Abstract
Biofilms consist of surface-adhered bacterial communities encased in an extracellular matrix composed of DNA, exopolysaccharides, and proteins. Extracellular DNA (eDNA) has a structural role in the formation of biofilms, can bind and shield biofilms from aminoglycosides, and induces antimicrobial peptide resistance mechanisms. Here, we provide evidence that eDNA is responsible for the acidification of Pseudomonas aeruginosa planktonic cultures and biofilms. Further, we show that acidic pH and acidification via eDNA constitute a signal that is perceived by P. aeruginosa to induce the expression of genes regulated by the PhoPQ and PmrAB two-component regulatory systems. Planktonic P. aeruginosa cultured in exogenous 0.2% DNA or under acidic conditions demonstrates a 2- to 8-fold increase in aminoglycoside resistance. This resistance phenotype requires the aminoarabinose modification of lipid A and the production of spermidine on the bacterial outer membrane, which likely reduce the entry of aminoglycosides. Interestingly, the additions of the basic amino acid l-arginine and sodium bicarbonate neutralize the pH and restore P. aeruginosa susceptibility to aminoglycosides, even in the presence of eDNA. These data illustrate that the accumulation of eDNA in biofilms and infection sites can acidify the local environment and that acidic pH promotes the P. aeruginosa antibiotic resistance phenotype.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic pathogen that causes persistent infections in immunocompromised patients, such as those with cystic fibrosis (CF). The capacity of P. aeruginosa to form a bacterial biofilm is thought to directly promote chronic P. aeruginosa infection of the CF lung. Biofilms consist of communities of bacteria encased in an extracellular matrix that resist host immune clearance or eradication using available antimicrobial therapies. The extracellular biofilm matrix components contribute to the robust resistance exhibited by biofilm communities. Therefore, understanding how biofilm polymers contribute to fostering bacterial resistance to antibiotics is paramount to expanding the treatment options available to patients with chronic biofilm infections.
The biofilm matrix consists predominantly of polysaccharides, proteins, and nucleic acids (1–4). Despite macromolecule heterogeneity, most research has focused on the role of bacterially produced exopolysaccharides (EPSs) in biofilm establishment and maturation. The integral role of extracellular DNA (eDNA) in biofilm formation was first identified in P. aeruginosa (2), but eDNA has since been shown to be a ubiquitous biofilm matrix polymer across most Gram-positive and Gram-negative bacterial species (3, 5–7). In fact, within P. aeruginosa biofilms, eDNA is the most abundant matrix polymer (8, 9).
Extracellular DNA has multiple functions in biofilm formation and stability: it promotes biofilm formation by aiding in initial bacterium-surface adhesion by modulating charge and hydrophobicity interactions between the bacteria and the abiotic surface (10, 11). In addition to mediating cell-substrate interactions, eDNA also influences 3-dimensional biofilm architecture and stability by acting as a cell-cell interaction polymer (2, 3, 12–14). Recent elegant research furthers our understanding of the integral role of eDNA in biofilm organization, where the polymer ensures proper cellular alignment within the biofilm, facilitating efficient movement of bacterial cells to the microcolony periphery and allowing biofilm expansion (15). In addition to adhesion and organization, eDNA has a central role in the structural maturation exhibited by flow cell-cultivated, mushroom-shaped P. aeruginosa microcolonies. Flow cell microcolonies demonstrate that eDNA localizes to the surface and stalk of the microcolonies and when absent arrests microcolony maturation (3, 16).
The origin of eDNA in the matrix of in vitro-grown, single-species biofilms is genomic DNA released from cell lysis, prophage-mediated cell death, quorum sensing (QS) release, or DNA-containing outer membrane vesicles (OMVs) (3, 17–21). However, in human sites of infection, such as the CF lung, the eDNA detected is almost entirely derived from human polymorphonuclear leukocytes (PMNs) that are recruited heavily to quell infection (22). This abundance of neutrophil DNA likely results from a combination of the PMN immune response of NETosis and leukocyte lysis mediated by bacterial virulence factors (23–28).
Given the abundance and importance of eDNA in P. aeruginosa biofilm structures, current research has sought to assess how this matrix polymer influences bacterial persistence in the presence of sustained antibiotic treatment. Initial research highlighted that eDNA derived from CF lung sputum was capable of binding to and sequestering aminoglycosides (29–33). Interestingly, more recent research has illustrated that PMN-derived genomic DNA can be incorporated into P. aeruginosa biofilms and confer increased bacterial resistance to aminoglycoside treatment (16). An unidentified component of the biofilm matrix is required to limit the diffusion of tobramycin to the interior of the microcolony, temporarily protecting the matrix-encapsulated bacteria from exposure to the antibiotic (34).
In addition to acting as a passive shield, eDNA is a highly efficient divalent metal cation chelator, activating the PhoPQ and PmrAB two-component systems (TCS) that detect limiting Mg2+. Cation chelation by eDNA leads the expression of genes controlled by PhoPQ and PmrAB, including the pmr operon (PA3552-PA3559), which is required for the addition of aminoarabinose to the phosphates of the lipopolysaccharide (LPS) lipid A moiety (35–37). DNA also induces expression of the PA4773-PA4774 genes (speDE homologs), which are required to produce spermidine on the outer surface of the bacterial membrane (38). Both eDNA-induced surface modifications function to mask the negative surface charges and are required to limit antimicrobial peptide binding, membrane damage and killing (35, 38).
In this report, we describe a novel function of extracellular DNA in acidifying culture media and biofilm cultures. We also show that acidic pH is an environmental signal that activates the PhoPQ and PmrAB TCS, leading to significant increases in aminoglycoside resistance under acidic conditions. Further, we demonstrate that acidic culture conditions produced by eDNA can be neutralized by l-arginine or sodium bicarbonate to restore aminoglycoside sensitivity, highlighting a possible treatment strategy for acidic sites of chronic infection by P. aeruginosa, including the CF lung.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids employed in this study are listed in Table 1. For the experiments presented, P. aeruginosa was cultured in unbuffered LB or HEPES-buffered BM2 medium (38) at 37°C. Pseudomonas aeruginosa PAO1 and lux-tagged PAO1::p16Slux were used as wild-type (wt) strains. The pH values of all the media used in this study were measured and are shown in Table 2.
TABLE 1.
Strains and plasmid used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strain name or relevant genotype | ||
| PAO1 | Wild-type P. aeruginosa | R. E. Hancock |
| PAO1::p16Slux | PAO1 expressing lux reporter to16S rRNA gene | 64 |
| ΔbmfR | Deletion of bfmR regulator, DNA-overproducing mutant of PAO1 | 44 |
| PA3553::lux | Transcriptional lux fusion due to mini-Tn5-lux insertion in PA3553 (arnC); aminoarabinose modification of lipid A phosphates | 51 |
| PA4773::lux | Transcriptional lux fusion due to mini-Tn5-lux insertion between PA4773 and PA4774 spermidine synthesis genes | 51 |
| oprH-lux | PAO1 with oprH-luxCDABE reporter integrated in the chromosome with CTX | 65 |
| pqsB::lux | Mini-Tn5-lux insertion mutant in pqsB | 51 |
| phoP::xylE | phoP insertion mutant with xylE-Gmr cassette | 66 |
| phoQ::xylE | phoQ insertion mutant with xylE-Gmr cassette | 66 |
| pmrA::xylE | pmrA insertion mutant with xylE-Gmr cassette | 36 |
| pmrB::xylE | pmrB insertion mutant with xylE-Gmr cassette | 36 |
| PA4773::lux, phoP | PA4773::lux fusion in phoP::xylE background | 36 |
| PA4773::lux, phoQ | PA4773::lux fusion in phoQ::xylE background | 36 |
| PA4773::lux, pmrA | PA4773::lux fusion in pmrA::xylE background | 36 |
| PA4773::lux, pmrB | PA4773::lux fusion in pmrB::xylE background | 36 |
| Plasmid p3552-lux | PA3552 promoter-lux transcriptional fusion in broad-host-range pUCP23 plasmid | 36 |
TABLE 2.
