Abstract
Background
To identify mutant genes with high-frequency-risk-expression between lung adenocarcinoma and normal samples.
Methods
The ribonucleic acid RNA-Seq data GSE34914 and GSE37765 were downloaded from the Gene Expression Omnibus database, including 12 lung adenocarcinoma samples and six controls. All RNA-Seq reads were processed and the gene-expression level was calculated. Single nucleotide variation (SNV) was analyzed and the locations of mutant sites were recorded. In addition, the frequency and risk-level of mutant genes were calculated. Gene Ontology (GO) functional analysis was performed. The reported cancer genes were searched in tumor suppressor genes, Cancer Genes, and the Catalogue of Somatic Mutations in Cancer (COSMIC) database.
Results
The SNV annotations of somatic mutation sites showed that 70% of mutation sites in the exon region occurred in the coding sequence (CDS). Thyroid hormone receptor interactor (TRIP)12 was identified with the highest frequency. A total of 118 mutant genes with high frequency and high-risk were selected and significantly enriched into several GO terms. No base mutation of cyclin C (CCNC) or RAB11A was recorded. At fragments per kilobase per million reads (FPKM) ≥ 56.5, reported tumor suppressor genes catenin (cadherin-associated protein), delta (CTNND)1, dual specificity phosphatase (DUSP)6, malate dehydrogenase (MDH)1 and RNA binding motif protein (RBM)5, were identified. Notably, signal transducer and activator of transcription 2 (STAT2) was the only transcription factor (TF) with high-risk mutation and its expression was detected.
Conclusion
For the mutant genes with high-frequency-risk-expression, CTNND1, DUSP6, MDH1 and RBM5 were identified. TRIP12 might be a potential cancer-related gene, and expression of TF STAT2 with high-risk was detected. These mutant gene candidates might promote the development of lung adenocarcinoma and provide new diagnostic potential targets for treatment.
Keywords: High expression, high frequency, high-risk, lung adenocarcinoma, mutant gene, RNA-Seq
Introduction
Lung cancer is the most common cancer with high-incidence and high-mortality worldwide and is classified into two groups based on histological type: small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC).1 Adenocarcinoma, a subtype of NSCLC, originates in peripheral lung tissue, accounting for nearly 40% of lung cancers.2 Although recent examinations are advanced, early detection of lung cancer is still a challenge for clinicians, because the biomarkers for early diagnosis are still lacking and there are multiple molecular pathways that mediate lung carcinogenesis.3,4 A deep understanding of the molecular mechanism is necessary for lung adenocarcinoma prevention and treatment.
Similar to other cancers, lung adenocarcinoma can be activated by oncogenes and inactivated by tumor suppressor genes. To date, two major pathways of mediated lung adenocarcinoma have been verified: an epidermal growth factor receptor (EGFR)-dependent pathway in never-smokers and a Kirsten rat sarcoma oncogene (KRAS)-dependent signaling module in smokers.5,6 EGFR mutations distribute throughout the four kinase domain (exons 18–21) and comprise of in-frame deletions, in-frame insertions/duplications and point mutations.7 Most KRAS mutations involve replacement of glycine 12 and glycine 13, and activate mitogenic and proliferative signaling through the RAF-MEK- extracellular-signal-regulated-kinase (ERK) cascade.8 Human epidermal growth factor receptor (HER)2/NEU9 oncogenes exclusively occur in lung adenocarcinoma. Other oncogenes include anaplastic lymphoma kinase (ALK) (fusion),10 MET,11 kinase insert domain receptor (KDR),12 EPHA, and MAP2K1.13
Meanwhile, methylation, focal DNA deletion, and mutation of tumor suppressor genes have been described in lung adenocarcinoma.4 Tumor protein (TP)53 is the most frequently mutated tumor suppressor gene in lung adenocarcinoma.14 Others include CDKN2A,15 STK116 and LRP1B.17 In addition, increased gene dosage of thyroid transcriptional factor−1/NK2 homeobox 1 (TITF-1/NKX2-1) is prevalent in lung adenocarcinoma, which may act as a lineage-specific gene in lung cancer.18 However, most of the mutations that occur in lung adenocarcinoma are still unknown. Few studies have verified successful target-specific treatment strategies for lung adenocarcinoma with mutations.
