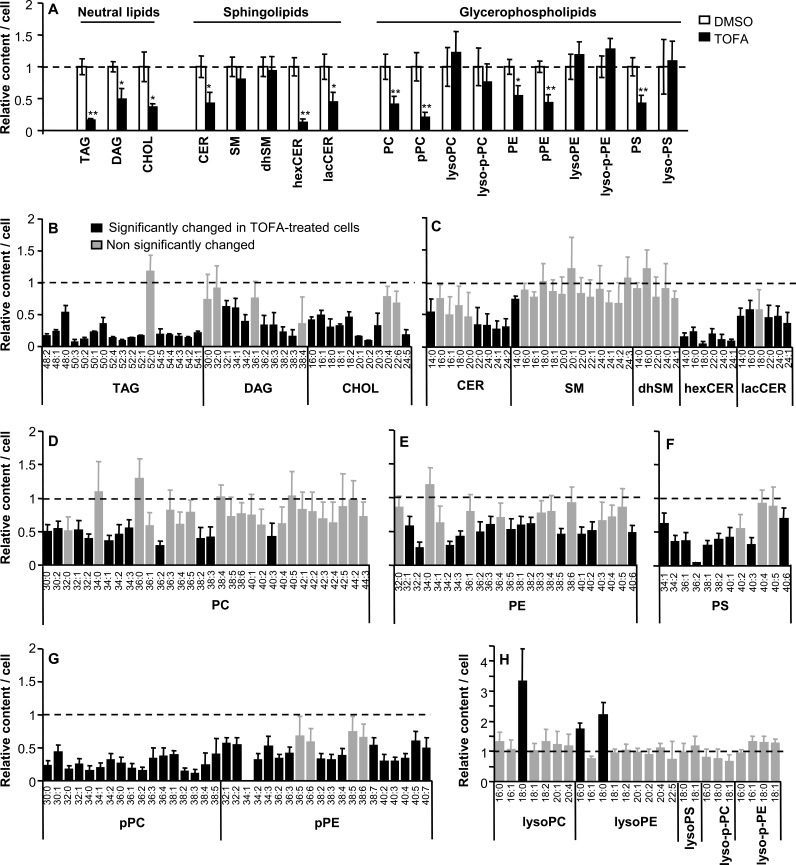
TOFA is an inhibitor of ACC that has been used to evaluate the involvement of this enzyme in infections by a variety of viruses (18, 31–34). To determine its effect on the cellular lipid content, Vero cells were treated with TOFA for 24 h and the lipid content was analyzed by mass spectrometry and compared to that of control cells treated in parallel with the same amount of drug solvent (Fig. 1). These experiments were performed in serum-free medium to avoid possible interference of serum lipids in the analysis. Eighteen lipid classes (3 neutral lipids [NLs], 10 glycerophospholipids [GPLs], and 5 sphingolipids [SLs]) were included in the analysis (Fig. 1A). A significant decrease in the amount of NLs (triacyclglycerol [TAG], diacylglycerol [DAG], and cholesteryl esters [CHOL]) was observed in TOFA-treated cells (Fig. 1A). Among SLs, ceramide (CER), monohexosylceramide (hexCER), lactosylceramide (lacCER), sphingomyelin (SM), and dihydroSM (dhSM) were analyzed. The amount of CER and its derivatives containing sugar moieties (hexCER and lacCER) was significantly reduced in TOFA-treated cells. In contrast, the content of SM and dhSM was not significantly reduced by treatment with TOFA (Fig. 1A). To evaluate the effect of TOFA on the cellular GPLs, we analyzed the content of phosphatidylcholine (PC), 1-alkenyl-2-acyl-glycero-3-phosphocholine (referred to as plasmalogen-PC [p-PC; plasmenylcholine]), 1-acyl-glycero-3-phosphocholine (lyso-PC), lyso-plasmenylcholine (lyso-p-PC), phosphatidylethanolamine (PE), 1-alkenyl-2-acyl-glycero-3-phosphoethanolamine (p-PE), 1-acyl-glycero-3-phosphoethanolamine (lyso-PE), lyso-p-PE, phosphatidylserine (PS), and 1-acyl-glycero-3-phosphoserine (lyso-PS). A significant decrease in the amount of PC, p-PC, PE, p-PE, and PS was noted in TOFA-treated cells, whereas no significant change in the lysolipids analyzed (lyso-PC, lyso-p-PC, lyso-PE, lyso-p-PE, and lyso-PS) was induced by treatment with TOFA. The lack of reduction in the amount of lysolipids can be explained because lysolipids are produced by the action of phospholipases (35) rather than by a biosynthetic process. Among the 18 lipid classes analyzed, a total of 208 different molecular species could be identified (Fig. 1B to H). From these species, 89 remained unaltered in TOFA-treated cells (42.8%) and a significant increase was observed in only three of the identified species (1.4%) that corresponded to lysolipids (Fig. 1H). Such an increase could likely correspond to a stress response including the activation of phospholipases (35). On the other hand, on 116 lipid species (55.8%) a significant reduction was noted, confirming that inhibition of ACC using TOFA has a major impact on lipid synthesis, reducing the content of a wide variety of cellular lipids.
FIG 1.
