Abstract
Background
Francisella tularensis, a gram-negative bacterium replicates intracellularly within macrophages and efficiently evades the innate immune response. It is able to infect and replicate within Kupffer cells, specialized tissue macrophages of the liver, and to modulate the immune response upon infection to its own advantage. Studies on Francisella tularensis liver infection were mostly performed in animal models and difficult to extrapolate to the human situation, since human infections and clinical observations are rare.
Results
Using a human co-culture model of macrophages and hepatocytes we investigated the course of infection of three Francisella tularensis strains (subspecies holarctica – wildtype and live vaccine strain, and mediasiatica - wildtype) and analyzed the immune response triggered upon infection. We observed that hepatocytes support the intracellular replication of Franciscella species in macrophages accompanied by a specific immune response inducing TNFα, IL-1β, IL-6 and fractalkine (CX3CL1) secretion and the induction of apoptosis.
Conclusions
We could demonstrate that this human macrophage / hepatocyte co-culture model reflects strain-specific virulence of Francisella tularensis. We developed a suitable tool for more detailed in vitro studies on the immune response upon liver cell infection by F. tularensis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0621-3) contains supplementary material, which is available to authorized users.
Keywords: Francisella tularensis, Human macrophage, Hepatocyte, Co-culture, Liver
Background
Francisella tularensis (F. tularensis) is a gram-negative bacterium that causes the zoonosis tularemia [1]. Due to its high infectivity it is considered as a class A bioweapon agent [2]. F. tularensis primarily infects and persists in macrophages and is thus used as model bacterium to study adopted strategies to evade primary immune detection [3]. As human infections with F. tularensis and related human clinical trials are rare, most of the data available on mechanisms of F. tularensis replication are derived from mouse models [4]. However, due to interspecies differences in the immune response and opposing evolutionary strategies for resistance vs. tolerance or resilience to infection [5, 6] it remains difficult to extrapolate results obtained in animal models to the human situation [7].
Currently, four subspecies (ssp.) of F. tularensis are generally accepted: ssp. tularensis, holarctica, mediasiatica, and novicida. F. tularensis ssp. tularensis is highly virulent in hares and the cause of Type A tularemia, whereas F. tularensis ssp. holarctica is less virulent and causes Type B tularemia; F. tularensis subsp. mediasiatica is a rare pathogen with unique biochemical characteristics that has only been isolated in Kazakhstan and Turkmenistan in Central Asia and exhibits virulence in hares similar to Type B organisms [8]. Since the infectious dose of F. tularensis wild type strains is very low, the attenuated type B live vaccine strain (LVS) thus has often been used as surrogate for the virulent strains. The LVS strain is able to induce diseases in mice similar to those seen in humans, but possesses practically no risk for laboratory personnel [9–11]. Both, virulent and attenuated strains of F. tularensis, survive macrophage phagocytosis. They escape into the cytoplasm by preventing acidification and maturation of the phagosome [12–14].
Kupffer cells represent specialized tissue macrophages within the liver [15] and account for 80 – 90 % of the total macrophage pool of the body [16]. In response to F. tularensis infection, several morphological alterations of the liver tissue have been observed [17]. In this context, it has been supposed that hepatocytes as well as dendritic cells may support the intracellular replication of F. tularensis without undergoing pyroptosis or apoptosis. Moreover, F. tularensis is assumed to delay induction of cell death in host cells to its own advantage until exit from its intracellular environment [2].
So far, there is only little knowledge about the infection and replication cycles of F. tularensis within human liver tissue. To investigate these processes in vitro, we co-cultured primary human monocyte-derived macrophages with the recently developed hepatocyte cell line HepaRG, which differentiates into cells with a hepatocyte phenotype and into cells exhibiting a biliary epithelial cell phenotype [18]. In contrast to other hepatic cells lines, i.e. HepG2 or Hep2/C3A, HepaRG cells remain functionally stable during prolonged culture, self-organize with functional bile canaliculi-like structures and respond to inflammatory cytokines [18]. Using this human macrophage / hepatocyte co-culture approach we characterized the infection and replication of F. tularensis ssp. holarctica, spp. mediasiatica and the attenuated LVS strain.
Results
To characterize the impact of hepatocytes on the intracellular replication rate of F. tularensis in mono-cell cultures of macrophages or hepatocytes as well as in macrophage/hepatocyte co-cultures, cells were infected with F. tularensis and cultured up to 72 h. The F. tularensis replication rate was measured by flow cytometry using an anti- F. tularensis lipopolysaccharide (LPS) antibody to detect intracellular bacteria. Macrophages were discriminated from hepatocytes in the flow cytometric analyses by combined gating of FSC/SSC scattering and fluorescence measurement of FITC-labeled antibody directed against the leukocyte marker protein CD45. F. tularensis ssp. holarctica and LVS strain were reliably detected in macrophages and hepatocytes. Although it has been concluded from previous studies that all three F. tularensis ssp. possess a common LPS lipid A structure [19], we were unable to detect intracellular LPS after infection with F. tularensis ssp. mediasiatica. However, presence and replication of intracellular viable bacteria in the hepatocyte / macrophages co-culture of all three F. tularensis strains were confirmed by colony forming unit (CFU) assays from lysates of infected cells (Fig. 1, Additional file 1: Figure S1).
Fig. 1.
