Abstract
The nucleotide sequences of three IncU plasmids from Aeromonas spp. isolated from ornamental fish are described. They had a typical IncU backbone for plasmid replication and maintenance functions, but conjugative transfer modules were disrupted. The gene qnrS2 was inserted into mpR as a mobile insertion cassette. Novel Tn3 family transposons carrying putative toxin-antitoxin and plasmid stability genes were identified. The study demonstrates high plasticity of IncU plasmids from aquatic environments.
TEXT
Aeromonas is a ubiquitous microorganism distributed in water ecosystems and includes species causing various kinds of infections in fish and humans. Antibiotic resistance in Aeromonas in aquatic environments is associated especially with plasmids of the IncU family. Most IncU plasmids described in this genus to date share a highly conserved backbone represented by genes for plasmid replication, stability, and conjugative transfer and one variable region for resistance genes to antibiotics, such as tetracyclines, sulfonamides, trimethoprim, streptomycin, and chloramphenicol (1–6). Quinolones are broad-spectrum antimicrobial agents widely used in human and veterinary medicine, and they are among the most widely used antibiotics in aquacultures (7). Aeromonas isolates harboring quinolone resistance (PMQR) genes on IncU plasmids have been recently reported in river water, lake water, and fish (2, 3, 6, 8), highlighting the role of this plasmid family in dissemination of clinically relevant resistance mechanisms in a wide range of aquatic environments.
In our previous study, IncU plasmids carrying qnrS2 and aac(6′)-Ib-cr genes in Aeromonas spp. from ornamental fish originating from various geographic locations were identified (8). The aim of this study was to elucidate the structure and features of three representative IncU plasmids carrying PMQR genes by sequencing and bioinformatic analysis. At present, complete nucleotide sequences of nine IncU plasmids are available in the GenBank database. Four of them, plasmids pRA3 (GenBank accession no. DQ401103), pFBAOT6 (GenBank accession no. CR376602), pAC3 (GenBank accession no. KM204147), and pP2G1 (GenBank accession no. HE616910), originate from Aeromonas spp., and only the last two plasmids carry PMQR genes (updated on 22 July 2015).
The three sequenced IncU plasmids pAH6, pAH227, and pASCH21 were nonconjugative and varied in size (20 to 40 kb). Plasmid pAH6 (∼20 kb) originated from Aeromonas hydrophila from a koi carp bred in the Czech Republic. It has been identified as a highly diffused plasmid in isolates of A. hydrophila and Aeromonas sobria from Czech koi carps and tropical fish imported from Asia (8). The other two plasmids, pAH227 (∼40 kb) and pASCH21 (∼35 kb), carrying qnrS2, originated from A. hydrophila from a koi carp from the Czech Republic and A. sobria from a tropical fish from Thailand, respectively. Plasmids were transferred to chemically competent Escherichia coli DH5α cells by transformation followed by selection of the transformants on Luria-Bertani agar with ciprofloxacin (0.05 mg/liter) as described previously (8). Plasmid DNAs were extracted from transformants using the PureLink HiPure plasmid filter midiprep kit (Invitrogen, Life Technologies, Germany) and sequenced using the 454 Genome Sequencer Junior system (Roche, Prague, Czech Republic) on a standard DNA fragment library following the manufacturer's protocol. One, one, and three contigs for plasmids pAH6, pAH227, and pASCH21, respectively, ranging from 250 to 41,761 bp in size with at least a 444-fold coverage, were obtained using GS De Novo Assembler v2.6 software. Gaps between contigs were filled by PCR, and the obtained amplicons were sequenced using the Sanger method to link the contigs (Applied Biosystems, Foster City, CA, USA). Geneious 7.0.6 software (Biomatters, Auckland, New Zealand) was used to combine the data from the 454 Genome Sequencer Junior system and Sanger sequencing. The BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST) was used for further data analysis. The plasmids were mapped against IncU plasmids pRA3 and pFBAOT6 to verify the obtained sequences. pRA3 and pFBAOT6 were used as reference plasmids to annotate the plasmid sequences.
