Abstract
CA-MA is a hybrid antimicrobial peptide (AMP) derived from two naturally occurring AMPs, cecropin A and magainin 2. CA-MA shows strong antimicrobial activity against Gram-negative and Gram-positive bacteria but also exhibits cytotoxicity toward mammalian cells. Our objective was to identify CA-MA analogues with reduced cytotoxicity by systematic replacement of amino acids with positively charged R groups (His and Lys), aliphatic R groups (Leu), or polar R groups (Glu). Among the CA-MA analogues studied (CMA1 to -6), CMA3 showed the strongest antimicrobial activity, including against drug-resistant Escherichia coli and Pseudomonas aeruginosa strains isolated from hospital patients. CMA3 appeared to act by inducing pore formation (toroidal model) in the bacterial membrane. In cytotoxicity assays, CMA3 showed little cytotoxicity toward human red blood cells (hRBCs) or HaCaT cells. Additionally, no fluorescence was released from small or giant unilamellar vesicles exposed to 60 μM CMA3 for 80 s, whereas fluorescence was released within 35 s upon exposure to CA-MA. CMA3 also exerted strong lipopolysaccharide (LPS)-neutralizing activity in RAW 264.7 cells, and BALB/c mice exposed to LPS after infection by Escherichia coli showed improved survival after administration of one 0.5-mg/kg of body weight or 1-mg/kg dose of CMA3. Finally, in a mouse model of septic shock, CMA3 reduced the levels of proinflammatory factors, including both nitric oxide and white blood cells, and correspondingly reduced lung tissue damage. This study suggests that CMA3 is an antimicrobial/antiendotoxin peptide that could serve as the basis for the development of anti-inflammatory and/or antimicrobial agents with low cytotoxicity.
INTRODUCTION
When microorganisms infect vertebrate or invertebrate animals, the secretion of cytokines, chemokines, and other peptides is an important component of the animals' innate immune response (1, 2). Among those other peptides are antimicrobial peptides (AMPs), which play key roles in the innate immune responses of mammals, amphibians, and insects, among others (3, 4). Indeed, more than 2,300 different AMPs have been identified and isolated from a wide range of organisms, including bacteria (n = 322), protozoa (n = 6), fungi (n = 12), plants (n = 306), and animals (n = 1,801). Most AMPs contain at least 10 amino acid residues, have a net charge ranging from −3 to +20, and exhibit <60% hydrophobicity (5). Together, these features underlie the bactericidal activity of AMPs, enabling them to bind and form damaging pores in the membranes of Gram-negative and Gram-positive bacteria. To induce pore formation, AMPs have an α-helical structure and the ability to interact with anionic components of the cytoplasmic membrane, resulting in the formation of toroidal, carpet, or barrel stave pores (6). Additionally, many AMPs can potentially prevent septic shock via their antiendotoxic activity through electrostatic interaction with lipopolysaccharides (LPS) and lipoteichoic acid in the bacterial membrane (7–9).
CA-MA is a hybrid AMP synthesized from residues 1 to 8 from cecropin A, a naturally occurring AMP isolated from the hemolymph of Hyalophora cecropia (one of the giant silk moths) and residues 1 to 12 of magainin 2 from the skin of Xenopus laevis (African clawed frog) (10, 11). Like most naturally occurring AMPs, CA-MA targets the bacterial cytoplasmic membrane and appears to have strong activity against both Gram-negative and Gram-positive bacteria (12). Unfortunately, however, CA-MA also shows cytotoxicity against normal mammalian cells. A recent study characterized a set of CA-MA analogues in which amino acid residues were systematically replaced at locations carrying positively charged R groups (His and Lys), aliphatic R groups (Leu), or polar R groups (Glu) (13). Analogues CMA1 to -6, especially CMA6 peptide, reportedly showed the strongest binding affinity for the bacterial cell membrane (12–15).
In the present study, we assessed the antimicrobial activities of CA-MA and CMA1 to -6 against Gram-negative and Gram-positive drug-resistant bacterial strains, as well as their cytotoxic activities against cultured HaCaT cells. In addition, we identified a possible mechanism of action against the Escherichia coli cell membrane and detected anti-inflammatory activity against E. coli 0111:B4 LPS, as well as endotoxin-neutralizing activity in BALB/c mice injected with E. coli 0111:B4 LPS.
MATERIALS AND METHODS
Microorganisms.
Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 15692) were obtained from the American Type Culture Collection. The strains of Staphylococcus aureus (strains 3518, 1870, 1630, 4716, 3551, and 5155) and P. aeruginosa (strains 434, 5018, 4891, 3399, 341, 138, 778, 4007, 3344, 4076, 3552, 3543, 3290, and 4891) used are ampicillin- and gentamicin-resistant strains isolated from hospital patients with otitis media at Chonnam National University, Republic of Korea.
Peptide synthesis and purification.
The peptides were synthesized using the 9-fluorenylmethoxycarbonyl (Fmoc) solid-phase method on Rink amide 4-methyl benzhydrylamine resin (0.55 mmol/g; Novabiochem) with a Liberty microwave peptide synthesizer (CEM Co., Matthews, NC). To obtain N-terminal fluorescently labeled peptides, the resin-bound peptides were treated with 20% (vol/vol) piperidine in dimethylformamide (DMF) to remove the protective Fmoc group from the N-terminal amino acid residue. The peptides were then cleaved from the corresponding resins, precipitated with ether, and extracted. The resultant crude peptides were purified by reversed-phase preparative high-performance liquid chromatography (RP-HPLC) on a Jupiter C18 column (250 by 21.2 mm, 15 μM, 300 Å) using a 0-to-60% acetonitrile gradient in water also containing 0.05% trifluoroacetic acid. The purity of the extracted peptides (more than 95%) was then confirmed using analytical reversed-phase HPLC on a Jupiter proteo C18 column (250 by 4.6 mm, 90 Å, 4 μM). The molecular mass of the peptides was confirmed using a matrix-assisted laser desorption ionization (MALDI) mass spectrometer (MALDI II; Kratos Analytical Instruments) (16).
Antimicrobial activity.
Bacterial cells were cultured at 37°C in appropriate culture media (tryptic soy broth [TSB], Luria broth [LB], and 0.05% NaCl with nutrient broth [NB]), and the antimicrobial activities of the AMPs were determined using microdilution assays and/or the NCCLS method. Briefly, twofold serial dilutions of each peptide (CA-MA and its analogues; 0.5 to 64 μM) or conventional antibiotics (ampicillin and gentamicin; 6.25 to 800 μM) were added in duplicate to bacterial cell-containing medium (5 × 105 CFU/ml) during the mid-logarithmic phase of growth. The samples were then incubated for 24 h at 37°C, after which the MICs of the peptides were determined by measuring the absorbance level at 650 nm. The lowest peptide concentration that completely inhibited growth was defined as the MIC (16, 51).
Hemolytic activity.
Fresh human red blood cells (hRBCs) from healthy donors were centrifuged at 800 × g and washed with phosphate-buffered saline (PBS) until the supernatant was clear. Twofold serial dilutions of the peptides in PBS were added to a 96-well plate, after which the hRBCs were added to obtain a final concentration of 8% (vol/vol). The samples were then incubated with mild agitation for 1 h at 37°C. The samples were then centrifuged at 800 × g for 10 min, and the absorbance of the supernatant was measured at 414 nm. All the measurements were performed in triplicate (11, 12), and the percentages of hemolysis were calculated using equation 1:
| (1) |
where 100% hemolysis was defined as the absorbance measured from hRBCs exposed to 1% Triton X-100 and zero hemolysis was characterized using hRBCs alone in PBS (16).
Cytotoxicity.
To examine the cytotoxic effects of the peptides, HaCaT (human keratinocyte) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and 10% fetal calf serum at 37°C in a humidified chamber under a 5% CO2 atmosphere. Growth inhibition was evaluated using 3-(4,5-dimethylthizol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays to measure cell viability. After cells were seeded into a 96-well plate at a density of 2 × 104/well and incubated for 24 h, twofold serial dilutions of each peptide in DMEM were added to the wells, and the cells were incubated for an additional 24 h at 37°C. Thereafter, 10 μl of MTT (5 mg/ml) was added to each well, and the plate was incubated for 4 h. The supernatants were then removed, and 50 μl of DMSO was added to each well to dissolve any remaining precipitate. Finally, the absorbance at 570 nm was measured using a microtiter reader (16).
Binding affinity of peptides to SUV liposomes.
