Abstract
We employed an endpoint genotyping method to update the prevalence rate of positivity for the TR34/L98H mutation (a 34-bp tandem repeat mutation in the promoter region of the cyp51A gene in combination with a substitution at codon L98) and the TR46/Y121F/T289A mutation (a 46-bp tandem repeat mutation in the promoter region of the cyp51A gene in combination with substitutions at codons Y121 and T289) among clinical Aspergillus fumigatus isolates obtained from different regions of Iran over a recent 5-year period (2010 to 2014). The antifungal activities of itraconazole, voriconazole, and posaconazole against 172 clinical A. fumigatus isolates were investigated using the European Committee on Antimicrobial Susceptibility Testing (EUCAST) broth microdilution method. For the isolates with an azole resistance phenotype, the cyp51A gene and its promoter were amplified and sequenced. In addition, using a LightCycler 480 real-time PCR system, a novel endpoint genotyping analysis method targeting single-nucleotide polymorphisms was evaluated to detect the L98H and Y121F mutations in the cyp51A gene of all isolates. Of the 172 A. fumigatus isolates tested, the MIC values of itraconazole (≥16 mg/liter) and voriconazole (>4 mg/liter) were high for 6 (3.5%). Quantitative analysis of single-nucleotide polymorphisms showed the TR34/L98H mutation in the cyp51A genes of six isolates. No isolates harboring the TR46/Y121F/T289A mutation were detected. DNA sequencing of the cyp51A gene confirmed the results of the novel endpoint genotyping method. By microsatellite typing, all of the azole-resistant isolates had genotypes different from those previously recovered from Iran and from the Dutch TR34/L98H controls. In conclusion, there was not a significant increase in the prevalence of azole-resistant A. fumigatus isolates harboring the TR34/L98H resistance mechanism among isolates recovered over a recent 5-year period (2010 to 2014) in Iran. A quantitative assay detecting a single-nucleotide polymorphism in the cyp51A gene of A. fumigatus is a reliable tool for the rapid screening and monitoring of TR34/L98H- and TR46/Y121F/T289A-positive isolates and can easily be incorporated into clinical mycology algorithms.
INTRODUCTION
Azole resistance in Aspergillus fumigatus is a global and evolving public health threat which translates into treatment failure (1). Surveillance studies indicate that the incidence of azole resistance is increasing (2–6), with the TR34/L98H mutation (a 34-bp tandem repeat mutation in the promoter region of the cyp51A gene in combination with a substitution at codon L98) emerging in multiple European countries and in the Middle East, Asia, and Africa and with a new resistance mechanism, the TR46/Y121F/T289A mutation (a 46-bp tandem repeat mutation in the promoter region of the cyp51A gene in combination with substitutions at codons Y121 and T289), emerging more recently in Europe and India (2–6). We also previously reported the occurrence of the TR34/L98H mutation in 3.2% of clinical Aspergillus fumigatus isolates obtained from patients in Iran to the end of 2009 (5).
The trend of increases in the rates of azole resistance among A. fumigatus isolates in different regions and patient groups exemplifies the fact that knowledge of the (local) epidemiology of azole-resistant Aspergillus diseases is important for clinical mycology/microbiology reference laboratories (7–9). Moreover, rapid and specific molecular methods for the identification of the recently identified azole-resistant A. fumigatus strains can significantly influence a timely decision on patient management (10).
In our search for a novel, rapid, sensitive, accurate, and high-throughput method for detection and screening of azole resistance in A. fumigatus, we found that endpoint genotyping targeting a single-nucleotide polymorphism (SNP) in the cyp51A gene could provide an option. The quantitative analysis of SNPs has been a reliable method in diagnostic microbiology for identification of a single nucleotide in the genomes of humans (11–15), viruses (16–20), and bacteria (18). In this assay, an extension probe can be simply designed to anneal to the template in a position that places the mutation site immediately adjacent to the 3′ end of the probe, and the use of dideoxynucleoside triphosphates (ddNTPs) allows the extension of only 1 nucleotide from the 3′ end of the probe. Labeling of each ddNTP with a different fluorescent dye allows the differentiation of the genotype at the SNP by the color of the extended probes (11–20).
In the current study, we therefore evaluated the prevalence of TR34/L98H- and TR46/Y121F/T289A-positive isolates among clinical Aspergillus fumigatus isolates obtained from patients with Aspergillus diseases in Iran over a recent 5-year period (2010 to 2014), using PCR sequencing and the novel endpoint genotyping assay targeting SNPs in the cyp51A gene of A. fumigatus.
MATERIALS AND METHODS
Fungal isolates.
