Abstract
Alterations in resting‐state networks (RSNs) are often associated with psychiatric and neurologic disorders. Given this critical linkage, it has been hypothesized that RSNs can potentially be used as endophenotypes for brain diseases. To validate this notion, a critical step is to show that RSNs exhibit heritability. However, the investigation of the genetic basis of RSNs has only been attempted in the default‐mode network at the region‐of‐interest level, while the genetic control on other RSNs has not been determined yet. Here, we examined the genetic and environmental influences on eight well‐characterized RSNs using a twin design. Resting‐state functional magnetic resonance imaging data in 56 pairs of twins were collected. The genetic and environmental effects on each RSN were estimated by fitting the functional connectivity covariance of each voxel in the RSN to the classic ACE twin model. The data showed that although environmental effects accounted for the majority of variance in wide‐spread areas, there were specific brain sites that showed significant genetic control for individual RSNs. These results suggest that part of the human brain functional connectome is shaped by genomic constraints. Importantly, this information can be useful for bridging genetic analysis and network‐level assessment of brain disorders. Hum Brain Mapp 36:3959–3972, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: resting‐state networks, heritability, twin study
INTRODUCTION
Our understanding of large‐scale human brain networks has been revolutionized by the inception of resting‐state functional magnetic resonance imaging (rsfMRI) [Biswal et al., 1995; Fox and Raichle, 2007]. rsfMRI measures functional connectivity between brain regions based on the temporal synchronization of spontaneously fluctuating rsfMRI signals. This technique has led to a remarkable discovery that during a state of rest, the human brain is organized into multiple large‐scale networks including the auditory, visual, sensorimotor, executive control, attention, default‐mode, basal ganglia, and cerebellum networks [Allen et al., 2011; Beckmann et al., 2005; De Luca et al., 2006; Smith et al., 2009]. These resting‐state networks (RSNs) are consistent with the functional networks derived from task‐based fMRI studies of various experimental paradigms and are implicated in specific brain functions [Smith et al., 2009]. Furthermore, the intrinsic connectivity architecture of RSNs constitutes the human brain “functional connectome” [Biswal et al., 2010].
Although the spatial patterns of RSNs are generally consistent across healthy individuals with modest‐to‐high reliability [Shehzad et al., 2009], appreciable inter‐individual variability still exists [Damoiseaux et al., 2006; Shehzad et al., 2009]. Importantly, the inter‐individual variability between twins or pedigrees enables the examination of genetic and environmental influences on RSNs and the functional connectome [Fornito et al., 2011; Glahn et al., 2010; Korgaonkar et al., 2014; van den Heuvel et al., 2013]. For instance, Fornito et al. [2011] investigated topological changes in network dynamics of adult twins and demonstrated that the global cost‐efficiency is genetically heritable, whereas the genetic effects on regional cost‐efficiency are heterogeneous across different brain regions [Fornito et al., 2011]. In another study with twin children, the normalized path length, which reflects the global integration of brain networks, was found to be under genetic control, but the level of connectivity and normalized clustering coefficient, which measures the segregation of brain networks, were not [van den Heuvel et al., 2013].
The genetic basis of the default‐mode network is also under interrogation. In a pedigree study, the posterior cingulate/precuneus within the default‐mode network showed the highest heritability in functional connectivity and gray‐matter density [Glahn et al., 2010]. In another twin study using seed‐based correlational analysis, the functional connectivity between posterior cingulate cortex and right inferior parietal cortex in the default‐mode network was found to be genetically heritable [Korgaonkar et al., 2014]. However, these two studies were both focused on the genetic control over regions of interest (ROIs), whereas the variability of genetic effects across voxels within each ROI remains unknown. More importantly, genetic influences on RSNs other than the default‐mode network have not been studied yet.
Therefore, to enhance our understanding of the genetic basis of RSNs, in the present study we have used a twin design to investigate the genetic and environmental contributions to individual RSNs. RSN spatial maps were obtained using the group independent component analysis (ICA) approach [Calhoun et al., 2001b]. The genetic and environmental effects on each RSN were estimated by fitting the functional connectivity covariance of each voxel in the RSN to the classic ACE twin model [Neale and Cardon, 1992]. This model tests the hypothesis that the variance of a given phenotype (i.e., the functional connectivity of a voxel within a RSN) can be decomposed into the contributions of additive genetic (A), common environmental (C) and unique environmental (E) factors, and relies on the assumption that monozygotic (MZ) twins share 100% of their genetic information, while dizygotic (DZ) twins on average share 50% of their genetic information [Neale and Cardon, 1992]. Our results showed that there was large variability in genetic influences across different RSNs. Sensory networks tend to be under stronger genetic control while the heritability of cognitive networks was generally weaker.