Small unilamellar vesicles (SUVs) were prepared using the sonication method (17, 18). The desired phospholipid mixture (phosphatidylcholine-sphingomyelin [PC-SM], 2:1, wt/wt) was dissolved in chloroform, dried in a glass tube under nitrogen, and then lyophilized overnight to remove any residual solvent. Dry lipid films were then added to 1 to 2 ml of 10 mM HEPES buffer (pH 7.4), and the resulting mixtures were vortexed. To prepare the SUVs, the lipid dispersions were sonicated in a bath-type sonicator for 30 min, until the turbidity had cleared. After preparation of the vesicles, the suspensions were extruded 15 times through polycarbonate membranes with 0.05 and 0.2 μM pores, which ensured a homogeneous SUV population. The concentration was determined using a standard phosphate assay (19). Circular dichroism (CD) spectra were recorded at 25°C on a Jasco 810 spectropolarimeter (Jasco, Easton, MD, USA) equipped with a temperature control unit. A 0.1-cm-path-length quartz cell was used with 30 μM and 60 μM CA-MA/CMA3 in 300 μM SUVs in 10 mM HEPES buffer (pH 7.4). At least three scans were acquired and averaged to improve the signal-to-noise ratio at 250 to 190 nm (20).
Preparation and visualization of GUV liposomes.
Giant unilamellar vesicles (GUVs) were prepared using the electroformation method. Briefly, the desired phospholipid mixture (PC-SM, 2:1, wt/wt) was prepared in chloroform, after which 100-μl aliquots of the lipid mixture were deposited on indium tin oxide (ITO)-coated glass slides (25 by 35 by 1.1 mm; Sigma-Aldrich, St. Louis, MO), which were then spin coated at 600 rpm for 5 min. Any residual organic solvent was removed by evacuation in a vacuum for at least 2 h. Two ITO glass slides, one with a lipid film, were then placed such that they were separated only by a poly(dimethylsiloxane) spacer so as to form an electroformation chamber (25 by 25 by 1 mm). The chamber was filled with 0.1 M sucrose in 10 mM HEPES buffer (pH 7.4) through a hole in the poly(dimethylsiloxane) spacer. A 1.7-Vpp (peak to peak voltage), 10 Hz alternating current (AC) field was then applied to the ITO slides using a function generator (Agilent 33220A; Agilent Technology, USA). After 1.5 h, the electric field was changed to 4 Vpp, 4 Hz for 10 min to detach the liposomes that formed on the slides. The liposome solution was then gently removed from the electroformation chamber and diluted in 10 mM HEPES buffer containing 0.1 M glucose. Aliquots of the resultant GUV suspension were deposited on microscope slides and allowed to settle for 1 min. This settling was due to the density difference between the sugar solutions inside and outside the liposomes. The liposomes on the slides were examined under a microscope (IX71; Olympus, Tokyo, Japan), and images were recorded using a digital charge-coupled device (CCD) camera (DP71; Olympus) and video recorder and were analyzed using software provided by the manufacturer (21).
CD analysis.
CD spectra were recorded at 25°C on a Jasco 810 spectropolarimeter (Jasco, Easton, MD, USA) equipped with a temperature control unit. A 0.1-cm-path-length quartz cell containing 30 μM peptide solution was used along with 30 mM sodium dodecyl sulfate (SDS; wt/vol). At least five scans were acquired and averaged to improve the signal-to-noise ratio at 250 to 190 nm. Mean residue ellipticity ([θ], deg · cm2 · dmol−1) was calculated using the following equation (20):
| (2) |
where obs is the measured signal (ellipticity) in millidegrees, l is the optical path length of the cell in centimeters, and c is the molar concentration of a peptide (mean molar concentration of the residues: c = number of residues in the peptide × molar concentration of the peptide [mol/liter]).
Sytox green uptake analysis.
E. coli cells were grown to mid-logarithmic phase at 37°C, washed, and resuspended (2 × 107/ml) in 10 mM sodium phosphate buffer (pH 7.2). The cells were then incubated with 1 μM Sytox green for 15 min in the dark. After the addition of 2 to 64 μM peptide (CA-MA or CMA3), the time-dependent increases in fluorescence caused by the binding of the cationic dye to intracellular DNA were monitored (excitation at 485 nm and emission at 520 nm) (20).
Membrane depolarization of intact bacterial cells.
The membrane-depolarizing activities of the peptides were evaluated in E. coli cells using the membrane potential-sensitive fluorescent dye DiSC3-5 (3,3′-dipropylthiadicarbocyanine iodide). Two batches of bacterial cells (experiment and control) were grown to mid-log phase at 37°C, harvested, and washed three times with buffer A (20 mM glucose, 5 mM HEPES [pH 7.2]). The cells were then resuspended to an A600 of 0.05 in buffer A containing 0.1 M KCl, after which DiSC3-5 was added to a final concentration of 0.1 μM and the mixture was incubated for ∼60 min until the fluorescence level stabilized. The peptides were then added to bacterial suspensions, and the changes in fluorescence were continuously recorded (excitation at 622 nm and emission at 670 nm) for 30 min (20).
CLSM.
To analyze the cellular distribution of the peptides, E. coli cells were incubated with tetramethyl rhodamine (TAMRA)-labeled peptide and monitored using confocal laser-scanning microscopy (CLSM). The peptides were added to the cells using the same procedure used for the antimicrobial assays (see above). TAMRA-labeled peptides at their respective MICs were added to 100 μl of cell suspension. After incubation for 30 min, the cells were pelleted by centrifugation at 4,000 × g for 5 min and washed three times with ice-cold PBS. The localization of the TAMRA-labeled peptides was then examined under an inverted LSM510 laser-scanning microscope (Carl Zeiss, Gottingen, Germany) equipped with a helium/neon laser (543-nm line). The resulting images were recorded digitally in a 512- by 512-pixel format (22).
SEM.
Morphological alteration of E. coli cells was assessed by comparing cells incubated in the absence or presence of CA-MA or CMA3. E. coli cells grown to mid-logarithmic phase at 37°C were resuspended in 10 mM sodium phosphate buffer (pH 7.4) to an optical density at 600 nm (OD600) of 0.15, after which 1/2 MIC of each peptide was added. After incubation for 10 min at 37°C, the cells were fixed, dehydrated through a 50-to-100% ethanol series (10 min at each step), and coated with platinum. The cells were then examined under a scanning electron microscope (SEM) (JSM-7100F; JEOL, Japan) (23).
LPS binding analysis.
CD spectra were recorded at 25°C on a Jasco 810 spectropolarimeter (Jasco, Easton, MD, USA) equipped with a temperature control unit. A 0.1-cm-path-length quartz cell containing a 50 μM peptide solution was used, along with 10 mM sodium phosphate buffer (pH 7.4) and 30 mM E. coli LPS. At least five scans were acquired and averaged to improve the signal-to-noise ratio at 250 to 190 nm (20).
LPS aggregation analysis.
To analyze LPS aggregation based on light scattering, several concentrations of TAMRA-labeled CMA3 (0, 5, 10, 20, 30, and 40 μg/ml) were mixed with 25-μg/ml E. coli LPS solution in 10 mM sodium phosphate buffer (pH 7.4) and incubated for 30 min at 37°C. The aggregation sample was collected by centrifugation at 12,000 × g and resuspended in 10 mM sodium phosphate buffer (pH 7.4). The dose-dependent increases in fluorescence caused by the aggregation of TAMRA-labeled CMA3 with E. coli LPS were then monitored (excitation at 570 nm and emission at 570 nm) (24).
Neutralization of LPS.
The neutralization of LPS by the peptides was assessed using a chromogenic limulus amebocyte lysate assay. A constant concentration of E. coli LPS (1 ng/ml) was incubated with various concentrations of the peptides (0 to 25 μM) at 37°C in the wells of a pyrogenic sterile microtiter plate. Fifty-microliter aliquots of the mixtures were then added to equal volumes of limulus amebocyte lysate reagent, and the mixtures were incubated for 10 min at 37°C. A yellow color developed upon the addition of 100 μl of a chromogenic substrate solution. The reaction was stopped by adding 25% acetic acid, after which the absorbance was measured at 405 nm (20).
Mammalian cell culture.
Raw 264.7 (mouse macrophage) cells were cultured in DMEM supplemented with antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin) and 10% fetal calf serum at 37°C in a humidified chamber under an atmosphere containing 5% CO2. For experimentation, the cells were plated in 12-well plates (105/well) and cultured for 24 h under the same conditions.
Nitric oxide assay.