One hundred seventy-two clinical A. fumigatus isolates obtained from 142 patients with Aspergillus diseases were investigated. These patients included 88 patients with chronic pulmonary aspergillosis (CPA; 61.97%), 23 patients with allergic bronchopulmonary aspergillosis (ABPA; 16.19%), 20 patients with aspergilloma (14.08%), and 11 patients with invasive pulmonary aspergillosis (7.75%). Patient-related data were collected in accordance with the applicable rules concerning the review of research ethics committees at the Tehran University of Medical Sciences, and informed consent was obtained from all patients. The isolates were stored in 10% glycerol broth at −80°C at the Tehran University Mycology Reference Centre in Iran (Tables 1 and 2).
TABLE 1.
Distribution of azole-resistant and azole-susceptible (wild-type) A. fumigatus isolates examined in this study according to year of isolation
| Yr of isolation | No. of isolates with each phenotype and resistance mechanism |
||
|---|---|---|---|
| Wild type | Resistant TR34/L98H mutant | Resistant TR46/Y121F/T289A mutant | |
| 2010 | 24 | 1 | |
| 2011 | 35 | 1 | |
| 2012 | 37 | 1 | |
| 2013 | 38 | 2 | |
| 2014 | 32 | 1 | |
| Total | 166 | 6 | 0 |
TABLE 2.
Underlying disease and in vitro susceptibilities of six clinical Aspergillus fumigatus isolates that grew on the 4-well platesa
| Azole-resistant Aspergillus fumigatus isolate | Underlying diseaseb | MIC (mg/liter) |
|||
|---|---|---|---|---|---|
| Amphothericin B | Itraconazole | Voriconazole | Posaconazole | ||
| T-IR-AF 1002 | CPA | 0.5 | ≥16 | 4.0 | 0.25 |
| T-IR-AF 1088 | CPA | 0.5 | ≥16 | 2.0 | 0.5 |
| T-IR-AF 1143 | CPA | 0.5 | ≥16 | 8.0 | 0.5 |
| T-IR-AF 1416 | CPA | 0.5 | ≥16 | 8.0 | 0.5 |
| T-IR-AF 1499 | ABPA | 0.5 | ≥16 | 4.0 | 0.5 |
| T-IR-AF 1521 | CPA | 0.5 | ≥16 | 8.0 | 0.5 |
All isolates were positive for the 34-bp tandem repeat in the promoter region of the cyp51A gene and the L98H amino acid substitution (nucleotides are numbered from the translation start codon ATG of cyp51A) in the cyp51A gene, and all patients had previously been exposed to azoles.
CPA, chronic pulmonary aspergillosis; ABPA, allergic bronchopulmonary aspergillosis.
The isolates were submitted to various fungus culture collections across Iran over the last 5 years (2010 to 2014) for species identification and antifungal susceptibility testing and were then submitted to the Mycology Reference Centre at the School of Hygiene & Institute of Public Health Research, Tehran University of Medical Sciences, Tehran, Iran. The isolates were subcultured on Sabouraud dextrose agar (SDA) supplemented with 0.02% chloramphenicol for 5 days at 35 to 37°C. The isolates were originally identified by experienced technicians on the basis of macroscopic colony morphology, the microscopic morphology of the conidia and conidium-forming structures, and the ability to grow at 48°C, and their identities were further confirmed by sequence-based analysis of parts of the β-tubulin and calmodulin genes, as described previously (21, 22). All isolates were plated onto a four-well agar plate containing one well each with 4 mg/liter of itraconazole, 1 mg/liter of voriconazole, and 0.5 mg/liter of posaconazole and a growth control well (23). The ability to grow on each well was assessed after 48 h. Any isolate that was able to grow on one of the azole-containing media was further investigated by antifungal susceptibility testing, PCR sequencing of the cyp51A gene, and the novel SNP endpoint genotyping technique.
In addition, a collection of wild-type and azole-resistant A. fumigatus strains (10 wild-type strains, 8 strains positive for the TR34/L98H mutation, and 6 strains positive for the TR46/Y121F/T289A mutation) were obtained from the culture collection of Radboud University Medical Centre, Nijmegen, the Netherlands. The genomic DNAs of these isolates were used as negative and positive controls for amplification and detection of the L98H and Y121F mutations by the novel quantitative PCR assays that we developed.
In vitro antifungal susceptibility testing.
In vitro antifungal susceptibility testing was performed using a broth microdilution method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines (24). Itraconazole, voriconazole, posaconazole, and amphotericin B were assayed over a 2-fold concentration range of from 16 to 0.016 mg/liter. Visual readings were performed with a reading mirror, and the MIC was defined as the lowest antifungal concentration that inhibited growth by 100% after 48 h compared with the growth of the drug-free well. Susceptibility tests were performed three times with each strain on different days. Paecilomyces variotii (ATCC 22319), Candida parapsilosis (ATCC 22019), and Candida krusei (ATCC 6258) were used for quality control in all experiments. The EUCAST breakpoints and epidemiological cutoff (ECOFF) values were used for the interpretation of the in vitro drug susceptibility testing results (25).