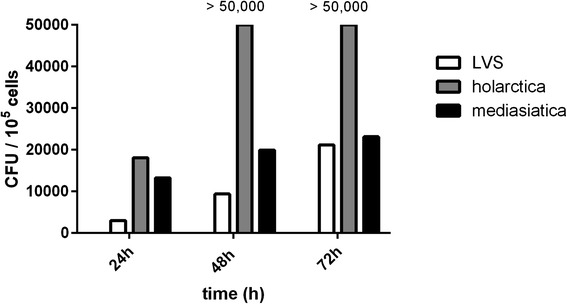
Quantification of colony forming units from cell lysates of macrophage (6 %)/hepatocyte (94 %) co-cultures 24, 48 and 72 h after infection with F. tularensis LVS, spp. holarctica or spp. mediasiatica plated on cysteine heart agar dishes. Data of one representative experiment out of a series of 5 independent experiments for each time point and F. tularensis strain is shown
Replication of F. tularensis LVS strain and F. tularensis ssp. holarctica was confirmed in monocultures of macrophages and in hepatocytes (Fig. 2a and b, left part) as well as in the co-culture of both cell types (Fig. 2a and b, right part). Interestingly, the highest LPS content was detected in co-cultures with the lowest macrophage content of 6 % in the co-culture (94 % hepatocytes) (Additional file 2: Figure S2A). An increase in the macrophage content in the cell cultures resulted in a diminished LPS signal reflecting first, F. tularensis preferentially enter macrophages compared to hepatocytes, and second, in consequence bacterial MOI per individual macrophage cell is reduced in the co-culture.
Fig. 2.
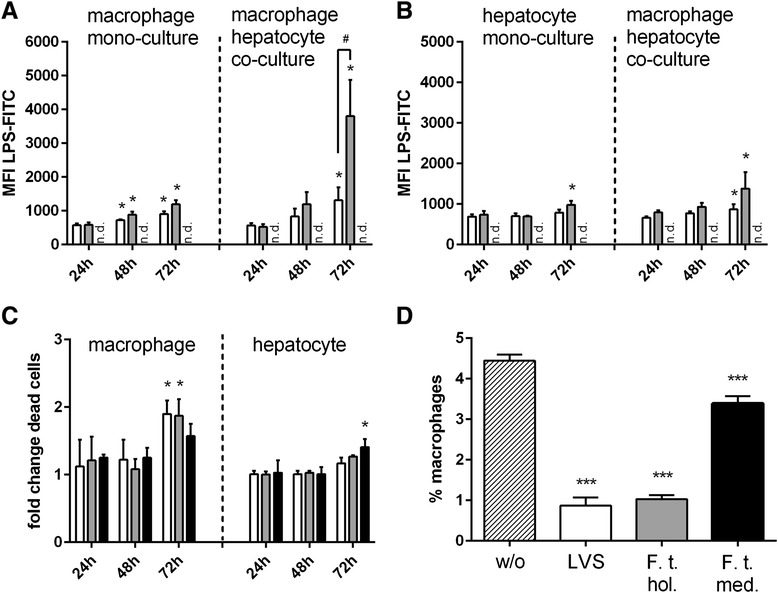
Detection of intracellular LPS and cell death in macrophage (6 %) / hepatocyte (94 %) co-cultures infected with LVS (open bars), spp. holarctica (grey filled bars) or spp. mediasiatica (black filled bars) and untreated control (hatched bars). a-b Flow cytometric (FACS) detection of intracellular Francisella tularensis by detection of LPS in a macrophages and b hepatocytes in the mono-cell culture (left side of dashed line) and co-culture (right side of dashed line) (MFI mean fluorescence intensity); c fold change of increase in cell death in macrophages (left side of dashed line) and hepatocytes (right side of dashed line) compared to untreated control in the co-culture; d percentage of detectable macrophages in the co-culture 72 h after infection without (w/o) and with incubation of LVS strain (LVS), F. tularensis holarctica (F. t. hol.) or F. tularensis mediasiatica (F. t. med.). Statistical significance was calculated using student’s t-test (a-d: * p < 0.05 compared to corresponding condition 24 h after infection with the respective strain; # p < 0.05, statistical significance between indicated conditions calculated with student’s t-test)
The strong increase in replication rate of F. tularensis ssp. holarctica in macrophages and hepatocytes in the co-culture compared with respective mono-cell cultures, indicates a supportive function of hepatocytes for bacterial replication (Fig. 2a, b). Furthermore, we observed an increased proportion of dead macrophages infected with the LVS strain and F. tularensis ssp. holarctica in the co-culture after 72 h, whereas at the same time point F. tularensis ssp. mediasiatica induced a significant onset of cell death preferentially in hepatocytes, but not in macrophages (Fig. 2c). An analysis of the total percentage of macrophages revealed that infection with all three F. tularensis strains resulted in a significantly diminished macrophage numbers after 72 h in the co-culture. A comparative analysis with different proportions of macrophages in the co-culture revealed that a decline of present macrophages correlated with the detected LPS signal confirming bacterial infection and related subsequent cell death (Additional file 2: Figure S2B). Thus, in the subsequent experiments co-cultures of 6 % macrophage and 94 % hepatocytes were used to enable sufficient bacterial infection rate and allowing an efficient study of the related cellular response. Under these conditions the observed loss of detectable macrophages in response to bacterial infection was most prominent in cell cultures infected with LVS and F. tularensis ssp. holarctica compared to F. tularensis ssp. mediasiatica (Fig. 2d).
Induction of apoptosis in macrophages in response to infection with all three F. tularensis strains was detected in the co-cultures with hepatocytes at the level of cleaved caspase-3. In these assays, infection with F. tularensis ssp. holarctica caused the highest rate of apoptosis induction in macrophages 24 h and 72 h post infection (Fig. 3b-d, b).
Fig. 3.