RAW 264.7 cells (5 × 105/well) were incubated for 9 h at 37°C with 1 μg/ml E. coli LPS in the presence or absence of 50 μg/ml CMA3 in DMEM supplemented with serum and antibiotics in a humidified chamber with a 5% CO2 atmosphere. The culture supernatants were then collected, mixed with Griess reagent (equal volume), and incubated for 10 min at room temperature. The NO produced was measured on a microplate reader at 540 nm. The changes evoked in NO levels were determined by comparison with data points obtained in the absence of CMA3 (25).
Effects of CMA3 on LPS-dependent TNF-α induction.
The expression level of a proinflammatory cytokine (tumor necrosis factor alpha [TNF-α]) was evaluated after the addition of E. coli LPS (1 μg/ml) to RAW 264.7 cells (5 × 105/well) in the presence or absence of CMA3 (50 μg/ml) in DMEM supplemented with serum and antibiotics. The cells were incubated for 9 h at 37°C in a humidified chamber with a 5% CO2 atmosphere. TNF-α secretion was measured in the culture supernatant using a mouse TNF-α enzyme-linked immunosorbent assay (ELISA) kit (Biotechnology Komabiotech) (22). The levels were quantified by measuring the absorbance at 450 nm on a VERSA max microplate reader.
Western blot analysis.
Western blot analysis was performed as previously described (22). Briefly, proteins extracted from mouse organs were separated by SDS-PAGE in a 15% polyacrylamide gel for 3 h and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, USA) for 1 h using 90 V. Each membrane was then incubated overnight at 4°C with anti-TNF-α or anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody (Santa Cruz Biotechnology) in a 5% skim milk solution (Abfrontier). The membranes were washed with Tris-buffered saline containing Tween 20 (0.05% TBST) and incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG. Finally, the membrane blots were developed using a Western blot detection kit (Abfrontier).
Immunocytochemistry (ICC) analysis.
RAW 264.7 cells (5 × 105/ml) were cultured in an 8-well plate for 12 h at 37°C under an atmosphere containing 5% CO2 and then stimulated for 9 h with E. coli LPS (1 μg/ml) in the presence or absence of CMA3 (50 μg/ml). Thereafter, the cells were washed with PBS, fixed in 4% paraformaldehyde (15 min), and permeabilized using 0.5% Triton X-100 in PBS. The cells were then washed again by gently shaking in PBS for 5 min three times, blocked in blocking buffer (5% bovine serum albumin in PBS at 200 μl/well) for 1 h with gentle shaking, and then incubated with a 1:100 dilution of primary antibody (anti-TNF-α antibody; Abfrontier) for 12 h at 4°C. Thereafter, they were washed three times in PBS (5 min/wash) and incubated with a 1:200 dilution of secondary antibody (fluorescein isothiocyanate [FITC]-conjugated goat anti-mouse IgG) for 1 h at 37°C. The nuclei were stained using DAPI (0.25 μg/ml; Life Technologies), and the staining was visualized using a confocal laser-scanning microscope (LSM510; Carl Zeiss, Gottingen, Germany) (26).
In vivo assays.
Studies were conducted according to the standards of the institutional ethics committee of Chosun University and to the checklist for ethical consideration of cytotoxicity studies (authorization no. CIACUC2015-A0011) (https://www.cre.or.kr/article/policy/1382313). Male BALB/c mice (aged 7 weeks, 23 ± 0.5 g) were injected intraperitoneally (i.p.) with 15 mg of E. coli 0111:B4 LPS (Sigma) per kg of body weight. The CMA3 peptide was injected i.p. into mice at a concentration of 0.5 or 1 mg/kg 30 min after the E. coli 0111:B4 LPS injection. Survival was followed for 40 h, after which mice were sacrificed, blood and lungs were collected, lungs were fixed, white blood cells (WBCs) were counted, and NO and histochemical assays were performed. Whole lungs were stained with hematoxylin and eosin (H&E) (27).
Mouse survival rate.
In the induced-inflammation and toxicity experiments, groups of 7-week-old male BALB/c mice (n = 10) were injected i.p. with 0.5 or 1 mg/kg CMA3 (100 μl) 30 min prior to i.p. injection of 100 μl PBS buffer and/or 15 mg/kg E. coli 0111:B4 LPS. Experimental mice were monitored for survival over the subsequent 60 h (27).
WBC staining and cell counts using Hoechst stain.
Whole-blood samples were extracted from mouse heart tissues soaked in heparin for 30 min. After complete separation of red and white blood cells (RBCs and WBCs, respectively) from the serum by centrifugation for 10 min at 800 × g, the cells were resuspended in PBS buffer (8%, vol/vol). The 8% RBCs were stained using 0.25 mg/ml Hoechst dye for 10 min at 37°C. The cells (10 μl) were then observed under a fluorescence microscope (IX71; Olympus, Tokyo, Japan), and the WBCs were counted (28).
Nitric oxide assay.
Whole-blood samples were collected following induction of inflammation using 15 mg/kg E. coli 0111:B4 LPS in the absence or presence of 1 mg/kg CMA3. Thirty minutes after LPS injection, the BALB/c mice were injected i.p. with either CMA3 (0.5 and 1 mg/kg) or PBS alone. Forty hours after treatment, whole-blood samples were extracted and centrifuged at 4,000 × g for 10 min at 4°C. The supernatant was then retained for NO assays. To determine NO levels, Griess reagent [N-(1-naphthyl) ethylenediamine dihydrochloride; 0.1% in distilled water] was added to the samples, which were then mixed with equal volumes of sulfanilic acid (1% in phosphoric acid). Aliquots of the resultant mixture (50 μl) were incubated for 30 min at 25°C with continuous mixing, and the absorbance at 548 nm was measured using a microplate reader. NO levels were calculated against a standard curve constructed using sodium nitrite (27).
H&E staining.
Whole-lung-tissue samples were removed, washed once in PBS buffer, and then transferred to 4% paraformaldehyde for 24 h. The samples were next dehydrated in 50% and then 100% ethanol (2 h each), followed by 3 washes in xylene substitute (1 h). They were then embedded in paraffin, cut into 4-μm-thick sections using a microtome (Thermo Scientific), and stained with H&E. The stained sections were evaluated using an inverted microscope (IX71; Olympus, Tokyo, Japan) (27).
RESULTS
Modification of the CA-MA hybrid AMP and bactericidal and cytotoxic activity.
We initially designed the CA-MA hybrid AMP. Analogue peptides were then produced with the aim of reducing the hydrophobicity and cytotoxicity of the peptides while maintaining the antimicrobial activity. We constructed six CA-MA analogues (CMA1 to -6) by replacing the amino acid residues in positions 13 to 17 with positive (H or K), aliphatic (L), or polar (Q) amino acids. The resultant peptides each had an approximate charge of +8 under physiological conditions (Table 1; Fig. 1A). With the exception of CMA6, all of the analogues had shorter retention times than CA-MA during RP-HPLC, indicating an increase in polar surface area (Table 1). When we examined the antimicrobial activities of the analogues against Gram-negative and Gram-positive bacteria, we found that, like CA-MA, the analogue peptides exhibited strong antimicrobial activities against Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 15692) at 2 to 4 μM (data not shown).
TABLE 1.
Sequences, chromatography retention times, and molecular weights of CA-MA and its analogues
| Peptide | Sequence | Mean retention time (min)a | Mol wt |
|
|---|---|---|---|---|
| Observed | Calculated | |||
| CA-MA | KWKLFKKIGIGKFLHSAKKF-NH2 | 23.6 | 2,404.3 | 2,401.0 |
| CMA1 | KWKLFKKIGIGKHFLSAKKF-NH2 | 20.9 | 2,405.0 | 2,402.2 |
| CMA2 | KWKLFKKIGIGKFLSAHKKF-NH2 | 22.3 | 2,405.0 | 2,404.2 |
| CMA3 | KWKLKKHIGIGKHFLSAKKF-NH2 | 17.8 | 2,394.9 | 2,393.2 |
| CMA4 | KWKHLKKIGIGKFLSAHKKF-NH2 | 19.1 | 2,394.9 | 2,390.8 |
| CMA5 | KWKHLKKIGIGKHFLSAKKF-NH2 | 17.6 | 2,394.9 | 2,392.6 |
| CMA6 | KWKLFKKIGIGKFLQSAKKF-NH2 | 23.9 | 2,396.0 | 2,394.3 |
Mean retention time during reversed-phase high-performance liquid chromatography (RP-HPLC).