DNA extraction.
DNA was isolated as described previously (26); in brief, the isolates were cultured on Sabouraud dextrose agar slants. Conidia were harvested and added to 200 μl of breaking buffer (100 mM NaCl, 10 mM Tris-HCl, pH 8, 2% Triton X-100, 1% sodium dodecyl sulfate, 1 mM EDTA, pH 8) with ∼0.1-g glass beads (diameters, 0.4 to 0.6 mm). After shaking by vortexing, the conidia were incubated at 70°C for 30 min while they were shaken. Then, 200 μl of phenol-chloroform-isoamyl alcohol (25:24:1) saturated with pH 8.0 aqueous buffer was added, and the samples were incubated for 5 min while they were shaken. After centrifugation for 5 min, the upper phase containing the DNA was transferred to a new tube. One microliter of DNA was used per PCR mixture.
Strain identification and cyp51A sequence analysis.
All isolates were identified using sequence-based analysis of the calmodulin and β-tubulin genes, as described previously (21, 22). The sequence of the promoter region and the full coding sequence of the cyp51A gene were determined by amplification and subsequent sequencing as described previously (26–28). To detect mutations, the sequences were compared with the cyp51A gene sequence with GenBank accession number AF338659 (29).
Endpoint genotyping.
Using a LightCycler 480 real-time PCR system, a novel endpoint genotyping analysis method was evaluated to detect the L98H and Y121F mutations in all of the 172 clinical A. fumigatus isolates, as described previously (J. Zoll, S. Seyedmousavi, W. J. Melchers, and P. E. Verweij, submitted for publication). The assay is based on the competition during annealing between probes detecting the wild type and the mutants. The use of locked nucleic acid (LNA) residues at the SNP and adjacent positions increases the discriminative properties of the probes. A fragment of the A. fumigatus cyp51A gene covering both the L98 and Y121 codons was amplified in the presence of TaqMan probes detecting L98, L98H, Y121, and Y121F. The primer and probe sequences used in the current study are shown in Table 3.
TABLE 3.
Sequences of primers and probes used for detection of L98H and Y121F mutations in cyp51A gene of Aspergillus fumigatus
| Assay | Primer or probe | Sequence (5′–3′)a |
|---|---|---|
| cyp51A amplification | Forward primer | GGCGTTCAGGGGAACGAG |
| cyp51A amplification | Reverse primer | CTTGATGAACTTTTTCTGCTCCATCAG |
| cyp51A L98 detection | L98 probe | 6FAM-AACGGCAAG+C+T+CAAGGATGTC-BBQ |
| cyp51A L98H detection | L98H probe | Cy5-CAACGGCAAG+C+A+CAAGGATGTCA-BBQ |
| cyp51A Y121 detection | Y121 probe | LC610-TTGGGACAATC+A+T+ACACCACGTCCG-BBQ |
| cyp51A Y121F detection | Y121F probe | 6HEX-TTGGGACAATC+A+A+ACACCACGTCCG-BBQ |
The following dyes were used as 5′fluorophores: 6-carboxyfluorescein (6FAM), LightCycler Red 610 dye (LC610), cyanine 5 (Cy5), and 3′-quencher BlackBerry quencher (BBQ). +X (where X indicates any nucleotide), an LNA residue.
The real-time PCR was performed using a Roche LC480 instrument II. The PCR mixture formulation was 350 nM either forward or reverse primer, 250 nM the TaqMan probe, and 1 μl sample DNA in the Roche LC480 probe master mix according the manufacturer's protocol. Thermal cycling was performed with an initial decontamination program for 10 min at 40°C, followed by hot-start activation and initial DNA denaturation for 10 min at 94°C. Template DNA was amplified in a two-step cycling program of 45 cycles consisting of denaturation for 10 s at 94°C and annealing and extension for 1 min at 60°C. A series of control samples was analyzed in parallel. Control samples consisted of DNA extracted from A. fumigatus with wild-type, TR34-L98H, and TR46-Y121F/T289A cyp51A. The cyp51A genotype was determined on the basis of the fluorescence ratios of the discriminative probes.
Microsatellite genotyping.
Genotyping was performed on all A. fumigatus isolates for which the MIC of itraconazole was ≥16 mg/liter, using an A. fumigatus short tandem repeat (STR) assay, as described previously (29–31). Briefly, six loci consisting of three trinucleotide repeat fragments (A. fumigatus STRs 3A, 3B, and 3C) and three tetranucleotide repeat fragments (A. fumigatus STRs 4A, 4B, and 4C) were amplified by using fluorescently labeled primers (29–31). The sizes of the fragments were determined by addition of the GeneScan LIZ500 marker and subsequent analysis of the fragments on an Applied Biosystems 3730 DNA analyzer. Assignment of repeat numbers in each marker was determined from the GeneScan data by using Peak Scanner (version 1.0) software (Applied Biosystems). The sizes of the fragments were determined on the basis of the size of the LIZ500 marker, and the repeat numbers of these isolates were compared to those of a collection of 20 Dutch TR34/L98H-positive isolates. Allele-sharing distance matrices were generated from the tandem repeat numbers and were used as input into the Neighbor program of the PHYLIP (version 3.6) software package to produce dendrograms (32–34).
