Abstract
Background
The echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) rearrangement is almost in mutual exclusion to epidermal growth factor receptor (EGFR) and v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations, with rare exceptions. This study aimed to search for the coexisting gene alterations in Chinese patients with lung adenocarcinoma (LAC).
Methods
We detected mutations in the EGFR, KRAS, and ALK gene rearrangements in samples from 131 Chinese patients with LAC. ALK rearrangements were identified by fluorescent in situ hybridization. Mutations in EGFR (exons 19 to 21) and KRAS (codons 12 and 13) were determined by real time polymerase chain reaction.
Results
All patients were classified into four distinct genotype groups: EGFR mutations (n = 63; 48.1%), ALK rearrangements (n = 9; 6.9%), KRAS mutations (n = 8; 6.1%), and the wild-type (unmutated) genotype of all three genes (WT/WT/WT) (n = 53; 40.5%). Interestingly, two never-smoking women (2/131, 1.5%) harbored coexisting ALK rearrangement and EGFR mutation. ALK rearrangement occurred more frequently in young patients (8/9) (P = 0.687), non-smokers (8/9) (P = 0.077), and those who had no family history of LAC (8/9) (P = 1.000); all KRAS mutations occurred in the EGFR wild type (P = 0.007). KRAS mutations were generally detected in young patients (6/8) (P = 0.658) and in those who had no family history (7/8) (P = 1.000); EGFR mutations correlated with gender (P = 0.001), and smoking status (P < 0.001).
Conclusions
Two patients harboring both EGFR mutation and EML4-ALK rearrangement were detected in this study. Our data was apparently inconsistent with the traditional view that the EML4-ALK fusion gene in patients is resistant to EGFR-TKIs.
Keywords: Coexisting, EGFR, EML4-ALK, K-RAS, lung cancer
Introduction
Non–small cell lung cancer (NSCLC) is increasingly recognized as a heterogeneous set of diseases at the molecular level and these differences can drive therapeutic decision making.1–3 These genes have been defined as “driver genes,” and such driver genes include epidermal growth factor receptor (EGFR), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), v-raf murine sarcoma viral oncogene homolog B (BRAF), phosphatidylinositol-4, 5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA), and echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4- ALK). The discovery of a number of these molecular alterations underlying lung cancer has led to uniquely targeted therapies with specific inhibitor drugs, such as erlotinib and gefitinib for mutations in the EGFR4 or crizotinib for the gene translocation resulting in the EML4-ALK oncogene.1
In 2007, the fusion of ALK with EML4 was identified in NSCLC. The EML4-ALK fusion gene arises as a result of an inversion in chromosome 2 that juxtaposed the 5 end of the EML4 gene with the 3 end of the ALK gene. The frequency of the fusion gene is approximately 6.7% in NSCLC.5 The clinical features of NSCLC that harbor EML4-ALK include light- or never-smokers, younger age and adenocarcinomas.6,7 These same features can also be observed in NSCLC patients likely to harbor EGFR mutations.8–10
Many studies focus on the relationships between EML4-ALK and EGFR gene mutation. Previous studies5,6,11–13 have proved that the EML4-ALK fusion gene and EGFR gene mutation is independent of molecular events; the EML4-ALK fusion gene can be another reason for acquired resistance to EGFR-tyrosine kinase inhibitors (TKIs), such as the KRAS mutation.6,14 To the best of our knowledge, the coexistence of the EML4-ALK fusion gene and EGFR gene mutation has only been reported in six patients.15–20
According to previous research, EGFR and ALK concomitant mutations occur most frequently in women, East Asians, non-smokers, and patients with adenocarcinomas. In this study, we investigated 131 Chinese patients who were diagnosed with lung adenocarcinoma (LAC). We analyze the relationship between EML4-ALK fusion genes and EGFR and KRAS mutations to further clarify the relationship between clinicopathologic features and these three gene mutations.
Material and methods
Tumor specimens
Tumor specimens were obtained from 131 consecutive patients who underwent surgery for pathologically proven LAC between November 2011 and November 2012 at Sun Yat-sen University Cancer Center. Detailed demographic and clinical information of these patients was available from the Sun Yat-sen University Cancer Center Surveillance System. No patient underwent either neoadjuvant chemoradiotherapy or target therapy. Tumors were staged according to the International Association of the Study for Lung Cancer (IASLC) tumor node metastasis (TNM) staging system.21 The tumor blocks were fixed in 10% buffered formaldehyde and embedded in paraffin. The blocks were cut in 4 μm consecutive sections and stained with hematoxylin and eosin (HE). Slides rich in viable tumor cells were submitted for fluorescent in situ hybridization (FISH) and real time polymerase chain reaction (RT-PCR) analysis.