Influence of the addition of extracellular DNA on the pH of bacterial growth media
| Medium | pH |
|---|---|
| LB | 7.02 |
| LB + 0.03% DNA | 6.51 |
| LB + 0.06% DNA | 6.4 |
| LB + 0.125% DNA | 6.12 |
| LB + 0.20% DNA | 5.75 |
| LB + 0.25% DNA | 5.52 |
| LB + 0.50% DNA | 4.81 |
| LB + 1.00% DNA | 4.22 |
| BM2 | 7 |
| BM2 + 0.03% DNA | 7.02 |
| BM2 + 0.06% DNA | 6.95 |
| BM2 + 0.125% DNA | 6.91 |
| BM2 + 0.25% DNA | 6.78 |
| BM2 + 0.50% DNA | 6.4 |
| BM2 + 1.00% DNA | 5.73 |
| LB (pH 5.5) | 5.5 |
| LB (pH 5.5) + l-Arg (0.4%) | 8.25 |
| LB + 0.20% DNA + 10 μM Mg2+ | 5.76 |
| LB + 0.20% DNA + 10 μM Mg2+ + 10 mM Tris | 7 |
| LB + 0.20% DNA + 10 mM NaHCO3 | 7.4 |
Gene expression assays.
Gene expression from lux-tagged strains was performed in a high-throughput format using 96-well microplates as previously described (38). Briefly, overnight cultures were grown in LB medium, diluted 1/1,000 into 150 μl of LB medium adjusted to pH 5.5 or pH 7.0 in black, 96-well clear bottom microtiter plates (Thermo Fisher, USA), and overlaid with 75 μl of mineral oil to prevent evaporation. Microplate planktonic cultures were incubated at 37°C in a Wallac Victor3 luminescence plate reader (PerkinElmer, USA), and optical density at 600 nm (OD600; growth) and luminescence (counts per second [CPS]; gene expression) readings were taken every 20 min throughout 18 h of growth. Data are shown from mid-log-phase, 7-h time points.
MIC assays.
P. aeruginosa strains were cultured overnight in LB alone, LB supplemented with 0.2% (wt/vol) salmon sperm DNA (USB, USA; UB14405S2), or acidic LB at pH 5.5. Overnight cultures were normalized and diluted 1,000-fold into fresh LB media of the same culturing conditions (LB alone, supplemented with DNA, or at pH 5.5) in 96-well microtiter trays (Thermo Fisher, USA; 100 μl/well) containing a 2-fold dilution series of antibiotic. Microplates were incubated for 24 h at 37°C to assess the MIC of antibiotic required to inhibit bacterial growth below an OD600 of 0.1. MIC experiments were also performed in LB medium containing 0.2% (wt/vol) salmon sperm DNA and 0.4% (wt/vol) l-arginine (Sigma-Aldrich, USA). Each MIC assay was performed at least three times, and representative values are presented in the tables as specified below.
lux assay of antibiotic killing.
Killing assays were carried out as previously described (35, 39). Briefly, mid-log-phase P. aeruginosa PAO1::p16Slux cultures from LB alone, LB supplemented with 0.2% (wt/vol) salmon sperm DNA, or acidic LB at pH 5.5 were washed and resuspended in LB alone and added to each well of a microtiter plate containing various concentrations of gentamicin or tobramycin (150 μl/well). Bacterial viability was assessed by measuring luminescence (CPS) in the Wallac Victor3 microplate reader every 2 to 5 min for 1 h at room temperature (20°C). Relative survival was calculated by comparing antibiotic-treated PAO1::p16Slux and untreated control bacterial suspensions. Each killing experiment was performed at least five times, and representative curves are shown.
Cultivation, imaging, and analysis of pH in flow cell biofilms.
Biofilms were cultivated in single-channel flow cells and stained with C-SNARF-4 to measure the pH of biofilms as previously described (40). Briefly, P. aeruginosa PAO1 was grown to mid-log phase at 37°C in LB supplemented with 30 μg/ml of gentamicin (OD600 = 0.6) and then syringe inoculated upstream of the sterilized flow chambers. After inoculation, flow was arrested to promote bacterial adhesion for 2 h. Biofilms were cultivated in 3 g/liter of dilute Trypticase soy broth (dTSB) with 5 μg/ml of gentamicin at a flow rate of 0.05 ml/min for 6 days. Once biofilms reached maturity, flow cell inputs were clamped and the flow chambers were injected with dTSB containing 10 μM C-SNARF-4 for biofilm imaging within 15 min of C-SNARF-4 staining. Biofilm microcolonies were visualized with a Leica DMIREB2 inverted microscope equipped with an ORCA-ER digital camera and Openlab software (Improvision) with the 100× objective. Z-stack slices of microcolony structure were captured from the biofilm substratum to the bulk fluid layer above using 0.4-μm increments. Captured images of C-SNARF-4 fluorescence/pixel intensity were analyzed using Quorum Angstrom Optigrid software. The pH of biofilm microenvironments was quantified as previously described (40). We calibrated 10 μM C-SNARF-4 in 1/10 TSB at pHs 5.6, 5.8, 6.0, 6.2, 6.4, 6.6, 6.8, 7.0, and 7.2 to assess the dye fluorescence profile at 488-nm excitation and emission in two channels, 525 nm and 605 nm (Em 525 and Em 605, respectively). This combination of emission channels was used to generate a standard curve of ratios that represent specific pH values as previously described (40). The pH microenvironments in cultivated biofilms were determined from a series of two-channel optical images by calculating the ratio of emission intensities between the two channels (Em 605/Em 525) and comparing this value to the standard curve generated as described above. Each biofilm cultivation experiment was conducted two times, and 10 fields of view, consisting of 15 to 40 Z-stack slices, were acquired under each condition. The C-SNARF-4 fluorescence ratio was analyzed in 10 separate locations of 10 μm2, in positions corresponding to the bulk fluid, biofilm periphery, and interior of the microcolonies. Representative images are presented.
Cultivation, imaging, and analysis of pH in coverslip biofilms.
Biofilms were cultivated on 22-mm by 22-mm glass coverslips (VWR, USA) submerged in 3 ml of LB medium in a six-well plate (Nunc, USA) at 37°C. After 24 h, conditioned medium was replaced, and after 48 and 72 h of incubation, coverslips were rinsed and stained (10 min) in the dark with 10 μM C-SNARF-4 (Life Technologies, USA) and 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI; Life Technologies). After excess stain was removed, coverslip biofilms were visualized with a Leica DMIREB2 inverted microscope equipped with an ORCA-ER digital camera and Openlab software (Improvision) with the 100× objective as described above. The pH of biofilm microenvironments was quantified as described above.