RESULTS
Prevalence of azole-resistant A. fumigatus isolates in Iran from 2010 to 2014.
The global distribution of azole-resistant and azole-susceptible (wild-type) A. fumigatus isolates examined in this study is shown in Table 1 according to the year of isolation. All isolates were identified to be A. fumigatus by sequence analysis of the ITS1, ITS2, and β-tubulin genes. Of the 172 A. fumigatus isolates, 6 isolates (recovered from separate patients) grew on the wells containing itraconazole and voriconazole, indicating a prevalence of 3.5%.
Antifungal resistance phenotypic analysis.
Table 2 shows the underlying disease of the patients and the in vitro susceptibilities of six clinical A. fumigatus isolates that grew on the 4-well plates. All six isolates showed a multiresistant phenotype, and the MICs of itraconazole (≥16 mg/liter) and voriconazole (≥2 mg/liter) for these isolates were high. Five of these isolates were recovered from patients with chronic pulmonary aspergillosis (CPA), and one was from a patient with allergic bronchopulmonary aspergillosis (ABPA).
Resistance mechanism.
As shown in Fig. 1, quantitative analysis of the single-nucleotide polymorphisms showed the presence of the TR34/L98H mutation in the cyp51A gene of the 6 out of the 172 A. fumigatus isolates for which the MICs of itraconazole and voriconazole were elevated. However, no A. fumigatus isolates harboring the TR46/Y121F/T289A mutation were detected. Sequence analysis of the cyp51A gene confirmed the presence of the TR34/L98H mutation in those 6 isolates, yet no other polymorphisms were identified in any of the 172 isolates tested.
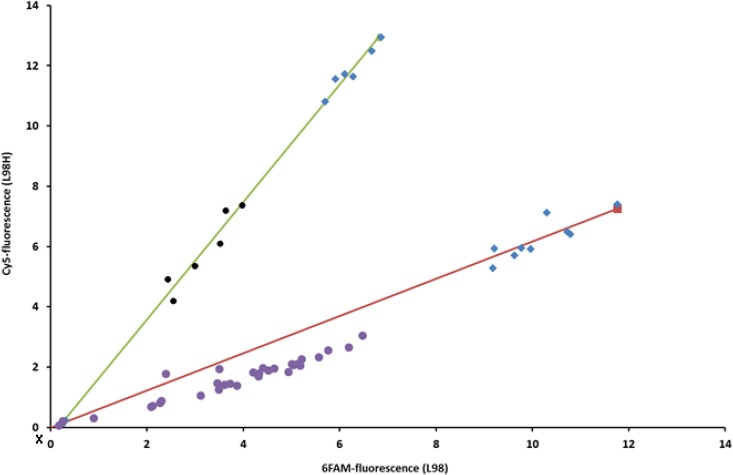
FIG 1.
Endpoint fluorescence plot of single-nucleotide variance for detection of the L98H mutation in clinical Aspergillus fumigatus isolates using a quantitative PCR assay. Relative L98 (6-carboxyfluorescein [6FAM]) and L98H (cyanine 5 [Cy5]) fluorescence levels are plotted on the y and x axes, respectively. Blue diamonds, control isolates; purple circles, clinical A. fumigatus isolates without a mutation in the cyp51A gene at L98; black circles, clinical A. fumigatus isolates harboring the L98H substitution in the cyp51A gene; X, nuclease-free water, which was used as a negative control.
Genotypic analysis.
Microsatellite typing of six STR loci showed identical patterns for two out of the six azole-resistant isolates, proving that the two isolates had a similar phylogenetic origin and, possibly, the same origin. Of note, the two patients from whom these isolates were recovered lived in the same geographical area. Comparison of genetic relatedness by the generation of dendrograms of the STR profiles showed that the 6 Iranian clinical isolates clustered apart from the 20 Dutch TR34/L98H control isolates and those previously isolated in Iran between 2003 and 2009 (5).