Alterations of cellular lipid content upon inhibition of ACC using TOFA. (A) Relative amounts of different classes of NLs, SLs, and GPLs in Vero cells treated with 10 μg/ml TOFA or not treated (DMSO) determined by mass spectrometry at 24 h after drug addition. The experiments were performed in serum-free medium. Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005. (B to H) Relative amounts of different lipid molecular species (NLs [B], SLs [C], PC [D], PE [E], PS [F], plasmalogens [G], and lysophospholipids [H]) in cells treated with TOFA as in panel A. Black bars denote molecular species significantly altered (P < 0.05) relative to control cells. Gray bars denote molecular species not significantly changed relative to untreated cells. Bars corresponding to untreated cells have been omitted from the figure for clarity. Dashed lines, mean value for each lipid in untreated cells. Data are presented as means ± SDs (three replicates).
TOFA inhibits infection by WNV and USUV.
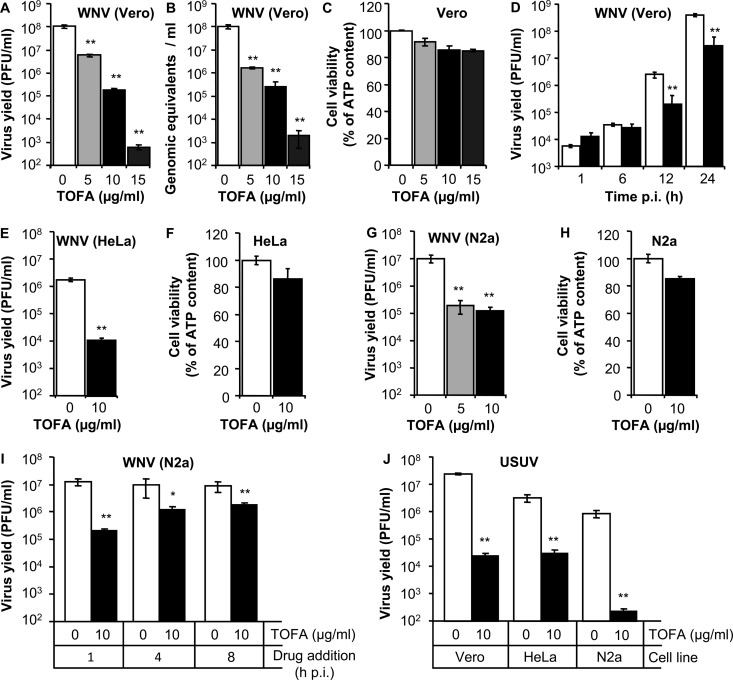
Vero cells were infected with WNV, and different concentrations of TOFA were added 1 h p.i. to avoid possible interference of the drug with virus entry. TOFA inhibited WNV multiplication in a dose-dependent manner (Fig. 2A and B). Both the production of infectious particles (Fig. 2A) and the release of genome-containing particles to the culture medium (Fig. 2B) were significantly reduced by TOFA. The toxicity of the drug was analyzed in parallel by determination of the cellular ATP content (Fig. 2C), confirming that the concentrations of TOFA tested inhibited WNV infection, exerting small effects on cell viability. A time course analysis of the production of infectious virus revealed that TOFA inhibited the multiplication of WNV also at time points as early as 12 h p.i. (Fig. 2D). Next, the effect of TOFA on WNV infection in other cell lines was tested. An inhibition similar to that observed on Vero cells (Fig. 2A and B) was noted in TOFA-treated HeLa cells (Fig. 2E and F). Considering that WNV is a neurotropic virus, the effect of TOFA was also assayed in the neuroblastoma cell line N2a. TOFA also inhibited the infection of WNV in N2a cells, showing also a minor impact on cell viability (Fig. 2G and H). When N2a cells were used to address the effect of TOFA addition to infected cultures at 1, 4, and 8 h p.i., a significant inhibition of WNV infection was observed in all cases (Fig. 2I), indicating that treatment with TOFA impaired WNV infection after virus entry. Even more, TOFA also significantly reduced the production of USUV, a related flavivirus, in Vero, HeLa, and N2a cells (Fig. 2J). These results suggest that the inhibition of ACC can be also effective against other related flaviviruses.
FIG 2.
TOFA inhibits WNV and USUV multiplication. (A) Reduction of WNV infectious particle production in Vero cells treated with different concentrations of TOFA. Cells were infected with WNV (MOI of 0.5 PFU/cell), and virus yield in culture supernatant was determined by plaque assay at 24 h p.i. (B) Quantification by quantitative RT-PCR of genome-containing particles in culture supernatant of Vero cells infected as in panel A. (C) Evaluation of the toxicity of TOFA on Vero cells by determination of cellular ATP content 24 h posttreatment. (D) Time course analysis of virus production in TOFA-treated cells. Vero cells were infected with WNV (MOI of 1 PFU/cell) and treated with 10 μg/ml TOFA. The amount of infectious virus in the supernatant of infected cultures was determined by plaque assay at different times p.i. (E) Reduction of WNV infectious particle production (24 h p.i.) in HeLa cells infected (MOI of 0.5 PFU/cell) and treated with TOFA. (F) Evaluation of the toxicity of TOFA on HeLa cells 24 h posttreatment. (G) Reduction of WNV infectious particle production (24 h p.i.) in N2a cells infected (MOI of 0.5 PFU/cell) and treated with TOFA. (H) Evaluation of the toxicity of TOFA on N2a cells 24 h posttreatment. (I) Inhibition of WNV multiplication in N2a cells by adding TOFA at different times p.i. N2a cells were infected with WNV (MOI of 0.5 PFU/cell), and 10 μg/ml TOFA was added at the indicated time points. Virus yield was determined by plaque assay at 24 h p.i. (J) Inhibition of USUV infection in different cell lines using TOFA. Vero, HeLa, and N2a cells were infected with USUV (MOI of 0.5 PFU/cell) and treated with TOFA. Virus yield was determined by plaque assay at 24 h p.i. Data are presented as means ± SDs (three replicates). Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005.