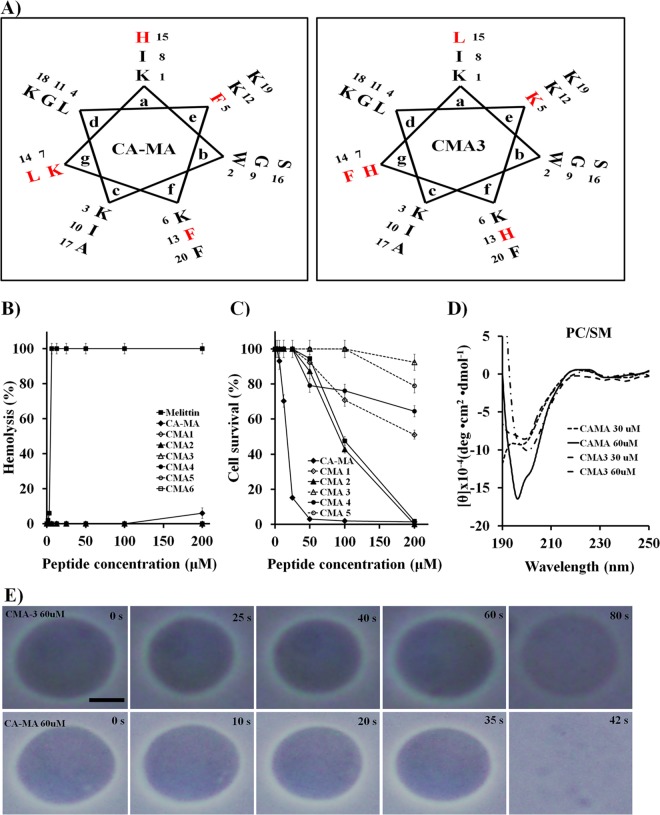
FIG 1.
Diagrams of CA-MA and CMA3. (A) Helical wheel projections of CA-MA and CMA3. Residues that were replaced in CMA3 are shown in red font in both projections. (B) Hemolytic activities of CA-MA and its analogue peptides against hRBCs, compared with that of melittin peptide. Error bars indicate standard deviations (SD) (P < 0.005). (C) Cytotoxicities of CA-MA and the indicated analogues toward HaCaT. Error bars indicate SD (P < 0.005). (D) CD spectra for CA-MA and CMA3 in the presence of small unilamellar vesicles (PC-SM, 2:1, wt/wt). Mean values are presented; n = 3. (E) Photomicrographs showing the effects of CA-MA and CMA3 on giant unilamellar vesicles (PC-SM, 2:1, wt/wt). The size bar represents 5 μm.
Antimicrobial activities against drug-resistant strains.
Given the results summarized above, we selected CMA3 for analysis of its activity against drug-resistant E. coli (CCARM strains 1229 and 1238) and P. aeruginosa strains isolated from patients with otitis media in Chonnam National University Hospital using National Committee for Clinical and Laboratory Standards (NCCLS) guidelines. CA-MA and CMA3 showed stronger activities against drug-resistant bacteria (E. coli and P. aeruginosa strains) than ampicillin or gentamicin did (Table 2). Furthermore, the hemolytic activity of CA-MA and its analogues was tested against hRBC cells (Fig. 1B). Incubation of the hRBC cells in the presence of 200 μM CA-MA peptide resulted in about 6% hemolysis, but no hemolytic activity was observed in the hRBC cells for higher peptide concentrations (200 μM) of its analogues. On the other hand, melittin, a honeybee venom toxin that has been reported to possess potent antimicrobial activity over a broader spectrum (29), exhibited 100% hemolytic activity at 3.13 μM (Fig. 1B).
TABLE 2.
Antimicrobial activities of AMPs and antibiotics against drug-resistant bacterial strains
| Straina | MIC (μM) of: |
|||||
|---|---|---|---|---|---|---|
| CA-MA in: |
CMA3 in: |
Ampicillin in MHB | Gentamicin in MHB | |||
| SPb | MHBc | SP | MHB | |||
| E. coli CCARM 1229 | 8 | 32 | 8 | 32 | 800 | 400 |
| P. aeruginosa 3552 | 8 | 4 | 8 | 4 | 800 | 400 |
| S. aureus 3518 | 2 | 64 | 2 | >64 | 800 | 800 |
| S. aureus 1870 | 2 | >64 | 2 | >64 | 800 | 400 |
| S. aureus 1630 | 2 | 64 | 2 | >64 | 800 | 800 |
| S. aureus 4716 | 4 | 64 | 4 | >64 | 800 | 400 |
| S. aureus 3551 | 1 | 64 | 1 | >64 | 800 | 800 |
The S. aureus and P. aeruginosa strains used are drug-resistant strains isolated from patients with otitis media in a hospital.
This assay was performed in 10 mM sodium phosphate (pH 7.4) with 10% Mueller-Hinton broth (MHB).
The procedure used was in accordance with NCCLS (National Committee for Clinical Laboratory Standards) methods.
Cytotoxic activity.
The cytotoxicities of CA-MA and its analogues were tested against HaCaT cells (Fig. 1C). Incubating HaCaT cells in the presence of 12.5 μM CA-MA resulted in about 85% cell lysis. However, CMA3 at a concentration of 100 μM exhibited no cytotoxicity toward HaCaT cells and showed minimal cytotoxicity (7.6%) at 200 μM. Among the six analogues, CMA3 had the strongest antimicrobial activity with no evident toxicity against mammalian cells, even at high concentrations (Fig. 1C).
We then investigated the cytotoxic mechanism of CA-MA and CMA3 by using small unilamellar vesicles (SUVs) and giant unilamellar vesicles (GUVs) constructed of phosphatidylcholine-sphingomyelin (PC-SM; 2:1, wt/wt) to approximate mammalian cell membrane components (17, 18, 21). Conformational changes to CA-MA and CMA3 were identified using CD spectroscopy. Our CD measurements indicated that CA-MA (60 μM) had strong binding affinity for PC-SM (2:1, wt/wt) SUV liposomes, whereas CMA3 peptide (60 μM) had much weaker affinity (Fig. 1D). Moreover, imaging of GUVs exposed to the peptides showed that at a concentration of 60 μM, CA-MA rapidly interacted with PC-SM (2:1, wt/wt) GUVs and disrupted the vesicles within about 42 s. In contrast, at the same concentration, CMA3 showed little interaction with GUVs, even after 80 s. These results indicate that CA-MA, but not CMA3, rapidly accumulates on PC-SM GUVs, leading to membrane disruption and vesicle lysis (Fig. 1E).
Helicity and membrane permeabilization by CA-MA and its analogues.
To analyze the structural features of CA-MA analogues and the effects of amino acid substitution, we first used far-UV CD spectra to evaluate their structures in aqueous solution (in 10 mM sodium phosphate buffer [pH 7.4]) (Fig. 2A) and in 30 mM SDS buffer (Fig. 2B). In aqueous buffer, the AMPs did not self-associate, and they assumed a random coil structure. In SDS solution, in contrast, all six CA-MA analogues assumed an α-helical structure, suggesting that the peptides' antimicrobial efficacy reflects that structure. We also assessed the mechanism by which CA-MA and CMA3 disrupted the cytoplasmic membrane of E. coli ATCC 25922 cells loaded with 1 μM Sytox green (Fig. 2C). There is no change in intracellular Sytox fluorescence without membrane disruption, since Sytox does not cross intact membranes. Upon the addition of CA-MA or CMA3 to E. coli cells, membrane disruption was evident from the dose-dependent increase in Sytox fluorescence intensity (Fig. 2C), as well as the rapid membrane depolarization at the MIC (2 μM CA-MA and 4 μM CMA3) (Fig. 2D). From these results, it appears that CA-MA and CMA3 disrupt both the inner and outer cytoplasmic membranes of E. coli cells.
FIG 2.
Mechanism of CA-MA and its analogue peptides. Circular dichroism (CD) spectra for CA-MA analogues in aqueous solution (A) and SDS solution (B). (C) Time-dependent influx of Sytox green into E. coli cells. The cells were incubated with 1 μM Sytox green until the basal fluorescence reached a steady state. CA-MA or CMA3 was then added, and the fluorescence was measured at the indicated time points (excitation at 485 nm and emission at 520 nm). Error bars indicate SD (P < 0.005). (D) Time-dependent depolarization of E. coli cells by CA-MA and CMA3. The E. coli concentration in mid-logarithmic phase was adjusted to an OD600 of 0.05 and preequilibrated for 10 min with DisC3-5. CA-MA (black) or CMA3 (gray) was then added, each at its MIC (2 μM and 4 μM, respectively), and changes in fluorescence were monitored continuously (excitation at 622 nm and emission at 670 nm) and plotted in arbitrary units. The dramatic second change in fluorescence was elicited by the addition of Triton X-100 (0.1%) to determine the maximum response. Mean values are presented; n = 3.