With the advent of various technologies, such as single nucleotide polymorphism (SNP) arrays, and the second-generation sequencing and tumor-sequencing project, the mutational spectrum of lung adenocarcinoma has substantially increased.4 Ribonucleic acid (RNA)-Seq is a powerful new technology for transcriptome analysis, which provides both gene expression and single nucleotide variation (SNV) information.19,20 Currently, point mutations in expressed exons of the human genome can be identified by using RNA-seq data.
In the present study, somatic mutations of transcriptional genes in 12 lung adenocarcinoma samples were analyzed based on RNA-seq data. Mutations were screened according to the frequency, risk-level and expression level. Thus, mutant genes with high frequency, high risk-level, and high expression level were identified. The discovery of new oncogenes and tumor suppressor mutations in lung adenocarcinoma may provide potential targets for personalized therapeutic strategies.
Materials and methods
Data acquisition
The two expression profiling data by high throughput sequencing were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/ ID: GSE34914 21 and GSE3776522), including 12 lung adenocarcinoma and six normal samples. A total of 18 samples included six lung adenocarcinoma tumors with wild-type KRAS (GSE34914), primary lung adenocarcinoma tumors, and adjacent normal tissues of six Korean female never-smoker patients (GSE37765). All of the six lung adenocarcinoma tumors with wild-type KRAS were grade I or II and were obtained by surgical resection. The primary lung adenocarcinoma tumors and adjacent normal tissues of the six Korean female never-smoker patients, with an average age of 55.5 years, were grade T1N0M0 or T2N0M0. All patients had first-time (i.e. non-recurrent) lung cancers. None of the patients had distant metastasis or a family history of lung cancer. No preoperative chemotherapy/radiotherapy was performed.
Total RNA was isolated using miRNeasy Mini Kit columns according to the manufacturer's protocol (Qiagen, Hilden, Germany). All samples showed RNA Integrity Numbers (RIN) > 7.0, as determined using the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Sequencing was conducted on the Illumina Genome Analyzer (Illumina Inc., San Diego, CA, USA) following the manufacturer standard protocol. Reads were generated in a paired-end orientation.
Read alignment, transcript assembly, and differential expression
All RNA-Seq reads were mapped to the reference human genome (hg19) of UCSC (University of California Santa Cruz) by using TopHat23 software. For read alignment, mismatches (two bases by default) were permitted in one read. Other parameters were set up according to the default settings of TopHat. Transcripts were then assembled using Cufflinks24 (http://cufflinks.cbcb.umd.edu/) based on Refseq gene annotation, as well as the alignment results of TopHat. Cuffdiff, a part of the Cufflinks package, calculated gene expression levels of transcripts by using the FPKM (fragments per kilobase per million reads) method.25 Mutant genes with FPKM value ≥1 were selected.
Identification of single nucleotide variation (SNV)
After remove of polymerase chain reaction (PCR) duplicates, the SNVs of the aligned reads were called using the Sequence Alignment/Map (SAM) tool.26 In order to minimize the risk of false-positive SNV callings, the Phred score of each base must be more than 28. Meanwhile, coverage of each credible SNV was > 50 x and the minimum quality score of high reliability SNV was 50. In addition, all variants were analyzed based on the reported SNP of 1000 Genome databases and dbSNP137. To effectively excise the interference of RNA editing in transcriptome, SNV callings were optimized by combining the RNA-Seq data of six normal controls.
Somatic mutations and high-risk assessment
VarioWatch27 software was used to annotate the SNVs in coding sequence and then analyze the functional impact of gene products based on risk assessment software. For mutation sites in coding sequence (CDS) resulting in transition, transversion, and indel, these mutant genes were defined as high-risk genes. SNVs with high functional risk levels were selected and their loci functions were further analyzed.