TOFA inhibits the replication of WNV genome.
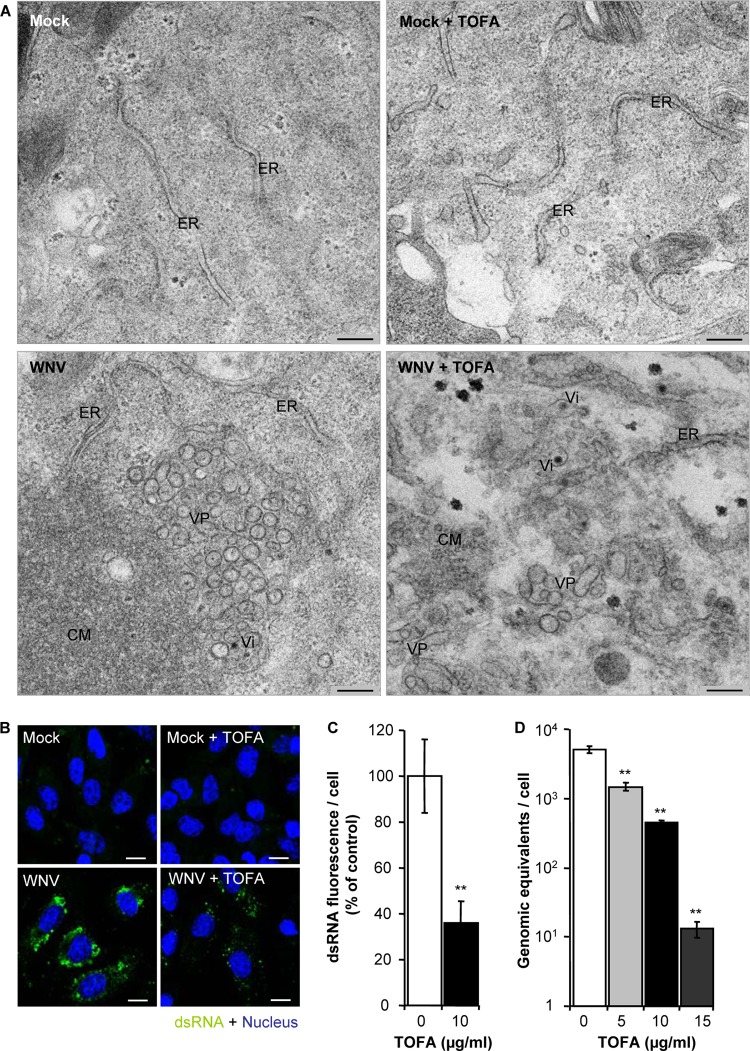
The infection with WNV induces marked changes in the intracellular membrane architecture to create an appropriate microenvironment for viral replication (7, 11). Hence, we analyzed the effect of TOFA on Vero cells infected with WNV by means of transmission electron microscopy (Fig. 3A). Mock-infected cells treated with TOFA or not treated exhibited normal cellular architecture (Fig. 3A, upper panels). In contrast, control infected cells not treated with TOFA exhibited characteristic infection-associated ultrastructural alterations, such as convoluted membranes (CM), vesicle packets (VPs), and electron-dense virions (Vi) (Fig. 3A, lower left panel). In infected cells treated with TOFA, Vi could be recognized but properly arranged CM or VPs, such as those observed in control cells, could not be detected. Instead, disordered VP-like materials and small cumuli of membranous materials resembling CM were observed (Fig. 3A, lower right panel). Since the replication of the flavivirus genome takes place in the VP (4, 36), our observations could indicate that the alteration of cellular lipids induced by TOFA impaired the development of WNV replication complexes. To support this hypothesis, the amount of double-stranded RNA (dsRNA) intermediates (a well-characterized marker of flavivirus replication complex [4, 7]) was analyzed by immunofluorescence in infected cells treated or not treated with TOFA (Fig. 3B). As expected, no dsRNA was detected in uninfected cells, treated or not treated with TOFA (Fig. 3B, upper panels). In contrast, dsRNA was observed in both control and TOFA-treated cells infected with WNV. However, the intensity of the dsRNA fluorescence was lower in infected cells treated with TOFA than in control infected cells (Fig. 3B, lower panels). The quantification of this signal confirmed a statistically significant reduction in the cellular content of dsRNA intermediates in TOFA-treated cells (Fig. 3C). Furthermore, the analysis by quantitative RT-PCR revealed that treatment with TOFA induced a statistically significant dose-dependent reduction in the amount of cell-associated viral RNA (Fig. 3D). Taken together, these results suggest that the alteration of cellular lipid content induced by TOFA impairs WNV replication complex assembly and hence viral replication.
