Abstract
Background
Drug resistance significantly weakens the efficacy of cancer treatment, and the BIM (also known as the BCL2L11 gene) deletion polymorphism has been identified as a potential biomarker for drug resistance. In this retrospective study, we included a total of 290 patients with advanced non-small cell lung cancer (NSCLC) who received treatment with epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) and chemotherapy.
Methods
The BIM deletion polymorphism of each patient was detected by polymerase chain reaction. EGFR mutations were detected by denaturing high-performance liquid chromatography methods and the amplification refractory mutation system.
Results
The BIM deletion polymorphism was detected in 45/290 (15.5%) Chinese NSCLC patients. No associations were observed between the BIM deletion and clinic-pathologic characteristics of patients. The BIM deletion polymorphism was predictive of shorter progression-free survival in Chinese patients with EGFR-mutant adenocarcinoma and who were treated with EGFR-TKIs (7.30 vs. 9.53 months, P = 0.034). Additionally, we found that the BIM deletion polymorphism was an effective predictor of short progression-free survival in individuals with EGFR-mutant NSCLC and treated with chemotherapy containing pemetrexed (3.32 vs. 5.30, P = 0.012) or second-/beyond-line chemotherapy containing taxanes (1.53 vs. 2.61 months, P = 0.025). The BIM deletion was not correlated with overall survival.
Conclusion
The BIM deletion polymorphism occurs in 15.5% of Chinese NSCLC patients, and is a biomarker for resistance to TKIs and chemotherapy. However, BIM deletion was not a decisive factor in overall survival.
Keywords: Chemotherapy, EGFR tyrosine kinase inhibitor, non-small cell lung cancer, polymorphism
Introduction
Doublet chemotherapy is the standard regimen for treating non-small cell lung cancer (NSCLC). Several biomarkers including ERCC1,1 RRM1,2 and βIII-tubulin3 have emerged as potential predictive markers for the efficacy of chemotherapy in treating NSCLC. Epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs) can also greatly benefit patients with EGFR mutant tumors.4–8 However, drug resistance can limit the therapeutic outcome of EGFR-TKIs. Recent studies have shown that K-ras mutation and the echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) fusion gene may be associated with primary resistance of EGFR-TKIs.9,10 A mutation at T790M in EGFR, amplification of the MET oncogene, and the activation of PI3KCA, IGF-1R may all contribute to secondary drug resistance.
The apoptosis-regulating gene BIM (also known as BCL2L11, Bcl-2 Interacting Mediator of cell death) has emerged as another focus of drug resistance research. BIM contains a single apoptosis domain (BH3), which regulates the cellular life-or-death switch.11 BIM activity is mainly regulated by the extracellular signal-regulated kinase (ERK) and protein kinase B (AKT) pathways. ERK phosphorylates BIM and promotes BIM degradation post-transcriptionally by proteasomes. AKT suppresses BIM transcription factor FOXO3a, thus reducing the expression of BIM.12,13 BIM initiates the stress pathway (or mitochondrial pathway), which inactivates Bcl-2-like proteins, provokes Apaf-1, and, thus, induces apoptosis.
The BIM deletion polymorphism is a 2903-bp genomic deletion commonly found (12.3%) in the Asian population. It is located in intron-2 and regulates the expression of exon-3 and exon-4. Exon 4 encode for the BH3 domain, required for apoptotic function. After TKI exposure, cells with BIM deletion favour the expression of exon-3 that does not encode the BH3 domain, resulting in impaired apoptosis, and consequently, the cells develop a resistance. Clinical data have shown that the BIM deletion polymorphism is predictive for a shorter progression-free survival (PFS) in individuals with EGFR-mutant NSCLC and treated with EGFR-TKI therapy.14,15
In this study, we analyzed the BIM deletion polymorphism in 290 consecutive Chinese NSCLC patients with and without an EGFR mutation to examine the effect of this polymorphism on the efficacy of chemotherapy and EGFR-TKIs (either a single Erlotinib 150 mg qd or Gefitinib 250 mg qd), as well as overall survival (OS). We also examined the association between the BIM deletion polymorphism and clinical pathology characteristics of NSCLC.