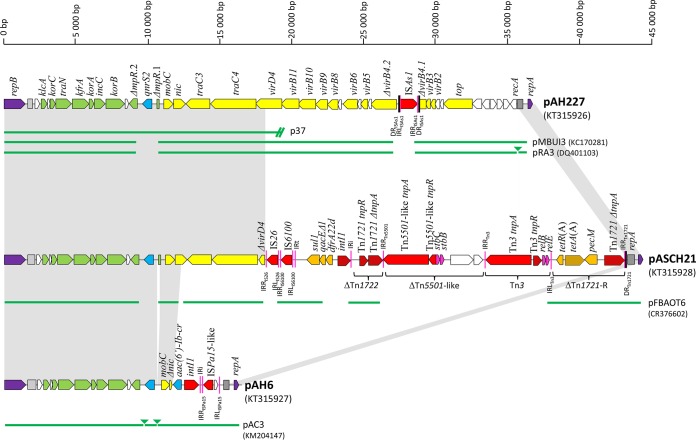
All three sequenced plasmids showed a highly conserved backbone. Genes involved in plasmid replication and maintenance showed significant sequence identity with the corresponding regions of other plasmids in the IncU family. The replication module was represented by the genes repA and repB and repetitive sequences (Fig. 1). RepB showed 99.8% and 100% sequence identity with the replication protein in pRA3 and pFBAOT6, respectively. RepA was 100% identical with the transcriptional repressor protein of IncU archetypes. The complete set of plasmid maintenance genes, found adjacent to the replication module, was disrupted by insertion of qnrS2 into the gene mpR coding for a putative zinc-metalloprotease as observed in other qnrS2-carrying IncU plasmids (2, 3, 6). Significant differences between the three sequenced plasmids were observed in the transfer modules involved in plasmid conjugation and in plasmid variable regions containing various antibiotic resistance genes and mobile elements.
FIG 1.
Linear maps of PMQR-carrying IncU plasmids pAH227, pASCH21, and pAH6 from Aeromonas. Open reading frames (ORFs) are shown as arrows, indicating the direction of transcription. Plasmid scaffold, including the plasmid replicons, maintenance genes, and transfer region, are shown in violet, green, and yellow, respectively. PMQR genes are shown in blue, while other resistance genes are indicated in orange. IS, intI1, tnpA, and tnpR are shown in red. Other genes are shown in white or pink. Dark gray boxes represent repetitive regions adjacent to rep genes. Inverted repeats (IRL, left inverted repeat; IRR, right inverted repeat) and direct repeats (DR) are shown in purple and black lines, respectively. The extent of transposons is indicated below the plasmid map. The green lines below the maps correspond to highly similar sequences from other fully and partially (p37) sequenced IncU plasmids. Homologous regions are indicated by gray shading.
Plasmid pAH227 was 36,559 bp in size and had 52.3% G+C content. It carried qnrS2 and showed significant similarity to pFBAOT6 and pRA3 (92% coverage, 99% identity). The majority of the plasmid sequence (33,947 bp) was composed of conserved IncU plasmid backbone. It contained a complete transfer region as seen in pRA3. However, the transfer region in pAH227 was disrupted by the insertion sequence ISAs1 inserted into the gene coding for conjugal transfer protein VirD4, likely resulting in the loss of conjugative abilities of the plasmid, as demonstrated in vitro (8). The ISAs1 was flanked by imperfect 22-bp inverted repeats (IRs) and showed 100% nucleotide identity to a transposase gene found in A. salmonicida (GenBank accession no. CP000645) and Aeromonas media (GenBank accession no. CP007567) genomes. Target site duplication of 10 bp (ACTGCATACC) at the ISAs1 boundaries indicated the insertion via transposition. In contrast to other IncU plasmids described in Aeromonas spp. to date, no accessory genes apart from qnrS2 were found on pAH227. However, plasmids pMBUI3 (KC170281) and pMBUI7 (KC170284) from an unknown bacterial species recently isolated from freshwaters in the United States show great similarity (91% to 92% coverage, 93% to 95% identity) to pAH227 and consist entirely of IncU plasmid backbone with no known accessory genes (9). These plasmids may represent IncU archetypes before the acquisition of resistance genes.
