Abstract
The kelch 13 (K13) propeller gene is associated with artemisinin resistance. In a previous work, there were no mutations found in 138 Plasmodium falciparum isolates collected in 2012 and 2013 from patients residing in Dakar, Senegal (M. Torrentino-Madamet et al., Malar J 13:472, 2014, http://dx.doi.org/10.1186/1475-2875-13-472). However, the N554H, Q613H, and V637I mutations were identified in the propeller region of K13 in 92 (5.5%) isolates in 2013 and 2014. There were five polymorphisms identified in the Plasmodium/Apicomplexa-specific domain (K123R, N137S, N142NN/NNN, T149S, and K189T/N).
TEXT
Malaria resistance to most antimalarial drugs has developed in Southeast Asia and has spread to Africa. The World Health Organization (WHO) has recommended artemisinin-based combination therapy (ACT) as the first-line treatment for malaria since 2005. ACT has been recommended by the Senegalese National Malaria Control Program as the first-line treatment for uncomplicated malaria since 2006. The emergence of Plasmodium falciparum resistance to artemisinin and its derivatives manifests as delayed parasite clearance following treatment with artesunate monotherapy or ACT and has recently developed in Southeast Asia (1, 2). This clinical resistance was correlated with in vitro resistance, which manifests as an increase in the ring-stage survival rate after contact with artemisinin (3). Therefore, the spread of artemisinin resistance from Asia to Africa may be a serious threat for malaria control and elimination.
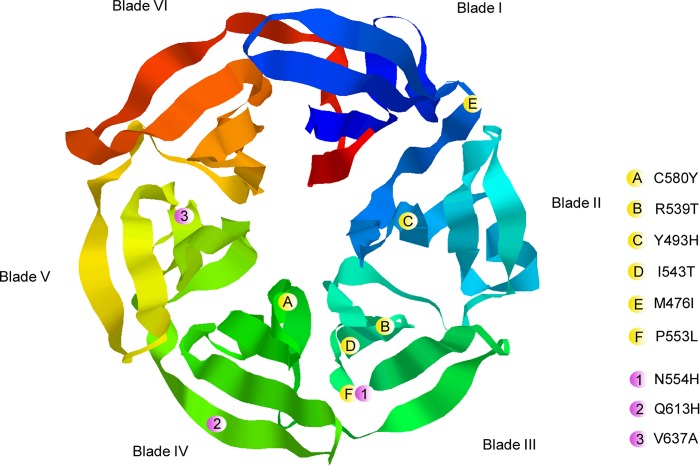
Mutations in the propeller domain of the kelch 13 (K13) gene (PF3D71343700) were recently associated with in vivo and in vitro resistance to artemisinin in Southeast Asia (4). The K13 propeller gene is located on chromosome 13 and encodes a kelch protein. There are 6 kelch motifs called blades at the C terminus (Fig. 1). Each propeller blade is composed of approximately 50 amino acids that form four antiparallel beta-sheet secondary structures arranged around a canal. The propeller domain harbors multiple protein-protein interaction sites (5). The Y493H, R539T, I543T, and C580Y mutations were also correlated with in vivo and in vitro artemisinin resistance in Southeast Asia (2, 4, 6–8). The C580Y mutation is located in the first beta-sheet of the fourth blade and is associated with delayed parasite clearance leading to in vivo resistance (4). A previous study used site-directed mutagenesis with zinc finger nucleases to modify the K13 propeller gene locus. The results confirmed the importance of the Y493H, R539T, I543T, and C580Y mutations in mediating in vitro artemisinin resistance (9). Although the four mutations are widely present in Southeast Asia, they are not observed in Africa (10–14). However, the P553L mutation, which is associated with delayed parasite clearance in Southeast Asia, was detected in Kenya and Malawi (12).
FIG 1.
Locations of the mutations in the predicted three-dimensional (3D) model of the K13 propeller domain (RasTop software, Phyre 2 server). The locations of the various mutations are indicated by spheres, in which yellow represents mutations correlated with artemisinin resistance reported by Ariey et al. (4) and pink represents mutations observed in Dakar, Senegal.