Materials and methods
Patient population
This study enrolled 290 consecutive NSCLC patients treated at the Peking University Cancer Hospital between July 2005 and March 2012. All enrolled patients had a pathologically confirmed stage of NSCLC, available lymphocytes, and were receiving treatment with an EGFR-TKI. The EGFR mutation status of the patients had been previously assessed using denaturing high-performance liquid chromatography (DHPLC) and the amplification refractory mutation system (ARMS). The patient population consisted of 191 EGFR-mutant patients and 99 EGFR-wild type patients; 162 females and 128 males; 103 smokers and 187 non-smokers. All patients treated with EGFR-TKIs were of locally advanced and advanced disease. All patient laboratory data was obtained and recorded independently and blinded from clinical review until analyses by a biostatistician. This study was reviewed and approved by the Institutional Ethics Committee of the Peking University Cancer Hospital, and all patients signed an informed consent for participation in the study and use of their biological samples (Table 1).
Table 1.
Baseline characteristics of the lung cancer patients
| BIM polymorphism (n = 45) | No BIM polymorphism (n = 245) | P-value | ||
|---|---|---|---|---|
| Mean age at diagnosis (years) (mean ± SD) | 59.98 ± 11.08 | 58.33 ± 11.73 | 0.383† | |
| Gender | Male (44.0%) | 23 | 105 | 0.305§ |
| Female (56.0%) | 22 | 140 | ||
| Smoking | Yes (35.1%) | 20 | 82 | 0.156§ |
| No (64.9%) | 25 | 163 | ||
| Pathology | Adenocarcinoma (82.1%) | 35 | 203 | 0.683¶ |
| Squamous (13.4%) | 8 | 31 | ||
| Large cell lung cancer (2.8%) | 2 | 6 | ||
| Others (1.7%) | 0 | 5 | ||
| Stage at diagnosis | Stage I (7.2%) | 2 | 19 | 0.507‡ |
| Stage II (2.1%) | 0 | 5 | ||
| Stage IIIa (4.5%) | 1 | 12 | ||
| Stage IIIb (7.6%) | 6 | 16 | ||
| Stage IV (78.7%) | 36 | 193 | ||
| Differentiation at diagnosis | Low (43.3%) | 24 | 102 | |
| Intermediate (28.5%) | 10 | 73 | 0.483¶ | |
| High (9.6%) | 3 | 25 | ||
| Unknown (18.6%) | 8 | 45 | ||
| EGFR mutation | Mutation (66.0%) | 30 | 161 | 0.901§ |
| Wild type (34.0%) | 15 | 84 | ||
| EGFR exon mutated | 19 (52.6%) | 10 | 90 | 0.071¶ |
| 21 (40.1%) | 17 | 60 | ||
| 19 & 21 (7.3%) | 3 | 11 | ||
t-test.
Fisher's Exact Test.
Pearson Chi-Square.
Continuity correction. EGFR, epidermal growth factor receptor; SD, standard deviation.
Evaluation of treatment response
Objective tumor response was determined using the Response Evaluation Criteria in Solid Tumors (RECIST 1.1). All patients underwent a computed tomography (CT) scan covering target lesions every six weeks (two cycles) during the course of chemotherapy, and every eight weeks during the EGFR-TKI treatment phase of therapy. Brain or bone metastases were evaluated every three or six months by magnetic resonance imaging or bone scintigraphy.
Patient response to therapy was categorized into the following four groups, based on the RECIST guidelines: (i) complete response (CR); (ii) partial response (PR); (iii) stable disease (SD); and (iv) progression of disease (PD). OS was calculated as the time from the beginning of therapy to death or the last follow-up visit. PFS was calculated from the date of the beginning of chemotherapy to the date of tumor progression or death.
Specimen collection and DNA extraction
Each patient contributed a single sample (4 mL) of anticoagulated venous blood anytime before or after treatment. The samples were stored at 4°C for four hours, and then centrifuged at 2500 rpm for 10 minutes at 4°C. Then, 2 mL of Hanks solution was added to the sediment, and the mixture was poured into a 15 mL tube containing 4 mL of separating medium. After centrifugation at 1800 rpm for 15 minutes at 4°C, the suspended lymphocytes were removed and added to 4 mL Hanks solution. This mixture was then centrifuged at 2500 rpm for 10 minutes at 4°C, and the sediment (lymphocytes) were stored at −80°C. Genomic DNA was extracted from patient lymphocytes using a blood DNA kit (DP120420, Tiangen Biotech, Beijing, China). DNA concentrations were detected by Nanodrop spectrophotometer (Utrospec III, Pharmacia, Stockholm, Sweden), and DNA samples were stored at −20°C.