Visualization of E. coli membrane disruption using CLSM and SEM.
To further investigate the effects of CA-MA and CMA3 on E. coli, TAMRA-labeled CA-MA and CMA3 were incubated with E. coli at their respective MICs. Examination of the cells using CLSM showed the peptides to be localized on the E. coli cell surface (Fig. 3A). Visualization of E. coli cells using SEM following incubation of the cells with CA-MA or CMA3 at their respective MICs revealed that the peptides caused pore formation in the cytoplasmic membrane that appeared consistent with the toroidal-pore model. This confirms that both CA-MA and CMA3 kill bacteria through the formation of damaging pores in the cytoplasmic membrane (Fig. 3B).
FIG 3.
Morphological analysis of E. coli membrane with CA-MA and CMA3 peptides. (A) Localization of TAMRA-labeled CA-MA and CMA3 in E. coli. The cells were incubated for 10 min with the MIC of TAMRA–CA-MA (top) or TAMRA-CMA3 (bottom) and then examined using a confocal laser-scanning microscope. The scale bar is 5 μm. DIC, differential interference contrast. (B) Morphological analysis of E. coli cells in the absence and presence of CA-MA and CMA3 using SEM. CA-MA and CMA3 were incubated with E. coli cells at the MIC in 10 mM sodium phosphate buffer (containing 10% medium). The scale bar is 1.0 μm.
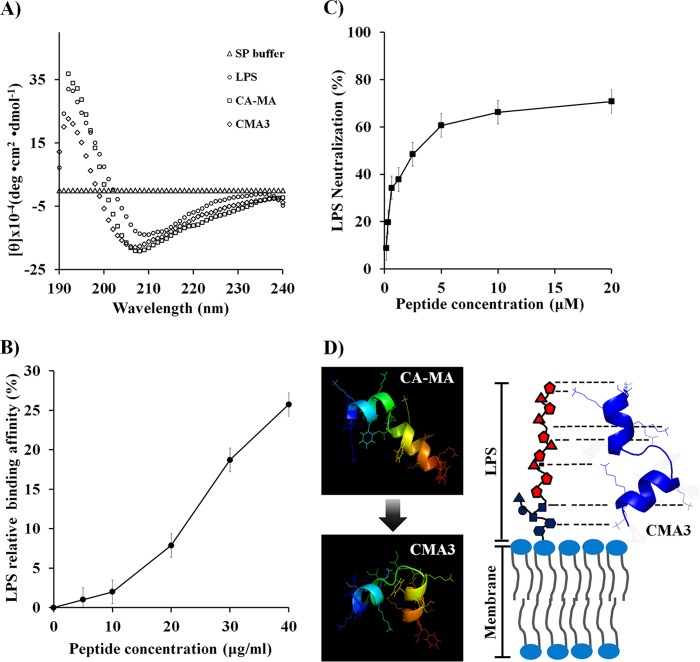
LPS binding activity.
Because CA-MA and CMA3 have a charge of +8 under physiological conditions, we anticipated that they would bind to LPS. We therefore used CD spectra to analyze the binding of CA-MA and CMA3 (50 μM) to E. coli LPS (30 mM), which we found shifted the peak minimum from 210 nm to 190 nm (Fig. 4A). When TAMRA-labeled CMA3 (0, 5, 10, 20, 30, or 40 μg/ml) was incubated with E. coli LPS (25 μg/ml) in 10 mM sodium phosphate buffer for 30 min, we observed dose-dependent increases in the fluorescence intensity, reflecting aggregation of the TAMRA-labeled peptide with LPS. At 40 μg/ml, the peptide showed strong aggregation (25.8%) with the E. coli LPS at 25 μg/ml (Fig. 4B). Furthermore, using the limulus amebocyte lysate method with 0.156 to 20 μM CMA3 and 1 ng/ml LPS, we determined that CMA3 also dose dependently neutralized LPS, showing 70.78% neutralization at 20 μg/ml (Fig. 4C). The computer modeling shown in Fig. 4D revealed that bending of the CMA3 peptide (at lysine residues) results in it having strong affinity for LPS, enabling it to bind to and neutralize the endotoxin.
FIG 4.
CMA3 binding to E. coli LPS, as evaluated using CD (A), TAMRA-labeled peptide (B), and a limulus amebocyte lysate chromogenic endotoxin quantitation kit (C). (A) Far-UV CD spectra confirming that structural changes are induced upon mixing 30 μM CMA3 with 30 mM LPS. (B) Relative binding of the TAMRA-labeled CMA3 in the presence of 25 μg/ml LPS at 37°C (fluorescence spectrometry). Mean values are presented; n = 3. Error bars indicate SD (P < 0.005). (C) LPS neutralization by CMA3 determined using an endotoxin quantitation kit. Mean values are presented; n = 3. Error bars indicate SD (P < 0.005). (D) Diagram of CMA3 binding to and neutralizing LPS in the bacterial membrane. The peptide structures were calculated by using PEP-FOLD server (http://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD/).
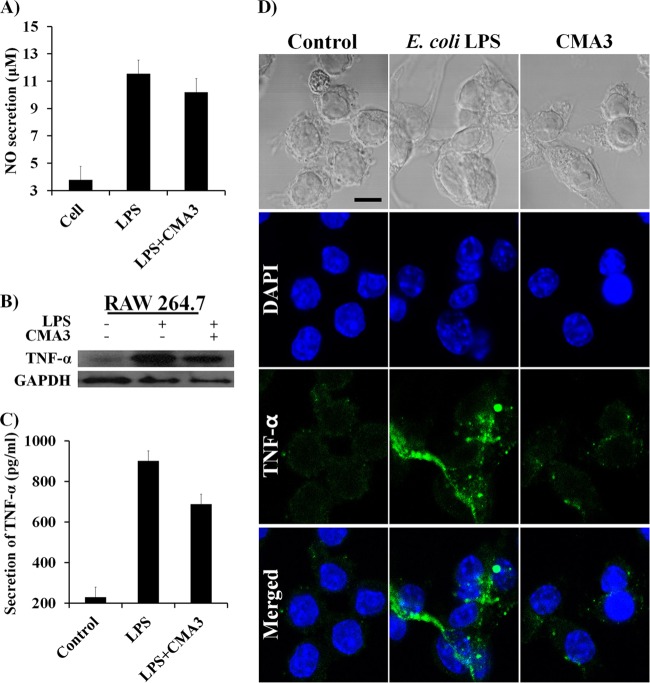
Anti-inflammatory effect of CMA3 on RAW 264.7 cells.
The ability of 50 μg/ml CMA3 to neutralize 1 μg/ml LPS prompted us to ask whether CMA3 could suppress the inflammatory response elicited by LPS. To address that question, we assessed the effect of CMA3 on E. coli LPS (1 μg/ml)-induced secretion of nitric oxide (NO) and the proinflammatory cytokine TNF-α from RAW 264.7 cells. As shown by the results in Fig. 5A, LPS significantly increased the NO secretion from RAW 264.7 cells compared with the NO secretion from control cells (11.45 μM versus 3.78 μM, respectively), but that response was diminished (to 10.19 μM) after treatment with 50 μg/ml CMA3. Furthermore, Western blot analysis and CLSM showed that LPS stimulates the expression of TNF-α in RAW 264.7 cells and that this response too was suppressed by CMA3 (Fig. 5B and D). Correspondingly, TNF-α ELISA analysis of culture supernatants showed that CMA3 significantly reduced the secretion of TNF-α in RAW 264.7 cells compared with its secretion in cells induced with LPS only (656.75 pg/ml versus 904.75 pg/ml, respectively). In the CMA3-treated cells, the peptide concentration of 50 μg/ml reduced TNF-α secretion to 248 pg/ml (Fig. 5C).
FIG 5.
Endotoxin activity of CMA3 antimicrobial peptide against induced inflammation in RAW 264.7 cells. (A) Effect of CMA3 on E. coli LPS-induced NO production in RAW 264.7 cells. All values represent the mean results ± SD from three individual experiments (P < 0.005). (B) Western blot showing intracellular TNF-α levels in RAW 264.7 cells stimulated with E. coli LPS with and without CMA3. (C) TNF-α levels in culture supernatants of RAW 264.7 cells stimulated with LPS with and without CMA3. TNF-α level were determined using an ELISA. Error bars indicate ±SD (P < 0.005). (D) Immunohistochemical analysis of the effects of CMA3 on LPS-induced TNF-α expression in RAW 264.7 cells. Shown are confocal micrographs of RAW 264.7 cells stained for TNF-α (green); the nuclei were labeled with DAPI (blue). The scale bar is 10 μm.