We collected 103 samples from falciparum malaria patients attending the Hôpital Principal de Dakar, Senegal, from November 2013 to January 2014 and August 2014 to December 2014 (59 from 2013 and 44 from 2014). Sixty-four percent of the patients were recruited from the emergency department. The other patients were recruited from the intensive care unit (12%), pediatric department (7%), infectious diseases department (5%), maternity department (3%), and other units (9%). There was no information available on antimalarial treatment prior to admission. Despite the recommendations of the WHO, the patients were treated with quinine until November 2014 and with artesunate or artemether-lumefantrine at the Hôpital Principal de Dakar. Informed verbal consent was obtained from the patients or their parents/guardians before blood collection. The study was approved by the ethics committee of the Hôpital Principal de Dakar.
Venous blood samples were collected in Vacutainer acid citrate dextrose (ACD) tubes prior to patient treatment. The total genomic DNA of each isolate was extracted using the QIAamp DNA minikit, according to the manufacturer's recommendations (Qiagen, Germany). A malaria diagnosis was confirmed using a thin blood smear, rapid diagnosis test, and real-time quantitative PCR.
The K13 propeller gene was amplified using a PCR and nested-PCR method described previously (7). The following primers were used for PCR: 3′-GGG AAT CTG GTG GTA ACA GC-5′ and 3′-CGG AGT GAC CAA ATC TGG GA-5′, and 3′-GCC TTG TTG AAA GAA GCA GA-5′ and 3′-GCC AAG CTG CCA TTC ATT TG-5′. The K13 propeller gene was successfully sequenced in 58 PCR and 92 nested-PCR amplicons, and the results were compared to the reference P. falciparum 3D7 strain. The K13 mutations were confirmed three times by sequencing the products of three different PCRs.
The polymorphisms identified in this study are reported in Table 1. The mutations associated with in vitro resistance in Southeast Asia, such as Y493H, R539T, I543T, and C580Y (2, 4–6), were not observed. The M476I mutation obtained in vitro on the F32 Tanzanian strain after artemisinin pressure was not identified (4).
TABLE 1.
Nonsynonymous mutations observed in the K13 gene in P. falciparum isolates from Dakar, Senegal, collected from November 2013 to January 2014 and August 2014 to December 2014a
| Amino acid change or insertion and location | Bladed | Referent sequence | Mutant sequenceb | No. of isolates/total isolates (%) |
|---|---|---|---|---|
| K123Rc | — | AAA | AGA | 1/58 (1.7) |
| N137Sc | — | AAT | AGT | 1/58 (1.7) |
| N142NN | — | AAT | 7/58 (12.1) | |
| N142NNN | — | AAT AAT | 1/58 (1.7) | |
| T149S | — | ACT | TCT | 1/58 (1.7) |
| K189T | — | AAA | ACA | 18/58 (31.0) |
| K189Nc | — | AAA | AAT | 1/58 (1.7) |
| N554H | 3 | AAT | CAT | 1/92 (1.1) |
| Q613Hc | 4 | CAA | CAT | 1/92 (1.1) |
| V637I | 5 | GTT | ATT | 3/92 (3.3) |
Mutations described were not found in the same isolates.
Mutated bases are in bold type.
Novel mutation.
—, absence of blade numbered only in the propeller domain.
The two novel mutations K123R and N137S in the N-terminal domain were each reported in 1.7% (1/58) of the isolates. One or two asparagine (N or NN) insertions at codon 142 were found in 13.8% (8/58) of the isolates. These insertions at codon 142 were observed in 10.9% of the samples collected in the Hôpital Principal de Dakar in 2012 and 2013 (10). The T149S mutation was reported in 4.7% (4/64) of cases in 2012 and 2013 in Dakar (10); in this study, the mutation was found in 1.7% (1/58) of the isolates. The isolates with the T149S mutation (unknown location of collection) previously found were associated with a parasite clearance half-life of <5 h (2). There were two mutations at codon 189. We found that 31.0% (18/58) of isolates presented the K189T mutation, and 1.7% (1/58) presented the K189N mutation. In recent studies, the K189T mutation was found in 42.2% of the samples in Dakar in 2012 and 2013 (10) and in 34.5% (10/29) of the samples in Uganda (15). The mutation was also identified in one isolate in Bangladesh (8). Furthermore, the K189N mutation was reported in 3.4% (1/29) of the cases in Uganda (15). Among the 20 isolates previously found (unknown collection location), only one (in Bangladesh) was associated with a parasite clearance half-life of <5 h (2).