Fluorescent in situ hybridization (FISH) analysis of anaplastic lymphoma kinase (ALK) rearrangements
Bacterial artificial chromosomes, RP11-667I6 and RP11-100C1 (Children's Hospital Oakland Research Institute), were used as break-apart probes for the EML4 and ALK genes, respectively. Bacterial artificial chromosome DNA was labeled with either spectrum red 2-deoxyuridine, 5-triphosphate (dUTP) or spectrum green 11-dUTP by nick translation (Vysis) according to the manufacturer's recommendations. Slides for metaphase FISH from the cell lines were prepared using standard cytogenetic methodologies. The probes were hybridized and washed according to standard FISH procedures. A positive FISH result for ALK rearrangement was defined as >15% of tumor cells with a split signal and was confirmed by immunohistochemistry.
Real-time polymerase chain reaction (RT-PCR) analysis of epidermal growth factor receptor (EGFR) and KRAS mutations
We used an EGFR kit (GP Medical Technologies Ltd, Beijing, China) to detect a deletion in exon 19 (delE746A750) and a mutation in exon 21 (L858R) with RT-PCR. Details of sequencing and diagnosis criteria have been described previously.22 Genotyping of KRAS exon 2 (codons 12 and 13) was performed by RT-PCR detection according to standard protocols.23
Statistical analyses
Statistical analysis was performed using SPSS software (standard version 16.0, SPSS, Chicago, IL). The Chi-square or Fisher's exact test was performed to determine the associations between the presence of three major oncogenic alterations and patients' characteristics. A two-sided probability value of less than 0.05 was considered statistically significant.
Results
Patient characteristics
A total of 57 women and 74 men, aged from 31 to 85 years (median, 58.0 years), were included in the study. Of the 131 patients, 57 were smokers, and 74 were non-smokers. The other clinical-pathologic characteristics of the 131 patients are listed in Table 1.
Table 1.
Relationships between clinical-pathological characteristics and these three genes in patients with lung adenocarcinoma
| Variable | Total | EML4-ALK rearrangement | EGFR mutation | KRAS mutation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 131 | n | % | P-value | n | % | P-value | n | % | P-value | |
| Gender | 0.502 | 0.001 | 0.728 | |||||||
| Male | 74 | 4 | 5.41 | 26 | 35.14 | 4 | 5.41 | |||
| Female | 57 | 5 | 8.77 | 37 | 64.91 | 4 | 7.02 | |||
| Age | 0.687 | 1.000 | 0.658 | |||||||
| ≤65 | 105 | 8 | 7.61 | 51 | 48.57 | 6 | 5.71 | |||
| >65 | 26 | 1 | 3.84 | 12 | 46.15 | 2 | 7.69 | |||
| Smoking status | 0.077 | <0.001 | 0.728 | |||||||
| Never | 74 | 8 | 10.81 | 46 | 62.16 | 4 | 5.41 | |||
| Ever | 57 | 1 | 1.75 | 17 | 29.82 | 4 | 7.02 | |||
| Family history† | 1.000 | 0.215 | 1.000 | |||||||
| No | 112 | 8 | 7.14 | 51 | 45.54 | 7 | 6.25 | |||
| Yes | 19 | 1 | 5.26 | 12 | 63.16 | 1 | 5.26 | |||
| PTNM stage | 0.293 | 0.060 | 0.691 | |||||||
| IA | 29 | 3 | 10.34 | 13 | 44.83 | 1 | 3.45 | |||
| IB | 32 | 2 | 6.25 | 15 | 46.88 | 3 | 9.38 | |||
| IIA | 11 | 1 | 9.09 | 2 | 18.18 | 1 | 9.09 | |||
| IIB | 5 | 1 | 20.00 | 2 | 40.00 | 1 | 20.00 | |||
| IIIA | 38 | 1 | 2.63 | 21 | 55.26 | 2 | 5.26 | |||
| IIIB | 3 | 1 | 33.33 | 0 | 0.00 | 0 | 0.00 | |||
| IV | 13 | 0 | 0.00 | 10 | 76.92 | 0 | 0.00 | |||
Family history: whether the three generations of immediate family members of the patients had malignant tumor. EGFR, epidermal growth factor receptor; EML4-ALK, echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase; KRAS, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; PTNM, pathological tumor node metastasis stage.