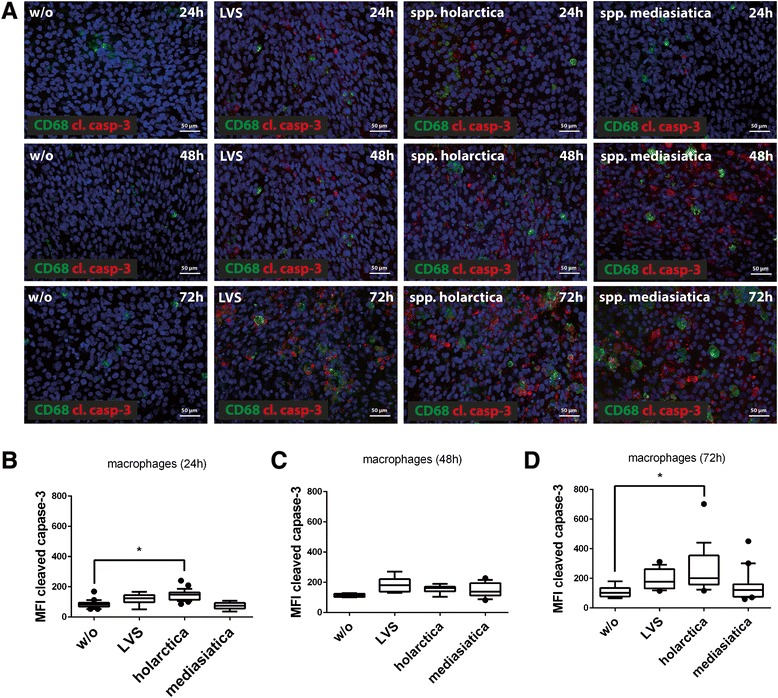
a Immunofluorescence staining of macrophages marker protein CD68 (green) and apoptosis marker cleaved caspase-3 (cl. casp-3, red) in co-culture of 6 % macrophages and 94 % hepatocytes. Nuclei are stained with DAPI (blue). Cells were infected with LVS, spp. holarctica and spp. mediasiatica and subsequently co-cultured for 24, 48 and 72 h, respectively. b-d Quantification of the mean fluorescence intensity (MFI) of labeled antibody for detection of the apoptosis marker cleaved caspase-3 (cCasp-3) in macrophages b) 24h, c) 48h and d) 72h post infection. Whisker dot plots show boxes ranging from 10th (lower box boundary) to 90th (upper box boundary). Vertical line inside the box marks the median. Outliers are shown as filled circles. Statistical significance was calculated using one-way ANOVA with Dunnett's multiple comparisons (* p < 0.05 for indicated conditions)
It has been demonstrated in mice that after 16 h of intravenous infection with F. tularensis infectious foci in liver tissue are formed and accompanied by infiltration of both neutrophils and monocytes [20]. Infected mice developed typical signs of hepatitis within 24–72 h post infection [21] and during this period, pro-inflammatory cytokines, including interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF) α were secreted at the active sites of infection [11, 22–24]. To characterize the inflammatory response in our humanized co-culture model the secretion of these pro-inflammatory cytokines in response to F. tularensis infection was measured in the supernatant of the cell culture by cytometric bead arrays (CBA). In addition, the release of fractalkine (FKN, CX3CL1), a chemokine that controls the survival of invading monocytes and their differentiation into functionally diverse macrophage subsets upon liver injury [25] was analyzed by CBA. In chronic liver injury FKN elicits liver protective functions via its receptor CX3CR1 by promoting hepatic macrophage survival and restriction of pro-inflammatory macrophage polarization [26, 27].
We observed a robust and sustained inflammatory response upon infection with all three F. tularensis strains up to 72 h after infection. TNFα and IL-1β levels were highest 24 h after infection and subsequently declined in the further course of culture (Fig. 4a, b). A release of IL-6 was similarly triggered by infection with all Francisella strains after 24 h. Whereas IL-6 levels declined in F. tularensis ssp. holarctica and ssp. mediasiatica infected co-cultures up to 72 h, IL-6 levels remained stable upon infection with the LVS strain (Fig. 4c). In contrast to the release of pro-inflammatory cytokines TNFα, IL-1β and IL-6, the release of FKN increased from 24 h up to 72 h post infection (Fig. 4d). Interestingly, FKN release negatively correlated with pro-inflammatory cytokine release in a time-dependent as well as strain-specific manner, indicating that the release of the FKN chemokine negatively correlates with F. tularensis-mediated inflammation response. Among all three F. tularensis strains tested, F. tularensis ssp. mediasiatica infection induced the secretion of only low FKN levels but high amounts of TNFα and IL-1β were secreted relative to the LVS strain and F. tularensis ssp. holarctica.
Fig. 4.