FIG 3.
Effect of TOFA on replication of WNV. (A) Intracellular membrane rearrangements in WNV-infected Vero cells observed by transmission electron microscopy 24 h p.i. Upper panels show representative images of uninfected cells untreated (left) or treated with 10 μg/ml TOFA (right). Lower panels display representative images of infected cells (MOI, 10 PFU/cell) untreated (left) or treated with TOFA (right). VP, vesicle packet; Vi, virion; CM, convoluted membranes; ER, endoplasmic reticulum. Bars, 200 nm. (B) Visualization of intracellular dsRNA accumulation in cells infected with WNV (MOI, 10 PFU/cell) and treated with 10 μg/ml TOFA at 24 h p.i. Cells were fixed and processed for immunofluorescence using monoclonal antibody to dsRNA J2 and a secondary antibody coupled to Alexa Fluor 488 (green). Nuclei were stained with ToPro-3 (blue). Mock-infected cells processed in parallel are also included as a control. Bars, 10 μm. (C) Quantification of the fluorescence intensity of dsRNA in cells infected and treated with TOFA as shown in panel B. (D) Amount of cell-associated viral RNA in cell cultures infected with WNV (MOI of 0.5 PFU/cell) and treated with TOFA determined by quantitative RT-PCR at 24 h p.i. Data are presented as means ± SDs (three replicates). Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005.
TOFA reduces the lipid content of WNV-infected cells.
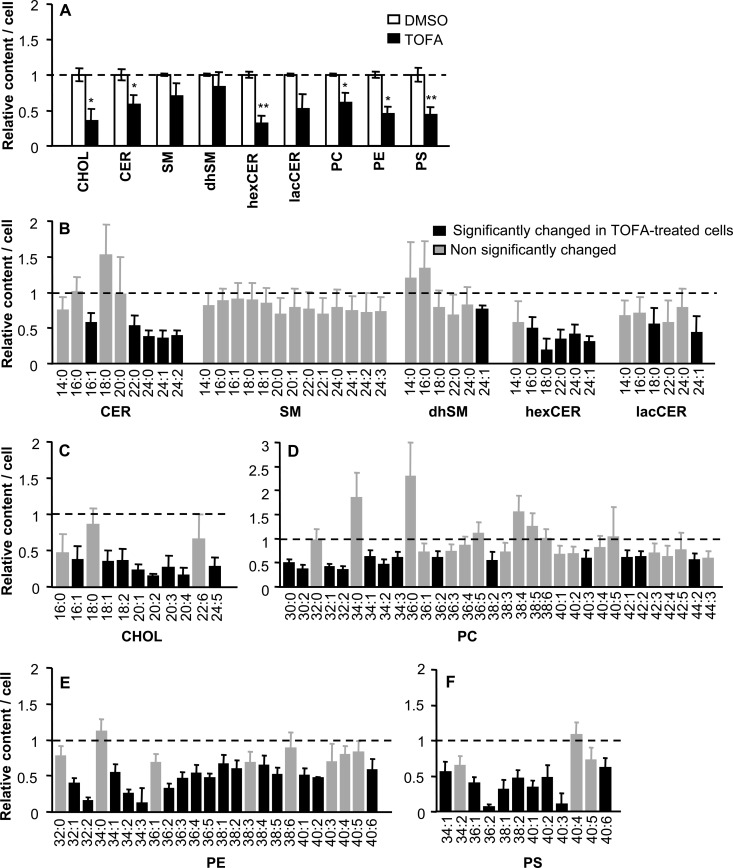
The amount of lipids in WNV-infected cells was analyzed by mass spectrometry (Fig. 4). In this case, in order to simplify the analysis, we focused on 9 lipid classes, including representative members of NLs (CHOL), SLs (CER, SM, dhSM, hexCER, and lacCER), and GPLs (PC, PE, and PS). These lipid classes included important membrane components, some of which had been previously associated with different features of virus infection (8–10, 37, 38). Relative to infected cells not treated with the drug, a significant reduction in the content of CHOL, CER, hexCER, PC, PE, and PS was observed in infected cells treated with TOFA (Fig. 4A). Overall, these results supported the idea that, as observed in uninfected cells (Fig. 1), TOFA inhibited lipid synthesis in WNV-infected cells. Among the 9 lipid classes compared, a total of 119 different molecular species could be identified and analyzed (Fig. 4B to F). A significant reduction of 59 molecular species was noted in infected cells treated with TOFA (49.6% of species analyzed), 48 of which (81% of species analyzed) were also significantly reduced in uninfected cells treated with TOFA (Fig. 1). These findings indicated that the lipid changes induced by TOFA were similar in both WNV-infected and uninfected cells.
FIG 4.