MATERIALS AND METHODS
Participants
Subjects were recruited from local schools of Chongqing, China through posters and flyers. After a complete description of the study, written informed consent was obtained from all subjects and subjects’ guardians. Subjects with any psychiatric disorders, nervous system diseases or severe physical diseases were excluded from the study. One hundred and twelve (112) healthy twins participated in this study. There were 32 pairs of MZ twins (64 individuals) between the ages of 12 and 18 (mean ± sd = 15.7 ± 1.5 years, 53% female) and 24 pairs of DZ twins (48 individuals) between the ages of 13 and 19 (mean ± sd = 16.0 ± 1.5 years, 54% female). All twins were right‐handed and reared together. The IQ of the subject was assessed using the Chinese version of Wechsler intelligence scale for children (C‐WISC). Zygosity was determined based on the features of short tandem repeats and amelogenin [Yang et al., 2006].
MRI Image Acquisition
MRI image acquisition was performed on a 3T scanner (Signa, GE Medical Systems, Waukesha, WI) at The First Affiliated Hospital of Chongqing Medical University. Subjects were instructed to lie in the scanner with eyes closed while keeping their head still. High‐resolution T1 structural MRI images were acquired using the 3DT1 pulse sequence with the parameters as follows: repetition time (TR) = 6.2 ms, echo time (TE) = 2.8 ms, matrix size = 256 × 256, field of view (FOV) = 24 cm × 24 cm, slice number = 166, slice thickness = 1.2 mm. rsfMRI data were acquired using the gradient‐echo echo planar imaging (EPI) pulse sequence with the parameters as follows: TR = 3000 ms, TE = 30 ms, flip angle = 90°, matrix size = 64 × 64, FOV = 24 cm × 24 cm, slice number = 33, slice thickness = 3.5 mm. For each rsfMRI run, 240 volumes were acquired. At the end of the rsfMRI scan, subjects were asked to respond to ensure they did not fall asleep during the scan.
Image Preprocessing
Data preprocessing was carried out using the Data Processing Assistant for Resting‐State fMRI toolkit (http://rfmri.org/DPARSF) [Yan and Zhang, 2010]. The first five volumes of each rsfMRI run were discarded to allow magnetization to reach steady state. Slice‐timing correction was performed using the last slice (33rd) as the reference frame and the slice order of ascending odd number slices first followed by ascending even number slices. Head motion was corrected using the realignment function in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/), which corrects motion based on a rigid body model with six movement parameters (three translational and three rotational movement parameters). Functional images were subsequently spatially normalized to the Montreal Neurological Institute (MNI) space using an EPI template [Ashburner and Friston, 1999], and resliced to 3 × 3 × 3 mm3 voxels. Finally, all functional images were spatially smoothed using a Gaussian filter with a full‐width‐half‐maximum (FWHM) of 4 mm. A relatively small smoothing kernel size was used to make sure we will be able to detect small spatial variations of genetic and environmental effects within each ROI.
Group ICA
Group ICA was carried out on preprocessed data of all 112 subjects using the GIFT toolbox (http://mialab.mrn.org/software/gift/). The optimal number of independent components was first estimated to be 63 using the Minimum Description Length criterion. ICA was performed using the Infomax algorithm. Subject‐specific spatial maps and time courses were calculated using the GICA back reconstruction method [Calhoun et al., 2001b]. Component images of each subject were scaled to z‐scores. Each mean component image was thresholded using the feature selection method based on a Normal‐Gamma‐Gamma model developed in [Allen et al., 2011]. Specifically, the distribution of t values of all gray matter voxels for each component was modeled as a mixture of a Normal distribution and two Gamma distributions. The threshold of a given component was selected based on the estimated mean (μ) and variance (σ) of the Normal distribution at t > μ + 4σ.
Out of 63 ICA components, 26 nonartifactual components were identified, in which 22 components spatially matched the ICA component templates previously reported [Allen et al., 2011] based on the goodness‐of‐fit and the template‐matching method described in [Greicius et al., 2004]. The goodness‐of‐fit was calculated by the average t value of voxels falling within the template minus the average t value of voxels falling outside the template, and the template‐matching method selected the component with the greatest goodness‐of‐fit for each template [Greicius et al., 2004] (mean ± std of goodness‐of‐fit from all 22 components selected = 5.28 ± 1.64). Four additional ICA components were included due to the correspondence of their spatial locations to RSNs reported in [Smith et al., 2009] (two components in the visual network, one component in the default‐mode network and one component in the cerebellum network). These 26 components were used to generate eight RSNs including the default‐mode, auditory, basal ganglia, executive control, sensorimotor, cerebellum, attention, and visual networks described in [Allen et al., 2011] and [Smith et al., 2009].
Heritability Analysis
Univariate twin analysis was used for assessing the genetic and environmental influences on individual RSNs. Specifically, the classic ACE twin model was applied to estimate the contributions of three latent factors—additive genetic factor (A), shared environmental factor (C), and unique environmental factor (E)—to the variance of z‐scores of each voxel in an ICA component map across all subjects. For each twin pair, the observed z‐scores (z 1 from Twin 1; z 2 from Twin 2) were modeled using a linear combination of these three latent factors:
| (1) |
As a result, the covariance matrix Σ for (z 1, z 2) can be written in the following equation [Neale and Cardon, 1992]:
| (2) |
Based on the assumption that MZ twins share 100% of their genetic information and DZ twins on average share 50% of their genetic information, we have g = 1 for MZ twins and g = 0.5 for DZ twins in Eq. (2). Therefore, coefficients a, c, and e can be estimated by fitting the right side of Eq. (2) to the covariance matrix of (z 1, z 2) (i.e., Σ) using the maximum likelihood estimation, and the values of a2, c2, and e2, respectively, represent the relative contributions of A, C, and E to the variance of z‐scores.