RESULTS
P. aeruginosa biofilms contain acidic microdomains.
Biofilms are bacterial aggregates that contain heterogeneous microenvironments due to the limited diffusion of oxygen, nutrients, and metabolic waste products through the exopolysaccharide and DNA matrix polymer structure (41). Several studies have employed ratiometric dyes and sensors to measure the pH gradients occurring in microenvironments within biofilms (40, 42, 43). Although these studies indicate that P. aeruginosa biofilms contain acidic microdomains, the source and effects of the acidic conditions on P. aeruginosa remain unknown. We hypothesized that the accumulation of extracellular DNA (eDNA) in the biofilm matrix may contribute to the formation of acidic microenvironments. We first quantified how the addition of eDNA alters the pH of common bacterial growth media. Addition of eDNA rapidly acidified nonbuffered LB medium and was still able to acidify HEPES-buffered BM2 defined medium, but to a lesser extent (Table 2). The acidification is likely due to the phosphates on the phosphodiester backbone acting as a source of protons.
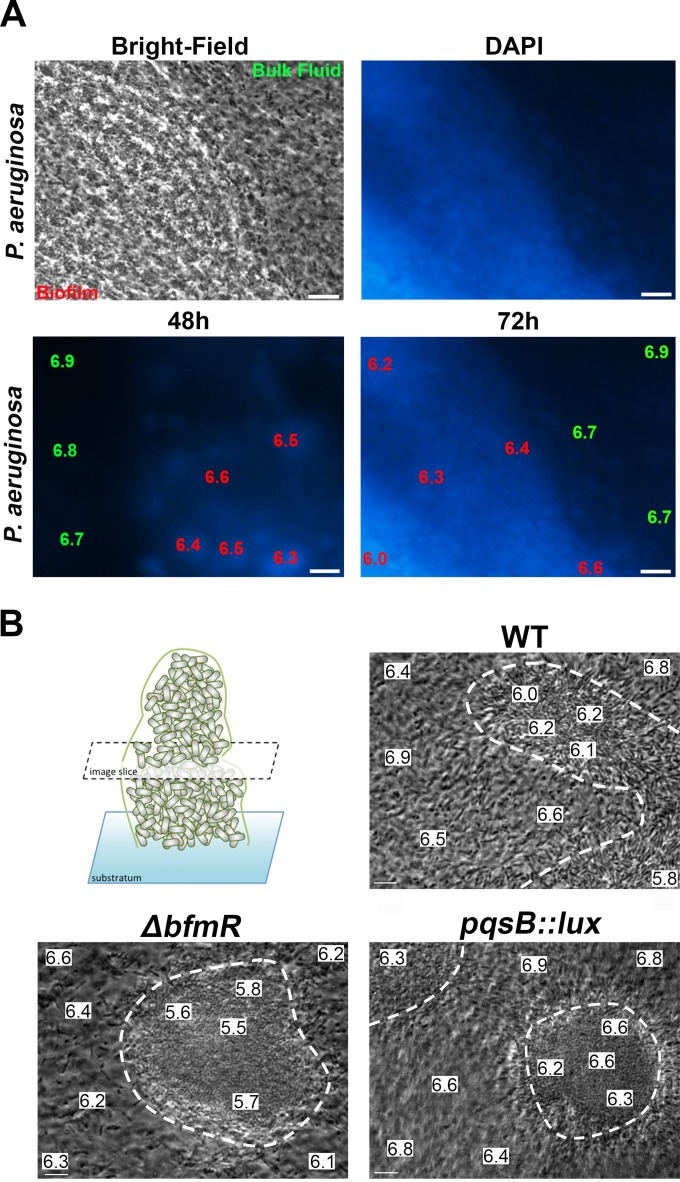
In order to determine whether eDNA influences the acidification of P. aeruginosa biofilms, we assessed the pH of structurally simple, glass slide-adhered biofilms formed by the wild-type PAO1 strain. DAPI stain was used to show the accumulation of eDNA in biofilm aggregates but not in the planktonic cells present in the bulk fluid (Fig. 1A). The biofilm pH was determined using the pH-sensitive ratiometric dye C-SNARF-4 as previously described (40). After 2 days of biofilm cultivation, the DNA-staining biofilm regions attained a reduced pH of 6.3 to 6.5, compared to the neutral pH of 6.7 to 6.9 in the bulk fluid (Fig. 1A). After 3 days of cultivation, the biofilm regions became increasingly acidic and the pH was reduced to 6 (Fig. 1A). Interestingly, supplementation of the growth media with 10 mM HEPES (pH 7) buffer neutralized the interior of the biofilms over the course of 3 days (data not shown). These results demonstrate that DNA-staining regions of biofilms accumulate a reduced pH compared to that of the bulk fluid.
FIG 1.
Visualization of acidic microdomains in glass-adhered and flow chamber-cultivated Pseudomonas aeruginosa biofilms. (A) Bright-field image (left) of DAPI-stained, glass-adhered biofilm (right). pH measurements were made using the ratiometric pH dye C-SNARF in P. aeruginosa wild-type biofilms that were costained with DAPI. Representative micrographs were captured at 48 and 72 h of biofilm cultivation. The data shown are the pH values in a 10-μm2 area of either the bulk fluid, the biofilm periphery, or biofilm interior. Scale bar, 10 μm. (B) Bright-field image slices of P. aeruginosa wild-type, ΔbfmR (eDNA overproducer), and pqsB::lux (eDNA underproducer) microcolonies after 6 days of flow chamber cultivation. pH measurements using C-SNARF in P. aeruginosa wild-type, ΔbfmR, and pqsB::lux microcolonies were determined in the bulk fluid, the biofilm-fluid interface, and biofilm core in the middle depth of the microcolony. The white dotted lines delineate the adhered microcolony edges. Representative micrographs are presented, and data are the pH values within a 10-μm2 area. Scale bar, 10 μm.
To further examine the relationship between eDNA production and biofilm pH, we cultivated flow chamber biofilms of wild-type PAO1, as well as bacterial strains that were previously shown to overproduce or underproduce eDNA as a secreted matrix polymer. The P. aeruginosa bfmR gene encodes an essential regulator of biofilm development, where mutation of this gene leads to biofilm structures with increased bacterial cell lysis and eDNA release (44). Conversely, the Pseudomonas quinolone signal (pqs) operon is part of a biosynthetic pathway involved in Pseudomonas aeruginosa quorum signaling, and mutation of this pathway leads to biofilm structures that possess markedly less eDNA and are more sensitive to detergent-mediated disruption (3). Flow chamber cultivation of these bacterial strains for 6 days allowed us to assess the pH within bacterial microcolonies in a steady state (40). Within the flow chamber biofilms, we determined the pH in distinct regions of the microcolony edges, the microcolony centers, and the bulk fluid regions (Fig. 1B). Wild-type P. aeruginosa PAO1 microcolonies attained a reduced biofilm pH of 6 to 6.2, while the biofilm pH in the ΔbfmR mutant had a more acidic interior of pH 5.5 to 5.8. We also demonstrated that biofilms formed by the pqsB::lux mutant, which releases less eDNA than wild-type cultures, produced microcolonies with a relatively neutral pH, 6.2 to 6.6 (Fig. 1B). In general, the pH values in the bulk fluid and near the periphery of microcolonies tended to remain more neutral than in the interior of biofilm microcolonies, except for the acidic bulk fluid from the ΔbfmR strain, which may be due to the increased release of eDNA (44). Together, these results suggest a process whereby the accumulation of bacterial eDNA contributes to the formation of acidic microenvironments within biofilms.
Exogenous DNA acidifies planktonic cultures to induce P. aeruginosa resistance to aminoglycosides.