Reduction of cellular lipid content in WNV-infected cells treated with TOFA. (A) Relative amounts of different lipid classes in Vero cells infected with WNV (MOI of 50 PFU/cell) and treated with 10 μg/ml TOFA or not (DMSO), determined by mass spectrometry at 24 h p.i. The experiments were performed in serum-free medium. Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005. (B to F) Relative amounts of different lipid molecular species (SLs [B], CHOL [C], PC [D], PE [E], and PS [F]) in cells infected with WNV and treated with TOFA as in panel A. Black bars denote molecular species significantly altered (P < 0.05) relative to untreated infected cells. Gray bars denote molecular species not significantly changed relative to untreated infected cells. Bars corresponding to untreated infected cells have been omitted from the figure for clarity. Dashed lines, mean value for each lipid in untreated infected cells. Data are presented as means ± SDs (three replicates).
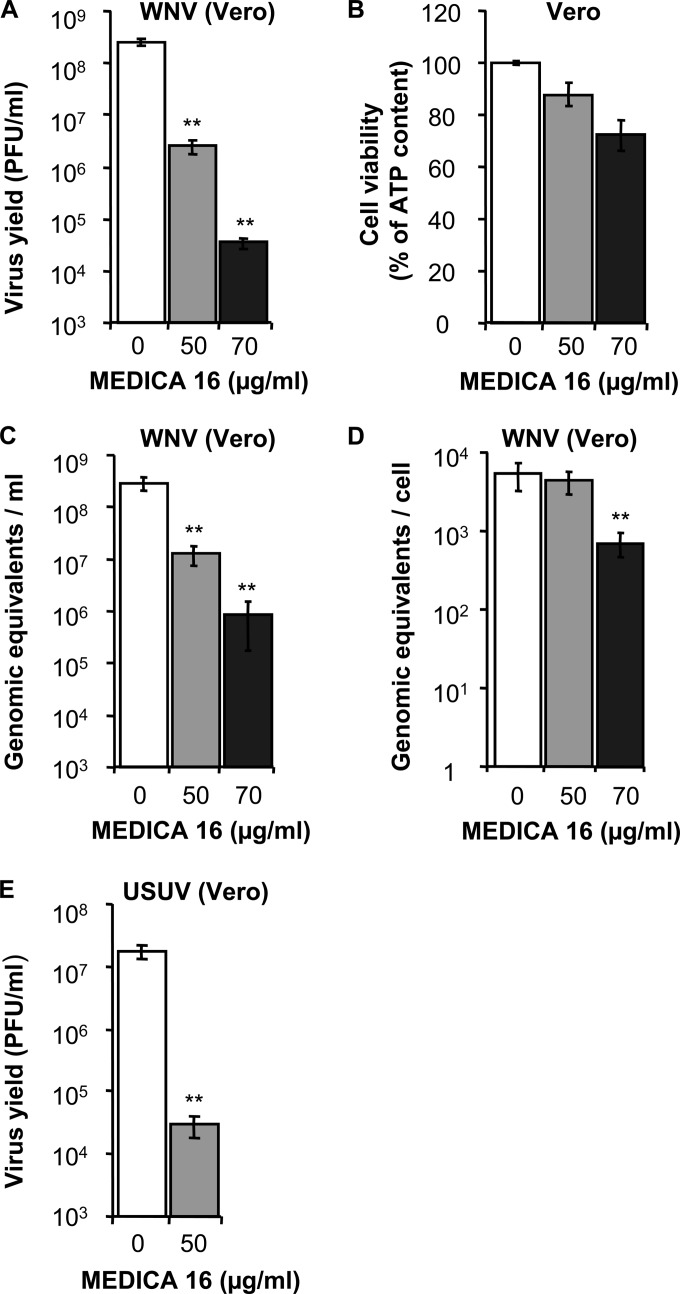
MEDICA 16 inhibits the infection of WNV and USUV.
Treatment of Vero cells with MEDICA 16, an ACC inhibitor different from TOFA (39), also resulted in a dose-dependent inhibition of the production of infectious WNV (Fig. 5A) at concentrations that did not severely impair the viability of the cells (Fig. 5B). The quantification of genome-containing particles by quantitative RT-PCR revealed that MEDICA 16 reduced the release of these viral particles to the culture medium (Fig. 5C). Remarkably, the extent of this inhibition was lower than the reduction produced in virus yield (compare Fig. 5A and C). This could indicate that treatment with MEDICA 16 was inducing the formation of noninfectious genome-containing WNV particles, a phenomenon that was not observed in infected cells treated with TOFA, in which the decrease in the production of infectious particles was similar to the reduction in the release of genome-containing particles (Fig. 2). Moreover, the concentration of 50 μg/ml MEDICA 16 that inhibited the production of infectious WNV and the release of genome-containing particles did not inhibit accumulation of cell-associated viral RNA within infected cells. However, a significant reduction of viral replication was observed at a higher concentration of 70 μg/ml (Fig. 5D). Nevertheless, MEDICA 16 strongly inhibited the infection with USUV (Fig. 5E), confirming that this hypolipidemic agent can also inhibit the multiplication of WNV-related flaviviruses.
FIG 5.