DISCUSSION
In the present study, we found a 3.5% prevalence of resistance to triazoles resulting from the TR34/L98H mutation in A. fumigatus isolates obtained from patients with underlying Aspergillus disease in Iran over a recent 5-year period (2010 to 2014). There was not a significant increase in the prevalence of azole-resistant A. fumigatus harboring the TR34/L98H mutation. Of note, five out of six azole-resistant isolates were recovered from CPA patients. The significant predilection to CPA is in agreement with the findings of recent studies in Iran which demonstrated that CPA is the most common clinical presentation of aspergillosis in individuals with healed tuberculosis (35). Importantly, patients with CPA require long-term maintenance antifungal therapy to improve symptoms and prevent the recurrence of infection (36, 37).
The acquisition of azole resistance in A. fumigatus is an emerging public health problem which definitely needs continued surveillance at the national and international levels (9). The main molecular mechanism of azole resistance in A. fumigatus is explained by several mutations in the cyp51A gene (38). Two common genetic variants associated with resistance to azoles are the TR34/L98H mutation and the TR46/Y121F/T289A mutation (1, 2). Both mutations are predominantly found in the environment, show a strong tendency to migrate, and have now been reported in many countries from several continents (3, 23, 40–42). In addition, the clinical implications of infection due to A. fumigatus isolates harboring these mutations are significant, as they cause both diagnostic challenges and azole treatment failure (8, 43).
Since effective monitoring of azole resistance requires effective detection methods, rapid diagnostic tools are warranted to obtain a better understanding of the scale of this emerging problem and to detect the emergence of new resistance mechanisms early (7, 8). In the current study, we employed for the first time a rapid and simple one-step endpoint genotyping quantitative PCR assay (11–20) to detect the L98H and Y121F mutations in TR34/L98H- and TR46/Y121F/T289A-positive azole-resistant A. fumigatus isolates. Interestingly, all of the A. fumigatus isolates in which the L98H mutation was confirmed by PCR sequencing of the cyp51A gene were correctly found to be mutated from the endpoint fluorescence plots (Fig. 1). The quantitative SNP assay used in the current study is based on the competition between probes detecting the wild type and the mutants (11, 16–18, 44). Endpoint measurements of the fluorescent signal for the mutant probe versus that for the wild-type probe were used for target detection and SNP discrimination (16–20). Importantly, this is a rapid method that is technically simple to perform and can easily be employed in clinical diagnostic laboratories.
Of note, molecular techniques are a promising tool to rapidly provide information about the azole resistance genotype of A. fumigatus isolates. Mellado et al. used PCR amplification of the cyp51A region followed by DNA sequencing (45); PCR assays were performed using primers generated from the unique sequence of the A. fumigatus cyp51A gene, and the A. fumigatus cyp51A gene was further evaluated by consecutive DNA sequence analysis to detect and identify point or tandem repeat mutations (45). Using real-time quantitative PCR, Klaassen et al. applied a mixed-format assay and analyzed the melting curves obtained with specific probe primers to detect clinically related mutations at positions Gly54, Leu98, Gly138, and Met220 of the cyp51A gene of A. fumigatus (38). The L98H and TR34 mutations have also been detected using specific PCR assays targeting each mutation (46), a nested PCR assay followed by DNA sequencing (47), and a PCR-restriction fragment length polymorphism (RFLP) assay (39). In addition, PCR-based assays were also tested to detect cyp51A gene mutations directly in clinical samples (48, 49). Moreover, two commercial multiplex real-time PCR assays which are able to differentiate susceptible from resistant A. fumigatus strains and identify various mutations of the cyp51A gene directly in serum and bronchoalveolar lavage fluid samples were recently introduced (50, 51).
In conclusion, our data show that there was not a significant increase in the prevalence of azole-resistant A. fumigatus isolates harboring the TR34/L98H resistance mechanism over a recent 5-year period in Iran. The quantitative assay detecting a single-nucleotide polymorphism in the cyp51A gene of A. fumigatus is a powerful surveillance method with high epidemiological and clinical relevance to determine whether A. fumigatus isolates have acquired the TR34/L98H and or TR46/Y121F/T289A mutations and can easily be incorporated into clinical mycology algorithms.
ACKNOWLEDGMENTS
S.S. has received research and travel grants from Astellas Pharma B.V. and a travel grant from Gilead Sciences. P.E.V. has served as a consultant and has received research grants from Astellas, Basilea, Gilead Sciences, Merck, and Pfizer. None of the other authors has a conflict of interest.
This publication was prepared as a collaborative study between the Department of Medical Mycology and Parasitology, School of Hygiene & Institute of Public Health Research, Tehran University of Medical Sciences, Tehran, Iran (research project fund no. 93-02-27-23062), and the Department of Medical Microbiology, Radboud University Medical Centre, Nijmegen, The Netherlands.