Determination of the BIM polymorphic deletion
Two separate polymerase chain reactions (PCR) were performed to determine the presence or absence of the deletion polymorphism in the DNA extracted from patient blood samples. The non-deletion allele was genotyped using the forward primer 5′-CCACCAATGGAAAAGGTTCA-3′ and the reverse primer 5′-CTGTCATTTCTCCCCACCAC-3.′ The deletion allele was genotyped using the forward primer 5′-CCACCAATGGAAAAGGTTCA-3′ and the reverse primer 5′-GGCACAGCCTCTATGGAGAA-3.′ PCR reactions were performed a using PCR instrument (2720 Thermal Cycler, Applied Biosystems, Grand Island, NY, USA) under the following conditions: 95°C for five minutes, (95°C for 30 seconds, 58°C for 50 seconds, and 72°C for one minute) × 35 cycles, and 72°C for 10 minutes. The PCR products for the deletion (284 bp) and the non-deletion (362 bp) alleles were analyzed on a 1.5% agarose gel. The first two PCR products were sequenced, and then used as positive controls in later studies.
Epidermal growth factor receptor (EGFR) mutation detection
The EGFR exon 19 deletion or exon 21 substituted mutations were detected by DHPLC, using a previously reported method.16 ARMS, a more sensitive method, was used to re-evaluate the cases with EGFR wild type by DHPLC.
Statistical analyses
Statistical analysis was performed using SPSS 16.0 software. The relationships between the BIM deletion polymorphism and relevant factors such as gender, age, smoking history, tumor stage, and histological type were examined using the χ2 test, with P < 0.05 representing a bilateral significant difference. The relationship between the BIM deletion polymorphism and response to treatment was analyzed by the χ2 test, and the relationships between the BIM deletion polymorphism and PFS and OS were analyzed using Kaplan-Meier survival curves. The prognostic factors for response to treatment, including age, gender, smoking history, tumor stage, differentiation, and BIM deletion polymorphism were examined by logistic analysis. The prognostic factors for PFS and OS following EGFR-TKI treatment were analyzed by Cox-regression analysis.
Results
BIM deletion polymorphism and clinic-pathologic parameters
Forty-five (15.5%) BIM deletions were detected in 290 lymphocyte samples. The rates of BIM deletion in the EGFR mutation group and EGFR wild type group were 15.7% (30/191) and 15.2% (15/99), respectively. Patients with or without the deletion polymorphism did not differ with respect to age, gender, smoking status, tumor differentiation, pathology, tumor stage or EGFR mutation status.
Association between BIM deletion polymorphism and EGFR-tyrosine kinase inhibitors (TKI) response/progression-free survival (PFS)
Among the 290 NSCLC patients, 103 received an EGFR-TKI as first-line treatment, and 187 received it as second-line treatment or beyond. As of 10 October 2013, 247 of the 290 patients showed signs of disease progression, and 43 of those patients were still receiving an EGFR-TKI.
The median PFS time for patients in the BIM deletion group was 4.60 months compared with 6.17 months for patients without the deletion (P = 0.549). Considering the different responses in patients with different pathology and EGFR-mutant status, we subdivided patients into EGFR-mutant adenocarcinoma (n = 159), EGFR-mutant squamous cell lung cancer (n = 25) and EGFR-wild type lung cancer (n = 99) groups. In subgroup analysis, the BIM deletion polymorphism was predictive for a shorter PFS in individuals with EGFR-mutant adenocarcinoma when treated with an EGFR-TKI (7.30 vs. 9.53 months, P = 0.034). However, the BIM deletion polymorphism was not a good predictor for PFS in EGFR-mutant squamous cell lung cancer (1.07 vs. 4.20 months, P = 0.646) and EGFR-wild type patients (2.07 vs. 2.06 months, P = 0.190). In EGFR-mutant adenocarcinoma cases, patients who received an EGFR-TKI as first-line treatment and as a second-line treatment or more, the median difference in PFS between groups with and without the BIM deletion was not statistically significant (7.87 vs. 11.43 months, P = 0.068 and 7.07 vs. 7.77 months, P = 0.283, respectively).