Gene functional enrichment analysis
For functional analysis of the obtained genes, DAVID (Database for Annotation, Visualization and Integrated Discovery)28 was performed for GO (Gene Ontology)29 enrichment analysis. GO categories were classified into Biological Process (BP), Molecular Function (MF) and Cellular Component (CC) GO-terms. We used the DAVID to identify over-represented GO categories based on hypergeometric distribution with a P-value less than 0.05.
The mutant genes with transcriptional regulation were selected, labeled, and input into the TS genes30 database and the Cancer Genes31 database to screen the cancer-related genes for further analysis based on the Catalogue of Somatic Mutations in Cancer (COSMIC)32 database.
Results
Somatic mutations and SNV in lung adenocarcinoma
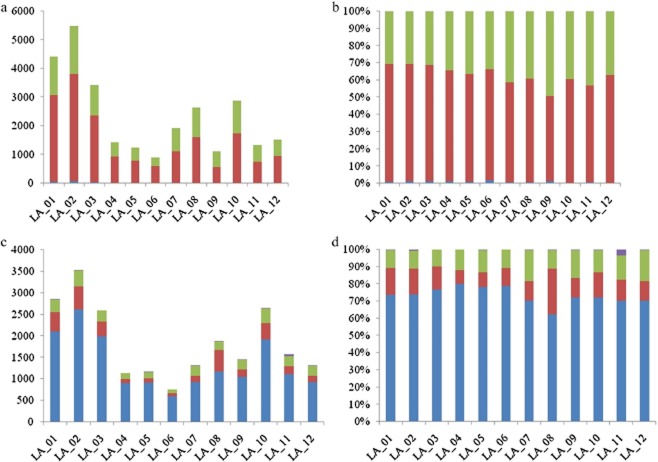
According to the data processing and sub-filtering of RNA-seq in 12 lung adenocarcinoma samples, potential somatic mutations of each sample were obtained (Fig. 1). In different lung adenocarcinoma samples, the number of somatic base mutations in transcriptional genes was significantly different. The highest quantity variance of mutation site among cancer samples was up to approximately six times. In each cancer sample, transition was the main mutation type (more than 70%), transversion was nearly 30%, and the frequency of indel was only 0.4 % (Fig. 1b).
Figure 1.
Statistics of different somatic mutations in 12 lung adenocarcinoma samples. (a and b) The number and percentage of different types of somatic mutations in 12 lung adenocarcinoma samples. (c and d) The number and percentage of different locations of mutationd in exon region. (a) ( ) all_transversion, (
) all_transversion, ( ) all_transition, (
) all_transition, ( ) all_indel. (b) (
) all_indel. (b) ( ) all_transversion, (
) all_transversion, ( ) all_transition, (
) all_transition, ( ) all_indel. (c) (
) all_indel. (c) ( ) GT-AG, (
) GT-AG, ( ) 5′ untranslated region (UTR), (
) 5′ untranslated region (UTR), ( ) 3′UTR, (
) 3′UTR, ( ) CDS. (d) (
) CDS. (d) ( ) GT-AG, (
) GT-AG, ( ) 5′UTR, (
) 5′UTR, ( ) 3′UTR, (
) 3′UTR, ( ) coding sequence (CDS).
) coding sequence (CDS).
The SNV annotations of somatic mutation sites showed that about 70% of mutation sites located in the exon region occurred in CDS, while 3′ untranslated region (UTR) and 5′ UTR each contributed about 10% (Fig. 1c,d). There was little difference among the samples and there were very few mutations located in GT-AG splicing sites.
High-frequency mutation sites
The frequency of each mutation site in cancer samples was calculated (Table 1). A total of 19278 mutation sites were founded. The mutation sites in different cancer samples were highly diverse and 96.97% of them were only detected in single samples. At the frequency ≥ 5 and mutation rate > 42%, 14 mutation sites were selected. The remaining mutation sites displayed individual differences among different cancer samples.
Table 1.