MEDICA 16 inhibits WNV and USUV multiplication. (A) Reduction of WNV infectious particle production in Vero cells treated with MEDICA 16. Cells were infected with WNV (MOI of 0.5 PFU/cell), and virus yield in culture supernatant was determined by plaque assay at 24 h p.i. (B) Evaluation of the toxicity of MEDICA 16 on Vero cells by determination of cellular ATP content 24 h posttreatment. (C) Determination by quantitative RT-PCR of the amount of genome-containing particles in culture supernatant of Vero cells infected with WNV (MOI of 0.5 PFU/cell) at 24 h p.i. (D) Amount of cell-associated viral RNA in cell cultures infected with WNV (MOI of 0.5 PFU/cell) and treated with MEDICA 16 determined by quantitative RT-PCR at 24 h p.i. (E) Inhibition of USUV infection in Vero cells using MEDICA 16. Vero cells were infected with USUV (MOI of 0.5 PFU/cell) and treated with MEDICA 16, and virus yield was determined by plaque assay at 24 h p.i. Data are presented as means ± SDs (three replicates). Statistically significant differences are indicated: *, P < 0.05; **, P < 0.005.
DISCUSSION
Lipids play multifaceted roles during virus infection. In such scenarios, the pharmacological manipulation of cellular lipids can reduce virus infection by different mechanisms. These mechanisms include (i) the impairment of membrane rearrangements associated with viral replication, (ii) the generation of an unfavorable metabolic environment for virus replication, or (iii) the alteration of the viral envelope composition and the impairment of viral particle assembly (8, 17, 18, 31).
Treatment with TOFA reduced the synthesis of multiple cellular lipids, including those previously described to be involved in WNV infection, such as GPLs, SLs, and CHOL (8, 10, 15). Our electron microscopy results showed that TOFA induced a reduction in the amount of membrane rearrangements associated with WNV replication. In addition, quantitative RT-PCR analysis indicated that this drug reduced the synthesis of viral RNA, which was compatible with the reduction of the accumulation of dsRNA intermediates observed by immunofluorescence in infected cells treated with TOFA. Taken together, all these results suggest that the inhibition of lipid synthesis exerted by TOFA could impair the membrane rearrangements necessary for the replication of WNV. This mechanism of action would be similar to that recently reported for hepatitis C virus in cells treated with an inhibitor of the ACC (34). An additional point to be considered is that although treatment with TOFA reduced the amount of multiple lipid species, it also increased the amount of some lysolipids. Interestingly, the production of these lysolipids could be related to an activation of a stress response in TOFA-treated cells (35). The possible contributions of this stress to the antiviral effect of TOFA still remain to be evaluated.
In the case of the other ACC inhibitor tested, MEDICA 16, the inhibition exerted by the lowest concentration of this drug assayed (50 μg/ml) seemed to rely mainly on a reduction of infectious virions rather than on RNA synthesis inhibition, although at a high concentration (70 μg/ml) this drug also inhibited RNA replication. Blockage of infectious virus production has been described for other viruses treated with drugs affecting lipid metabolism (31, 40). In this regard, it should be considered that replication and assembly of viral particles are coupled processes in flaviviruses (5). This may explain our results showing that the alteration of lipid metabolism could affect both processes. In contrast to what was described for other members of the Flaviviridae family (41), there is no evidence, to our knowledge, of palmitoylation of WNV proteins. Thus, palmitoylation does not seem to be an antiviral target of action for ACC inhibitors in WNV infection.
Although the strategy of manipulating a major cellular metabolic pathway such as lipid biosynthesis could induce multiple side effects in vivo, it has to be considered that current antivirals target other major biosynthetic pathways, such as nucleotide biosynthesis (for a discussion, see reference 18). Indeed, the clinical value of lipid-lowering agents such as the statins, which are cholesterol synthesis inhibitors broadly used against cardiovascular diseases, supports the feasibility of therapeutic interventions targeting the lipid metabolism (42). Consistently, hypolipidemic drugs targeting the ACC are being evaluated for the treatment of multiple human disorders from certain cancers to obesity and diabetes (20, 21) and even viral infections (18, 34, 43). In addition, the results obtained with hypolipidemic agents (i.e., nordihydroguaiaretic acid) that target cellular factors other than ACC also support the feasibility of lipid-based antiviral approaches (17, 44). In this context, the potential of ACC inhibitors reported here to impair the replication of different flaviviruses, such as WNV and USUV, could contribute to the development of broad-spectrum antiflaviviral drugs.
In this work, we have provided further evidence supporting the idea that pharmacological manipulation of the lipid biosynthetic route can impair flavivirus infection. Specifically, we have identified the ACC as a druggable target to be considered for antiviral development against WNV and related flaviviruses.
ACKNOWLEDGMENTS
We thank M. Guerra and M. T. Rejas for help with electron microscopy and M. Calvo and Eva Dalmau for technical assistance. We thank E. García and J. Avila for N2a cells.
Funding Statement
This work was supported by grants RTA 00036-2011 (to J.-C.S.), BIO2011-24351 (to F.S.), AGL2014-52395-C2-1-R (to F.S.), E-RTA2013-0013 (to J.-C.S. and F.S.), S2013/ABI-2906 (to J.-C.S. and F.S.), AGL2014-56518-JIN (to M.A.M.-A.), and CTQ2014-54743-R (to J.C.). Work at CBMSO was also supported by Fundación Ramón Areces. A.V.-C. is a recipient of a “Contrato de formación postdoctoral” from MINECO. T.M.-R. is a recipient of a “Formación de Personal Investigador (FPI)” predoctoral fellowship from INIA.