REFERENCES
- 1.Seyedmousavi S, Verweij P. 2015. Azole resistance in Aspergillus fumigatus: mechanisms, route of resistance selection, and clinical implications, p 1–17. In Gotte M, Berghuis A, Matlashewski G, Wainberg M, Sheppard D (ed), Handbook of antimicrobial resistance. Springer, New York, NY. doi: 10.1007/978-1-4939-0667-3_22-1. [DOI] [Google Scholar]
- 2.Verweij PE, Chowdhary A, Melchers WJG, Meis JF. 20 October 2015. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiederhold NP, Gil VG, Gutierrez F, Lindner JR, Albataineh MT, McCarthy DI, Sanders C, Fan H, Fothergill AW, Sutton DA. 21 October 2015. First detection of TR34/L98H and TR46 Y121F T289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. J Clin Microbiol. doi: 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhary A, Sharma C, van den Boom M, Yntema JB, Hagen F, Verweij PE, Meis JF. 2014. Multi-azole-resistant Aspergillus fumigatus in the environment in Tanzania. J Antimicrob Chemother 69:2979–2983. doi: 10.1093/jac/dku259. [DOI] [PubMed] [Google Scholar]
- 5.Seyedmousavi S, Hashemi SJ, Zibafar E, Zoll J, Hedayati MT, Mouton JW, Melchers WJ, Verweij PE. 2013. Azole-resistant Aspergillus fumigatus, Iran. Emerg Infect Dis 19:832–834. doi: 10.3201/eid1905.130075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denning DW, Perlin DS. 2011. Azole resistance in Aspergillus: a growing public health menace. Future Microbiol 6:1229–1232. doi: 10.2217/fmb.11.118. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen E, Lagrou K, Verweij PE. 2013. Azole resistance in Aspergillus fumigatus: a growing public health concern. Curr Opin Infect Dis 26:493–500. doi: 10.1097/QCO.0000000000000005. [DOI] [PubMed] [Google Scholar]
- 8.Seyedmousavi S, Mouton JW, Melchers WJ, Bruggemann RJ, Verweij PE. 2014. The role of azoles in the management of azole-resistant aspergillosis: from the bench to the bedside. Drug Resist Updat 17:37–50. doi: 10.1016/j.drup.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Warris A. 2015. Azole-resistant aspergillosis. J Infect 71(Suppl 1):S121–S125. doi: 10.1016/j.jinf.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Perlin DS. 2009. Antifungal drug resistance: do molecular methods provide a way forward? Curr Opin Infect Dis 22:568–573. doi: 10.1097/QCO.0b013e3283321ce5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst CD, Zuiverloon TC, Hafner C, Zwarthoff EC, Knowles MA. 2009. A SNaPshot assay for the rapid and simple detection of four common hotspot codon mutations in the PIK3CA gene. BMC Res Notes 2:66. doi: 10.1186/1756-0500-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budowle SA, Gonzalez S, Budowle B, Eisenberg AJ, Grange RW. 2008. A novel SNaPshot assay to detect the mdx mutation. Muscle Nerve 37:731–735. doi: 10.1002/mus.21027. [DOI] [PubMed] [Google Scholar]
- 13.Li L, Li CJ, Zhang YJ, Zheng L, Jiang HX, Si-Tu B. 2011. Simultaneous detection of CYP3A5 and MDR1 polymorphisms based on the SNaPshot assay. Clin Biochem 44:418–422. doi: 10.1016/j.clinbiochem.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Lou C, Cong B, Li S, Fu L, Zhang X, Feng T, Su S, Ma C, Yu F, Ye J, Pei L. 2011. A SNaPshot assay for genotyping 44 individual identification single nucleotide polymorphisms. Electrophoresis 32:368–378. doi: 10.1002/elps.201000426. [DOI] [PubMed] [Google Scholar]
- 15.Murphy KM, Geiger T, Hafez MJ, Eshleman JR, Griffin CA, Berg KD. 2003. A single nucleotide primer extension assay to detect the APC I1307K gene variant. J Mol Diagn 5:222–226. doi: 10.1016/S1525-1578(10)60477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan S, Boltz DA, Li J, Oshansky CM, Marjuki H, Barman S, Webby RJ, Webster RG, Govorkova EA. 2011. Novel genotyping and quantitative analysis of neuraminidase inhibitor resistance-associated mutations in influenza A viruses by single-nucleotide polymorphism analysis. Antimicrob Agents Chemother 55:4718–4727. doi: 10.1128/AAC.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolotin S, Robertson AV, Eshaghi A, De Lima C, Lombos E, Chong-King E, Burton L, Mazzulli T, Drews SJ. 2009. Development of a novel real-time reverse-transcriptase PCR method for the detection of H275Y positive influenza A H1N1 isolates. J Virol Methods 158:190–194. doi: 10.1016/j.jviromet.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Vries E, Jonges M, Herfst S, Maaskant J, Van der Linden A, Guldemeester J, Aron GI, Bestebroer TM, Koopmans M, Meijer A, Fouchier RA, Osterhaus AD, Boucher CA, Schutten M. 2010. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J Clin Virol 47:34–37. doi: 10.1016/j.jcv.