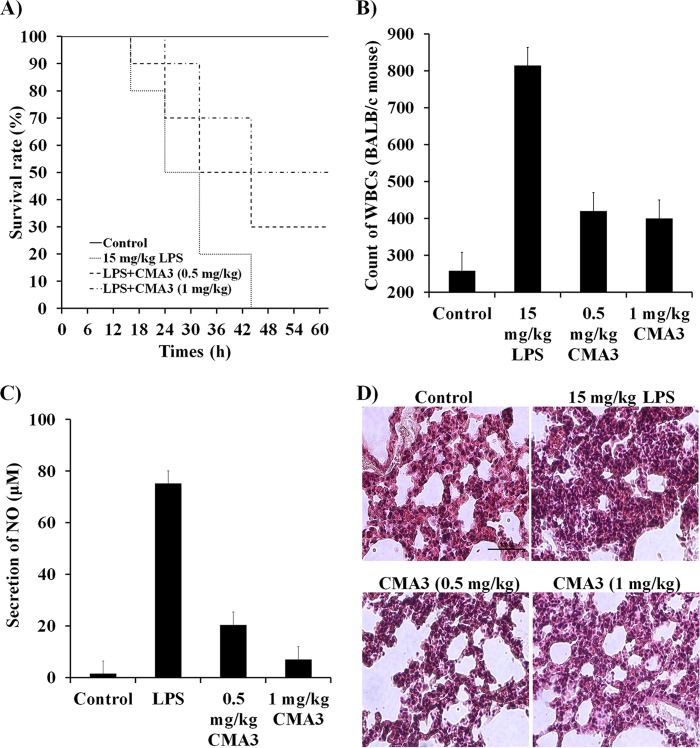
In vivo CMA3 activity.
To evaluate the therapeutic activity of CMA3 in a model of severe sepsis, BALB/c mice were injected with 0.5 mg/kg or 1 mg/kg CMA3 peptide prior to injection of E. coli 0111:B4 LPS (15 mg/kg). The CMA3 pretreatment increased the survival rate at 40 h posttreatment by more than 50% compared to the survival of mice treated only with LPS (Fig. 6A). In addition, the WBC counts were also significantly higher (814 WBCs) in E. coli 0111:B4 LPS-treated BALB/c mice than in untreated control mice (258 WBCs). However, pretreatment with CMA3 dose dependently reduced the WBC counts, to 420 WBCs with 0.5 mg/kg CMA3 and to 400 WBCs with 1 mg/kg CMA3 (Fig. 6B). Similarly, CMA3 also reduced the elevated serum NO levels otherwise seen following LPS administration, where the NO levels were 35 and 6.99 μM with 0.5 and 1 mg/kg CMA3 treatment, respectively, compared with 75.1 μM in serum induced with LPS only (Fig. 6C). Indeed, CMA3-treated mice showed no serious increase in NO levels following LPS administration. Consistent with those findings, lungs from E. coli 0111:B4 LPS-treated mice were significantly swollen and/or damaged, but CMA3 administration appeared to partially reverse that swelling and damage (Fig. 6D). These data indicate that the CMA3 antimicrobial peptide is also a strong neutralizer of endotoxin activity in BALB/c mice.
FIG 6.
CMA3 antimicrobial peptide effects on E. coli LPS 0111:B4-induced inflammation in the BALB/c mice. (A) CMA3 peptide increased survival rates in mice with E. coli 0111:B4 LPS (15 mg/kg)-induced septic shock. The CMA3 peptide prolonged survival in BALB/c mice injected with E. coli 0111:B4 LPS. In the E. coli 0111:B4 LPS treatment model, CMA3 (0.5 and 1 mg/kg) or PBS alone was injected intraperitoneally (i.p.) 30 min after administration of LPS. (B) CMA3 peptide inhibited white blood cell (WBC) infiltration. BALB/c mice were stimulated (10 μl) with E. coli 0111:B4 LPS in the presence or absence of CMA3 peptide. Error bars indicate ±SD (P < 0.005). (C) CMA3 peptide inhibited nitric oxide (NO) production. In BALB/c mice injected with E. coli 0111:B4 LPS, CMA3 (0.5 and 1 mg/kg) or PBS alone was injected i.p. after 30 min, and NO was analyzed in the whole-blood serum. Error bars indicate ±SD (P < 0.005). (D) Morphological changes in the lungs were analyzed 40 h after i.p. E. coli 0111:B4 LPS injection followed by treatment with CMA3 peptide (0.5 and 1 mg/kg) or PBS. The tissue samples were stained with H&E. The size bar represents 100 μm.
DISCUSSION
The immune response to bacterial infection involves the production of cytokines, chemokines, and other proteins, including AMPs (2, 30–34) such as cathelicidin/LL-37 (35, 36), by various immune cells (e.g., macrophages, monocytes, and NK cells). Bacterial infection also induces epithelial cells in the skin, lungs, gut, and breast to produce these peptides (37, 38), which also show anticancer activity against drug-resistant MCF-7 breast cancer cells (39–41).
In the present study, we examined CA-MA, a hybrid peptide synthesized from truncated versions of the α-helical structures of the AMPs cecropin A and magainin 2, two proteins that interact with the bacterial cytoplasmic membrane. Through the use of a set of substituted analogues, CA-MA was shown to act by binding to or inserting into the bacterial membrane (15). The parent peptides exert potent antimicrobial effects against both Gram-negative and Gram-positive bacteria but also exhibit hemolytic and cytotoxic activities against normal mammalian cells (10, 11). The CA-MA hybrid shows stronger antimicrobial activity than either of the parent peptides but does not induce hemolysis; however, it retains some cytotoxicity toward mammalian cells (12–14).
The purpose of the present study was to obtain additional information about the mechanism of action of a set of peptides derived from CA-MA through amino acid substitution. CA-MA has an α-helical structure when interacting with the cell membrane, and the source of CA-MA's cytotoxicity toward mammalian cells appears to be related in part to the interaction of the linear benzyl side chains of phenylalanine with the cell membrane (6). During the interaction, phenylalanines in positions 5, 13, and 20 of the peptide are aligned with one another in close proximity. It was therefore reasoned that cytotoxicity could be reduced by replacing those amino acids with alanine (16). In the present study, therefore, we eliminated or left the phenylalanine at position 5 and/or replaced the phenylalanine at position 13 with histidine in an effort to reduce cytotoxicity; also replaced were the lysine, histidine, and leucine residues, as summarized in Table 1 and Fig. 1A. Also, all of the analogue peptides were configured to have the same charge (+8). Among the analogues created, CMA3 was eluted earliest (17.3 min) during HPLC, indicating it to be the least hydrophobic. In that regard, we believe the hydrophobicity of AMPs contributes to their cytotoxicity (hRBC and HaCaT cells), which was therefore reduced by the replacement of hydrophobic amino acid residues (such as Phe). CA-MA had strong affinity for GUV membranes (PC-SM liposome), which were rapidly disrupted upon CA-MA binding, but CMA3 had much weaker affinity. The reason for the reduced cytotoxicity was the elimination (position 5) and replacement (positions 13 and 14) of the phenylalanine residues. Eliminating the phenylalanines from CA-MA was particularly important for reducing cytotoxicity toward mammalian cells (HaCaT cells).
Furthermore, we confirmed the antimicrobial activities of CA-MA and its analogues against standard and drug-resistant strains of Gram-negative and Gram-positive bacteria. CA-MA and CMA3 appeared to have the strongest antimicrobial activity against drug-resistant strains of P. aeruginosa in 10 mM sodium phosphate buffer with 10% Mueller-Hinton broth, though these peptides also showed good activity when the NCCLS method was used. The mechanism of action of CMA3 against drug-resistant strains remains to be tested, but in E. coli cells, CMA3, like CA-MA, appears to induce pore formation that disrupts both the inner and outer membranes. Moreover, under SEM, the pores formed appear to be consistent with the toroidal model (42). In contrast, two conventional antibiotic drugs, ampicillin and gentamicin, had no antimicrobial activity against drug-resistant strains, even at high concentrations.
The mortality and morbidity caused by infection with bacteria like S. aureus and P. aeruginosa are often related to the inflammatory response elicited; for example, the septic shock induced by LPS, a cell membrane component of Gram-negative bacteria, and lipoteichoic acid (LTA), a membrane component of Gram-positive bacteria (8, 43, 44). These responses reflect in large part the enhanced expression of inflammatory mediators, such as Toll-like receptor 4 (TLR4) and NO-related proteins (e.g., inducible NO synthase). Through its interaction with TLR4 on macrophage cells, LPS stimulates the production of NO and proinflammatory cytokines like TNF-α and interleukin 1β (IL-1β) (45, 46).