It is well known that multiple bacterial species are aminoglycoside resistant when grown under acidic pH conditions (45, 46). Since extracellular DNA acidifies culture media and biofilms (Table 2; Fig. 1), we wanted to decipher the contribution of acidification by eDNA to aminoglycoside resistance. P. aeruginosa cultures supplemented with eDNA (0.2%; 2 mg/ml) resulted in substantially higher MICs for several aminoglycosides (Table 3). The addition of 0.2% DNA reduced the pH of LB medium to ∼5.75. There was no effect of eDNA on the MIC for chloramphenicol or tetracycline, but interestingly, there was a 2- to 8-fold increase in resistance to carbenicillin (Table 3).
TABLE 3.
Acidification and extracellular DNA increases P. aeruginosa resistance to aminoglycosides
| Growth condition | MIC (μg/ml)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gm | Tm | Km | Ak | Sm | Nm | Cm | Tet | Cb | |
| LB (pH 7) | 4 | 1 | 4 | 4 | 50 | 60 | 80 | 20 | 40 |
| LB (pH 5.5) | 32 | 8 | 16 | 16 | 100 | >128 | 80 | 20 | 80 |
| LB + 0.2% DNA | 32 | 8 | 16 | 32 | >200 | >128 | 80 | 20 | 160 |
Gm, gentamicin; Tm, tobramycin; Km, kanamycin; Ak, amikacin; Sm, streptomycin; Nm, neomycin; Cm, chloramphenicol; Tet, tetracycline; Cb, carbenicillin.
P. aeruginosa also demonstrated increased aminoglycoside resistance under acidic (pH 5.5) conditions (Table 3), and there was a gradual increase in the MIC of tobramycin as the pH decreased. The MIC of tobramycin is 1 μg/ml at the neutral pH of 7 and increases to 2, 4, and 8 μg/ml with corresponding decreases in pH to 6.5, 6, and 5.5. To confirm that acidification by eDNA was the cause of increased aminoglycoside resistance, we attempted to neutralize the pH in the presence of eDNA by adding 10 mM Tris (pH 7.0) to the growth media, which partially restored the sensitivity of the aminoglycosides (Table 4). Similarly, quenching the cation chelation effects by addition of excess cations also partially restored aminoglycoside susceptibility (Table 4). A low concentration of magnesium was used (10 μM), since millimolar concentrations of cations antagonize aminoglycoside binding and increase resistance (47). When we combined neutralizing the pH of eDNA to 7 with Tris buffer with the addition of magnesium, aminoglycoside susceptibility was fully restored to the levels observed in the absence of eDNA (Table 4). These results suggest that the cation chelating and acidifying properties of eDNA act in a synergistic fashion to promote resistance of planktonic P. aeruginosa to aminoglycosides.
TABLE 4.
DNA- and pH-induced aminoglycoside resistance in P. aeruginosa PAO1 is blocked by neutralizing the pH
| Medium condition | pH | MIC (μg/ml) |
|
|---|---|---|---|
| Gm | Tm | ||
| eDNA | |||
| LB + DNA (0.2%) | 5.75 | 16 | 16 |
| LB | 7 | 4 | 2 |
| LB + DNA + 10 μM Mg2+ | 5.76 | 8 | 8 |
| LB + DNA + 10 mM Tris | 7.02 | 8 | 4 |
| LB + DNA + 10 μM Mg2+ + 10 mM Tris | 7 | 2 | 2 |
| LB + DNA + l-Arg (0.4%) | 8.25 | 4 | 2 |
| LB + DNA + 10 mM NaHCO3 | 7.4 | 2 | 1 |
| Acid pH | |||
| LB (pH 5.5) | 5.5 | 8 | 8 |
| LB | 7 | 1 | 1 |
| LB (pH 5.5) + l-Arg (0.4%) | 8.25 | 2 | 1 |
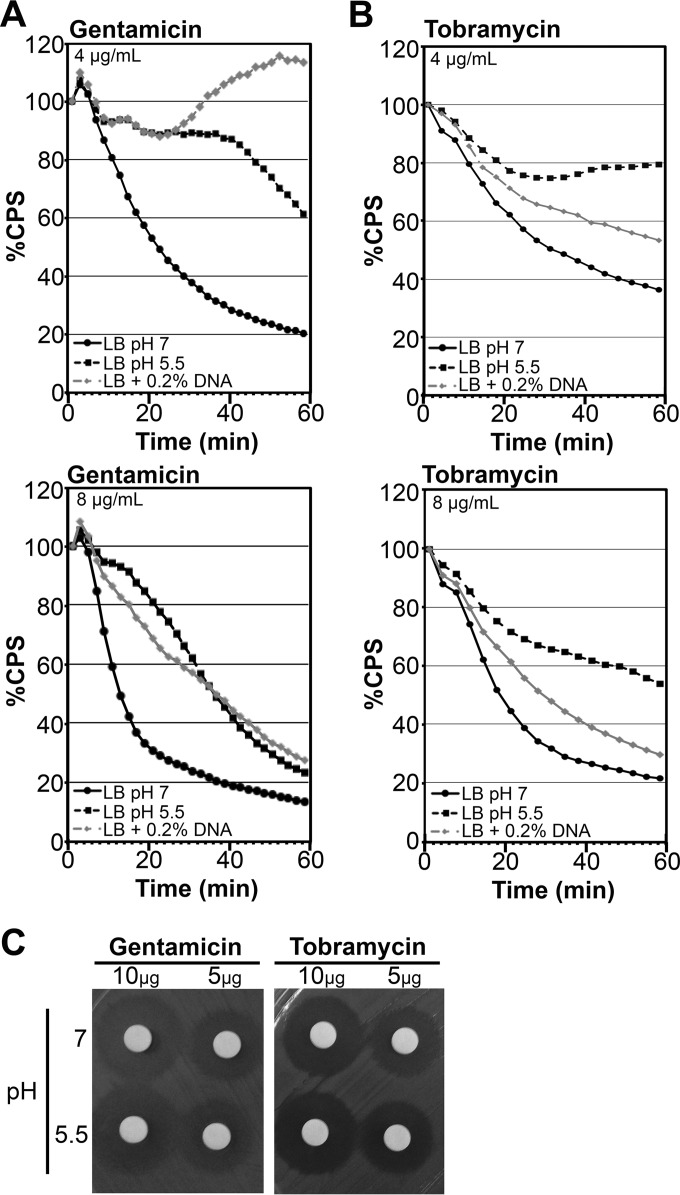
Under neutral pH with the addition of excess magnesium, it does not appear that eDNA has the ability to bind and sequester aminoglycosides (Table 4). However, previous research has highlighted that alginate, extracellular DNA, or other biofilm matrix components can act as a shield that binds to aminoglycosides and limits their penetration into the interior of biofilms (16, 34, 48). To confirm that eDNA was not sequestering aminoglycosides under these conditions, we performed kill curves of P. aeruginosa PAO1::p16Slux cultured in the presence and absence of 0.2% eDNA, resuspended in LB alone, and challenged with gentamicin or tobramycin for 1 h. Aminoglycosides quickly killed bacteria precultured in LB at pH 7, as measured by the rapid loss of luminescence over time (Fig. 2A and B). In contrast, PAO1::p16Slux precultured in the presence of eDNA demonstrated greater tolerance to aminoglycosides, which was also true of cells precultured at acidic pH (5.5) (Fig. 2A and B). Importantly, resistance was due to an active resistance mechanism and not a loss of antibiotic activity, since aminoglycosides incubated in acidic pH 5.5 maintained their antimicrobial activity in the disk diffusion assay (Fig. 2C).
FIG 2.