We next analyzed the association between the BIM deletion polymorphism and clinical outcomes. The overall response rates (ORR) and disease free survival rates (DCR) in patients with the BIM deletion polymorphism and treated with a EGFR-TKI were 33.3% and 71.1%, respectively, whereas the ORR and DCR in patients without the BIM deletion were 22.9% and 70.6%, (P = 0.946 and P = 0.133), respectively. Additionally, a subgroup analysis based on EGFR mutation, pathology, and treatment lines of TKI also failed to show significant differences in ORR and DCR between patients with or without the BIM deletion.
Association between BIM deletion polymorphism and chemotherapy response/PFS
Among the 290 patients enrolled in this study, 222 received chemotherapy. Because these patients were treated using a wide range of chemotherapy regimens, they were subdivided into the following five groups: (i) taxane chemotherapy (including docetaxel and paclitaxel); (ii) pemetrexed chemotherapy; (iii) gemcitabine chemotherapy; (iv) vinorelbine chemotherapy; and (v) platinum based chemotherapy. Although there was some patient overlap between the groups, each patient was included only once in each group, and only the first time they received a therapeutic regimen was taken into account.
At the time of the last follow-up visit on 10 April 2013, 202 of 222 patients had progressed, and 17 patients had suffered a serious adverse event (SAE), which caused the discontinuation of chemotherapy. SAEs occurred more often in patients with the BIM deletion (18.9%), compared to those without the BIM deletion (5.4%), and the difference between the two groups was statistically significant (P = 0.012).
The BIM deletion was predictive for a shorter PFS in individuals with EGFR-mutant NSCLC and treated with chemotherapy containing pemetrexed (3.32 vs. 5.30 months, P = 0.012), or platinum (4.07 vs. 4.73 months, P = 0.025). However, in the EGFR-wild type group, the predictive ability of the BIM deletion was not statistically significant.
Sixty-seven percent of patients received docetaxel as a second line therapy or beyond. In this group, we also found that the BIM deletion was a strong predictive factor for short response duration to taxane-containing chemotherapy (1.53 vs. 2.61 months, P = 0.025). Additionally, being homozygous rather than heterozygous for the BIM deletion was predictive for an even shorter PFS (0.75 vs. 2.70 months, P < 0.001).
The presence or absence of the BIM deletion polymorphism was not predictive for PFS in patients receiving chemotherapy containing gemcitabine or vinorelbine (P = 0.727 and P = 0.959, respectively), regardless of EGFR status (Table 2, Fig 1). Moreover, a subgroup analysis showed the BIM deletion had no affect on ORR (25.0% vs.19.6%, P = 0.481) or DCR (62.5% vs. 65.9%, P = 0.708) following chemotherapy. BIM deletion in EGFR-mutant non-squamous cell lung cancer is a negative predictive factor of chemotherapy (n = 166, 5.87 vs. 4.07, P = 0.010). However, the predictive ability was not seen in squamous cell lung cancer (n = 25).