Plasmid pASCH21 contained qnrS2 and was 44,649 bp in size, with an average G+C content of 56.5%. It was composed of a 17,694-bp IncU plasmid backbone and a 26,955-bp accessory region. The plasmid backbone was disrupted within the gene coding for VirD4 conjugal transfer protein by a continuous highly mosaic variable region, consisting of sets of resistance genes, transposable elements, and integrons. The sequencing data indicated that pASCH21 was a derivate of pFBAOT6 (65% coverage, 99% identity). Both plasmids contained plasmid backbone sharing 99% sequence similarity and a related genetic load represented by an In4-like class 1 integron and a Tn21-like unit transposon, Tn1721. In both plasmids, Tn1721 was separated into left (Tn1721-L, i.e., truncated region corresponding to Tn1722) and right [Tn1721-R, harboring tetA(A) resistance gene] ends. pFBAOT6 had a complete Tn1721 surrounded by direct repeats (DRs) at both ends, suggesting its acquisition by a single transposition event. On the other hand, the left end of Tn1721-L in pASCH21, including DR, IRLTn1721-L, and part of mcp (previously designated orfI) encoding methyl-accepting chemotaxis protein, has been removed, likely by insertion of the integron. Both plasmids contained insertion sequence IS6100 upstream of the class 1 integron, a typical feature of In4-like integrons. However, the integrons of pASCH21 and pFBAOT6 differed. pASCH21 contained a dfrA22d cassette coding for resistance to trimethoprim, while pFBAOT6 had the streptomycin resistance gene cassette aadA2. In addition, the integron of pFBAOT6 was inserted at the left end of an intact sequence of Tn1721-L, while in pASCH21, the integron was truncated Tn1721-L in mcp. Furthermore, the sequence of a remnant tni402 and a terminal IR typical for In4-like integrons were missing in pASCH21, likely as a result of insertion of IS26 adjacent to IS6100. Two Tn3 family transposons were found between Tn1721-L and Tn1721-R. Tn5501-like was inserted immediately downstream of ΔTn1721-L, truncating the gene tnpA of the transposon. This novel transposon is flanked by typical 38-bp left IRs and exhibits 90% to 91% identity at the amino acid level with Tn3 family transposons identified in IncP1 plasmids pGNB1 (10) and pB8 (11) isolated from unknown bacteria collected from activated sludge from wastewater treatment plants. Compared to the IncP1 plasmids, Tn5501-like in pASCH21 carried four genes, two of them coding for putative plasmid stabilization proteins showing significant similarity to proteins identified in plant-associated Pseudomonas savastanoi (12, 13). Insertion of another 4,717-bp Tn3 transposon downstream of Tn5501-like likely removed parts of Tn5501-like and Tn1721-R, including their corresponding IRs. This Tn3 transposon was flanked by 38-bp IRs; however, the insertion has not generated target site duplications; thus, it is unlikely that it has been acquired by transposition. The transposon carried toxin-antitoxin gene pairs, probably ensuring plasmid stability. These putative plasmid stability proteins exhibited 86% and 96% identity at the amino acid level with RelE and RelB proteins, respectively, encoded by small cryptic ColE2-type plasmids in A. salmonicida (14).