The above twin analysis was carried out on a voxel‐by‐voxel basis using OpenMx 1.4 (http://openmx.psyc.virginia.edu/) [Boker et al., 2011] in the R environment (R3.0.2, http://www.r-project.org/). A double‐entering method, in which each subject was entered twice, once with the label Twin 1 and the other time with the label Twin 2 [Plomin et al., 2013], was used to avoid any potential confounding effects due to the twin label and ensure the identical variance between the Twin 1 group and Twin 2 group. Before fitting, three head motion components used in [Couvy‐Duchesne et al., 2014] including mean translation, maximum translation and mean rotation, as well as age and gender were first regressed out from the observed variables as covariates. Subsequently, voxel‐wise twin analysis generated the A, C, and E maps for each nonartifactual ICA component. The A, C, and E maps for each RSN were then generated by correspondingly combining the A, C, and E maps of all components belonging to the same RSN. In the case when a RSN had more than one component and these components were spatially overlapping, the A, C, and E values for each overlapped voxel were then, respectively, calculated by weighted averaging the corresponding A, C, and E values across all overlapping components with the weight of the z‐scores in these components.
The statistical significance of the A and C values for each voxel was, respectively, tested by dropping the factor under test (A or C) from the ACE model and comparing the fitting results of the full model (ACE) and the reduced model (CE or AE) using the likelihood ratio test. Considering the non‐negativity constraint of variance components, P values were calculated using the method described in [Dominicus et al., 2006]. The result indicated that the C factor did not significantly account for the variance of any voxels in six of eight networks after multiple‐comparison correction using a false discovery rate of 0.05 [Genovese et al., 2002], with the exception of eight scattered voxels in the sensorimotor network and nine scattered voxels in the visual network. Insignificant contributions from the common environmental factor to RSNs have also been reported in numerous previous studies using the same model [Blokland et al., 2011; Fornito et al., 2011; Korgaonkar et al., 2014; van den Heuvel et al., 2013]. Therefore, the C factor was dropped and all results reported here were based on the AE model.
An endophenotype for each RSN was defined as a cluster of voxels that met two criteria: (1) at the cluster level, the significance of the genetic factor (i.e., A factor) must be at P < 0.05, and (2) at the voxel level, the heritability of individual voxels must be higher than 40% (i.e., A > 0.4 for all voxels in the cluster). The threshold of A > 0.4 was selected based on a recent publication by [Korgaonkar et al., 2014]. The P value for each endophenotypic cluster was estimated based on the Monte Carlo simulation with 20,000 iterations using Rest AlphaSim [Song et al., 2011] (http://www.restfmri.net), with the mask of the corresponding RSN map, FWHM = 4 mm, cluster connection radius = 3.5 mm, and individual voxel threshold probability = 0.05.
All data analysis procedures are summarized in Figure 1.
Figure 1.
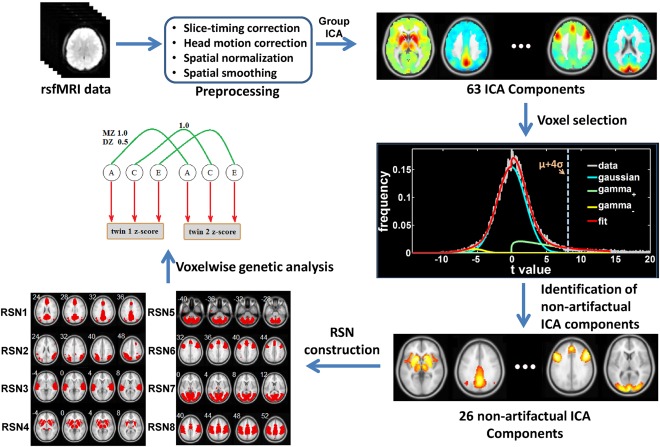
The workflow of data analysis.
RESULTS
Subjects between the MZ and DZ groups were comparable in age, gender, and IQ. There was no statistically significant difference in age (two‐sample t‐test, P = 0.22), gender (Chi‐square test, P = 0.91), or IQ (two‐sample t‐test, P = 0.34) between MZ and DZ twins. The demographic information of all subjects is summarized in Table 1.
Table 1.
Summary of subjects’ demographic information
| Zygosity | Number of twin pairs | Gender | Age (mean ± SD) | IQ (mean ± SD) | |
|---|---|---|---|---|---|
| Number of males | Number of females | ||||
| MZ | 32 | 30 | 34 | 15.7 ± 1.5 | 122.3 ± 14.7a |
| DZ | 24 | 22 | 26 | 16.0 ± 1.5 | 125.0 ± 14.6 |
IQ scores of one MZ twin pair were not measured.