Table 2.
BIM deletion polymorphism and response in different regimen
| BIM deletion | No deletion | BIM deletion | P-value | |||
|---|---|---|---|---|---|---|
| Median PFS (months) | 95%CI | Median PFS (months) | 95%CI | |||
| Taxanes (n = 115) | 13 (11.3%) | 2.61 | 1.90–3.77 | 1.53 | 0.63–2.43 | 0.025 |
| EGFR + (n = 60) | 6 | 2.02 | 0.89–3.17 | 1.78 | 0.77–2.29 | 0.129 |
| EGFR – (n = 55) | 7 | 2.22 | 1.68–3.79 | 1.27 | 1.01–1.52 | 0.470 |
| Pemetrexed (n = 112) | 21 (18.75%) | 4.17 | 3.82–5.72 | 2.80 | 1.99–5.68 | 0.111 |
| EGFR +(n = 71) | 14 | 5.30 | 3.82–8.04 | 3.32 | 2.31–5.36 | 0.012 |
| EGFR – (n = 41) | 7 | 3.51 | 2.62–4.64 | 1.67 | 0.32–5.86 | 0.461 |
| Gemcitabine (n = 72) | 15 (20.8%) | 3.93 | 3.75–4.51 | 2.27 | 2.92–7.54 | 0.727 |
| EGFR + (n = 42) | 11 | 3.35 | 2.78–4.12 | 1.77 | 1.19–6.27 | 0.880 |
| EGFR – (n = 29) | 4 | 4.90 | 2.47–7.87 | 5.23 | 3.63–16.50 | 0.960 |
| Vinorelbine (n = 18) | 3 (16.6%) | 4.27 | 2.47–6.07 | 5.00 | 3.88–6.12 | 0.959 |
| EGFR + (n = 15) | 2 | 4.07 | 1.86–6.45 | 5.04 | † | 0.842 |
| EGFR – (n = 3) | 1 | 4.49 | † | 5.00 | † | 0.225 |
| Platinum (n = 220) | 32 | 4.20 | 3.82–4.98 | 4.19 | 3.13–6.47 | 0.210 |
| EGFR + (n = 137) | 19 | 4.73 | 1.94–6.20 | 4.07 | 3.40–6.39 | 0.025 |
| EGFR – (n = 83) | 13 | 3.67 | 3.13–5.01 | 5.00 | 3.06–9.00 | 0.419 |
| EGFR-TKIs (n = 290) | 45 (15.5%) | 6.17 | 5.48–8.78 | 4.60 | 1.00–8.49 | 0.549 |
| EGFR + (n = 191) | 30 | 8.47 | 7.47–11.27 | 6.69 | 2.67–9.23 | 0.023 |
| EGFR – (n = 99) | 15 | 2.06 | 1.24–4.56 | 2.07 | 0.67–4.53 | 0.190 |
Cannot be calculated because of limited number of cases. CI, confidence interval; EGFR, epidermal growth factor receptor; PFS, progression-free survival; TKIs, tyrosine kinase inhibitors.
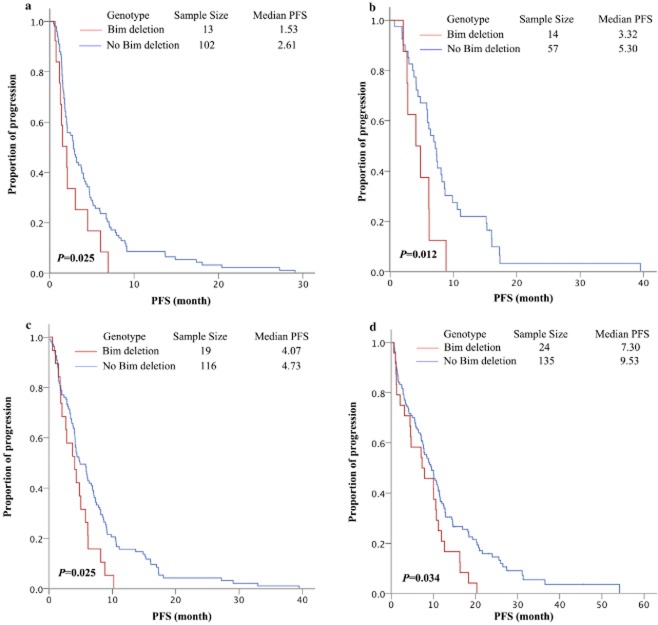
Figure 1.
Correlation with BIM deletion and progression-free survival of chemotherapy or epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs). (a) Taxanes contained chemotherapy for non-small cell lung cancer (NSCLC) as second-/beyond-line (1.53 vs. 2.61 months, P = 0.025). (b) EGFR-mutant NSCLC treated with pemetrexed contained chemotherapy (3.32 vs. 5.30 months, P = 0.012). (c) EGFR-mutant NSCLC platinum contained chemotherapy (4.07 vs. 4.73 months, P = 0.025). (d) EGFR-mutant adenocarcinoma treated with EGFR-TKIs (7.30 vs. 9.53 months, P = 0.034).
Association between BIM deletion polymorphism and patient survival
As of 10 April 2013, 182 (62.5%) of the 290 patients had died, 16 (5.5%) had been lost to follow-up, and 92 patients remained alive. The presence of a BIM deletion was not associated with OS (21.87 vs. 21.90 months, P = 0.627). Similarly, the OS rates of patients with the BIM deletion in the EGFR mutation and wild-type groups were 21.10 and 21.87 months, respectively, which were similar to those of patients without the BIM deletion (20.50 and 21.41 months, respectively), and the differences were not statistically significant.