Frequency of mutation sites, mutant genes and high-risk mutant genes in 12 lung adenocarcinoma samples
| Mutation Frequency | Mutation sites | Mutant genes | High-risk mutant genes | |||
|---|---|---|---|---|---|---|
| Counts | (%) | Counts | (%) | Counts | (%) | |
| 1 | 18693 | 96.97 | 3872 | 43.34 | 2681 | 54.56 |
| 2 | 429 | 2.23 | 2526 | 28.27 | 1409 | 28.66 |
| 3 | 111 | 0.57 | 1372 | 15.36 | 522 | 10.62 |
| 4 | 31 | 0.16 | 629 | 7.04 | 187 | 3.80 |
| ≥ 5 | 14 | 0.07 | 535 | 5.99 | 118 | 2.40 |
| Total | 19278 | 100 | 8934 | 100 | 4917 | 100 |
Based on the annotation of 14 high-frequency mutation sites (Table 2), 12 mutations occurred in CDS and two mutations in 5′ UTR. In addition, the base mutations in CDS included 11 missense mutations, which could cause changes in protein amino acid sequence and result in abnormal protein function.
Table 2.
High-frequency mutation sites in 12 lung adenocarcinoma samples
| Chromosome | Position | Reference | Mutation | Gene Symbol | Located Region | Mutation Type | Mutation Frequency |
|---|---|---|---|---|---|---|---|
| chr2 | 230670518 | G | C | TRIP12 | CDS | Missense | 9 |
| chr1 | 26212360 | T | C | STMN1 | CDS | Missense | 9 |
| chr22 | 18111371 | G | T | ATP6V1E1 | CDS | Missense | 6 |
| chr20 | 44536542 | T | C | PLTP | CDS | Missense | 6 |
| chr17 | 73038276 | C | A | ATP5H | CDS | Missense | 6 |
| chr17 | 73750053 | G | C | ITGB4 | CDS | Missense | 5 |
| chr12 | 95645847 | A | G | VEZT | CDS | Splicing regulation | 5 |
| chr12 | 89743338 | T | C | DUSP6 | CDS | Missense | 5 |
| chr12 | 51636114 | C | G | DAZAP2 | CDS | Missense | 5 |
| chr5 | 177576837 | G | A | NHP2 | CDS | Missense | 5 |
| chr4 | 106367542 | G | T | PPA2 | CDS | Missense | 5 |
| chr2 | 54040210 | G | A | ERLEC1 | CDS | Missense | 5 |
| chr1 | 109968926 | G | T | PSMA5 | 5′ UTR | Unkown | 5 |
| chr19 | 39616558 | C | A | PAK4 | 5′ UTR | Unkown | 5 |
CDS, coding sequence; Chr: chromosome; DUSP, dual specificity phosphatase; ITGB, integrin beta; PLTP, phospholipid transfer protein; PSMA, proteasome prosome, macropain subunit, alpha type; STMN, stathmin gene; TRIP, thyroid hormone receptor interactor.
Two mutations with the highest frequency were located in the CDS of stathmin gene (STMN)1 and thyroid hormone receptor interactor (TRIP)12, respectively. According to the Cancer Genes database, STMN1 was a reported oncogene and TRIP12 was involved in protein ubiquitination. For the other 10 mutations located in CDS, dual specificity phosphatase (DUSP)6, a tumor suppression gene, was involved in the phosphorus metabolic process based on GO analysis, which was functionally similar to three other genes (ATP5H, ATP6V1E1 and PPA2).
Gene mutation with high-frequency and high-risk
The frequency of mutant genes was calculated (Table 1). At the frequency ≥ 5, 535 mutant genes were identified, accounting for six percent of all mutant genes, the proportion of which were higher than mutant sites. The base mutations of TRIP12 and STMN were detected in 11 and nine cancer samples, respectively.
The frequency of high-risk mutant genes was calculated (Table 1). A total of 4917 mutant genes with high-risk were identified; 118 of these were detected in more than five cancer samples and belonged to high-risk-frequency mutant genes.