We first quantified the genetic and environmental influences on eight well‐known RSNs: the visual, basal ganglia, sensorimotor, cerebellum, auditory, attention, executive‐control, and default‐mode networks. The first two rows of each panel in Figures 2 and 3, respectively, show the voxel‐wise spatial patterns of the relative contributions of the genetic (A) and environmental (E) factors to each individual RSN. Overall, the results indicate that environmental effects accounted for the majority of the variance in wide‐spread areas for all RSNs. However, there were clearly specific brain sites that showed significant genetic control, suggesting that the human brain RSNs are partially shaped by genomic constraints.
Figure 2.
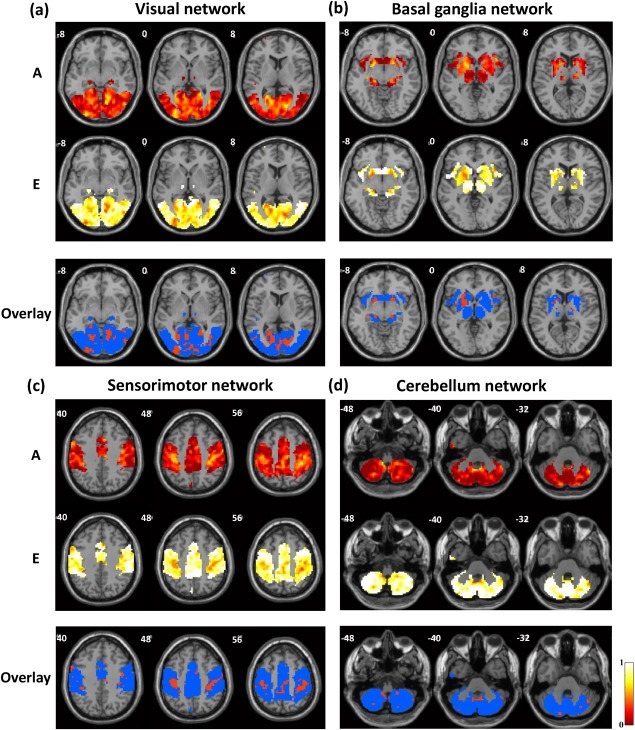
Genetic and environmental influences on the (a) visual network, (b) basal ganglia network, (c) sensorimotor network, and (d) cerebellum network. First two rows: the additive genetic (A, first row) and unique environmental (E, second row) maps. Third row: thresholded A map (A > 0.4, red) overlaid on the corresponding E map (blue).
Figure 3.
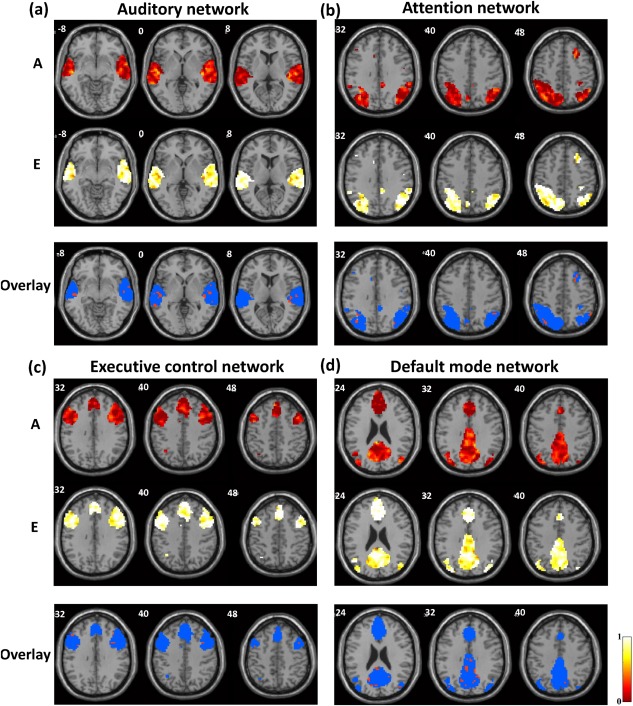
Genetic and environmental influences on the (a) auditory network, (b) attention network, (c) executive control network, and (d) default‐mode network. First two rows: the additive genetic (A, first row) and unique environmental (E, second row) maps. Third row: thresholded A map (A > 0.4, red) overlaid on the corresponding E map (blue).