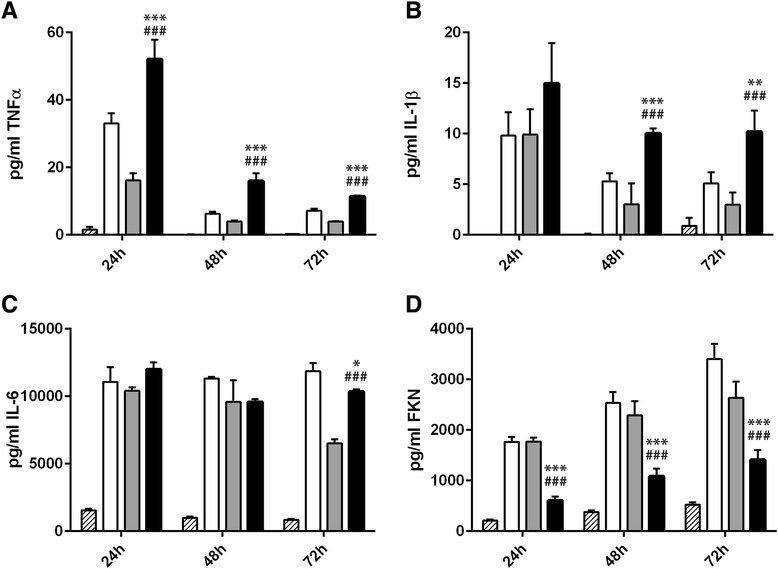
Cytokine secretion by macrophage/hepatocyte co-cultures of a TNFα, b IL-1β, c IL-6 and d fractalkine (FKN) in response to infection with LVS (open bars), spp. holarctica (grey filled bars) or spp. mediasiatica (black filled bars) or untreated control (hatched bars) after 24, 48 and 72 h of cell culture. Statistical significance was calculated using student’s t-test: *** p < 0.001 compared to identical time point after infection with LVS; ### p < 0.001 compared to identical time point after infection with spp. holarctica
F. tularensis attempts to evade the primary immune response by intracellular replication [3]. To study whether its presence in the cell culture is masked to suspended immune cells, we co-incubated infected cells with human white blood cells (WBC) seeded in a transwell filter above the co-cultures. In this setting we were not able to detect a significant number of migrating and adhesive leukocytes to the infected cell cultures. Only small amounts of less than 5 % migrating granulocytes were detectable (Fig. 5a) in naïve and infected cell cultures, but without significant differences in respect to cell numbers (Fig. 5a). Although secretion of TNFα and IL-1β was increased, we found only minor alterations of the time- or strain-dependent IL-6- and FKN-release (Fig. 5b-e) in presence of co-incubated WBC.
Fig. 5.
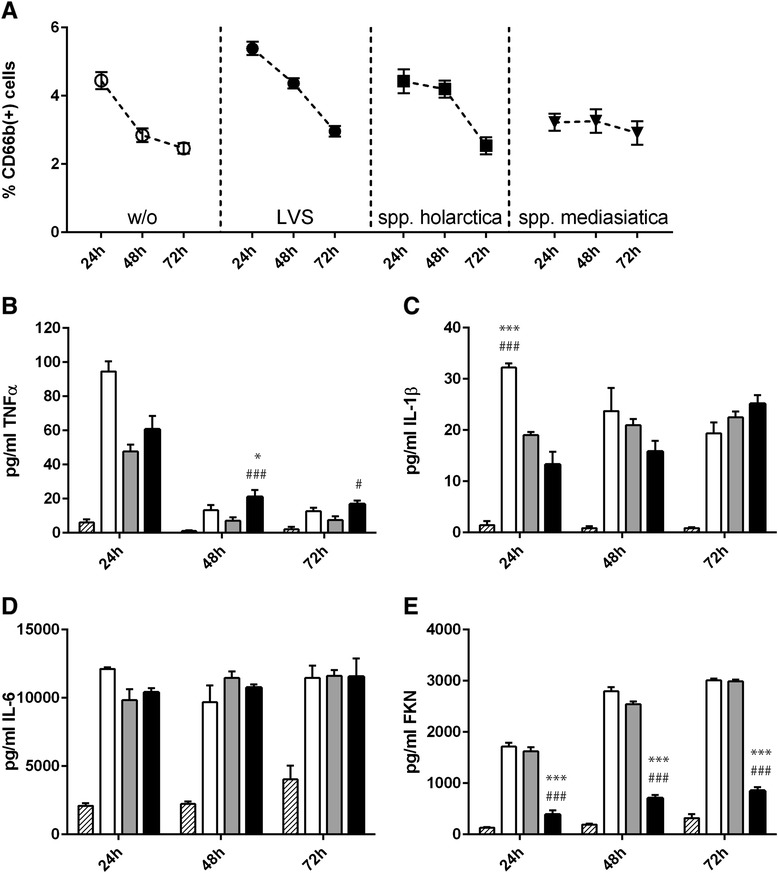
White blood cell (WBC) migration and cytokine secretion by macrophage (6 %) / hepatocyte (94 %) cultures co-incubated with 2 x105 WBC in a transwell filter insert. a Percentage of detected CD68b positive granulocytes by flow cytometry 24, 48 and 72 h post infection. Cytokine secretion of b TNFα, c IL-1β, d IL-6 and e fractalkine (FKN) in response to infection with LVS (open bars), spp. holarctica (grey filled bars) or spp. mediasiatica (black filled bars) or untreated control (hatched bars) after 24, 48 and 72 h of cell culture. Statistical significance was calculated using student’s t-test: * p < 0.05, *** p < 0.001 compared to identical time point after infection with LVS; # p < 0,05, ### p < 0.001 compared to identical time point after infection with spp. holarctica
Discussion
It has been suggested that hepatocytes as well as dendritic cells support the intracellular replication of F. tularensis without undergoing pyroptosis or apoptosis [2]. We could confirm this concept derived from the mouse models in the human cell culture model for the virulent subspecies F. tularensis mediasiatica but not for the subspecies holarctica or the attenuated LVS strain. In the human co-culture model we could detect a significant loss of macrophages infected with F. tularensis ssp. holarctica and the LVS strain, whereas the percentage of viable macrophages only slightly decreased after infection with F. tularensis ssp. mediasiatica.