pAH6 plasmid harbored qnrS2 and aac(6′)-Ib-cr genes and was found to be 15,886 bp in size, with an average G+C content of 54.9%. It shared extensive similarities with IncU plasmids pFBAOT6 (88% coverage, 100% identity) and pRA3 (87% coverage, 99% identity). The sequence is highly related to PMQR-carrying IncU plasmid pAC3 from Aeromonas sp. from a wastewater treatment plant in South Korea (99% coverage, 100% identity). pAH6 is one of the smallest IncU plasmids described so far. Recently, 8-kb IncU plasmid pPA-2 harboring blaKPC-2 and lacking genes involved in plasmid transfer and partitioning was identified in a clinical isolate of Pseudomonas aeruginosa (15), highlighting the ability of IncU plasmids to form smaller molecules. In pAH6, the conjugative transfer region was disrupted inside the gene nic encoding VirD2 relaxase by a class 1 integron harboring the gene cassette aac(6′)-Ib-cr. The 3′ part of the integron was removed, likely as a result of subsequent rearrangements after the integron acquisition. A complete In2 family integron with an aac(6′)-Ib-cr–blaOXA-1–catB3–arr-3 cassette array is present in other PMQR-carrying IncU plasmids in Aeromonas spp. isolated from river water in Spain and France (3, 6). Adjacent to the integron, pAH6 contained a 1,025-bp ISPa15-like element separated by a 114-bp sequence from the integron, as observed in IncU plasmid pFBAOT6 and IncP-6 plasmid Rms149 (4, 16). The In2-like integron and ISPa15-like element were suggested to form a composite transposon whose complete structure appears to be present in pFBAOT6 (4).
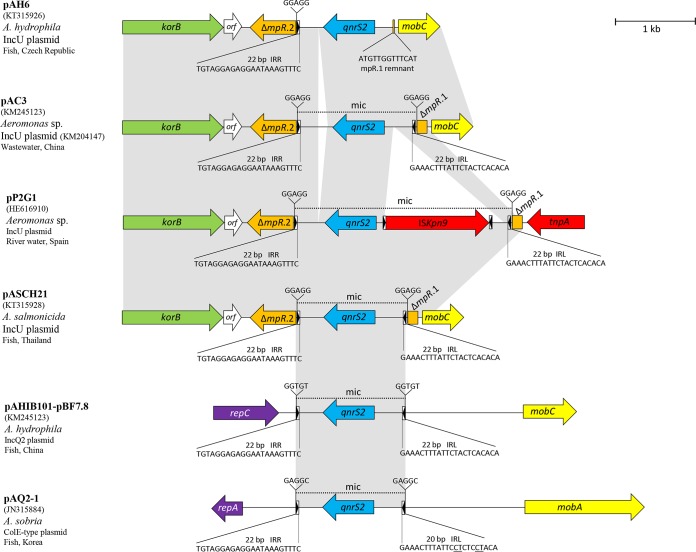
All three sequenced plasmids contained the qnrS2 gene inserted in the same position of the plasmid maintenance region into the gene mpR in the form of a mobile insertion cassette (mic). pAH227 and pASCH21 contained the intact 1,370-bp mic bracketed by 22-bp imperfect IRs and a 5-bp duplication of the target site (Fig. 2), suggesting the acquisition of the structure by transposition. The same insertion site of qnrS2 and structure of mic have been observed in other IncU plasmids (2, 3), highlighting the gene mpR as a hot spot for qnrS2 insertion in this plasmid group. The identical structure of mic seen in pCH21 and pAH227 was found in IncU plasmid p37 in Aeromonas punctata from river water in France (2) and in plasmids of other incompatibility groups, such as IncQ2 plasmid pAHIB101-pBF7.8 in A. hydrophila from a fish isolate in China (GenBank accession no. KM245123) and ColE-type plasmids pAQ2-1 and pAQ2-2 in A. sobria from fish in South Korea (17). This suggests that this qnrS2-carrying mic represents a promiscuous element moving between different replicons. In IncQ and ColE-type plasmids, the mic was inserted in plasmid backbones between genes encoding replication and mobilization proteins, generating target site duplication of 5 bp (Fig. 2). Other qnrS2-carrying IncU plasmids from Aeromonas spp. described so far contain various insertions of extra sequences inside the mic. Plasmid pP2G1 from river water in Spain carries ISKpn9 inserted at the 5′ end of qnrS2, while pAC3 and p42 from China and France, respectively, contain a 106-bp insert at the 3′ end of qnrS2 (6). In pAH6, IRLs and DRs of mic upstream of qnrS2 were missing, likely due to subsequent plasmid rearrangements after the acquisition of the qnrS2-carrying mic. This resulted in the loss of mobilization abilities of the gene.