Endophenotypic clusters defined by spatially connected voxels exhibiting statistically significant genetic control (P < 0.05 at the cluster level and A > 0.4 for all voxels [Korgaonkar et al., 2014]) were identified for each individual RSN. Specifically, for the visual network, two endophenotypic clusters located at left inferior occipital gyrus and left cuneus were found. Two clusters within the basal ganglia network displayed the characteristics of an endophenotype, with their peak voxels located at left amygdala and subcallosal gyrus, respectively. With respect to the sensorimotor network, four endophenotypic clusters were located at the right precentral gyrus, left postcentral gyrus, left precentral gyrus, as well as left median cingulate and paracingulate gyri, respectively. For the cerebellum network, two endophenotypic clusters were identified at Cerebellum_8_R and Cerebellum_9_L, respectively. Two endophenotypic clusters within the auditory network were found at the left middle temporal gyrus and right middle temporal gyrus, respectively. In terms of the attention network, one cluster at the opercular part of left inferior frontal gyrus exhibited the characteristics of an endophenotype. One endophenotypic cluster was found in the executive control network. The peak voxel was located at the triangular part of the right inferior frontal gyrus. For the default‐mode network, two endophenotypic clusters were identified, located at right posterior cingulate gyrus and left fusiform gyrus, respectively. More detailed information of each endophenotypic cluster including the cluster size, peak A value, anatomical location (in the definition of Automated Anatomical Labeling [Tzourio‐Mazoyer et al., 2002]) and MNI coordinates, as well as P value is summarized in Table 2. The spatial maps of all these clusters were shown in Supporting Information (Figure 1).
Table 2.
Summary of endophenotypic clusters for eight RSNs
| RSN | Cluster size (A > 0.4) | Peak A value | Peak A voxel location | Peak A voxel MNI coordinates | Cluster P value |
|---|---|---|---|---|---|
| Visual | 797 | 0.74 | Left inferior occipital gyrus | −27 −93 −9 | <0.00005 |
| 41 | 0.55 | Left cuneus | −9 −96 21 | 0.023 | |
| Basal ganglia | 262 | 0.69 | Left amygdala | −24 −3 −15 | <0.00005 |
| 73 | 0.77 | Subcallosal gyrus | 18 6 −15 | <0.00005 | |
| Sensorimotor | 573 | 0.83 | Right precentral gyrus | 63 6 24 | <0.00005 |
| 220 | 0.74 | Left precentral gyrus | −27 −18 72 | <0.00005 | |
| 207 | 0.79 | Left postcentral gyrus | −51 −12 18 | <0.00005 | |
| 71 | 0.64 | Left median cingulate and paracingulate gyri | −3 −42 54 | 0.00015 | |
| Cerebellum | 113 | 0.70 | Cerebellum_8_R | 33 −63 −54 | <0.00005 |
| 77 | 0.72 | Cerebellum_9_L | −6 −57 −39 | <0.00005 | |
| Auditory | 38 | 0.65 | Left middle temporal gyrus | −51 −15 −6 | 0.0048 |
| 31 | 0.60 | Right middle temporal gyrus | 51 −39 3 | 0.021 | |
| Attention | 65 | 0.61 | Opercular part of left inferior frontal gyrus | −54 12 6 | 0.00005 |
| Executive control | 47 | 0.70 | Triangular part of right inferior frontal gyrus | 48 27 24 | 0.00075 |
| Default mode | 338 | 0.79 | Right posterior cingulate gyrus | 6 −39 15 | <0.00005 |
| 38 | 0.75 | Left fusiform gyrus | −36 −36 −21 | 0.0183 |
To highlight the differential contributions of the genetic and environmental factors, the third row of each panel in Figures 2 and 3 displays the thresholded A map (A > 0.4, red) overlaid on the corresponding E map (blue) for each RSN. The results again show that all RSNs were to a certain extent genetically heritable, but the voxel‐wise heritability considerably varied across different RSNs.
For the purpose of comparing the level of genetic control across RSNs, we examined the distribution of A values for each RSN. Figure 4 displays the histograms of A values from all voxels in individual RSNs. Kolmogorov–Smirnov tests were applied to compare the distributions of A values between every two RSNs. The P values of these comparisons were listed in Table 3. The results indicate that, after Bonferroni correction, the relative A value distributions were significantly different across all RSNs except between the auditory and attention networks, as well as between the auditory and default‐mode networks.
Figure 4.
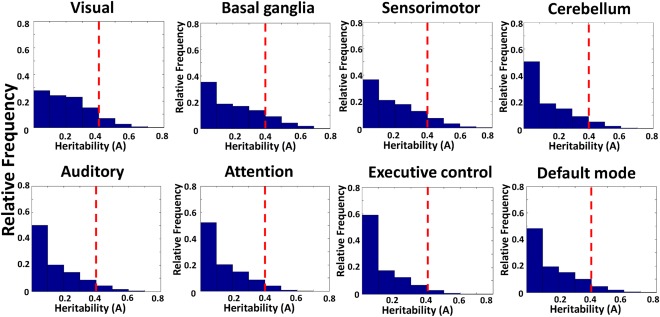
The histograms of A values of all RSNs. Dashed line indicates A = 0.4. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 3.