The 118 mutant genes obtained were significantly enriched into several GO terms, including purine nucleotide metabolic process, proteasomal ubiquitin-dependent-protein catabolic process and RNA processing, and were mainly functioned as ATPase, cytoskeleton protein, and RNA binding (Table 3). Although 10 high-risk-frequency mutant genes were enriched into cell death, the enrichments were not significant. The cellular localizations of protein products were not regular and were focused on vesicle, cytoskeleton, and ribonucleoprotein complex.
Table 3.
Gene Ontology function enrichment analysis of mutation genes with high frequency and high-risk
| GO Term | Counts | P-value | |
|---|---|---|---|
| BP | GO:0006163∼purine nucleotide metabolic process | 7 | 2.68E-03 |
| BP | GO:0043161∼proteasomal ubiquitin-dependent protein catabolic process | 4 | 0.04 |
| BP | GO:0006396∼RNA processing | 9 | 0.05 |
| BP | GO:0008219∼cell death | 10 | 0.09 |
| MF | GO:0008092∼cytoskeletal protein binding | 9 | 0.03 |
| MF | GO:0003723∼RNA binding | 11 | 0.04 |
| MF | GO:0042626∼ATPase activity, coupled to transmembrane movement of substances | 4 | 0.05 |
| CC | GO:0042470∼melanosome | 6 | 4.46E-04 |
| CC | GO:0031982∼vesicle | 14 | 9.77E-04 |
| CC | GO:0030529∼ribonucleoprotein complex | 12 | 1.15 E-03 |
| CC | GO:0005856∼cytoskeleton | 19 | 8.87 E-03 |
| CC | GO:0045177∼apical part of cell | 6 | 9.40 E-03 |
| CC | GO:0000502∼proteasome complex | 4 | 9.61 E-03 |
BP, biological process; CC, cellular component; GO, Gene Ontology; MF, molecular function.
For the 118 high-risk-frequency mutant genes, six reported tumor suppressor genes and two oncogenes were identified (Table 4). According to the COSMIC database, base mutations of beclin (BECN)1, catenin (cadherin-associated protein), delta (CTNND)1, DUSP6, malate dehydrogenase (MDH)1, RNA binding motif protein (RBM)5 and RAP1B had been reported. However, there was no record referring to the base mutation of cyclin C (CCNC) or RAB11A.
Table 4.
Gene annotation of cancer related genes in high-risk-frequency mutant genes
| Gene | Description | Tumor Suppressor or Oncogene |
|---|---|---|
| BECN1 | beclin 1 (coiled-coil, myosin-like BCL2 interacting protein) | Tumor Suppressor |
| CCNC | cyclin C | Tumor Suppressor |
| CTNND1 | catenin (cadherin-associated protein), delta 1 | Tumor Suppressor |
| DUSP6 | dual specificity phosphatase 6 | Tumor Suppressor |
| MDH1 | malate dehydrogenase 1, NAD (soluble) | Tumor Suppressor |
| RBM5 | RNA binding motif protein 5 | Tumor Suppressor |
| RAB11A | member RAS oncogene family | Oncogene |
| RAP1B | member RAS oncogene family | Oncogene |
Expression level of mutant genes with high-risk and high-frequency
Mutant genes with high-risk and high frequency were classified into three groups based on their expression level (Table 5). At a FPKM ≥ 56.5, 46 genes were selected and defined as high-expressed genes, including four reported tumor suppressor genes recorded in the Cancer Genes database, such as CTNND1, DUSP6, MDH1 and RBM5.
Table 5.
Expression level of mutant genes with high-risk and high frequency in lung adenocarcinoma samples
| Expression Level | Gene Counts | Tumor Suppressor | Oncogene | TF |
|---|---|---|---|---|
| High | 46 | CTNND1, DUSP6, MDH1, RBM5 | RAP1B | |
| Medium | 110 | BECN1, CCNC | RAB11A | STAT2 |
| Low | 14 |
BECN, beclin; CCNC, cyclin C; CTNND, catenin (cadherin-associated protein), delta 1; DUSP, dual specificity phosphatase; MDH, malate dehydrogenase; RBM, RNA binding motif protein; RNA, ribonucleic acid; STAT, signal transducer and activator of transcription; TF, transcription factor.