REFERENCES
- 1.Martin-Acebes MA, Saiz JC. 2012. West Nile virus: a re-emerging pathogen revisited. World J Virol 1:51–70. doi: 10.5501/wjv.v1.i2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinton MA. 2013. Replication cycle and molecular biology of the West Nile virus. Viruses 6:13–53. doi: 10.3390/v6010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suthar MS, Diamond MS, Gale M Jr. 2013. West Nile virus infection and immunity. Nat Rev Microbiol 11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 4.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apte-Sengupta S, Sirohi D, Kuhn RJ. 2014. Coupling of replication and assembly in flaviviruses. Curr Opin Virol 9:134–142. doi: 10.1016/j.coviro.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J Virol 88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillespie LK, Hoenen A, Morgan G, Mackenzie JM. 2010. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol 84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackenzie JM, Khromykh AA, Parton RG. 2007. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2:229–239. doi: 10.1016/j.chom.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Perera R, Riley C, Isaac G, Hopf-Jannasch AS, Moore RJ, Weitz KW, Pasa-Tolic L, Metz TO, Adamec J, Kuhn RJ. 2012. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog 8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Acebes MA, Merino-Ramos T, Blazquez AB, Casas J, Escribano-Romero E, Sobrino F, Saiz JC. 2014. The composition of West Nile virus lipid envelope unveils a role of sphingolipid metabolism in flavivirus biogenesis. J Virol 88:12041–12054. doi: 10.1128/JVI.02061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin-Acebes MA, Blazquez AB, Jimenez de Oya N, Escribano-Romero E, Saiz JC. 2011. West Nile virus replication requires fatty acid synthesis but is independent on phosphatidylinositol-4-phosphate lipids. PLoS One 6:e24970. doi: 10.1371/journal.pone.0024970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heaton NS, Randall G. 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe 8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heaton NS, Perera R, Berger KL, Khadka S, Lacount DJ, Kuhn RJ, Randall G. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A 107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carocci M, Hinshaw SM, Rodgers MA, Villareal VA, Burri DJ, Pilankatta R, Maharaj NP, Gack MU, Stavale EJ, Warfield KL, Yang PL. 2015. The bioactive lipid 4-hydroxyphenyl retinamide inhibits flavivirus replication. Antimicrob Agents Chemother 59:85–95. doi: 10.1128/AAC.04177-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aktepe TE, Pham H, Mackenzie JM. 2015. Differential utilisation of ceramide during replication of the flaviviruses West Nile and dengue virus. Virology 484:241–250. doi: 10.1016/j.virol.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 16.Martin-Acebes MA, Vazquez-Calvo A, Caridi F, Saiz JC, Sobrino F. 2013. Lipid involvement in viral infections: present and future perspectives for the design of antiviral strategies, p 291–321. In Valenzuela R. (ed), Lipid metabolism. InTech, Rijeka, Croatia. [Google Scholar]
- 17.Soto-Acosta R, Bautista-Carbajal P, Syed GH, Siddiqui A, Del Angel RM. 2014. Nordihydroguaiaretic acid (NDGA) inhibits replication and viral morphogenesis of dengue virus. Antiviral Res 109:132–140. doi: 10.1016/j.antiviral.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. 2008. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol 26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nchoutmboube JA, Viktorova EG, Scott AJ, Ford LA, Pei Z, Watkins PA, Ernst RK, Belov GA. 2013. Increased long chain acyl-Coa synthetase activity and fatty acid import is linked to membrane synthesis for development of picornavirus replication organelles. PLoS Pathog 9:e1003401. doi: 10.1371/journal.ppat.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zu X, Zhong J, Luo D, Tan J, Zhang Q, Wu Y, Liu J, Cao R, Wen G, Cao D. 2013. Chemical genetics of acetyl-CoA carboxylases. Molecules 18:1704–1719. doi: 10.3390/molecules18021704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourbeau MP, Bartberger MD. 2015. Recent advances in the development of acetyl-CoA carboxylase (ACC) inhibitors for the treatment of metabolic disease. J Med Chem 58:525–536. doi: 10.1021/jm500695e. [DOI] [PubMed] [Google Scholar]
- 22.Ashraf U, Ye J, Ruan X, Wan S, Zhu B, Cao S. 2015. Usutu virus: an emerging flavivirus in Europe. Viruses 7:219–238. doi: 10.3390/v7010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Nido J, Serrano L, Mendez E, Avila J. 1988. A casein kinase II-related activity is involved in phosphorylation of microtubule-associated protein MAP-1B during neuroblastoma cell differentiation. J Cell Biol 106:2057–2065. doi: 10.1083/jcb.106.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merino-Ramos T, Blazquez AB, Escribano-Romero E, Canas-Arranz R, Sobrino F, Saiz JC, Martin-Acebes MA. 2014. Protection of a single dose West Nile virus recombinant subviral particle vaccine against lineage 1 or 2 strains and analysis of the cross-reactivity with Usutu virus. PLoS One 9:e108056. doi: 10.1371/journal.pone.0108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin-Acebes MA, Saiz JC. 2011. A West Nile virus mutant with increased resistance to acid-induced inactivation. J Gen Virol 92:831–840. doi: 10.1099/vir.0.027185-0. [DOI] [PubMed] [Google Scholar]