FIG 2.
Mobile insertion cassette (mic) carrying qnrS2 gene and the insertion site on various plasmid backbones. Open reading frames (ORFs) are shown as arrows, indicating the direction of transcription. Plasmid scaffolds, including the plasmid replicons, maintenance genes, and transfer region, are shown in violet, green, and yellow, respectively. The gene qnrS2 is shown in blue. Inverted repeats (IRL, left inverted repeat; IRR, right inverted repeat) are shown in boxes, with a black arrow indicating the direction; their length and sequence are shown below the structures, and nucleotides that differed among the plasmids are underlined. Direct repeats (DR) flanking the mic are shown above the map. Homologous regions are indicated by gray shading.
Our study described complete sequences of IncU plasmids carrying PMQR determinants from A. hydrophila and A. sobria recovered from ornamental fish from distant geographic areas. pAH6-like IncU plasmids carrying qnrS2 and aac(6′)-Ib-cr genes have been found in various Aeromonas isolates from the Czech Republic, Thailand, Vietnam, and South Korea, suggesting the importance of this plasmid lineage in dissemination of PMQR genes among aquatic animals and the environment. None of the plasmids could be transferred to recipient bacteria by conjugation, most likely because of disruption of the conjugative transfer region. A general ability of Aeromonas environmental isolates to acquire free DNA was recently described (18), highlighting the role of transformation in the gene flow in natural environments, including water. Despite the loss of conjugative abilities, the IncU plasmids may be efficiently transferred in aquatic environments, most likely by the transformation of free plasmid DNA to naturally competent bacteria. IncU plasmids formed smaller molecules and acquired novel stability genes, which may give them the ability to pose a lower biological burden for host cells and to be maintained in antibiotic-free environments. Our study demonstrated higher variability in this plasmid family than previously anticipated, its rapid evolution, and an important role in the circulation of fluoroquinolone resistance in the environment.
Nucleotide sequence accession numbers.
The nucleotide sequences of plasmids pAH6, pAH227, and pASCH21 have been assigned the GenBank accession numbers KT315927, KT315926, and KT315928, respectively.
ACKNOWLEDGMENTS
This work was supported by a research project grant from the Czech Ministry of Agriculture (NAZV QJ1210237), Czech Science Foundation (15-14683Y/P502), CEITEC-Central European Institute of Technology (CZ.1.05/1.1.00/02.0068) from European Regional Development Fund and AdmireVet project (CZ.1.05/2.1.00/01.0006-ED0006/01/01) from the Czech Ministry of Education.
We thank Alois Cizek, Marie Slavikova, and Iva Kutilova from University of Veterinary and Pharmaceutical Sciences Brno for their assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Aoki T, Egusa S, Kimura T, Watanabe T. 1971. Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl Microbiol 22:716–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cattoir V, Poirel L, Aubert C, Soussy CJ, Nordmann P. 2008. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis 14:231–237. doi: 10.3201/eid1402.070677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picao RC, Poirel L, Demarta A, Silva CS, Corvaglia AR, Petrini O, Nordmann P. 2008. Plasmid-mediated quinolone resistance in Aeromonas allosaccharophila recovered from a Swiss lake. J Antimicrob Chemother 62:948–950. doi: 10.1093/jac/dkn341. [DOI] [PubMed] [Google Scholar]