Correlation between the clinic-pathological characteristics and three major oncogenic alterations
Of the 131 specimens, ALK rearrangement was detected in 9/131(6.9%). ALK rearrangement occurred more frequently in young patients (8/9) (P = 0.687), non-smokers (8/9) (P = 0.077), and those who had no family history of LAC (8/9) (P = 1.000). The rate of EGFR mutation in this study was 63/131 (48.1%), including exon 19 deletion mutations in 31 (23.7%) and exon 21 point mutations in 32 (24.4%) samples. There were significantly higher mutation rates in women (P = 0.001) and non-smokers (P < 0.001). KRAS was positive in eight (6.1%) of these 131 specimens, including codon 12 mutants in seven cases and one case with codon 13 mutant. The higher KRAS mutations occurred more frequently in young patients (6/8) and those who had no family history of LAC (7/8), although, there were no statistically significant results (Table 1). The frequency of no mutations (wild type [WT]) in any of the above three genes was 53/131 (40.5%) (Fig 1).
Figure 1.
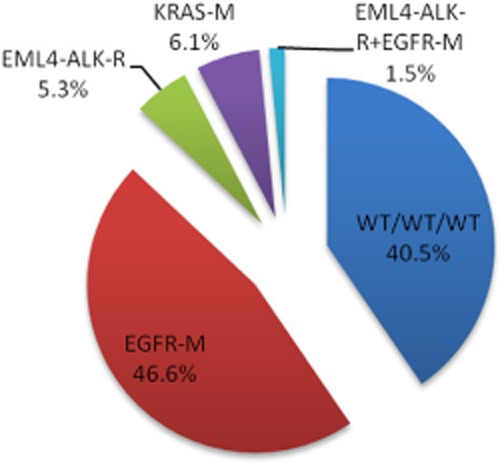
The proportions of major oncogenic alterations in 131 Chinese patients with lung adenocarcinoma. EGFR, epidermal growth factor receptor; EML4-ALK-R, echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase; KRAS, v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; M, mutation; R, rearrangement; WT/WT/WT, wild type (unmutated) of all 3 genes.
Relationship between the three major oncogenic alterations
In our data, ALK rearrangement was detected in 9/131 (6.9%), the rate of EGFR mutations was 63/131 (48.1%), and KRAS mutations were detected in 8/131 (6.1%) specimens. KRAS mutations occurred in EGFR wild type samples (P = 0.007) and similar results were seen in ALK rearrangement (P = 1.000), however, there were no statistically significant results (Table 2). ALK rearrangement was seen in the EGFR wild type, but two samples harbored both EGFR mutation and ALK rearrangement. Further analysis revealed that these two cases were female non-smokers and harbored mutations in exon 19 (delE746A750).
Table 2.
Relationships between these three genes in patients with lung adenocarcinoma
| Variable | Total | EGFR mutation | KRAS mutation | ||||
|---|---|---|---|---|---|---|---|
| n = 131 | n = 63 | % | P-value | n = 8 | % | P-value | |
| EML4-ALK status | 0.167 | 1.000 | |||||
| Rearrangement | 9 | 2 | 0 | ||||
| Wild | 122 | 61 | 8 | ||||
| KRAS status | 0.007 | 1.000 | |||||
| Wild | 123 | 63 | 0 | ||||
| Mutation | 8 | 0 | 8 | ||||
Discussion
While NSCLC was previously considered to be a single disease treated with standard cytotoxic chemotherapy, it is now becoming more appropriate to consider NSCLC as a collection of disease subtypes according to the driving oncogenic alteration, and to select treatment accordingly. Among many alterations, EGFR mutations, ALK rearrangements, and KRAS mutations are the three most frequently identified and clinically relevant genetic alterations in NSCLC.5,24,25
Two or more mutations of driver genes could exist concurrently in NSCLC. Lipson et al. identified 50 alterations in 21 genes, with at least one alteration being present in 83.0% (20 out of 24) of NSCLC patients (with a range of 1–7 alterations).26 Co-existence of multiple driver mutations has been taken into consideration by oncologists to obtain an in-depth understanding of cancer mechanism and therapeutic developments.27,28 In our research, about 1.5% of patients (2/131) harbored both EGFR mutation and ALK rearrangement, while further analysis revealed that these two cases were female non-smokers and harbored mutations in exon 19 (delE746A750).
EGFR is a 170 kilodaltons (kDa) membrane-bound protein encoded by 28 exons on chromosome 7p12, and has a tyrosine kinase activity after binding of several specific ligands to the extracellular domain. EGFR-tyrosine kinase has become an attractive target for the treatment of NSCLC, and agents targeting this receptor, including gefitinib, erlotinib, and cetuximab, are being investigated.29 Many studies report an association between mutations and some of the clinical features of NSCLC. EGFR mutation was suggested to be the best predictor for TKI sensitivity; it should be the patient selection criterion for EGFR targeting therapy. The rates of mutation in those studies were 44-55% in patients with adenocarcinoma, 51–68% in non-smokers, 42–62% in women, and 30–50% in Asian ethnicity, but, in contrast, the mutation rates were 10% in smokers, 14% in men, and 8% in adenocarcinoma patients from western countries.19,24,25,29 This was confirmed in our study, as we found significantly higher EGFR mutation rates in women (P = 0.001) and in non-smokers (P < 0.001).