- 26.Lanciotti RS, Kerst AJ, Nasci RS, Godsey MS, Mitchell CJ, Savage HM, Komar N, Panella NA, Allen BC, Volpe KE, Davis BS, Roehrig JT. 2000. Rapid detection of West Nile virus from human clinical specimens, field-collected mosquitoes, and avian samples by a TaqMan reverse transcriptase-PCR assay. J Clin Microbiol 38:4066–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blazquez AB, Saiz JC. 2010. West Nile virus (WNV) transmission routes in the murine model: intrauterine, by breastfeeding and after cannibal ingestion. Virus Res 151:240–243. doi: 10.1016/j.virusres.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Garanto A, Mandal NA, Egido-Gabas M, Marfany G, Fabrias G, Anderson RE, Casas J, Gonzalez-Duarte R. 2013. Specific sphingolipid content decrease in Cerkl knockdown mouse retinas. Exp Eye Res 110:96–106. doi: 10.1016/j.exer.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorrochategui E, Casas J, Perez-Albaladejo E, Jauregui O, Porte C, Lacorte S. 2014. Characterization of complex lipid mixtures in contaminant exposed JEG-3 cells using liquid chromatography and high-resolution mass spectrometry. Environ Sci Pollut Res Int 21:11907–11916. doi: 10.1007/s11356-014-3172-5. [DOI] [PubMed] [Google Scholar]
- 30.Fahy E, Sud M, Cotter D, Subramaniam S. 2007. LIPID MAPS online tools for lipid research. Nucleic Acids Res 35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greseth MD, Traktman P. 2014. De novo fatty acid biosynthesis contributes significantly to establishment of a bioenergetically favorable environment for vaccinia virus infection. PLoS Pathog 10:e1004021. doi: 10.1371/journal.ppat.1004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapadia SB, Chisari FV. 2005. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc Natl Acad Sci U S A 102:2561–2566. doi: 10.1073/pnas.0409834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaunt ER, Cheung W, Richards JE, Lever A, Desselberger U. 2013. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J Gen Virol 94:1310–1317. doi: 10.1099/vir.0.050146-0. [DOI] [PubMed] [Google Scholar]
- 34.Koutsoudakis G, Romero-Brey I, Berger C, Perez-Vilaro G, Perin PM, Vondran FW, Kalesse M, Harmrolfs K, Muller R, Martinez JP, Pietschmann T, Bartenschlager R, Bronstrup M, Meyerhans A, Diez J. 2015. Soraphen A: a broad-spectrum antiviral natural product with potent anti-hepatitis C virus activity. J Hepatol 63:813–821. doi: 10.1016/j.jhep.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Xu H, Valenzuela N, Fai S, Figeys D, Bennett SA. 2013. Targeted lipidomics—advances in profiling lysophosphocholine and platelet-activating factor second messengers. FEBS J 280:5652–5667. doi: 10.1111/febs.12423. [DOI] [PubMed] [Google Scholar]
- 36.Miorin L, Romero-Brey I, Maiuri P, Hoppe S, Krijnse-Locker J, Bartenschlager R, Marcello A. 2013. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol 87:6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. 2014. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol 88:12276–12295. doi: 10.1128/JVI.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carro AC, Damonte EB. 2013. Requirement of cholesterol in the viral envelope for dengue virus infection. Virus Res 174:78–87. doi: 10.1016/j.virusres.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Rose-Kahn G, Bar-Tana J. 1990. Inhibition of rat liver acetyl-CoA carboxylase by β,β′-tetramethyl-substituted hexadecanedioic acid (MEDICA 16). Biochim Biophys Acta 1042:259–264. doi: 10.1016/0005-2760(90)90018-S. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez-Calvo A, Saiz JC, Sobrino F, Martin-Acebes MA. 2011. Inhibition of enveloped virus infection of cultured cells by valproic acid. J Virol 85:1267–1274. doi: 10.1128/JVI.01717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welsch C, Albrecht M, Maydt J, Herrmann E, Welker MW, Sarrazin C, Scheidig A, Lengauer T, Zeuzem S. 2007. Structural and functional comparison of the non-structural protein 4B in flaviviridae. J Mol Graph Model 26:546–557. doi: 10.1016/j.jmgm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Opie LH. 2015. Present status of statin therapy. Trends Cardiovasc Med 25:216–225. doi: 10.1016/j.tcm.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Martinez JP, Hinkelmann B, Fleta-Soriano E, Steinmetz H, Jansen R, Diez J, Frank R, Sasse F, Meyerhans A. 2013. Identification of myxobacteria-derived HIV inhibitors by a high-throughput two-step infectivity assay. Microb Cell Fact 12:85. doi: 10.1186/1475-2859-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Syed GH, Siddiqui A. 2011. Effects of hypolipidemic agent nordihydroguaiaretic acid on lipid droplets and hepatitis C virus. Hepatology 54:1936–1946. doi: 10.1002/hep.24619. [DOI] [PMC free article] [PubMed] [Google Scholar]