Growth in the presence of extracellular DNA or under acidic conditions promotes aminoglycoside resistance in Pseudomonas aeruginosa. Mid-log-phase PAO1::p16Slux cells were grown in LB, LB at pH 5.5, or LB plus 0.2% DNA, resuspended in LB (5 × 107 CFU), and treated with various concentrations of gentamicin (A) or tobramycin (B). Luminescence (CPS) was measured as an indication of viability in lux kill-curve experiments throughout 1 h of antibiotic exposure. Cells were resuspended in LB buffer alone as a negative control, and all data are expressed as percent survival relative to the untreated control. Values shown are the means for at least 3 replicates. The standard deviation was within ±10% of the mean. (C) Aminoglycoside antibiotics were resuspended in LB medium with a pH of 7 or 5.5 for 18 h and then spotted at 10 μg/disk or 5 μg/disk and applied to the aminoglycoside-sensitive Escherichia coli in a disk diffusion assay.
Acidic pH induces the expression of protective surface modifications through PhoPQ and PmrAB signaling.
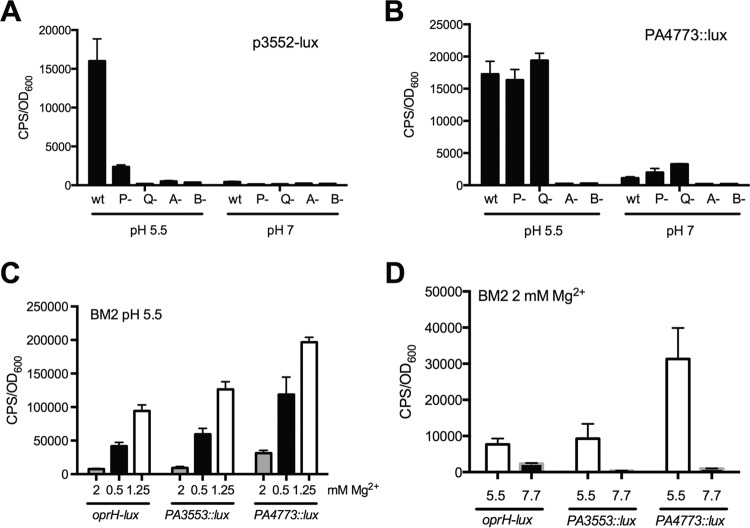
We have previously shown that eDNA binds and sequesters divalent cations, which mimics a Mg2+ limiting environment and strongly induces the expression of PhoPQ- and PmrAB-regulated genes (35, 38, 49). Since the accumulation of eDNA also leads to the acidification of biofilms (Fig. 1), we wanted to determine if acidic pH was itself an environmental condition that activates the PhoPQ and PmrAB systems. We monitored the expression of multiple transcriptional reporters under acidic pH conditions, including the spermidine synthesis PA4773::lux reporter, which is controlled solely by PmrAB (38), and the PA3553::lux reporter of the pmr operon, which is controlled by both PhoPQ and PmrAB (50). We also included an oprH-lux reporter of the outer membrane porin because PhoPQ solely regulates oprH (50). We first tested their expression in defined BM2 medium at neutral pH with limiting concentrations of Mg2+ (125 μM), and all three reporters were strongly induced (Fig. 3A). We also demonstrated that all three reporters of the PhoPQ and PmrAB systems were strongly induced in defined BM2 medium with acidic pH compared to neutral pH (Fig. 3B). We next tested the influence of acidic pH in rich LB medium, and interestingly, only PA3553::lux and PA4773::lux were highly induced (15- to 38-fold), while oprH showed a modest (<2-fold) increase (Fig. 3C). The effect of pH was not limited to pH 5.5, as both PA3553::lux and PA4773::lux reporters showed a progressive gene induction response as the pH was gradually decreased from neutral to 5.5 (Fig. 3D).
FIG 3.
Induction of PhoPQ- and PmrAB-controlled genes under acidic pH conditions. (A) Influence of limiting Mg2+ (125 μM) on the expression of oprH-lux, PA3553::lux, and PA4774::lux compared to that in BM2 defined medium at neutral pH 7 with 2 mM Mg2+. (B) Influence of acidic pH (5.5) on the expression of oprH-lux, PA3553::lux, and PA4774::lux compared to that in BM2 defined medium at neutral pH 7. (C) Influence of mildly acidic pH (5.5) in rich LB medium on the expression of oprH-lux, PA3553::lux, and PA4774::lux. (D) Effects of various pH values ranging between 5.5 and 7 on the expression of PA3553::lux and PA4774::lux. All values shown are the averages and standard deviations from 4 replicates. Gene expression is shown at ∼7 h of growth under the respective conditions.
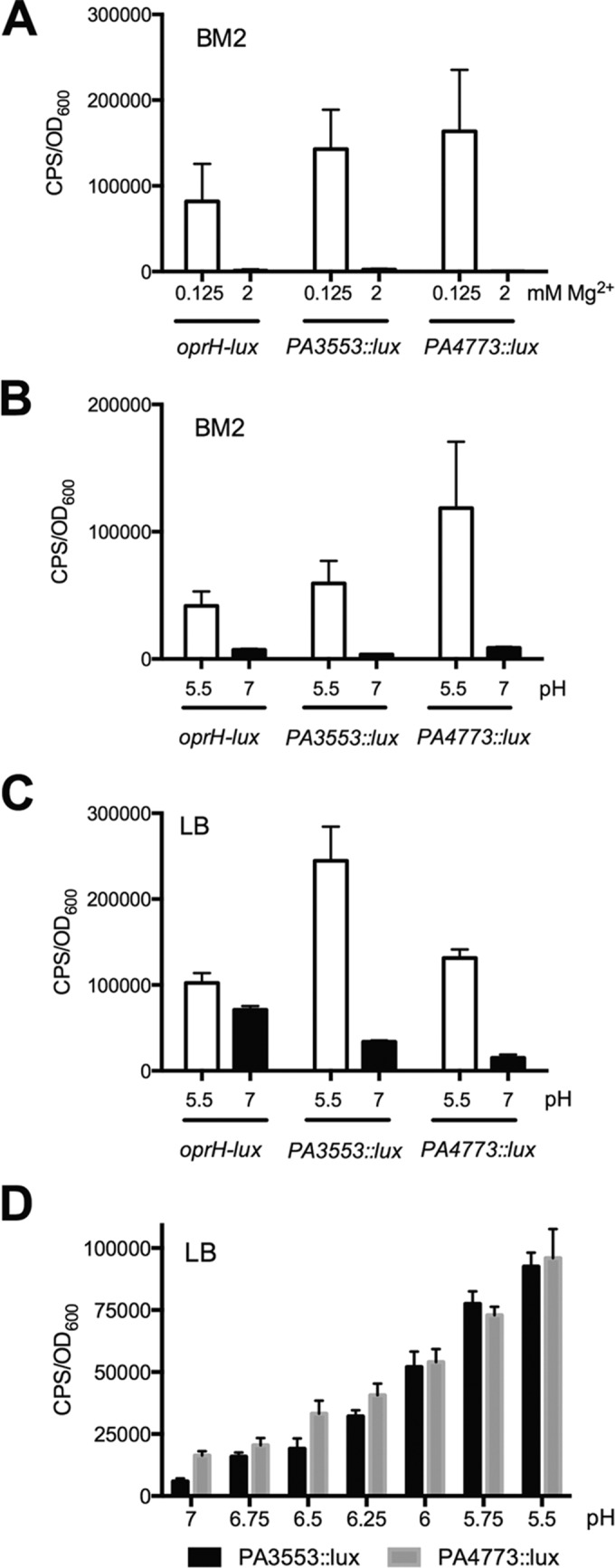
To confirm the ability of PhoPQ or PmrAB to sense acidic (pH 5.5) conditions in P. aeruginosa, we measured the gene expression of a plasmid-encoded PA3552 promoter-lux fusion introduced into wt, phoP::xylE, phoQ::xylE, pmrA::xylE or pmrB::xylE mutant backgrounds (Fig. 4A). The chromosomal PA3553::lux and plasmid pPA3352-lux reporters have similar expression patterns under the control of the promoter in front of PA3552. This analysis revealed that pPA3552-lux expression partially required PhoP but was strictly dependent on PhoQ and PmrAB for expression at pH 5.5 (Fig. 4A). We performed a similar experiment measuring the expression of the spermidine synthesis gene from a chromosomally encoded PA4773::lux fusion, as well as in double mutants in a phoP::xylE, phoQ::xylE, pmrA::xylE, or pmrB::xylE background. PmrAB was exclusively required to induce the spermidine synthesis genes under acidic (pH 5.5) conditions (Fig. 4B).