There were no previously reported mutations in the six K13 propeller blades in Dakar in 2012 and 2013 (10). In this study, we identified 5 isolates with mutations in blade 3, 4, or 5 (Fig. 1). The N554H mutation was found in 1.1% (1/92) of the isolates. The corresponding residue is located near the third beta-sheet structure of the 3rd blade and was found to already be mutated in African isolates. In Comoros (Grande Comore and Anjouan) in 2013, two isolates (6.9%) were found to carry the N554H and N554K mutations (16). A mutation in the corresponding codon (resulting in N554S) was found in one isolate from Ungoye, Kenya, in 2012 (11) and in one isolate from Mali in 2011 (17). A mutation in this codon has never been found in an isolate from Southeast Asia (18). This mutation is adjacent to the P553L mutation described in southern Asia and is associated with delayed parasite clearance following treatment with artesunate monotherapy in Asia (2). The P553L mutation was found in 0.53% of the isolates in Kisumu, Kenya, and in 0.59% in Machinga, Malawi (8, 12). The Q613H mutation was observed in 1.1% (1/92) of the isolates. It is located in the 4th beta-sheet of the 4th blade. The Q613E and Q613L mutations were already found in Africa (18). The Q613E mutation was previously identified in one isolate with a parasite clearance half-life of <5 h (2). There are previous reports of different single nucleotide polymorphisms (SNP) at codon 637, whose corresponding residue is located in the second beta-sheet of the 5th blade. The SNP was found to be V637A in 3.22% of the isolates from Bas-Congo, Democratic Republic of Congo (12). The V637D SNP was found in 0.8% of the isolates in Uganda (15). In this study, the V637I mutation was present in 3.3% (3/92) of the isolates. The mutation at codon 637 has not been yet observed in Southeast Asia (18).
The propeller is composed of six blades and harbors protein-protein interaction sites. Thus, mutations in the beta-sheet structures may affect protein interactions. The P. falciparum phosphatidylinositol-3-kinase (PI3K) is a putative binding partner of K13 and a target for artemisinin derivatives (19). Artemisinin binding leads to decreased PI3K activity, decreased phosphatidylinositol-3-phosphate (PI3P), and inhibition of parasite growth. Wild-type K13 binds PI3K and delivers it to ubiquitin ligase, which polyubiquitinates K13 and marks it for proteasomal degradation. Mutant K13 fails to bind PI3K. Further studies are needed to characterize the roles of these three mutations in artemisinin resistance. While no mutation was found in the propeller region of K13 in parasites from Dakar in 2012 and 2013, three mutations (5.5%) were identified in this domain in 2013 and 2014. As a result, surveillance of K13 polymorphisms must be implemented.
ACKNOWLEDGMENTS
We thank the patients and the staff of the Hôpital Principal de Dakar and Ndeye Fatou Diop and Maurice Gomis from the Hôpital Principal de Dakar for technical support.
We declare no conflicts of interest.
Funding Statement
This research was supported by the Schéma directeur Paludisme, Etat Major des Armées Françaises (grant LR 607a), and by the Ministère des Affaires Etrangères.