F. tularensis is presumed to delay induction of cell death in host cells to its own advantage until exit from its intracellular environment [2]. F. tularensis ssp. mediasiatica was found to efficiently adopt this strategy by preventing macrophage cell death in order to allow efficient replication rates. It was shown that infection of the macrophage-like J774 cell line with LVS results in apoptosis [28]. However, the number of bacterial burden was not affected. In this context, a role of the caspase-3-mediated cell death in favoring bacterial dissemination was assumed, but this mechanism is restricted to tissues in which macrophages play a central role in pathogen uptake and killing, such as the liver [2]. In mice, death of animals infected with F. tularensis appears to result from widespread sepsis and inflammation [29–31], and mortality was correlated with the extent of the inflammatory response [17] including release of pro-inflammatory IL-6, an early diagnostic marker of bacterial sepsis [32].
We observed the strongest pro-inflammatory response upon infection with F. tularensis ssp. mediasiatica that also induced the highest rate of apoptosis in the co-culture model of all tested F. tularensis strains. We further observed a reduced level of FKN, a chemokine released by cleavage from hepatocytes upon inflammation [33] and that is able to counteract liver inflammation [26] in response to F. tularensis ssp. mediasiatica infection. In mice, lethal infection with F. tularensis is associated with hypercytokinemia and biochemical markers for sepsis [29–31]. This sepsis is assumed to result from delayed cytokine up-regulation and insufficient recruitment of inflammatory cells [31]. Although we observed a slight increase of TNFα and IL-1β secretion upon WBC co-incubation, no effects on IL-6 release were observed. Based on the diminished immune response of co-incubated WBC we conclude that intracellular bacteria are mainly hidden from WBC, which impedes an efficient bacterial clearance.
On one hand, the observed high rate of apoptosis induction in macrophages infected with F. tularensis provides the host with the advantage of early elimination of infected cells and removal of microbial replication niches. On the other hand, the extensive cell death of tissue macrophages, which is a hallmark of virulent F. tularensis infection [2], and the reduced recruitment of phagocytes to the sites of infection favors the dissemination of bacteria and diminishes important innate immune responses [2, 29]. This double-edged sword needs to be tightly regulated by the host to ensure an efficient host defense against F. tularensis infection.
Conclusions
We could demonstrate that F. tularensis infection and related cellular immune response can be investigated in a human cell co-culture model of monocyte-derived macrophages and hepatocytes. This co-culture model depicts strain-specific virulence and the associated pathogenic potential of the tested F. tularensis strains. Using this model we could demonstrate that F. tularensis strains efficiently hide and replicate in macrophages depending on their virulent potential and thereby avoid detection by co-incubated WBC and subsequent aggravated immune response. We believe that the cell co-culture approach presented herein is a valuable tool for further, more detailed in vitro studies of F. tularensis pathophysiology. The potential of this co-culture models to study other intracellular persisting pathogens affecting the human liver will be characterized in further studies.
Methods
Cell isolation and culture
HepaRG hepatocytes
HepaRG cells were obtained from Biopredic International (Rennes, France). Undifferentiated HepaRG were seeded at a density of 2,7 × 104 cells/cm2 per well of a 12 well-plate and cultured in William’s Medium E (Biochrom, Berlin, Germany) containing 10 % (v/v) FCS (GIBCO, Darmstadt, Germany), 5 μg/ml insulin (Sigma-Aldrich, Steinheim, Germany), 2 mM glutamine (GIBCO), 5×10−5 M hydrocortisone-hemisuccinate (Sigma-Aldrich) and 100 U/ml penicillin/100 μg/ml streptomycin mixture (Pen/Strep) (GIBCO). The cells were cultured in a humidified cell incubator at 5 % CO2 and 37 °C for 14 days before differentiation. Medium was renewed every 3–4 days. Cell differentiation was induced for 14 days in the presence of 2 % (v/v) DMSO (Sigma-Aldrich) as described [34]. At day 8 before starting the infection with bacteria. Primary macrophages: PBMC were isolated from whole blood which was collected from healthy human donors that were informed about the aim of the study and gave written informed consent. The study was approved by the ethics committee of the Jena University Hospital. PBMCs were isolated by Ficoll density gradient centrifugation as described previously [35, 36] and monocytes isolated from PBMCs with the Dynabeads ® CD14 Isolation Kit according to the manufacturer’s protocol (Life Technologies, Darmstadt, Germany). For macrophages differentiation monocytes were seeded into each well of 12 well plates and cultured for 6 days in X-VIVO 15 medium (Lonza, Cologne, Germany) supplemented with 10 % (v/v) autologous human serum, 10 ng/ml human granulocyte macrophage colony-stimulating factor (GM-CSF) (PeproTech, Hamburg, Germany) without antibiotics. Medium was exchanged after three days. For co-culture of macrophages and hepatocytes, monocytes were plated and differentiated for 6 days to macrophages. HepaRG cells were subsequently added to the macrophage culture. Total percentage of macrophages in the co-culture was controlled by seeding different monocyte cell densities and filling up with hepatocytes to a resulting total cell density of 1.0 × 105 cell/cm2 used for all tested co-cultures.
White blood cell isolation
WBCs were isolated from 9 ml EDTA-blood using the erythrocyte lysis buffer of the QIAamp RNA Blood Mini Kit (Qiagen) followed by three washing steps in PBS containing 2 mM EDTA.
Francisella tularensis culture
Francisella tularensis ssp. holarctica (FLI isolate 06 T-0001 and live vaccine strain-LVS) and ssp. mediasiatica (F63) were provided by the cryobank of the German National Reference Laboratory for Tularemia, plated and cultured on cysteine heart agar (CHA) in a humidified incubator at 4,5 % CO2 and 37 °C for 72 h. Subsequently, colonies were suspended in PBS.