At the FPKM > 11.6 and < 56.5, 110 genes were identified and defined as medium-expressed genes, including two reported tumor suppressor genes (BECN1 and CCNC) and one reported oncogene (RAB11A). Notably, expression of transcription factor (TF) STAT2 was detected and it was the only TF with high-risk mutation. A total of 14 low-expressed genes were not associated with cancer.
Discussion
Compared with gene expression microarray, which relies on the design of prior probes and annotation of known transcripts, RNA-Seq is a revolutionary technology and can be used to analyze any transcriptome.20 RNA-Seq analysis can detect new transcripts, expand the depth of gene expression changes at a dynamic range, provide deep-insight into pathways and molecular events and, therefore, increase the annotation of the transcriptome.33 It has been reported that RNA-Seq can capture 81% of the exonic variants from well-expressed genes in the source sample.34 In this study, RNA-Seq data were used to analyse somatic mutations of transcriptional genes in lung adenocarcinoma samples compared with healthy controls.
STMN1 and TRIP12 were the selected mutations located in the CDS with the highest frequency. Based on the Cancer Genes database, STMN1 is a reported oncogene in NSCLC tissues and its mutation in lung adenocarcinoma may result in the disability of oncogenes. The far upstream sequence element-binding protein-1 (FBP-1) is a critical inducer of several stathmin family members.35 TRIP12 is involved in protein ubiquitination and may increase cancer risk by causing ubiquitin-mediated degradation of ARF (alternate reading frame of the INK4a/CDKN2A locus), the key activator of the tumor suppressor p53.36 TRIP12/ULF (a ubiquitin ligase) may be a novel sensor of oncogenic stress upstream of ARF and have pro-oncogenic activity if deregulated.37
Four reported tumor suppressor genes, CTNND1, DUSP6, MDH1, and RBM5, were identified as high-expressed mutant genes with high frequency and high-risk in lung adenocarcinoma. DUSP6, a cytoplasmic DUSP with high specificity for ERK, exerts antitumor effects via negative feedback regulation in NSCLC.38 RBM5, also called H37 gene or Luca15, is located in the 3p21.3 tumor suppressor region and its expression is decreased in 82% of primary NSCLCs.39 The tumor suppression model of H37 may be a post-transcriptional regulator for cell cycle/apoptotic–related proteins.40 MDH1 is overexpressed in muscle metastatic lesions of pancreatic adenocarcinoma41 and has redox activity in lung epithelial cells.42 The regulation of MDH1 in lung adenocarcinoma still needs to be confirmed in further studies. In NSCLC, CTNND1 can encode p120-catenin (p120ctn) and has been defined as a good candidate oncogene in a previous study.43 These differences may be related to the differences of samples, bioinformatic techniques, and cut-off criterion.
Meanwhile, mutant STAT2 was the only detected TF mutation with high frequency and high risk in lung adenocarcinoma. STAT2, a family of TF STATs, can mediate interferon (IFN)-effects and is phosphorylated on tyrosine 690.44 Current studies show that STAT3 plays a leading role of STATs in tumor inflammation and immunity by promoting pro-oncogenic inflammatory pathways. STAT2 as an important molecular target has been verified in skin squamous cell carcinoma cells. The potential function of STAT2 in lung adenocarcinoma should be verified with more evidence.
It should be highlighted that our study contains several limitations. In order to confirm our hypothesis, a larger number of samples are required for further investigation. Our study is a bioinformatics analysis and the potential functions of these mutant genes should be verified via experimental studies in future. Finally, no computational procedure is perfect and a specific computational method can identify specific DEGs that might be different from other methods.
Conclusion
In conclusion, based on high-throughput second-generation sequencing data, a dynamic gene expression profile combined with different biological information was used to analyze the potential factors that contribute to lung adenocarcinoma. According to the frequency, risk-level, and expression level, TRIP12, CTNND1, DUSP6, MDH1, RBM5, and STAT2 were separately identified, as their function in lung adenocarcinoma development had not been clearly elucidated. These candidate mutant genes may provide potential targets for the diagnosis and treatment of lung adenocarcinoma in future.