REFERENCES
- 1.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanish K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Seng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jttamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, et al. . 2015. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Meng Chuor C, Taylor WRJ, Suon S, Mercereau-Puijalon O, Fairhurst RM, Ménard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariey F, Witkowsky B, Amaratunga C, Beghain J, Langlois AC, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale JC, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams J, Kelso R, Cooley L. 2000. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol 10:17–24. doi: 10.1016/S0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- 6.Amaratunga C, Witkowski B, Khim N, Menard D, Fairhurst RM. 2014. Artemisinin resistance in Plasmodium falciparum. Lancet Infect Dis 14:449–450. doi: 10.1016/S1473-3099(14)70777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amaratunga C, Witkowski B, Dek D, Try V, Khim N, Miotto O, Ménard D, Fairhurst RM. 2014. Plasmodium falciparum founder populations in Western Cambodia have reduced artemisinin sensitivity in vitro. Antimicrob Agents Chemother 58:4935–4937. doi: 10.1128/AAC.03055-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takala-Harrison S, Jacob CG, Arze C, Cummings MP, Silva JC, Dondorp AM, Fukuda MM, Hien TT, Mayxay M, Noedl H, Nosten F, Kyaw MP, Thuy Nhien NT, Imwong M, Bethell D, Se Y, Lon C, Tyner SD, Saunders DL, Ariey F, Mercerau-Puijalon O, Menrad D, Newton PN, Khanthavong M, Hongvanthong B, Staezengruber P, Fuehrer HP, Swoboda P, Khan WA, Phyo AP, Nyunt MM, Nyunt MA, Brown TS, Adams M, Pepin CS, Bailey J, Tan JC, Ferdig MT, Clark TG, Miotto O, MacInnis B, Kwiatkowski DP, White NJ, Ringwald P, Plowe CV. 2014. Independent emergence of artemisinin resistance mutations among Plasmodium falciparum in Southeast Asia. J Infect Dis 211:670–679. doi: 10.1093/infdis/jiu491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. 2015. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torrentino-Madamet M, Fall B, Benoit N, Camara C, Amalvict R, Fall M, Dionne P, Ba Fall K, Nakoulima A, Diatta B, Diémé Y, Ménard D, Wade B, Pradines B. 2014. Limited polymorphisms in k13 gene in Plasmodium falciparum isolates from Dakar, Senegal in 2012–2013. Malar J 13:472. doi: 10.1186/1475-2875-13-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isozumi R, Uemura H, Kimata I, Ichinose Y, Logedi J, Omar AH, Kaneko A. 2015. Novel mutations in K13 propeller gene of artemisinin-resistant Plasmodium falciparum. Emerg Infect Dis 21:490–492. doi: 10.3201/eid2103.140898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor SM, Parobek CM, DeConti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga D, Martensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamau E, Campino S, Amenga-Etego L, Drury E, Ishengoma D, Johnson K, Mumba D, Kekre M, William Y, Mead D, Bouyou-Akotet M, Apinjoh T, Golassa L, Randrianarivelojosia M, Andagalu B, Maiga-Ascofare O, Amambua-Ngwa A, Tindana P, Ghansah A, McInnis B, Kwiatkowski D, Djimde AA. 2015. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J Infect Dis 211:1352–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escobar C, Pateira S, Lobo E, Lobo L, Teodosio R, Dias F, Fernandes N, Arez AP, Varandas L, Nogueira F. 2015. Polymorphisms in Plasmodium falciparum K13-propeller in Angola and Mozambique after the introduction of the ACTs. PLoS One 10:e0119215. doi: 10.1371/journal.pone.0119215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ. 2014. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouattara A, Kone A, Adams M, Fofana B, Maiga AW, Hampton S, Coulibaly D, Thera MA, Diallo N, Dara A, Sagara I, Gil JP, Bjorkman A, Takala-Harrison S, Doumbo OK, Plowe CV, Djimde AA. 2015. Polymorphisms in the K13-propeller gene in artemisinin-susceptible Plasmodium falciparum parasites from Bougoula-Hameau and Bandiagara, Mali. Am J Trop Med Hyg 92:1202–1206. doi: 10.4269/ajtmh.14-0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairhurst RM. 2015. Understanding artemisinin-resistant malaria: what a difference a year makes. Curr Opin Infect Dis 28:417–425. doi: 10.1097/QCO.0000000000000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torrentino-Madamet M, Collet L, Lepère JF, Benoit N, Amalvict R, Ménard D, Pradines B. 2015. K13-propeller polymorphisms in Plasmodium falciparum isolates from patients in Mayotte in 2013 and 2014. Antimicrob Agents Chemother, 59:7878–7881. doi: 10.1128/AAC.01251-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbengue A, Bhattacharjee S, Pandharkar T., Liu H, Estiu G, Stahelin RV, Rizk SS, Njimoh DL, Ryan Y, Chotivanich K, Nguon C, Ghorbal M, Lopez-Rubio JJ, Pfrender M, Emrich S, Mohandas N, Dondorp AM, Wiest O, Haldar K. 2015. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]