Infection
At the day of infection, bacterial suspensions were adjusted to an OD600 = 0.4, corresponding to 5 × 109 bacteria/ml. Cell cultures were incubated with F. tularensis bacteria (MOI 100:1) for 2 h in a humidified cell incubator at 4,5 % CO2 and 37 °C. Subsequently, the bacterial suspension was removed, washed with Williams medium E and further incubated in William’s Medium E containing 10 % (v/v) FCS, 5 μg/ml insulin, 2 mM glutamine, 5×10−5 M hydrocortisone-hemisuccinate and 10 μg/ml gentamicin (Sigma-Aldrich, Taufkirchen, Germany) for 30 min to kill extracellular bacteria. Infected cells with persisting Francisellae were than washed with PBS and cultured for indicated times in William’s Medium E containing 10 % (v/v) FCS, 5 μg/ml insulin, 2 mM glutamine, 5×10−5 M hydrocortisone-hemisuccinate without antibiotics.
White blood cell co-incubation
2 × 105 freshly isolated white blood cells were transferred into each cell culture inserts with 8 μm pores (Merck Millipore, Darmstadt) hanging above F.tularensis pre-inoculated co-cultures of HepaRG/macrophages. WBCs were incubated for up to 72 h with the infected co-cultures. Subsequently, inserts were discarded and cells in the subjacent well were harvested for flow cytometry. Granulocytes and T-cells were stained with fluorochrome-conjugated antibodies against CD66b and CD3 (BD Biosciences) and analyzed by flow cytometry.
Immunofluorescence staining
Cells were fixed with 4 % paraformaldehyde for 10 min at room temperature (RT). Staining was done with antibodies against CD68 (BD Biosciences, Heidelberg, Germany), cleaved caspase 3 (cCasp-3) (Cell Signaling Technology, Leiden, Netherlands), and goat-anti-rabbit-Cy3 (Dianova, Hamburg, Germany) and goat-anti-mouse-AlexaFluor488 (AF488) as secondary antibodies, and DAPI (Life Technologies, Karlruhe, Germany). Samples were embedded into fluorescent mounting medium (Dako, Hamburg, Germany). Subsequently, imaging was performed on an AXIO Observer Z1 fluorescence microscope equipped with Apotome 2 (Carl Zeiss AG, Jena, Germany).
Image analysis
Analysis of cleaved caspase-3 fluorescence signals in macrophages was done with ImageJ2 software. Specifically, macrophages were detected based on the expression of the cell-type-specific marker protein CD68 stained with AF488 coupled secondary antibody and cell borders of the cell marked as region of interest (ROI). ROI’s were than analyzed for fluorescence signals of cCasp-3 and mean fluorescence intensity (MFI) was calculated and plotted in a whisker box plot diagram.
Colony forming unit assay
After indicated incubation times, cells were lysed in 1 ml Aqua bidest. for 10 min at RT. Subsequently, 20–100 μl of the lysate were transferred and streaked on cysteine heart agar without antibiotics. Agar plates were cultivated for 2 days in a humidified incubator at 4,5 % CO2 and 37 °C. Afterwards colonies were counted.
Live/dead staining
Macrophages and hepatocytes were detached from cell culture dishes using 4 mg/ml Lidocaine and 5 mM EDTA (Sigma Aldrich) in PBS (Lonza,, Cologne, Germany), centrifuged at 300 x g at room temperature (RT) for 6 min, washed two times with PBS at RT and stained with LIVE/DEAD® Cell Viability assay (Life Technologies) for 30 min according to the manufacturer’s instructions.
FACS analysis
Macrophages were stained with antibody against CD45-APC-Cy7 (BD Bioscience, Heidelberg, Germany) and fixed with Inside Stain kit (Miltenyi Biotec, Bergisch Gladbach) according to manufacturer’s recommendations for permeabilization of cells thus allowing detection of F. tularensis LPS. Intracellular Francisellae were stained with 2,5 μg/ml FITC-conjugated antibody versus F. tularensis LPS (clone FB11 - HyTEST Ltd., Turku, Finland). Clone FB 11 does not recognize LPS of F. tularensis ssp. mediasiatica. Flow cytometry was performed on a BD FACS-Canto II (BD Biosciences) with FACSDiva software and analyzed using FlowJo X software (FlowJo LLC, Ashland, OR, USA).
Cytometric bead array (CBA)
Supernatants were collected after indicated time periods and immediately frozen at −80 °C. Cytokines were detected using CBA assay (BD Biosciences) according to the manufacturer’s protocol. Secretion of TNFα, IL-1β, IL-6, fractalkine and IFNα was analyzed using standard CBA flex sets. Analysis was performed on a BD FACS-Canto II cytometer with FACSDiva software. Data analysis was performed using FCAP Array V3 software (Softflow, Pecs, Hungary).
Statistics
All results are represented as mean of the performed experiments with standard deviation. Statistic tests were done with two-tailed, non-paired Student’s t-test or one-way ANOVA with Dunnett’s multiple comparisons test between indicated conditions. Statistical analysis was performed using GraphPad Prism 6.07 software (Graphpad Software, La Jolla, CA, USA). For CFU analysis data of one representative experiment out of a series of 5 independent experiments for each time point and F. tularensis strain is shown in Fig. 1 and Additional file 1: Figure S1.
Acknowledgements
We are grateful to the excellent technical work of Maria Franke and Margot Voigt. The authors would further like to acknowledge support of this work by grant of the German Research Platform for Zoonoses (FK 01KI1109).