Discussion
Earlier studies have suggested a link between the BIM deletion and resistance to targeted therapy with EGFR-TKIs.14,15,17 However, no data relevant to such findings has been collected for Chinese lung cancer patients. While an earlier study focused only on EGFR-mutant patients who received EGFR-TKIs, we found that the BIM deletion polymorphism was associated with a poor clinical outcome following EGFR-TKIs, as well as other types of chemotherapeutic agents, and may be useful as a negative predictive marker when selecting patients for such treatment.
In our study, the BIM deletion was associated with a poor outcome following chemotherapy with an EGFR-TKI in Chinese NSCLC patients with an EGFR mutation, which is accordant with results obtained by Ng et al.15 However, the predictive value of the BIM marker for EGFR-TKI treatment was not as strong as the predictive value of the EGFR mutation. Lynch et al.18 found that specific EGFR mutations were correlated with tumor response to gefitinib. EGFR-TKIs greatly increased PFS following EGFR-TKI therapy in cases with EGFR-mutant adenocarcinoma,4–8 and we found that the BIM deletion was predictive for an approximately two month decrease in PFS following TKI treatment (7.30 vs. 9.53 months, P = 0.034). Certain studies have identified the ERK and AKT pathways as being the two main systems downstream of EGFR that directly regulate activity of BIM.19–22 When activated, ERK phosphorylates BIM and promotes BIM degradation. AKT suppresses BIM transcription factor FOXO3a.23 When a BIM deletion occurs, apoptosis cannot be effectively initiated, even when the AKT and ERK pathways are repressed by EGFR-TKIs.
Subgroup analysis indicated that BIM deletion predicts shorter PFS only in EGFR-mutant adenocarcinoma lung cancer patients treated with EGFR-TKIs. A possible reason for a lack of a statistically significant association between BIM deletion and EGFR-TKIs in squamous cell lung cancer may be because of the limited number of patients in this subgroup (n = 25). As we have seen a difference in median PFS (1.07 vs. 4.20 months), larger sample clinical trials are needed to prove the negative predictive effects of BIM deletion in EGFR-mutant squamous cell lung cancer.
The BIM deletion is also predictive for a shorter PFS following chemotherapy with taxanes, platinum, and pemetrexed. Several researchers have proposed a link between paclitaxel resistance and BIM. Tan et al.20 reported that paclitaxel resistance was a result of BIM inactivation, and Li et al.22 suggested that BIM expression is related to paclitaxel treatment. Our clinical study confirmed these prior results using NSCLC cell lines. The mechanism of NSCLC resistance to platinum therapy mainly involves a failure to induce apoptosis. Wang et al. found that apoptosis induction by cisplatin in human NSCLC cells mainly occurs through the stress pathway.24 As BIM is an initiator of this pathway,23 it explains our clinical results. Regarding pemetrexed, very few studies have been conducted examining the relationship between BIM and the agent, and further investigations are needed.
In EGFR-wild type NSCLC patients, the BIM deletion was not predictive for PFS following treatment with EGFR-TKIs or certain other chemotherapy regimens, and we cannot fully explain the potential mechanisms contributing to this phenomenon. Based on the results described above, we speculate that BIM plays a significant role in triggering apoptosis, but only in patients with an EGFR mutation. While BIM mutations are present in both EGFR wild-type NSCLC cells and normal cell lines, they exist in an inactive precursor state.
Our study showed that the BIM deletion polymorphism is predictive of a negative outcome only in EGFR-mutant patients who receive pemetrexed and platinum based chemotherapy. But it was predictive of a shorter PFS in a taxane-based regimen with all patients enrolled. A possible explanation for this phenomenon is that most patients in this study received taxanes as second-line therapy or beyond. Because of intra-tumor genetic heterogeneity,25 chemotherapy may greatly influence EGFR mutation status. Therefore, the pre-treatment EGFR mutation status of a patient may not be completely consistent with the status following chemotherapy.
Eight large cell NSCLC patients were enrolled in our study, and two had the BIM polymorphic deletion. The BIM mutation rate in large cell NSCLC was higher than that in NSCLC (15.5% vs. 25.0%, P = 0.469). A larger prospective clinical study is required to determine whether a difference in BIM mutation rates may explain the poor response of large cell NSCLC to EGFR-TKIs.