FIG 4.
Role of PhoPQ and PmrAB in sensing acidic pH growth conditions. Gene expression of a plasmid-encoded pPA3552-lux reporter (A) and the chromosomally encoded PA4773::lux reporter (B) in rich LB medium adjusted to mildly acidic (pH 5.5) and neutral (pH 7) conditions is shown. Expression of both reporters is measured in wild-type and phoP::xylE (P-), phoQ::xylE (Q-), pmrA::xylE (A-), and pmrB::xylE (B-) mutant backgrounds. (C) Gene expression of oprH-lux, PA3553::lux, and PA4773::lux reporters in BM2 defined medium adjusted to pH 5.5 with a range of exogenous Mg2+ concentrations. (D) Despite the presence of repressing levels of 2 mM Mg2+, the oprH-lux, PA3553::lux, and PA4773::lux reporters are induced in BM2 defined medium adjusted to pH 5.5 compared to pH 7. All values are the averages and standard deviations from 4 replicates. Gene expression is shown at ∼7 h of growth under the respective conditions.
Since PhoPQ and PmrAB both respond to limiting cations as well as acidic pH, we wanted to determine if excess cations could suppress the acid-mediated induction of PhoPQ- and PmrAB-controlled genes. Indeed, the addition of increasing concentrations of excess Mg2+ did reduce the overall expression levels in acidic media (Fig. 4C). However, even in the presence of 2 mM Mg2+, which normally represses expression at neutral pH (Fig. 3A), there was still an acid pH-mediated induction of all three reporters of PhoPQ- and PmrAB-regulated genes (Fig. 4D). Collectively, these data indicate that the PhoPQ and PmrAB systems respond to acidic pH by inducing the expression of target genes and that there is complex interplay between the two independent environmental signals of acid pH and the cation concentration in the growth media.
Spermidine and aminoarabinose modifications protect planktonic P. aeruginosa from aminoglycoside killing.
Given our observations that acidic conditions increased P. aeruginosa resistance to aminoglycosides (Tables 3 and 4) and induced the expression of protective bacterial surface modifications (Fig. 3), we hypothesized that resistance to aminoglycosides could be due to a restricted outer membrane permeability mechanism. Therefore, we compared the MICs of wild-type P. aeruginosa and mutants with changes in the aminoarabinose modification of LPS and spermidine synthesis pathways cultured under mildly acidic (pH 5.5) conditions or in the presence of eDNA. Under both conditions, the aminoglycoside MICs were substantially lower for mutants with changes in the aminoarabinose-modified LPS or spermidine synthesis pathways than for the wild-type P. aeruginosa (Table 5). Acidic pH also promoted P. aeruginosa resistance to polymyxin B (Table 5), reinforcing the observation that these modifications protect the outer membrane from antimicrobial peptides in limiting Mg2+ or DNA-rich conditions (49, 51). These data highlight a novel P. aeruginosa response under acidic conditions, where spermidine production and the aminoarabinose-modified LPS contribute to reducing the outer membrane permeability and entry of aminoglycosides.
TABLE 5.
Spermidine and aminoarabinose cell surface modifications are required for P. aeruginosa resistance to aminoglycosides under acidic and DNA-enriched conditions
| Strain or relevant genotype | Growth condition | MIC (μg/ml) |
||
|---|---|---|---|---|
| Gm | Tm | PxnBa | ||
| PAO1 | LB (pH 5.5) | 32 | 16 | 8 |
| PA3553::lux | LB (pH 5.5) | 4 | 4 | 4 |
| PA4774::lux | LB (pH 5.5) | 8 | 4 | 4 |
| PAO1 | LB + DNA (0.2%) | 32 | 16 | 8 |
| PA3553::lux | LB + DNA (0.2%) | <2 | <2 | 2 |
| PA4774::lux | LB + DNA (0.2%) | 2 | 2 | 2 |
PxnB, polymyxin B.
l-Arginine and sodium bicarbonate neutralize the pH and restore P. aeruginosa sensitivity to aminoglycosides.
A previous report demonstrated that medium alkalinization upon addition of the amino acid l-arginine enhanced the effects of aminoglycosides on killing Gram-negative and Gram-positive bacteria in vitro and in vivo during an infection (52). Therefore, to determine whether l-arginine was capable of restoring aminoglycoside susceptibility in the presence of eDNA, we assessed the MIC of P. aeruginosa in the presence of gentamicin and tobramycin. The addition of 0.4% unbuffered l-arginine neutralized the pH and completely restored the aminoglycoside susceptibility of planktonic P. aeruginosa in the presence of eDNA (Table 4). l-Arginine addition also neutralized pH 5.5 conditions and restored sensitivity to gentamicin and tobramycin (Table 4). We also tested the ability of sodium bicarbonate to restore aminoglycoside sensitivity, as sodium bicarbonate production is deficient in CF airways, which results in acidic airway surfaces. Similar to l-arginine, the addition of 10 mM sodium bicarbonate (NaHCO3) neutralized the pH and restored aminoglycoside sensitivity in the presence of eDNA (Table 4). These results demonstrate possible treatment strategies to enhance aminoglycoside killing of Pseudomonas aeruginosa, especially for DNA-rich infection sites, like the CF lung.
DISCUSSION
Biofilm structures are comprised of heterogeneous microenvironments and bacterial growth states due to the limited diffusion of nutrients and oxygen, as well as the accumulation of waste products (41). Previous research illustrated that secreted EPS matrix polymers are responsible for the creation of distinct exopolysaccharide microdomains within biofilms (53). Here, we provide evidence that the accumulation of bacterial eDNA in the extracellular matrix contributes to the generation of acidic microdomains within P. aeruginosa biofilms. Depending on the amount of eDNA released, biofilms can achieve pH values ranging anywhere between pH 5.5 and 6.6 (Fig. 1).
We also demonstrate that acidic conditions act as a signal that is perceived by and activates the P. aeruginosa PhoPQ and PmrAB two-component systems. Previous research indicated that mildly acidic conditions (pH 5.8) did not affect the expression of the P. aeruginosa pmrB::xylE transcriptional reporter (36). However, we now realize that due to the sensor inactivation in the pmrB::xylE reporter strain, this strain was incapable of responding to pH 5.8 (36) (Fig. 4). P. aeruginosa responds to acidic pH similarly to the Salmonella enterica PhoPQ and PmrAB homologs (54–56). It is not yet clear why these Pseudomonas TCS appear to have redundant functions in sensing magnesium limitation and acidic pH, but the Salmonella PhoQ and PmrB sensors are well known to detect multiple independent signals (55, 56). Despite this similarity, these organisms employ distinct pathogenesis strategies as extracellular and intracellular pathogens and therefore encounter acidic conditions in different niches. Salmonella enterica survives transit through the acidic mammalian stomach and tolerates the acidified phagosome of professional phagocytes (54, 55). In contrast, P. aeruginosa likely encounters acidic pH when growing in DNA-rich biofilms and infection sites, such as the CF lung (49, 57, 58).