P values of Kolmogorov–Smirnov tests for examining the difference in the A value distributions between each two RSNs
| RSN | Auditory | Basal ganglia | Cerebellum | Default mode | Executive | Sensorimotor | Visual |
|---|---|---|---|---|---|---|---|
| Attention | 5.3 E−02 | 4.2 E−65 | 3.1 E−05 | 1.4 E−11 | 1.2 E−09 | 1.5 E−112 | 1.3 E−244 |
| Auditory | 2.3 E−34 | 3.7 E−05 | 3.7 E−03 | 4.9 E−12 | 4.0 E−39 | 5.7 E−95 | |
| Basal ganglia | 6.6 E−38 | 1.2 E−33 | 3.1 E−64 | 2.0 E−06 | 8.9 E−14 | ||
| Cerebellum | 2.9 E−07 | 1.6 E−12 | 3.8 E−53 | 2.6 E−130 | |||
| Default mode | 5.1 E−20 | 7.7 E−56 | 1.5 E−150 | ||||
| Executive | 8.0 E−89 | 7.3 E−162 | |||||
| Sensorimotor | 2.6 E−39 |
Figure 5 shows the portion of the voxels with A values >0.4 for all eight RSNs. The data indicate larger portions of voxels exhibiting relative strong genetic basis for the visual, basal ganglia, sensorimotor networks than the default‐mode, executive control and attention networks. Interestingly, visual, sensorimotor, and basal ganglia networks all subserve sensory and motor functions, whereas the default‐mode, executive control and attention networks are predominantly involved in cognitive functions. This result suggests that sensory networks may be under stronger genetic controls compared to cognitive networks.
Figure 5.
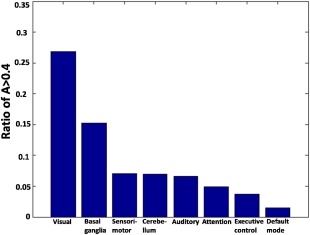
The ratio of the number of voxels with A > 0.4 for all RSNs. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
DISCUSSION
In the present study, we quantitatively estimated the genetic and environmental contributions to eight well‐organized RSNs in the human brain. The maps of genetic (A) and environmental (E) factors were generated (Figs. 2 and 3) and endophenotypic clusters of voxels were identified for each individual RSN (Table 2).
The present study is significant in several aspects. First, compared to the ROI‐based analysis used in previous studies [Glahn et al., 2010; Korgaonkar et al., 2014], our voxel‐wise approach provides more detailed spatial information regarding the genetic and environmental contributions to functional connectivity. Second, to our knowledge, this is the first study that examined the genetic effects on multiple RSNs including visual, basal ganglia, sensorimotor, cerebellum, auditory, executive control and attention networks. Considering the well‐known brain functions these RSNs are involved in, understanding the heritability of RSNs will help establish a genetic basis underlying fundamental brain functions [Thompson et al., 2013]. Third, since abnormal resting‐state functional connectivity (RSFC) is often associated with various neuropsychiatric and neurologic disorders [Broyd et al., 2009; Cocchi et al., 2012; Zalesky et al., 2012], endophenotypic clusters identified for each RSN might help link genetic and systems‐level circuit abnormalities in brain diseases [Meyer‐Lindenberg, 2009]. Fourth, given the increasing availability of animal RSFC data [Liang et al., 2011; Liang et al., 2012a, 2012b, 2014, 2013, 2015; Lu et al., 2012; Pawela et al., 2008; Sforazzini et al., 2014; Zhang et al., 2010], similar analyses can be readily carried out in animal studies. In combination with other well‐established preclinical tools, such studies will have great potentials to shed light on the genetic and molecular mechanisms underlying RSFC.
The significance of our findings also extends to clinical settings in which reliably identifying endophenotypes could help facilitate future studies aiming at improving diagnoses and treatment of psychiatric disorders. Neuropsychiatric diseases are complex, making them particularly difficult to diagnose and treat. Unlike in the simplistic case of classic Mendelian inheritance, where a given genotype leads to a definitive phenotype, neuropsychiatric diseases are produced by a combination of genetic and environmental factors that give rise to a diverse set of phenotypes. This gap between genotype and phenotype has been greatly reduced by the introduction of the “endophenotype,” which is defined as an intermediate phenotype that has a direct genetic linkage [Gottesman and Gould, 2003]. Thus, establishing RSNs as an endophenotype might be a convenient approach to understanding complex neuropsychiatric diseases because they more directly relate to gene regulation and expression than the larger overt phenotypes.
Establishing RSNs as an Endophenotype
Based on [Glahn et al., 2014; Gottesman and Gould, 2003], for a trait to be considered as an endophenotype, it must be (1) reproducibly measurable, (2) associated with the disease, (3) independent of clinical states, (4) presenting more in unaffected relatives than in the general population, and (5) heritable. RSNs are known to meet the first four criteria mentioned above. Specifically, RSNs are (1) measurable with the use of rsfMRI and are generally reproducible across healthy individuals [Beckmann et al., 2005; Damoiseaux et al., 2006; Smith et al., 2009; Zuo et al., 2010]; (2) alterations in RSNs have been tightly associated with various psychiatric and neurological disorders [Anand et al., 2005; Carter et al., 2012; Greicius et al., 2007, 2004; Hunter et al., 2012; Kennedy et al., 2006; Lowe et al., 2002; Lustig et al., 2003; Mayer et al., 2011; Tian et al., 2006; van Meer et al., 2012; Whitfield‐Gabrieli et al., 2009]; (3) as RSNs represent intrinsic neural network organization, they manifest in patients (and healthy subjects) independent of clinical states; and (4) abnormality of RSNs has been identified in unaffected relatives of schizophrenia and other mental disorders [Meda et al., 2012].