The BIM deletion was not correlated with OS, even in the EGFR-mutant group. However, the BIM deletion may still serve as a prognostic factor, because of its ability to predict negative outcomes for both targeted therapy and chemotherapy. Also, identification of the BIM deletion, when used in conjunction with characterization of the tumor stage,26 differentiation, pathology, tumor driver gene,27 and other factors may be useful for predicting OS rates of NSCLC patients.
Our current study had several limitations. First, this was a retrospective study conducted at a single medical center, and a prospective study is needed to verify the results. Second, the patient cohort sizes were small and the mutation rate was low, especially in subgroup analyses, which may result in some bias in the data analyzed. Third, the chemotherapy regimens and doses used in this study varied among patients, and may have biased the results.
Conclusion
In summary, results of our study suggest that the BIM deletion polymorphism can be used as a marker to predict a negative outcome when treating EGFR-mutant NSCLC patients with either EGFR-TKIs or chemotherapy. A prospective, multicenter study is required to validate our results.
Acknowledgments
We thank all patients for their participation in this study. We thank Dr. Ning Wang and Liping Qi, radiologists from Radiology Department of Beijing Cancer Hospital & Institute, for their contribution to response assessment.
Disclosure
No authors report any conflict of interest.
References
- Olaussen KA, Mountzios G, Soria JC. ERCC1 as a risk stratifier in platinum-based chemotherapy for nonsmall-cell lung cancer. Curr Opin Pulm Med. 2007;13:284–289. doi: 10.1097/MCP.0b013e32816b5c63. [DOI] [PubMed] [Google Scholar]
- Rosell R, Scagliotti G, Danenberg KD, et al. Transcripts in pretreatment biopsies from a three-arm randomized trial in metastatic non-small-cell lung cancer. Oncogene. 2003;22:3548–3553. doi: 10.1038/sj.onc.1206419. [DOI] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, et al. Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol. 2005;23:5900–5909. doi: 10.1200/JCO.2005.02.857. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S, Adams JM. The Bcl2 family: regulators of the cellular life-or-death switch. Nat Rev Cancer. 2002;2:647–656. doi: 10.1038/nrc883. [DOI] [PubMed] [Google Scholar]
- Yano T, Ito K, Fukamachi H, et al. The RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells undergoing transforming growth factor beta-induced apoptosis. Mol Cell Biol. 2006;26:4474–4488. doi: 10.1128/MCB.01926-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner A, Barrett T, Flavell RA, Davis RJ. Multisite phosphorylation regulates Bim stability and apoptotic activity. Mol Cell. 2008;30:415–425. doi: 10.1016/j.molcel.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- Liu JW, Chandra D, Tang SH, Chopra D, Tang DG. Identification and characterization of Bimgamma, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res. 2002;62:2976–2981. [PubMed] [Google Scholar]
- Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- Tan TT, Degenhardt K, Nelson DA, et al. Key roles of BIM-driven apoptosis in epithelial tumors and rational chemotherapy. Cancer Cell. 2005;7:227–238. doi: 10.1016/j.ccr.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, et al. BH3-only proteins Puma and Bim are rate-limiting for gamma-radiation- and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood. 2005;106:4131–4138. doi: 10.1182/blood-2005-04-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Moudgil T, Ross HJ, Hu HM. Apoptosis of non-small-cell lung cancer cell lines after paclitaxel treatment involves the BH3-only proapoptotic protein Bim. Cell Death Differ. 2005;12:292–303. doi: 10.1038/sj.cdd.4401554. [DOI] [PubMed] [Google Scholar]
- Akiyama T, Dass CR, Choong PF. Bim-targeted cancer therapy: a link between drug action and underlying molecular changes. Mol Cancer Ther. 2009;8:3173–3180. doi: 10.1158/1535-7163.MCT-09-0685. [DOI] [PubMed] [Google Scholar]
- Wang L, Chanvorachote P, Toledo D, et al. Peroxide is a key mediator of Bcl-2 down-regulation and apoptosis induction by cisplatin in human lung cancer cells. Mol Pharmacol. 2008;73:119–127. doi: 10.1124/mol.107.040873. [DOI] [PubMed] [Google Scholar]
- Bai H, Wang Z, Chen K, et al. Influence of chemotherapy on EGFR mutation status among patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:3077–3083. doi: 10.1200/JCO.2011.39.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–180. doi: 10.1016/S1470-2045(10)70087-5. [DOI] [PubMed] [Google Scholar]