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakauchi M, Ujike M, Obuchi M, Takashita E, Takayama I, Ejima M, Oba K, Konomi N, Odagiri T, Tashiro M, Kageyama T, Influenza Virus Surveillance Group of Japan. 2011. Rapid discrimination of oseltamivir-resistant 275Y and -susceptible 275H substitutions in the neuraminidase gene of pandemic influenza A/H1N1 2009 virus by duplex one-step RT-PCR assay. J Med Virol 83:1121–1127. doi: 10.1002/jmv.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takayama I, Nakauchi M, Fujisaki S, Odagiri T, Tashiro M, Kageyama T. 2013. Rapid detection of the S247N neuraminidase mutation in influenza A(H1N1)pdm09 virus by one-step duplex RT-PCR assay. J Virol Methods 188:73–75. doi: 10.1016/j.jviromet.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2008. Aspergillus section Fumigati: antifungal susceptibility patterns and sequence-based identification. Antimicrob Agents Chemother 52:1244–1251. doi: 10.1128/AAC.00942-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balajee SA, Borman AM, Brandt ME, Cano J, Cuenca-Estrella M, Dannaoui E, Guarro J, Haase G, Kibbler CC, Meyer W, O'Donnell K, Petti CA, Rodriguez-Tudela JL, Sutton D, Velegraki A, Wickes BL. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: where are we and where should we go from here? J Clin Microbiol 47:877–884. doi: 10.1128/JCM.01685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Linden JW, Arendrup MC, Warris A, Lagrou K, Pelloux H, Hauser PM, Chryssanthou E, Mellado E, Kidd SE, Tortorano AM, Dannaoui E, Gaustad P, Baddley JW, Uekotter A, Lass-Florl C, Klimko N, Moore CB, Denning DW, Pasqualotto AC, Kibbler C, Arikan-Akdagli S, Andes D, Meletiadis J, Naumiuk L, Nucci M, Melchers WJ, Verweij PE. 2015. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus Emerg Infect Dis 21:1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing. 2008. EUCAST technical note on the method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia-forming moulds. Clin Microbiol Infect 14:982–984. doi: 10.1111/j.1469-0691.2008.02086.x. [DOI] [PubMed] [Google Scholar]
- 25.Arendrup MC, Cuenca-Estrella M, Lass-Florl C, Hope WW. 2013. Breakpoints for antifungal agents: an update from EUCAST focussing on echinocandins against Candida spp. and triazoles against Aspergillus spp. Drug Resist Updat 16:81–95. doi: 10.1016/j.drup.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Camps SM, van der Linden JW, Li Y, Kuijper EJ, van Dissel JT, Verweij PE, Melchers WJ. 2012. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrob Agents Chemother 56:10–16. doi: 10.1128/AAC.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother 54:2425–2430. doi: 10.1128/AAC.01599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol 39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snelders E, van der Lee HA, Kuijpers J, Rijs AJ, Varga J, Samson RA, Mellado E, Donders AR, Melchers WJ, Verweij PE. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med 5:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Valk HA, Meis JF, Curfs IM, Muehlethaler K, Mouton JW, Klaassen CH. 2005. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J Clin Microbiol 43:4112–4120. doi: 10.1128/JCM.43.8.4112-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snelders E, Huis In't Veld RA, Rijs AJ, Kema GH, Melchers WJ, Verweij PE. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl Environ Microbiol 75:4053–4057. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balajee SA, Tay ST, Lasker BA, Hurst SF, Rooney AP. 2007. Characterization of a novel gene for strain typing reveals substructuring of Aspergillus fumigatus across North America. Eukaryot Cell 6:1392–1399. doi: 10.1128/EC.00164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein J. 2005. PHYLIP (phylogeny inference package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA. [Google Scholar]
- 34.Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL. 1994. High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457. doi: 10.1038/368455a0. [DOI] [PubMed] [Google Scholar]
- 35.Hedayati MT, Azimi Y, Droudinia A, Mousavi B, Khalilian A, Hedayati N, Denning DW. 2015. Prevalence of chronic pulmonary aspergillosis in patients with tuberculosis from Iran. Eur J Clin Microbiol Infect Dis 34:1759–1765. doi: 10.1007/s10096-015-2409-7. [DOI] [PubMed] [Google Scholar]
- 36.Al-Shair K, Atherton GT, Harris C, Ratcliffe L, Newton PJ, Denning DW. 2013. Long-term antifungal treatment improves health status in patients with chronic pulmonary aspergillosis: a longitudinal analysis. Clin Infect Dis 57:828–835. doi: 10.1093/cid/cit411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denning DW, Riniotis K, Dobrashian R, Sambatakou H. 2003. Chronic cavitary and fibrosing pulmonary and pleural aspergillosis: case series, proposed nomenclature change, and review. Clin Infect Dis 37(Suppl 3):S265–S280. doi: 10.1086/376526. [DOI] [PubMed] [Google Scholar]