Abbreviations
- CHA
Cysteine heart agar
- CFU
Colony forming unit
- CX3CL1
Fractalkine
- DMEM
Dulbecco’s modified eagle medium
- ECM
Endothelial cell medium
- FKN
Fractalkine
- F. tularensis
Francisella tularensis
- GM-CSF
Granulocyte-macrophage colony stimulating factor
- IL
Interleukin
- LPS
Lipopolysaccharide
- LVS
Live vaccine strain
- MOI
Multiplicity of infection
- Pen/Strp.
Penicillin/Streptomycin
- RT
Room temperature
- spp.
Subspecies
- WBC
White blood cell
- v/v
Volume/Volume
Additional files
Colony forming units assay from cell lysates of macrophage (6 %)/hepatocyte (94 %) co-cultures 24, 48 and 72 h after infection with F. tularensis LVS, spp. holarctica or spp. mediasiatica plated on cysteine heart agar dishes. Data of one representative experiment out of a series of 5 independent experiments for each time point and F. tularensis strain is shown. (TIF 77516 kb)
Detection of intracellular LPS in macrophage / hepatocyte co-cultures infected with LVS (open bars), spp. holarctica (grey filled bars) or spp. mediasiatica (black filled bars) and untreated control (hatched bars). A) Different amounts of macrophages in the co-culture were tested (6, 12 and 22 % of macrophages on total cell count). Flow cytometric detection of intracellular LPS in macrophages (MFI mean fluorescence intensity); B-D) percentage of remaining detectable macrophages after infection of the co-cultures with B) 6 % macrophages/94 % hepatocytes, C) 12 % macrophages/ 88 % hepatocytes and D) 22 % macrophages/ 88 % hepatocytes 72 h post infection. (TIF 32735 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KR performed the experiments. PO, HF, HT contributed reagents/materials/analysis tools. KR, PO, ASM analyzed the data. OH, HT, ASM wrote the manuscript. HT, ASM designed the study and supervised experiments. All authors read and approved the final version of the manuscript.
Contributor Information
Knut Rennert, Email: knut.rennert@med.uni-jena.de.
Peter Otto, Email: gps.otto@t-online.de.
Harald Funke, Email: harald.funke@med.uni-jena.de.
Otmar Huber, Email: otmar.huber@med.uni-jena.de.
Herbert Tomaso, Email: herbert.tomaso@fli.bund.de.
Alexander S. Mosig, Phone: +49-9396418, Email: alexander.mosig@med.uni-jena.de
References
- 1.McLendon MK, Apicella MA, Allen LA. Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol. 2006;60:167–85. doi: 10.1146/annurev.micro.60.080805.142126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parmely MJ, Fischer JL, Pinson DM. Programmed cell death and the pathogenesis of tissue injury induced by type A Francisella tularensis. FEMS Microbiol Lett. 2009;301:1–11. doi: 10.1111/j.1574-6968.2009.01791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Barrio L, Sahoo M, Lantier L, Reynolds JM, Ceballos-Olvera I, Re F. Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with Francisella tularensis LVS. PLoS Pathog. 2015;11:1–20. doi: 10.1371/journal.ppat.1004706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snoy PJ. Establishing efficacy of human products using animals: the US food and drug administration’s “animal rule”. Vet Pathol. 2010;47(5):774–8. doi: 10.1177/0300985810372506. [DOI] [PubMed] [Google Scholar]
- 5.Råberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–4. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- 6.Schneider DS, Ayres JS. Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol. 2008;8:889–95. doi: 10.1038/nri2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keim P, Johansson A, Wagner DM. Molecular epidemiology, evolution, and ecology of Francisella. Ann N Y Acad Sci. 2007;1105:30–66. doi: 10.1196/annals.1409.011. [DOI] [PubMed] [Google Scholar]
- 9.De Pascalis R, Chou AY, Ryden P, Kennett NJ, Sjostedt A, Elkins KL. Models derived from in vitro analyses of spleen, liver, and lung leukocyte functions predict vaccine efficacy against the Francisella tularensis Live Vaccine Strain (LVS). MBio. 2014;5:e00936-00913–3. [DOI] [PMC free article] [PubMed]
- 10.Anthony LS, Kongshavn PA. Experimental murine tularemia caused by Francisella tularensis, live vaccine strain: a model of acquired cellular resistance. Microb Pathog. 1987;2(1):3–14. doi: 10.1016/0882-4010(87)90110-0. [DOI] [PubMed] [Google Scholar]
- 11.Bokhari SM, Kim KJ, Pinson DM, Slusser J, Yeh HW, Parmely MJ. NK cells and gamma interferon coordinate the formation and function of hepatic granulomas in mice infected with the Francisella tularensis live vaccine strain. Infect Immun. 2008;76(4):1379–89. doi: 10.1128/IAI.00745-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asare R, Abu Kwaik Y. Molecular complexity orchestrates modulation of phagosome biogenesis and escape to the cytosol of macrophages by Francisella tularensis. Environ Microbiol. 2010;12(9):2559–86. doi: 10.1111/j.1462-2920.2010.02229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sjostedt A. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 2006;8(2):561–7. doi: 10.1016/j.micinf.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72(6):3204–17. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouwens L, Baekeland M, De Zanger R, Wisse E. Quantitation, tissue distribution and proliferation kinetics of Kupffer cells in normal rat liver. Hepatology. 1986;6(4):718–22. doi: 10.1002/hep.1840060430. [DOI] [PubMed] [Google Scholar]
- 16.Ishibashi H, Nakamura M, Komori A, Migita K, Shimoda S. Semin Immunopathol. vol. 31. Berlin: Springer; 2009. Liver architecture, cell function, and disease; pp. 399–409. [DOI] [PubMed] [Google Scholar]