ALK rearrangement, which results from a small inversion within chromosome 2p, is a newly identified driver oncogene in NSCLC.5,30 Results from a recent clinical trial of crizotinib, an inhibitor of the met proto-oncogene (hepatocyte growth factor receptor [MET]) and ALK, demonstrated a remarkable response rate and progression-free survival benefit in ALK-positive patients. In an unselected NSCLC population, the frequency of the ALK fusion gene ranged from 1.5% to 7.5%.5,19,31The frequency of ALK rearrangements (6.9%) in our study was similar to that reported for LAC in previous reports. ALK rearrangement occurred more frequently in young patients (8/9) (P = 0.687), non-smokers (8/9) (P = 0.077), and those who had no family history of LAC (8/9) (P = 1.000), but there was no statistical significance.
KRAS mutations are oncogenic missense mutations that occur more frequently in adenocarcinomas.32 Because tumors with KRAS mutations display primary resistance to EGFR-TKIs, molecular evaluation of KRAS is important to predict clinical treatment outcomes and to decide on a therapeutic option when treating NSCLC.24,33 In our study, KRAS mutations occurred more often in young patients (6/8) and in those who had no family history of LAC (7/8), although there were no statistically significant results.
Clinicopathologic features of NSCLC patients with EML4-ALK rearrangement and patients with (EGFR) mutations and KRAS mutations, including younger age, adenocarcinoma, and never or light smoking, are similar, but EML4-ALK rearrangement and EGFR and KRAS mutations have been shown to be mutually exclusive.11,13,34 However, there has been a rare case report regarding NSCLC patients who harbored the EML4-ALK fusion gene and EGFR gene mutations (in Table 3).
Table 3.
The clinical-pathological characteristics of non-small cell lung cancer harbored coexisting echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase (EML4-ALK) rearrangement and epidermal growth factor receptor (EGFR) mutation
| Patient No. | Gender | Age (years) | Smoking status | Histology | Ethnicity | EGFR mutation status | EML4-ALK fusion | EGFR-TKI response |
|---|---|---|---|---|---|---|---|---|
| 118 | Female | 56 | Never smoker | AC | Chinese | Exon19 deletion | Variant 3b | NA |
| 219 | Male | 72 | 60 pack year | AC+SCLC | Japanese | Exon19 deletion | Variant 1 | No use |
| 316 | Male | 48 | Never smoker | ASC | Italian | Exon19 deletion | Variant 1 | PD |
| 415 | Male | 72 | Never smoker | AC | Chinese | Exon21 L585 | Variant 1 | PR |
| 517 | Female | 65 | Never smoker | AC | English | Exon19 deletion | Unknown | CR |
| 620 | Female | 39 | Light smoker | AC | Japanese | Exon21 L585 | Variant 3b | PD |
| Present case 1 | Female | 54 | Never smoker | AC | Chinese | Exon19 deletion | Variant 1 | No use |
| Present case 2 | Female | 61 | Never smoker | AC | Chinese | Exon19 deletion | Variant 1 | No use |
AC, adenocarcinoma; ASC, adenosquamous carcinoma; CR, complete response; EGFR, epidermal growth factor receptor; EML4-ALK, echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase; PD, progressive disease; PR, partial response; SCLC, small cell lung cancer; TKI, tyrosine kinase inhibitor.
Of the eight patients in our study, the majority are female (5/8), Asian (6/8), never smokers (6/8), and with adenocarcinoma (6/8); the median age was 58 years old; the EGFR mutations are deletions in exon 19 (6/8) and can also be exon 21 (2/8), but EML4-ALK is the main variable one fusion. Of particular note, five patients received EGFR-TKI therapy (Table 3). One case showed partial response and one case showed complete response. Our data are apparently inconsistent with the traditional view that the EML4-ALK fusion gene in patients is resistant to EGFR-TKIs.
Our study was limited by the small sample size from a single institution. Our findings need to be proven by larger samples in further studies.
Conclusion
EML4-ALK rearrangement and EGFR and KRAS mutations have been shown to be mutually exclusive. In our research, 1.5% harbored coexisting ALK rearrangement and EGFR mutation. ALK status should be investigated in unexplained cases of TKI-resistance of EGFR mutated NSCLC.