Acidic pH has long been known to confer resistance to aminoglycosides (45, 46). One possible mechanism indicated that acidic conditions attenuate the inner membrane proton motive force (PMF), thus reducing the active uptake of positively charged antibiotics (45). Our results provide another mechanism involving reduced binding and entry of aminoglycosides across the outer membrane. Aminoglycosides are known to bind to the same negative surface charges as antimicrobial peptides and use the self-promoted uptake pathway to enter Gram-negative cells (47). Therefore, it is not surprising that masking these binding sites with aminoarabinose or spermidine contributes to aminoglycoside resistance. This is the first report linking aminoglycoside resistance to acidic or DNA-rich conditions with the PhoPQ- and PmrAB-controlled surface modifications. The accumulating evidence suggests that aminoarabinose-modified LPS and spermidine production act as a generic shield to counter multiple outer membrane threats, including protection from aminoglycosides and antimicrobial peptides (49) and from the antimicrobial, membrane-targeting action of DNA within neutrophil extracellular traps (NETs) (59).
The CF patient lung can contain up to 20 mg/ml of DNA (2%), which increases sputum viscosity and impedes sputum clearance (22, 32). The importance of eDNA in CF disease progression is underscored by the use of inhaled DNase (Pulmozyme) that reduces sputum viscosity, inflammation, and exacerbations and improves lung function and patient survival (57, 58). CF patient sputum is enriched in eDNA, which is largely derived from neutrophils and, to a lesser extent, bacterial sources (22). Recent evidence suggests that neutrophil extracellular traps are deployed in the CF lung, indicating that DNA accumulation is not solely due to cellular death but is also derived from a NETosis immune response (26–28, 60). Multiple studies have reported that the airway pH, as determined by measuring the pH of exhaled breath condensate, is more acidic in CF patients than control measurements from healthy individuals without CF (61, 62). Other chronic respiratory conditions also result in acidic pH measurements from the lung, including bronchiectasis, chronic obstructive pulmonary disease, and asthma (62, 63). Therefore, the accumulation of eDNA may contribute to acidification of sputum and aminoglycoside resistance in CF patients. An increased understanding of the diverse functions of extracellular DNA helps to explain the antibiotic resistance phenotypes exhibited by biofilms and during chronic CF lung infections. The use of simple pH-buffering agents alongside aminoglycosides may be a novel treatment strategy to combat chronic infection in the acidic, DNA-enriched lungs of cystic fibrosis patients.
ACKNOWLEDGMENTS
We thank Lori Johnson and Matthew Tse for technical assistance and preliminary experiments.
This work was funded by a Cystic Fibrosis Canada operating grant (www.cysticfibrosis.ca). M.W. is supported by a CF Canada Postdoctoral Fellowship.
REFERENCES
- 1.Moreau-Marquis S, Stanton BA, O'Toole GA. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm Pharmacol Ther 21:595–599. doi: 10.1016/j.pupt.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 3.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 4.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, Qu D. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 6.Abee T, Kovács ÁT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol 22:172–179. doi: 10.1016/j.copbio.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 7.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 8.Matsukawa M, Greenberg EP. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol 186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okshevsky M, Meyer RL. 2013. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit Rev Microbiol 41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 10.Busscher HJ, Norde W, van der Mei HC. 2008. Specific molecular recognition and nonspecific contributions to bacterial interaction forces. Appl Environ Microbiol 74:2559–2564. doi: 10.1128/AEM.02839-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. 2010. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol 76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. 2007. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology 153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 13.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 14.Yang L, Nilsson M, Gjermansen M, Givskov M, Tolker-Nielsen T. 2009. Pyoverdine and PQS mediated subpopulation interactions involved in Pseudomonas aeruginosa biofilm formation. Mol Microbiol 74:1380–1392. doi: 10.1111/j.1365-2958.2009.06934.x. [DOI] [PubMed] [Google Scholar]
- 15.Gloag ES, Turnbull L, Huang A, Vallotton P, Wang H, Nolan LM, Mililli L, Hunt C, Lu J, Osvath SR, Monahan LG, Cavaliere R, Charles IG, Wand MP, Gee ML, Prabhakar R, Whitchurch CB. 2013. Self-organization of bacterial biofilms is facilitated by extracellular DNA. Proc Natl Acad Sci U S A 110:11541–11546. doi: 10.1073/pnas.1218898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiang WC, Nilsson M, Jensen PO, Hoiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. 2013. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother 57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberger RE, Holden PA. 2004. Macromolecular composition of unsaturated Pseudomonas aeruginosa biofilms with time and carbon source. Biofilms 1:37–47. doi: 10.1017/S1479050503001066. [DOI] [Google Scholar]
- 18.Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb JS, Lau M, Kjelleberg S. 2004. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol 186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirov SM, Webb JS, O'May CY, Reid DW, Woo JK, Rice SA, Kjelleberg S. 2007. Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 153:3264–3274. doi: 10.1099/mic.0.2007/009092-0. [DOI] [PubMed] [Google Scholar]
- 21.Schooling SR, Beveridge TJ. 2006. Membrane vesicles: an overlooked component of the matrices of biofilms. J Bacteriol 188:5945–5957. doi: 10.1128/JB.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lethem MI, James SL, Marriott C, Burke JF. 1990. The origin of DNA associated with mucus glycoproteins in cystic fibrosis sputum. Eur Respir J 3:19–23. [PubMed] [Google Scholar]
- 23.Walker TS, Tomlin KL, Worthen GS, Poch KR, Lieber JG, Saavedra MT, Fessler MB, Malcolm KC, Vasil ML, Nick JA. 2005. Enhanced Pseudomonas aeruginosa biofilm development mediated by human neutrophils. Infect Immun 73:3693–3701. doi: 10.1128/IAI.73.6.3693-3701.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alhede M, Bjarnsholt T, Jensen PØ, Phipps RK, Moser C, Christophersen L, Christensen LD, van Gennip M, Parsek M, Høiby N, Rasmussen TB, Givskov M. 2009. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155:3500–3508. doi: 10.1099/mic.0.031443-0. [DOI] [PubMed] [Google Scholar]
- 25.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen A, Andersen CB, Givskov M, Tolker-Nielsen T. 2011. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Rep Reg 19:387–391. doi: 10.1111/j.1524-475X.2011.00681.x. [DOI] [PubMed] [Google Scholar]
- 26.Papayannopoulos V, Staab D, Zychlinsky A. 2011. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving DNase therapy. PLoS One 6:e28526. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manzenreiter R, Kienberger F, Marcos V, Schilcher K, Krautgartner WD, Obermayer A, Huml M, Stoiber W, Hector A, Griese M, Hannig M, Studnicka M, Vitkov L, Hartl D. 2012. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J Cyst Fibros 11:84–92. doi: 10.1016/j.jcf.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Dwyer M, Shan Q, D'Ortona S, Maurer R, Mitchell R, Olesen H, Thiel S, Huebner J, Gadjeva M. 2014. Cystic fibrosis sputum DNA has NETosis characteristics and neutrophil extracellular trap release is regulated by macrophage migration-inhibitory factor. J Innate Immun 6:765–779. doi: 10.1159/000363242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis SD, Burns WT. 1978. Effects of sputum from patients with cystic fibrosis on the activity in vitro of 5 antimicrobial drugs on Pseudomonas aeruginosa. Am Rev Respir Dis 117:176–178. [DOI] [PubMed] [Google Scholar]