The most important features of AMPs, affecting their antimicrobial activity, cytotoxicity, and ability to neutralize LPS, are their hydrophobicity and net positive charge (47). For example, AMPs must have the proper net positive charge (+7 to +9) to bind LPS or LTA and exert neutralizing activity (48). In the present study, CMA3 retained the same net charge (+8) as CA-MA but with reduced hydrophobicity. This combination conferred strong antimicrobial activity with minimal cytotoxicity and significantly increased the LPS binding affinity. These features are thought to reflect the strong electrostatic interaction between lysine residues in CMA3 and lipid A in LPS. We also found that E. coli LPS-stimulated production of both NO and TNF-α in a cultured macrophage cell line (RAW 264.7) was suppressed by CMA3, perhaps as modeled in Fig. 7. In addition, in vivo studies showed that CMA3 has strong endotoxin-neutralizing activity. LPS is located on the Gram-negative bacterial membrane, strongly induces inflammation (49), and is responsible for the physiological manifestations of various diseases, including pneumonia (50). Here, we show that serum levels of the inflammatory marker NO are significantly reduced when mice administered E. coli-derived LPS are pretreated with CMA3. Treatment with CMA3 also increased the survival rate among the mice and reduced lung damage. For these reasons, we suggest that CMA3 is able to bind to and neutralize endotoxin LPS (in the lipid A subunit). And because CMA3 also shows strong antimicrobial activity against drug-resistant P. aeruginosa strains, we suggest that treatment with CMA3 may relieve the inflammation caused by LPS from Gram-negative bacteria while exerting antimicrobial effects.
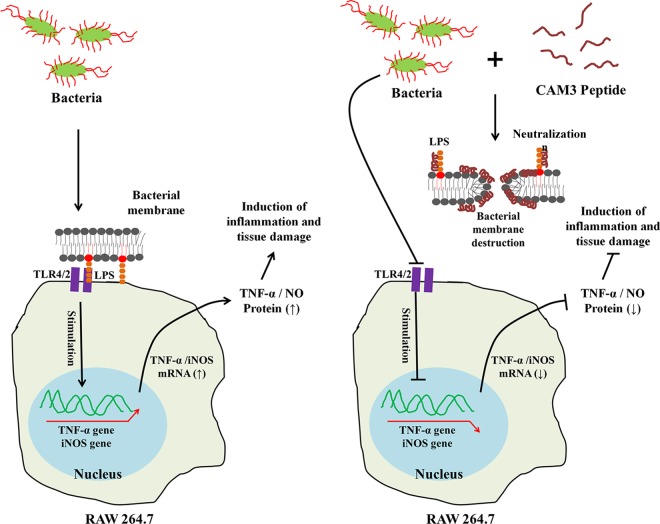
FIG 7.
Schematic diagram of a potential mechanism by which CMA3 suppresses LPS-induced inflammatory responses in RAW 267.4 macrophages. Left, LPS alone; right, LPS plus CMA3. CMA3 disrupts the E. coli cell membrane via toroidal pore formation and binds LPS, neutralizing it. →, stimulation; ⊣, inhibition.
Our results show that the AMP CMA3 has potent antimicrobial activity with little or no cytotoxicity toward hRBC and HaCaT cells, even at high concentrations. In addition, CMA3 shows strong endotoxin-neutralizing activity against LPS both in vivo and in vitro. CMA3 therefore appears to be an excellent antibiotic candidate.
ACKNOWLEDGMENTS
This work was supported by a Global Research Laboratory (GRL) grant (grant no. NRF-2014K1A1A2064460), the Global Collaborative R&D program (grant no. N0001229), and Chosun University, 2014.
REFERENCES
- 1.Miller LS, Cho JS. 2011. Immunity against Staphylococcus aureus cutaneous infections. Nat Rev Immunol 11:505–518. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai Y, Gallo RL. 2009. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defence. Trends Immunol 30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frew L, Stock SJ. 2011. Antimicrobial peptides and pregnancy. Reproduction 141:725–735. doi: 10.1530/REP-10-0537. [DOI] [PubMed] [Google Scholar]
- 4.Xie GH, Chen QX, Cheng BL, Fang XM. 2014. Defensins and sepsis. Biomed Res Int 2014:180109. doi: 10.1155/2014/180109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haney EF, Hancock RE. 2013. Peptide design for antimicrobial and immunomodulatory applications. Biopolymers 100:572–583. doi: 10.1002/bip.22250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bahar AA, Ren D. 2013. Antimicrobial peptides. Pharmaceuticals (Basel) 6:1543–1575. doi: 10.3390/ph6121543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray A, Cot M, Puzo G, Gilleron M, Nigou J. 2013. Bacterial cell wall macroamphiphiles: pathogen-/microbe-associated molecular patterns detected by mammalian innate immune system. Biochimie 95:33–42. doi: 10.1016/j.biochi.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 8.López-Abarrategui C, Del Monte-Martínez A, Reyes-Acosta O, Franco OL, Otero-González AJ. 2013. LPS inmobilization on porous and non-porous supports as an approach for the isolation of anti-LPS host-defense peptides. Front Microbiol 17:389. doi: 10.3389/fmicb.2013.00389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takagi S, Bai L, Ozeki T, Miyagi H, Kuroda K, Hayashi S, Yoneyama H, Ando T, Isogai E. 2014. A bovine myeloid antimicrobial peptide (BMAP-28) kills methicillin-resistant Staphylococcus aureus but promotes adherence of the bacteria. Anim Sci J 85:342–346. doi: 10.1111/asj.12109. [DOI] [PubMed] [Google Scholar]
- 10.Zasloff M. 1987. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc Natl Acad Sci U S A 84:5449–5453. doi: 10.1073/pnas.84.15.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiner H. 1982. Secondary structure of the cecropins: antibacterial peptides from the moth Hyalophora cecropia. FEBS Lett 137:283–287. doi: 10.1016/0014-5793(82)80368-2. [DOI] [PubMed] [Google Scholar]
- 12.Oh D, Shin SY, Lee S, Kang JH, Kim SD, Ryu PD, Hahm KS, Kim Y. 2000. Role of the hinge region and the tryptophan residue in the synthetic antimicrobial peptides, cecropin A(1-8)-magainin 2(1-12) and its analogues, on their antibiotic activities and structures. Biochemistry 39:11855-64. doi: 10.1021/bi000453g. [DOI] [PubMed] [Google Scholar]
- 13.Mereuta L, Roy M, Asandei A, Lee JK, Park Y, Andricioaei I, Luchian T. 2014. Slowing down single-molecule trafficking through a protein nanopore reveals intermediates for peptide translocation. Sci Rep 4:3885. doi: 10.1038/srep03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SY, Kang JH, Lee MK, Kim SY, Kim Y, Hahm KS. 1998. Cecropin A-magainin 2 hybrid peptides having potent antimicrobial activity with low hemolytic effect. Biochem Mol Biol Int 44:1119–1126. [DOI] [PubMed] [Google Scholar]
- 15.Shin SY, Kang JH, Jang SY, Kim Y, Kim KL, Hahm KS. 2000. Effects of the hinge region of cecropin A (1-8)-magainin 2(1-12), a synthetic antimicrobial peptide, on liposomes, bacterial and tumor cells. Biochim Biophys Acta 1463:209–218. doi: 10.1016/S0005-2736(99)00210-2. [DOI] [PubMed] [Google Scholar]
- 16.Lee JK, Park SC, Hahm KS, Park Y. 2013. Antimicrobial HPA3NT3 peptide analogs: placement of aromatic rings and positive charges are key determinants for cell selectivity and mechanism of action. Biochim Biophys Acta 1828:443–454. doi: 10.1016/j.bbamem.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Lee DG, Kim HN, Park Y, Kim HK, Choi BH, Hahm KS. 2002. Design of novel analogue peptides with potent antibiotic activity based on the antimicrobial peptide, HP (2-20), derived from N-terminus of Helicobacter pylori ribosomal protein L1. Biochim Biophys Acta 1598:185–194. doi: 10.1016/S0167-4838(02)00373-4. [DOI] [PubMed] [Google Scholar]
- 18.Mayer LD, Hope MJ, Cullis PR. 1986. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim Biophys Acta 858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 19.Stewart JCM. 1980. Colorimetric determination of phospholipids with ammonium ferrothiocyanate. Anal Biochem 104:10–14. doi: 10.1016/0003-2697(80)90269-9. [DOI] [PubMed] [Google Scholar]
- 20.Kim JY, Park SC, Yoon MY, Hahm KS, Park Y. 2011. C-terminal amidation of PMAP-23: translocation to the inner membrane of Gram-negative bacteria. Amino Acids 40:183–195. doi: 10.1007/s00726-010-0632-1. [DOI] [PubMed] [Google Scholar]