- 17.Chiavolini D, Alroy J, King CA, Jorth P, Weir S, Madico G, et al. Identification of immunologic and pathologic parameters of death versus survival in respiratory tularemia. Infect Immun. 2008;76:486–96. doi: 10.1128/IAI.00862-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillouzo A, Guguen-Guillouzo C. Evolving concepts in liver tissue modeling and implications for in vitro toxicology. Expert Opin Drug Metab Toxicol. 2008;4(10):1279–94. doi: 10.1517/17425255.4.10.1279. [DOI] [PubMed] [Google Scholar]
- 19.Hajjar AM, Harvey MD, Shaffer SA, Goodlett DR, Sjöstedt A, Edebro H, et al. Lack of in vitro and in vivo recognition of Francisella tularensis subspecies lipopolysaccharide by Toll-like receptors. Infect Immun. 2006;74:6730–8. doi: 10.1128/IAI.00934-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conlan JW, North RJ. Early pathogenesis of infection in the liver with the facultative intracellular bacteria Listeria monocytogenes, Francisella tularensis, and Salmonella typhimurium involves lysis of infected hepatocytes by leukocytes. Infect Immun. 1992;60(12):5164–71. doi: 10.1128/iai.60.12.5164-5171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White JD, Rooney JR, Prickett PA, Derrenbacher EB, Beard CW, Griffith WR. Pathogenesis of experimental respiratory tularemia in monkeys. J Infect Dis. 1964;114:277–83. doi: 10.1093/infdis/114.3.277. [DOI] [PubMed] [Google Scholar]
- 22.Cole LE, Elkins KL, Michalek SM, Qureshi N, Eaton LJ, Rallabhandi P, et al. Immunologic consequences of Francisella tularensis live vaccine strain infection: role of the innate immune response in infection and immunity. J Immunol. 2006;176:6888–99. doi: 10.4049/jimmunol.176.11.6888. [DOI] [PubMed] [Google Scholar]
- 23.Wickstrum JR, Bokhari SM, Fischer JL, Pinson DM, Yeh HW, Horvat RT, et al. Francisella tularensis induces extensive caspase-3 activation and apoptotic cell death in the tissues of infected mice. Infect Immun. 2009;77:4827–36. doi: 10.1128/IAI.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golovliov I, Sandstrom G, Ericsson M, Sjostedt A, Tarnvik A. Cytokine expression in the liver during the early phase of murine tularemia. Infect Immun. 1995;63(2):534–8. doi: 10.1128/iai.63.2.534-538.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimmermann HW, Trautwein C, Tacke F. Functional role of monocytes and macrophages for the inflammatory response in acute liver injury. Front Physiol. 2012;3:56. doi: 10.3389/fphys.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoyama T, Inokuchi S, Brenner DA, Seki E. CX3CL1-CX3CR1 interaction prevents carbon tetrachloride-induced liver inflammation and fibrosis in mice. Hepatology. 2010;52(4):1390–400. doi: 10.1002/hep.23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karlmark KR, Zimmermann HW, Roderburg C, Gassler N, Wasmuth HE, Luedde T, et al. The fractalkine receptor CX(3)CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology. 2010;52(5):1769–82. doi: 10.1002/hep.23894. [DOI] [PubMed] [Google Scholar]
- 28.Lai XH, Golovliov I, Sjostedt A. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect Immun. 2001;69(7):4691–4. doi: 10.1128/IAI.69.7.4691-4694.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma J, Mares CA, Li Q, Morris EG, Teale JM. Features of sepsis caused by pulmonary infection with Francisella tularensis Type A strain. Microb Pathog. 2011;51(1–2):39–47. doi: 10.1016/j.micpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma J, Li Q, Mishra BB, Pena C, Teale JM. Lethal pulmonary infection with Francisella novicida is associated with severe sepsis. J Leukoc Biol. 2009;86(3):491–504. doi: 10.1189/jlb.1208728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mares CA, Ojeda SS, Morris EG, Li Q, Teale JM. Initial delay in the immune response to Francisella tularensis is followed by hypercytokinemia characteristic of severe sepsis and correlating with upregulation and release of damage-associated molecular patterns. Infect Immun. 2008;76(7):3001–10. doi: 10.1128/IAI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin S, Huang Z, Wang M, Weng Z, Zeng D, Zhang Y, et al. Interleukin-6 as an early diagnostic marker for bacterial sepsis in patients with liver cirrhosis. J Crit Care. 2015;30(4):732–8. doi: 10.1016/j.jcrc.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 33.Turner SL, Mangnall D, Bird NC, Blair-Zajdel ME, Bunning RA. Effects of pro-inflammatory cytokines on the production of soluble fractalkine and ADAM17 by HepG2 cells. J Gastrointestin Liver Dis. 2010;19(3):265–71. [PubMed] [Google Scholar]
- 34.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, Cannie I, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc Natl Acad Sci U S A. 2002;99(24):15655–60. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosig S, Rennert K, Krause S, Kzhyshkowska J, Neunubel K, Heller R, et al. Different functions of monocyte subsets in familial hypercholesterolemia: potential function of CD14+ CD16+ monocytes in detoxification of oxidized LDL. FASEB J. 2009;23(3):866–74. doi: 10.1096/fj.08-118240. [DOI] [PubMed] [Google Scholar]
- 36.Wallert M, Mosig S, Rennert K, Funke H, Ristow M, Pellegrino RM, et al. Long-chain metabolites of alpha-tocopherol occur in human serum and inhibit macrophage foam cell formation in vitro. Free Radic Biol Med. 2014;68:43–51. doi: 10.1016/j.freeradbiomed.2013.11.009. [DOI] [PubMed] [Google Scholar]