Acknowledgments
This research was supported by National and Provincial Science and Technology Projects (No. 2012AA021502 and No.2012B031800295).
Disclosure
No authors report any conflict of interest.
References
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- Jänne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol. 2013;14:38–47. doi: 10.1016/S1470-2045(12)70489-8. [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA. 2003;290:2149–2158. doi: 10.1001/jama.290.16.2149. [DOI] [PubMed] [Google Scholar]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- Wong DW, Leung EL, So KK, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115:1723–1733. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol. 2010;17:889–897. doi: 10.1245/s10434-009-0808-7. [DOI] [PubMed] [Google Scholar]
- Solomon B, Varella-Garcia M, Camidge DR. ALK gene rearrangements: a new therapeutic target in a molecularly defined subset of non-small cell lung cancer. J Thorac Oncol. 2009;4:1450–1454. doi: 10.1097/JTO.0b013e3181c4dedb. [DOI] [PubMed] [Google Scholar]
- Mao C, Qiu LX, Liao RY, et al. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung Cancer. 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Kuo YW, Wu SG, Ho CC, Shih JY. Good response to gefitinib in lung adenocarcinoma harboring coexisting EML4-ALK fusion gene and EGFR mutation. J Thorac Oncol. 2010;5:2039–2040. doi: 10.1097/JTO.0b013e3181f43274. [DOI] [PubMed] [Google Scholar]
- Tiseo M, Gelsomino F, Boggiani D, et al. EGFR and EML4-ALK gene mutations in NSCLC: a case report of erlotinib-resistant patient with both concomitant mutations. Lung Cancer. 2011;71:241–243. doi: 10.1016/j.lungcan.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Popat S, Vieira de Araújo A, Min T, et al. Lung adenocarcinoma with concurrent exon 19 EGFR mutation and ALK rearrangement responding to erlotinib. J Thorac Oncol. 2011;6:1962–1963. doi: 10.1097/JTO.0b013e31822eec5e. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer. 2010;9:188. doi: 10.1186/1476-4598-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokawa G, Taguchi K, Ohba T, et al. First case of combined small-cell lung cancer with adenocarcinoma harboring EML4-ALK fusion and an exon 19 EGFR mutation in each histological component. J Thorac Oncol. 2012;7:e39–41. doi: 10.1097/JTO.0b013e3182762bcb. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Hayashi A, Morimoto T, et al. A case of lung adenocarcinoma harboring EGFR mutation and EML4-ALK fusion gene. BMC Cancer. 2012;12:558. doi: 10.1186/1471-2407-12-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- Zhang LJ, Cai L, Li Z, et al. Relationship between epidermal growth factor receptor gene mutation and copy number in Chinese patients with non-small cell lung cancer. Chin J Cancer. 2012;31:491–499. doi: 10.5732/cjc.011.10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcombe D, Theaker J, Guy SP. Brown T, Little S Detection of PCR products using self-probing amplicons and fluorescence. Nat Biotechnol. 1999;17:804–807. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Lin L, Takahashi T, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res. 2008;14:5731–5734. doi: 10.1158/1078-0432.CCR-08-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Res. 2012;22:375–385. doi: 10.1101/gr.120477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelzang NJ, Benowitz SI, Adams S, et al. Clinical cancer advances 2011: Annual Report on Progress Against Cancer from the American Society of Clinical Oncology. J Clin Oncol. 2012;30:88–109. doi: 10.1200/JCO.2011.40.1919. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Horn L, Pao W. EML4-ALK: honing in on a new target in non-small-cell lung cancer. J Clin Oncol. 2009;27:4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamura K, Takeuchi K, Togashi Y, et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol. 2008;3:13–17. doi: 10.1097/JTO.0b013e31815e8b60. [DOI] [PubMed] [Google Scholar]
- Gealy R, Zhang L, Siegfried JM, Luketich JD, Keohavong P Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomarkers Prev. 1999;8:297–302. [PubMed] [Google Scholar]
- Riely GJ, Marks J, Pao W. KRAS mutations in non-small cell lung cancer. Proc Am Thorac Soc. 2009;6:201–205. doi: 10.1513/pats.200809-107LC. [DOI] [PubMed] [Google Scholar]
- Kim HR, Shim HS, Chung JH, et al. Distinct clinical features and outcomes in never-smokers with nonsmall cell lung cancer who harbor EGFR or KRAS mutations or ALK rearrangement. Cancer. 2012;118:729–739. doi: 10.1002/cncr.26311. [DOI] [PubMed] [Google Scholar]