- 4.Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, Huys G, Swings J, Pickup RW. 2004. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl Environ Microbiol 70:7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorum H, L'Abee-Lund TM, Solberg A, Wold A. 2003. Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother 47:1285–1290. doi: 10.1128/AAC.47.4.1285-1290.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marti E, Balcazar JL. 2012. Multidrug resistance-encoding plasmid from Aeromonas sp. strain P2G1. Clin Microbiol Infect 18:E366–E368. doi: 10.1111/j.1469-0691.2012.03935.x. [DOI] [PubMed] [Google Scholar]
- 7.Quesada SP, Paschoal JA, Reyes FG. 2013. Considerations on the aquaculture development and on the use of veterinary drugs: special issue for fluoroquinolones–a review. J Food Sci 78:R1321–R1333. doi: 10.1111/1750-3841.12222. [DOI] [PubMed] [Google Scholar]
- 8.Dobiasova H, Kutilova I, Piackova V, Vesely T, Cizek A, Dolejska M. 2014. Ornamental fish as a source of plasmid-mediated quinolone resistance genes and antibiotic resistance plasmids. Vet Microbiol 171:413–421. doi: 10.1016/j.vetmic.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Brown CJ, Sen D, Yano H, Bauer ML, Rogers LM, Van der Auwera GA, Top EM. 2013. Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl Environ Microbiol 79:7684–7695. doi: 10.1128/AEM.02252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schluter A, Krahn I, Kollin F, Bonemann G, Stiens M, Szczepanowski R, Schneiker S, Puhler A. 2007. IncP-1-beta plasmid pGNB1 isolated from a bacterial community from a wastewater treatment plant mediates decolorization of triphenylmethane dyes. Appl Environ Microbiol 73:6345–6350. doi: 10.1128/AEM.01177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schluter A, Heuer H, Szczepanowski R, Poler SM, Schneiker S, Puhler A, Top EM. 2005. Plasmid pB8 is closely related to the prototype IncP-1beta plasmid R751 but transfers poorly to Escherichia coli and carries a new transposon encoding a small multidrug resistance efflux protein. Plasmid 54:135–148. doi: 10.1016/j.plasmid.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Qi M, Wang D, Bradley CA, Zhao Y. 2011. Genome sequence analyses of Pseudomonas savastanoi pv. glycinea and subtractive hybridization-based comparative genomics with nine pseudomonads. PLoS One 6:e16451. doi: 10.1371/journal.pone.0016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Palenzuela P, Matas IM, Murillo J, Lopez-Solanilla E, Bardaji L, Perez-Martinez I, Rodriguez-Moskera ME, Penyalver R, Lopez MM, Quesada JM, Biehl BS, Perna NT, Glasner JD, Cabot EL, Neeno-Eckwall E, Ramos C. 2010. Annotation and overview of the Pseudomonas savastanoi pv. savastanoi NCPPB 3335 draft genome reveals the virulence gene complement of a tumour-inducing pathogen of woody hosts. Environ Microbiol 12:1604–1620. doi: 10.1111/j.1462-2920.2010.02207.x. [DOI] [PubMed] [Google Scholar]
- 14.Boyd J, Williams J, Curtis B, Kozera C, Singh R, Reith M. 2003. Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid 50:131–144. doi: 10.1016/S0147-619X(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 15.Naas T, Bonnin RA, Cuzon G, Villegas MV, Nordmann P. 2013. Complete sequence of two KPC-harbouring plasmids from Pseudomonas aeruginosa. J Antimicrob Chemother 68:1757–1762. doi: 10.1093/jac/dkt094. [DOI] [PubMed] [Google Scholar]
- 16.Haines AS, Cheung M, Thomas CM. 2006. Evidence that IncG (IncP-6) and IncU plasmids form a single incompatibility group. Plasmid 55:210–215. doi: 10.1016/j.plasmid.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Han JE, Kim JH, Choresca CH Jr, Shin SP, Jun JW, Chai JY, Park SC. 2012. First description of ColE-type plasmid in Aeromonas spp. carrying quinolone resistance (qnrS2) gene. Lett Appl Microbiol 55:290–294. doi: 10.1111/j.1472-765X.2012.03293.x. [DOI] [PubMed] [Google Scholar]
- 18.Huddleston JR, Brokaw JM, Zak JC, Jeter RM. 2013. Natural transformation as a mechanism of horizontal gene transfer among environmental Aeromonas species. Syst Appl Microbiol 36:224–234. doi: 10.1016/j.syapm.2013.01.004. [DOI] [PubMed] [Google Scholar]