- 30.Mendelman PM, Smith AL, Levy J, Weber A, Ramsey B, Davis RL. 1985. Aminoglycoside penetration, inactivation, and efficacy in cystic fibrosis sputum. Am Rev Respir Dis 132:761–765. [DOI] [PubMed] [Google Scholar]
- 31.Ramphal R, Lhermitte M, Filliat M, Roussel P. 1988. The binding of anti-Pseudomonal antibiotics to macromolecules from cystic fibrosis sputum. J Antimicrob Chemother 22:483–490. doi: 10.1093/jac/22.4.483. [DOI] [PubMed] [Google Scholar]
- 32.Smith AL, Redding G, Doershuk C, Goldmann D, Gore E, Hilman B, Marks M, Moss R, Ramsey B, Rubio T. 1988. Sputum changes associated with therapy for endobronchial exacerbation in cystic fibrosis. J Pediatr 112:547–554. doi: 10.1016/S0022-3476(88)80165-3. [DOI] [PubMed] [Google Scholar]
- 33.Hunt BE, Weber A, Berger A, Ramsey B, Smith AL. 1995. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob Agents Chemother 39:34–39. doi: 10.1128/AAC.39.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng BS, Zhang W, Harrison JJ, Quach TPSJL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. 2013. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol 15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulcahy H, Charron-Mazenod L, Lewenza S. 2008. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog 4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McPhee JB, Lewenza S, Hancock RE. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 37.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson L, Mulcahy H, Kanevets U, Shi Y, Lewenza S. 2012. Surface-localized spermidine protects the Pseudomonas aeruginosa outer membrane from antibiotic treatment and oxidative stress. J Bacteriol 194:813–826. doi: 10.1128/JB.05230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilpert K, Hancock RE. 2007. Use of luminescent bacteria for rapid screening and characterization of short cationic antimicrobial peptides synthesized on cellulose using peptide array technology. Nat Protoc 2:1652–1660. doi: 10.1038/nprot.2007.203. [DOI] [PubMed] [Google Scholar]
- 40.Hunter RC, Beveridge TJ. 2005. Application of a pH-sensitive fluoroprobe (C-SNARF-4) for pH microenvironment analysis in Pseudomonas aeruginosa biofilms. Appl Environ Microbiol 71:2501–2510. doi: 10.1128/AEM.71.5.2501-2510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 42.de los Ríos A, Wierzchos J, Sancho LG, Ascaso C. 2003. Acid microenvironments in microbial biofilms of antarctic endolithic microecosystems. Environ Microbiol 5:231–237. doi: 10.1046/j.1462-2920.2003.00417.x. [DOI] [PubMed] [Google Scholar]
- 43.Hidalgo G, Burns A, Herz E, Hay AG, Houston PL, Wiesner U, Lion LW. 2009. Functional tomographic fluorescence imaging of pH microenvironments in microbial biofilms by use of silica nanoparticle sensors. Appl Environ Microbiol 75:7426–7435. doi: 10.1128/AEM.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova OE, Schurr JR, Schurr MJ, Sauer K. 2011. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol 81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlessinger D. 1988. Failure of aminoglycoside antibiotics to kill anaerobic, low-pH, and resistant cultures. Clin Microbiol Rev 1:54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gudmundsson A, Erlendsdottir H, Gottfredsson M, Gudmundsson S. 1991. Impact of pH and cationic supplementation on in vitro postantibiotic effect. Antimicrob Agents Chemother 35:2617–2624. doi: 10.1128/AAC.35.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hancock REW. 1984. Alterations in outer membrane permeability. Annu Rev Microbiol 38:237–264. [DOI] [PubMed] [Google Scholar]
- 48.Nichols WW, Dorrington SM, Slack MP, Walmsley HL. 1988. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother 32:518–523. doi: 10.1128/AAC.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewenza S. 2013. Extracellular DNA-induced antimicrobial peptide resistance mechanisms in Pseudomonas aeruginosa. Front Microbiol 4:21. doi: 10.3389/fmicb.2013.00021;. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, Brinkman FS, Hancock RE. 2006. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J Bacteriol 188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FS, Hancock RE. 2005. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res 15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lebeaux D, Chauhan A, Létoffé S, Fischer F, de Reuse H, Beloin C, Ghigo J. 2014. pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J Infect Dis 210:1357–1366. doi: 10.1093/infdis/jiu286. [DOI] [PubMed] [Google Scholar]
- 53.Lawrence JR, Swerhone GD, Kuhlicke U, Neu TR. 2007. In situ evidence for microdomains in the polymer matrix of bacterial microcolonies. Can J Microbiol 53:450–458. doi: 10.1139/W06-146. [DOI] [PubMed] [Google Scholar]
- 54.Bearson BL, Wilson L, Foster JW. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response. J Bacteriol 180:2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perez JC, Groisman EA. 2007. Acid pH activation of the PmrA/PmrB two-component system of Salmonella enterica. Mol Microbiol 63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prost LR, Miller SI. 2008. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Microbiol 10:576–582. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- 57.Jones AP, Wallis C. 2010. Dornase alfa for cystic fibrosis. Cochrane Database Syst Rev 3:CD001127. doi: 10.1002/14651858.CD001127.pub2. [DOI] [PubMed] [Google Scholar]
- 58.Konstan MW, Ratjen F. 2012. Effect of dornase alfa on inflammation and lung function: potential role in the early treatment of cystic fibrosis. J Cyst Fibros 11:78–83. doi: 10.1016/j.jcf.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Halverson TW, Wilton M, Poon KK, Petri B, Lewenza S. 2015. DNA is an antimicrobial component of neutrophil extracellular traps. PLoS Pathog 11(1):e1004593. doi: 10.1371/journal.ppat.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcos V, Zhou Z, Yildirim AO, Bohla A, Hector A, Vitkov L, Wiedenbauer EM, Krautgartner WD, Stoiber W, Belohradsky BH, Rieber N, Kormann M, Koller B, Roscher A, Roos D, Griese M, Eickelberg O, Doring G, Mall MA, Hartl D. 2010. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat Med 16:1018–1023. doi: 10.1038/nm.2209. [DOI] [PubMed] [Google Scholar]
- 61.Tate S, MacGregor G, Davis M, Innes JA, Greening AP. 2002. Airways in cystic fibrosis are acidified: detection by exhaled breath condensate. Thorax 57:926–929. doi: 10.1136/thorax.57.11.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ojoo JC, Mulrennan SA, Kastelik JA, Morice AH, Redington AE. 2005. Exhaled breath condensate pH and exhaled nitric oxide in allergic asthma and in cystic fibrosis. Thorax 60:22–26. doi: 10.1136/thx.2003.017327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kostikas K, Papatheodorou G, Ganas K, Psathakis K, Panagou P, Loukides S. 2002. pH in expired breath condensate of patients with inflammatory airway diseases. Am J Respir Crit Care Med 165:1364–1370. doi: 10.1164/rccm.200111-068OC. [DOI] [PubMed] [Google Scholar]
- 64.Riedel CU, Casey PG, Mulcahy H, O'Gara F, Gahan CG, Hill C. 2007. Construction of p16Slux, a novel vector for improved bioluminescent labeling of Gram-negative bacteria. Appl Environ Microbiol 73:7092–7095. doi: 10.1128/AEM.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. 2008. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog 4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macfarlane EL, Kwasnicka A, Ochs MM, Hancock RE. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol 34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]