Acknowledgments
We wish to express our warm thanks to Guiyuan Li, Shengming Yi, Fan Yang, Yongxin Zhou, Qiang Ji, Jianzhi Cai, Yunqing Mei.
Disclosure
No authors report any conflict of interest.
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. (Published erratum appears in CA Cancer J Clin 2011; 61: 134) [DOI] [PubMed] [Google Scholar]
- Hong WK, Hait WN, Kufe DW, Pollock RE. Holland-Frei Cancer Medicine Eighth Edition. Shelton, CT: PMPH-USA; 2010. [Google Scholar]
- Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadara H, Kabbout M, Wistuba II. Pulmonary adenocarcinoma: a renewed entity in 2011. Respirology. 2012;17:50–65. doi: 10.1111/j.1440-1843.2011.02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Tam IY, Chung LP, Suen WS, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non–small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- Mounawar M, Mukeria A, Le Calvez F, et al. Patterns of EGFR, HER2, TP53, and KRAS mutations of p14arf expression in non–small cell lung cancers in relation to smoking history. Cancer Res. 2007;67:5667–5672. doi: 10.1158/0008-5472.CAN-06-4229. [DOI] [PubMed] [Google Scholar]
- Yatabe Y. EGFR mutations and the terminal respiratory unit. Cancer Metastasis Rev. 2010;29:23–36. doi: 10.1007/s10555-010-9205-8. [DOI] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Xia N, An J, Jiang QQ, Li M, Tan J, Hu CP. Analysis of EGFR, EML4-ALK, KRAS, and c-MET mutations in Chinese lung adenocarcinoma patients. Exp Lung Res. 2013;39:328–335. doi: 10.3109/01902148.2013.819535. [DOI] [PubMed] [Google Scholar]
- Seto T, Higashiyama M, Funai H, et al. Prognostic value of expression of vascular endothelial growth factor and its flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung Cancer. 2006;53:91–96. doi: 10.1016/j.lungcan.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Yu J, Bulk E, Ji P, et al. The kinase defective EPHB6 receptor tyrosine kinase activates MAP kinase signaling in lung adenocarcinoma. Int J Oncol. 2009;35:175. doi: 10.3892/ijo_00000326. –179. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Onozato R, Kuwano H, Mitsudomi T. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol. 2009;4:22–29. doi: 10.1097/JTO.0b013e3181914111. [DOI] [PubMed] [Google Scholar]
- Chan EC, Lam SY, Fu KH, Kwong YL. Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: relationship with aberrant promoter methylation of the CDKN2A and RARB genes. Cancer Genet Cytogenet. 2005;162:10–20. doi: 10.1016/j.cancergencyto.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Li HX, Lei DS, Wang XQ, Skog S, He Q. Serum thymidine kinase 1 is a prognostic and monitoring factor in patients with non-small cell lung cancer. Oncol Rep. 2005;13:145–149. [PubMed] [Google Scholar]
- Liu CX, Musco S, Lisitsina NM, Yaklichkin SY, Lisitsyn NA. Genomic organization of a new candidate tumor suppressor gene, LRP1B. Genomics. 2000;69:271–274. doi: 10.1006/geno.2000.6331. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Yanagisawa K, Shinjo K, et al. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lin L, Jiang P, Wang D, Xing Y. A comparison of RNA-Seq and high-density exon array for detecting differential gene expression between closely related species. Nucleic Acids Res. 2011;39:578–588. doi: 10.1093/nar/gkq817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari KR, Rossell D, Necela BM, et al. Deep sequence analysis of non-small cell lung cancer: integrated analysis of gene expression, alternative splicing, and single nucleotide variations in lung adenocarcinomas with and without oncogenic KRAS mutations. Front Oncol. 2012;2:1–16. doi: 10.3389/fonc.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Jung Y, Park J, et al. A high-dimensional, deep-sequencing study of lung adenocarcinoma in female never-smokers. PLoS ONE. 2013;8:e55596. doi: 10.1371/journal.pone.0055596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beane J, Vick J, Schembri F, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res. 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC HsiaoFC, Yeh EC, et al. VarioWatch: providing large-scale and comprehensive annotations on human genomic variants in the next generation sequencing era. Nucleic Acids Res. 2012;40:W76–W81. doi: 10.1093/nar/gks397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. doi: 10.1038/nprot.2008.211. 2009; [DOI] [PubMed] [Google Scholar]