Therefore, to fully establish RSNs as an endophenotype, we need to understand the genetic influences and heritability of RSNs [Gottesman and Gould, 2003]. The literature studies have shown that the default‐mode network is heritable [Glahn et al., 2010; Korgaonkar et al., 2014]. In the present study, we examined the genetic basis of other RSNs, and the results showed that all RSNs were to a certain extent genetically heritable. Consistent with this notion, it has been shown that certain genotypes are associated with the functional connectivity within RSNs. For instance, one study compared low and high activity of monoamine oxidase A (MAOA) genotype and found that individuals with high levels of MAOA had increased functional connectivity within the posterior cingulate of the default‐mode network [Clemens et al., 2014]. Interestingly, the same brain region exhibited the highest heritability in the default‐mode network as revealed in the present study and the literature [Glahn et al., 2010]. In addition, another study demonstrated that the APOE genotype, a major genetic risk factor for Alzheimer's disease, influences functional connectivity of the lingual gyrus within the visual network [Trachtenberg et al., 2012]. In our study, the right lingual gyrus also displayed high heritability in the visual network (MNI coordinates: (12, −57, −6), A = 0.67, inside the cluster with the peak voxel at the left inferior occipital gyrus). This result may suggest a possible association between the APOE genotype and Alzheimer's disease that manifests via the endophenotype of RSFC in lingual gyrus. This hypothesis can be further tested using molecular genetic methods in future investigations. Taken together, given the significant genetic heritability and other features aforementioned for all RSNs, we believe RSNs can be established as an endophenotype.
Different Genetic Influences Across RSNs
An interesting observation of the present study is that the genetic influences appear to be stronger on sensory networks like the visual, sensorimotor and basal ganglia networks, reflected by larger ratios of voxels with high A values (Fig. 5), than cognition‐related networks such as default‐model, executive control and attention networks. We conjecture that this difference may be related to different developmental trajectories for these two types of networks. It has been well‐established that the development of sensory functions is completed at very early ages. For example, the critical period for proper neural development of vision in the cortex is the first year after birth [Antonini and Stryker, 1993; Wiesel and Hubel, 1963]. Meanwhile, the development of cognitive functions and related neural networks is long lasting throughout the entire adolescent period and into early adulthood [Fair et al., 2008]. We speculate that the protracted development of cognitive functions allows the environment to shape cognitive networks to a greater degree than sensory networks, which reach maturity at early ages. Ultimately, these findings may reflect a general principle of neural development in which phylogenetically “newer” brain functions, such as cognition, are associated with functional networks that have longer developmental trajectories. Moreover, the timing of the development of functional networks may be a reflection of the biological demand for that brain function. As described above, brain regions associated with vision mature faster than those related to cognitive functions, and one could argue that the ability to see is more vital to our survival than the ability to make decisions. Therefore, a stronger genetic control on the development of sensory networks might play an important evolutionary role. Notably, an exception to this notion was the auditory network. For unknown reasons, the genetic contribution to the auditory cortex was relatively low, even though it is also a major sensory network and matures early in life.
An alternative explanation to less genetic influences on cognitive RSNs relative to sensory RSNs is the age group of the twin sample used in the present study. It has been reported that the genetic influences over cognition increases with age from born to adulthood [Briley and Tucker‐Drob, 2013]. Therefore, given that the twin sample recruited in the present study is relatively young (age: 12–19), it is likely that the heritability of cognitive RSNs has not yet reached a maximum.
Technical Considerations and Potential Limitations
There are a few limitations in the present study. First, although motion‐related variances were considered in model fitting, other imaging‐associated variances were not taken into account and could be a confounding factor to our results. To thoroughly evaluate all imaging‐associated variances, a test‐retest study will be needed, which is unfortunately not pragmatic. Nonetheless, we believe it is unlikely that other imaging‐related variances could significantly affect the final results for two reasons. First, we found that the motion‐related variance, which typically represent a large portion of imaging‐associated variance, minimally contributed to A and E maps for all RSNs. In Supporting Information (Figure 2) we, respectively, compared the A and E maps between the cases with and without considering motion‐related variances in two example (visual and default‐mode) networks. For both networks, the spatial patterns of A and E maps were virtually identical between the two cases, suggesting that the motion variance had little effect on the relative A and E contributions to these two RSNs. The same result was observed in all other RSNs. Second, the ICA approach applied in the present study made the functional connectivity of RSNs “not significantly affected by structured noise over a relatively large range,” as suggested by a study comparing ICA and seed‐based correlational analysis [Ma et al., 2007]. As a result, the imaging‐related variance unlikely represented a major source of variance in our data, and thus will not significantly affect the final results.