- 38.Klaassen CH, de Valk HA, Curfs-Breuker IM, Meis JF. 2010. Novel mixed-format real-time PCR assay to detect mutations conferring resistance to triazoles in Aspergillus fumigatus and prevalence of multi-triazole resistance among clinical isolates in the Netherlands. J Antimicrob Chemother 65:901–905. doi: 10.1093/jac/dkq041. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad S, Khan Z, Hagen F, Meis JF. 2014. Simple, low-cost molecular assays for TR34/L98H mutations in the cyp51A gene for rapid detection of triazole-resistant Aspergillus fumigatus isolates. J Clin Microbiol 52:2223–2227. doi: 10.1128/JCM.00408-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Astvad KM, Jensen RH, Hassan TM, Mathiasen EG, Thomsen GM, Pedersen UG, Christensen M, Hilberg O, Arendrup MC. 2014. First detection of TR46/Y121F/T289A and TR34/L98H alterations in Aspergillus fumigatus isolates from azole-naive patients in Denmark despite negative findings in the environment. Antimicrob Agents Chemother 58:5096–5101. doi: 10.1128/AAC.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fischer J, van Koningsbruggen-Rietschel S, Rietschel E, Vehreschild MJ, Wisplinghoff H, Kronke M, Hamprecht A. 2014. Prevalence and molecular characterization of azole resistance in Aspergillus spp. isolates from German cystic fibrosis patients. J Antimicrob Chemother 69:1533–1536. doi: 10.1093/jac/dku009. [DOI] [PubMed] [Google Scholar]
- 42.Ozmerdiven GE, Ak S, Ener B, Agca H, Cilo BD, Tunca B, Akalin H. 2015. First determination of azole resistance in Aspergillus fumigatus strains carrying the TR34/L98H mutations in Turkey. J Infect Chemother 21:581–586. doi: 10.1016/j.jiac.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 43.Verweij PE, Ananda-Rajah M, Andes D, Arendrup MC, Bruggemann RJ, Chowdhary A, Cornely OA, Denning DW, Groll AH, Izumikawa K, Kullberg BJ, Lagrou K, Maertens J, Meis JF, Newton P, Page I, Seyedmousavi S, Sheppard DC, Viscoli C, Warris A, Donnelly JP. 2015. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resist Updat 21-22:30–40. doi: 10.1016/j.drup.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Hurst SF, Kidd SE, Morrissey CO, Snelders E, Melchers WJ, Castelli MV, Mellado E, Simmon K, Petti CA, Richardson S, Zhang S, Romanelli AM, Wickes BL, de Valk HA, Klaassen CH, Balajee SA. 2009. Interlaboratory reproducibility of a single-locus sequence-based method for strain typing of Aspergillus fumigatus. J Clin Microbiol 47:1562–1564. doi: 10.1128/JCM.00124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodriguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camps SM, Rijs AJ, Klaassen CH, Meis JF, O'Gorman CM, Dyer PS, Melchers WJ, Verweij PE. 2012. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. J Clin Microbiol 50:2674–2680. doi: 10.1128/JCM.00335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bader O, Weig M, Reichard U, Lugert R, Kuhns M, Christner M, Held J, Peter S, Schumacher U, Buchheidt D, Tintelnot K, Gross U, MykoLabNet D Partners . 2013. cyp51A-based mechanisms of Aspergillus fumigatus azole drug resistance present in clinical samples from Germany. Antimicrob Agents Chemother 57:3513–3517. doi: 10.1128/AAC.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denning DW, Park S, Lass-Florl C, Fraczek MG, Kirwan M, Gore R, Smith J, Bueid A, Moore CB, Bowyer P, Perlin DS. 2011. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clin Infect Dis 52:1123–1129. doi: 10.1093/cid/cir179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Linden JW, Snelders E, Arends JP, Daenen SM, Melchers WJ, Verweij PE. 2010. Rapid diagnosis of azole-resistant aspergillosis by direct PCR using tissue specimens. J Clin Microbiol 48:1478–1480. doi: 10.1128/JCM.02221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White PL, Posso RB, Barnes RA. 2015. Analytical and clinical evaluation of the PathoNostics AsperGenius assay for detection of invasive aspergillosis and resistance to azole antifungal drugs during testing of serum samples. J Clin Microbiol 53:2115–2121. doi: 10.1128/JCM.00667-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chong GL, van de Sande WW, Dingemans GJ, Gaajetaan GR, Vonk AG, Hayette MP, van Tegelen DW, Simons GF, Rijnders BJ. 2015. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. J Clin Microbiol 53:868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]