- Hulsegge I, Kommadath A, Smits MA. Globaltest and GOEAST: two different approaches for gene ontology analysis. BMC Proc. 2009;3:S10. doi: 10.1186/1753-6561-3-S4-S10. (Suppl.): [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Sun J, Zhao Z. TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35:D721–726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick BA, Phadke DP, Auerbach SS, et al. RNA-Seq profiling reveals novel hepatic gene expression pattern in aflatoxin B1 treated rats. PLoS ONE. 2013;8:e61768. doi: 10.1371/journal.pone.0061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli ET, Singh A, Shianna KV, Ge D, Smith JP, Maia JM. Research Screening the human exome: a comparison of whole genome and whole transcriptome sequencing. Genome Biology. 2010;11:R57. doi: 10.1186/gb-2010-11-5-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S, Malz M, Herpel E, et al. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non–small cell lung cancer. Cancer Res. 2009;69:2234–2243. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
- Hein R, Flesch-Janys D, Dahmen N, et al. A genome-wide association study to identify genetic susceptibility loci that modify ductal and lobular postmenopausal breast cancer risk associated with menopausal hormone therapy use: a two-stage design with replication. Breast Cancer Res Treat. 2013;138:529–42. doi: 10.1007/s10549-013-2443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Serrano M. The TRIP from ULF to ARF. Cancer Cell. 2010;17:317–318. doi: 10.1016/j.ccr.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Kobayashi S, Borczuk AC, et al. Dual specificity phosphatase 6 (DUSP6) is an ETS-regulated negative feedback mediator of oncogenic ERK signaling in lung cancer cells. Carcinogenesis. 2010;31:577–586. doi: 10.1093/carcin/bgq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JJ, West AR, Fishbein MC, Slamon DJ. A candidate tumor suppressor gene, H37, from the human lung cancer tumor suppressor locus 3p21.3. Cancer Res. 2002;62:3207–3213. [PubMed] [Google Scholar]
- Oh JJ, Razfar A, Delgado I, et al. 3p21.3 tumor suppressor gene H37/Luca15/RBM5 inhibits growth of human lung cancer cells through cell cycle arrest and apoptosis. Cancer Res. 2006;66:3419–3427. doi: 10.1158/0008-5472.CAN-05-1667. [DOI] [PubMed] [Google Scholar]
- Chaika NV, Yu F, Purohit V, et al. Differential expression of metabolic genes in tumor and stromal components of primary and metastatic loci in pancreatic adenocarcinoma. PLoS ONE. 2012;7:e32996. doi: 10.1371/journal.pone.0032996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess PC, Deng B, Hondal RJ, Matthews DE, van der Vliet A. Proteomic profiling of acrolein adducts in human lung epithelial cells. J Proteomics. 2011;74:2380–2394. doi: 10.1016/j.jprot.2011.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo SD, Angulo B, Suarez-Gauthier A, et al. Gene amplification of the transcription factor DP1 and CTNND1 in human lung cancer. J Pathol. 2010;222:89–98. doi: 10.1002/path.2732. [DOI] [PubMed] [Google Scholar]
- Clifford JL, Yang X, Walch E, Wang M, Lippman SM. Dominant negative signal transducer and activator of transcription 2 (STAT2) protein: stable expression blocks interferon α action in skin squamous cell carcinoma cells. Mol Cancer Ther. 2003;2:453–459. [PubMed] [Google Scholar]