- 21.Estes DJ, Mayer M. 2005. Electroformation of giant liposomes from spin-coated films of lipids. Colloids Surf B Biointerfaces 42:115–123. doi: 10.1016/j.colsurfb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK, Park SC, Hahm KS, Park Y. 2014. A helix-PXXP-helix peptide with antibacterial activity without cytotoxicity against MDRPA-infected mice. Biomaterials 35:1025–1039. doi: 10.1016/j.biomaterials.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 23.Hartmann M, Berditsch M, Hawecker J, Ardakani MF, Gerthsen D, Ulrich AS. 2010. Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy. Antimicrob Agents Chemother 54:3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park SC, Kim JY, Jeong C, Yoo S, Hahm KS, Park Y. 2011. A plausible mode of action of pseudin-2, an antimicrobial peptide from Pseudis paradoxa. Biochim Biophys Acta 1808:171–182. doi: 10.1016/j.bbamem.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Takada K, Hirose F, Yamabe S, Uehara Y, Mizuta H. 2013. Endoplasmic reticulum stress mediates nitric oxide chondrocyte apoptosis. Biomed Rep 1:315–319. doi: 10.3892/br.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J, Zhang YY, Guo L, Li H, Chen DF. 2013. Bupleurum polysaccharides attenuates lipopolysaccharide-induced inflammation via modulating Toll-like receptor 4 signaling. PLoS One 8:e78051. doi: 10.1371/journal.pone.0078051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papareddy P, Kalle M, Kasetty G, Mörgelin M, Rydengård V, Albiger B, Lundqvist K, Malmsten M, Schmidtchen A. 2010. C-terminal peptides of tissue factor pathway inhibitor are novel host defense molecules. J Biol Chem 285:28387–28398. doi: 10.1074/jbc.M110.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, Guo X, Xie W, Li Y, Ma M, Yuan T, Luo B. 2015. Resveratrol exerts an anti-apoptotic effect on human bronchial epithelial cells undergoing cigarette smoke exposure. Mol Med Rep 11:1752–1758. doi: 10.3892/mmr.2014.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habermann E, Jentsch J. 1967. Sequence analysis of melittin from tryptic and peptic degradation products. Hoppe-Seylers Z Physiol Chem 348:37–50. doi: 10.1515/bchm2.1967.348.1.37. [DOI] [PubMed] [Google Scholar]
- 30.Clark R, Kupper T. 2005. Old meets new: the interaction between innate and adaptive immunity. J Investig Dermatol 125:629–637. doi: 10.1111/j.0022-202X.2005.23856.x. [DOI] [PubMed] [Google Scholar]
- 31.Hotchkiss RS, Monneret G, Payen D. 2013. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brogden KA, Johnson GK, Vincent SD, Abbasi T, Vali S. 2013. Oral inflammation, a role for antimicrobial peptide modulation of cytokine and chemokine responses. Expert Rev Anti Infect Ther 11:1097–1113. doi: 10.1586/14787210.2013.836059. [DOI] [PubMed] [Google Scholar]
- 33.Cruz J, Ortiz C, Guzmán F, Fernández-Lafuente R, Torres R. 2014. Antimicrobial peptides: promising compounds against pathogenic microorganisms. Curr Med Chem 21:2299–2321. doi: 10.2174/0929867321666140217110155. [DOI] [PubMed] [Google Scholar]
- 34.Duplantier AJ, van Hoek ML. 2013. The human cathelicidin antimicrobial peptide LL-37 as a potential treatment for polymicrobial infected wounds. Front Immunol 4:143. doi: 10.3389/fimmu.2013.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandal SM, Silva ON, Franco OL. 2014. Recombinant probiotics with antimicrobial peptides: a dual strategy to improve immune response in immunocompromised patients. Drug Discov Today 19:1045–1050. doi: 10.1016/j.drudis.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 36.De Smet K, Contreras R. 2005. Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol Lett 27:1337–1347. doi: 10.1007/s10529-005-0936-5. [DOI] [PubMed] [Google Scholar]
- 37.Henzler Wildman KA, Lee DK, Ramamoorthy A. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 42:6545–6558. doi: 10.1021/bi0273563. [DOI] [PubMed] [Google Scholar]
- 38.Majchrzykiewicz JA, Kuipers OP, Bijlsma JJ. 2010. Generic and specific adaptive responses of Streptococcus pneumoniae to challenge with three distinct antimicrobial peptides, bacitracin, LL-37, and nisin. Antimicrob Agents Chemother 54:440–451. doi: 10.1128/AAC.00769-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.To KK, Ren SX, Wong CC, Cho CH. 2013. Reversal of ABCG2-mediated multidrug resistance by human cathelicidin and its analogs in cancer cells. Peptides 40:13–21. doi: 10.1016/j.peptides.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 40.Bhattacharjya S. 2010. De novo designed lipopolysaccharide binding peptides: structure based development of antiendotoxic and antimicrobial drugs. Curr Med Chem 17:3080–3093. doi: 10.2174/092986710791959756. [DOI] [PubMed] [Google Scholar]
- 41.Gaspar D, Veiga AS, Castanho MA. 2013. From antimicrobial to anticancer peptides. A review. Front Microbiol 4:294. doi: 10.3389/fmicb.2013.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melo MN, Ferre R, Castanho MA. 2009. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat Rev Microbiol 7:245–250. doi: 10.1038/nrmicro2095. [DOI] [PubMed] [Google Scholar]
- 43.Nell MF, Tiabringa GS, Wafelman AR, Verrijk R, Hiemstra PS, Drijfhout JW, Grot JJ. 2006. Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activity for therapeutic application. Peptides 27:649–660. doi: 10.1016/j.peptides.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 44.Heinbockel L, Sánchez-Gómez S, Martinez de Tejada G, Dömming S, Brandenburg J, Kaconis Y, Hornef M, Dupont A, Marwitz S, Goldmann T, Ernst M, Gutsmann T, Schürholz T, Brandenburg K. 2013. Preclinical investigations reveal the broad-spectrum neutralizing activity of peptide Pep19-2.5 on bacterial pathogenicity factors. Antimicrob Agents Chemother 57:1480–1487. doi: 10.1128/AAC.02066-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lei M, Jiao H, Liu T, Du L, Cheng Y, Zhang D, Hao Y, Man C, Wang F. 2011. siRNA targeting mCD14 inhibits TNF-α, MIP-2, and IL-6 secretion and NO production from LPS-induced RAW2647 cells. Appl Microbiol Biotechnol 92:115–124. doi: 10.1007/s00253-011-3371-7. [DOI] [PubMed] [Google Scholar]
- 46.Belkaid Y, Bouladoux N, Hand TW. 2013. Effector and memory T cell responses to commensal bacteria. Trends Immunol 34:299–306. doi: 10.1016/j.it.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenfeld Y, Lev N, Shai Y. 2010. Effect of the hydrophobicity to net positive charge ratio on antibacterial and antiendotoxin activities of structurally similar antimicrobial peptides. Biochemistry 49:853–861. doi: 10.1021/bi900724x. [DOI] [PubMed] [Google Scholar]
- 48.Jacob B, Park IS, Bang JK, Shin SY. 2013. Short KR-12 analogs designed from human cathelicidin LL-37 possessing both antimicrobial and antiendotoxic activities without mammalian cell toxicity. J Pept Sci 19:700–707. doi: 10.1002/psc.2552. [DOI] [PubMed] [Google Scholar]
- 49.Liu Y, Bao L, Xuan L, Song B, Lin L, Han H. 2015. Chebulagic acid inhibits the LPS-induced expression of TNF-α and IL-1β in endothelial cells by suppressing MAPK activation. Exp Ther Med 10:263–268. doi: 10.3892/etm.2015.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao M, Zhu T, Zhang W, Wang T, Shen YC, Wan QF, Wen FQ. 2014. Emodin ameliorates LPS-induced acute lung injury, involving the inactivation of NF-κB in mice. Int J Mol Sci 15:19355–19368. doi: 10.3390/ijms151119355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park SC, Kim JY, Lee JK, Yoo S, Kim H, Seo CH, Nah JW, Hahm KS, Park Y. 2011. Synthetic diastereomeric-antimicrobial peptide: antibacterial activity against multiple drug resistant clinical isolates. Biopolymers 96:130–136. doi: 10.1002/bip.21446. [DOI] [PubMed] [Google Scholar]