Another limitation of the present study is that the threshold of heritability in the definition of endophenotype is somewhat arbitrary. Considering that there is no consensus on the degree of the genetic contribution (i.e., A value) when defining an endophenotype, we used the threshold of A value based on a previous publication [Korgaonkar et al., 2014]. A similar degree of heritability has also been used to define endophenotypes in others studies. For instance, using a twin design Chen and Li showed that the additive genetic factor accounted for 31% (i.e., A = 0.31) of variance in the endophenotype of adolescent dysfunctional attitude [Chen and Li, 2014]. In another study using a mother‐offspring design, the heritability of delusional‐like experiences was estimated to be 0.35 (standard error 0.04), and these experiences were considered to represent a quantitative endophenotype for genetic studies of common mental disorders [Varghese et al., 2013]. Therefore, the degree of heritability chosen in the presented study should provide a reasonable threshold for defining endophenotype. Importantly, the genetic contributions in all clusters reported were statistically significant, confirming meaningful heritability of these endophenotypes.
In the present study, we dropped the full ACE model and adopted the AE model because of the insignificance of the common environmental (i.e., C) factor. Although insignificant effects from the C factor to RSNs have been reported in almost all previous studies using the same model [Blokland et al., 2011; Fornito et al., 2011; Korgaonkar et al., 2014; van den Heuvel et al., 2013], it has to be noted that the effects of the C factor may still be significant in studies with much larger statistical power. Neale and Cardon found that to be 80% sure about the common environmental factor accounting for 20% of the total variance, 692 twin pairs for each twin type (DZ or MZ) are required [Neale and Cardon, 1992]. Due to the moderate sample size of imaging genetics studies, the undetected effects of the C factor may be due to the lack of sufficient statistical power. In addition, like the most majority of these previous studies, we used the likelihood ratio test for selecting between the full ACE model and reduced models [Couvy‐Duchesne et al., 2014; Dominicus et al., 2006; Korgaonkar et al., 2014; Neale and Cardon, 1992]. It needs to be kept in mind that alternative statistical approaches are also available and might provide advantages over the likelihood ratio test. For instance, nonparametric permutation approaches can establish the exact empirical null distributions of A, C, and E factors, particularly under the circumstance that the sample size is not very large.
Also due to the moderate sample size of the present study, we included both DZ same‐sex and DZ opposite‐sex twins in the analysis. While there was no significant difference in the number of males and females in the DZ group, it is possible that opposite‐sex twin pairs can decrease the DZ correlation, thereby “artificially” increasing heritability. To address this issue, we calculated intra‐class voxelwise correlations for MZ, DZ same‐sex (12 pairs), and DZ opposite‐sex (12 pairs) groups, as well as the differences in mean, variance, and covariance of z‐scores between DZ same‐sex twins and DZ opposite‐sex twins for each ICA component in two RSNs that were under the strongest genetic control (i.e., visual and basal ganglia networks, Supporting Information Figure 3). The data show that the corresponding correlation maps between DZ same‐sex and DZ opposite‐sex groups were overall comparable. In addition, means, variances, and covariances can generally be equated between DZ same‐sex and DZ opposite‐sex groups, as reflected from the histograms of voxelwise difference between DZ same‐sex and DZ opposite‐sex twins for each nonartifactual ICA component in the two RSNs. Virtually all histograms show distributions with centers at zero. Based on this result, we believe that although including DZ opposite‐sex twin pairs may affect the estimation of heritability, this influence should be minimal. We will further investigate this issue with a larger sample size.
Nonstationarity could be another potential limitation in the present study. Nonstationarity due to local smoothness variation of noise requires a stringent cluster‐forming threshold and high spatial smoothing to control for false positive rates [Silver et al., 2011]. The measurements of resting‐state activity are unavoidably to include a few nonstationary components resulting from motion and physiological noise and, therefore, may affect the genetic analysis in the present study. However, because the group ICA approach is able to recover stationary sources even when nonstationary sources are present in the data, as suggested by previous studies [Calhoun et al., 2001a], the potential effects of nonstationarity should be minimized in the present study. Indeed, all ICA components used for the genetic analysis in the present study were consistent with the literature [Allen et al., 2011] owing to the template‐matching method applied [Greicius et al., 2004] and the well‐recognized templates used [Allen et al., 2011], suggesting that these ICA components were dominated by stationary sources. Therefore, the effects of nonstationarity on the genetic analysis should be minimal.
Conclusions
In conclusion, the present study has mapped the genetic and environmental influences on functional brain networks, and has identified specific endophenotypic clusters for individual RSNs. Our findings suggest that part of the human functional connectome is shaped by genetic constraints and the genetic control is heterogeneous across different RSNs. This study may be useful for determining the genetic basis of the human functional connectome.
Supporting information
Supporting Information
ACKNOWLEDGMENT
We would like to thank Dr. Elena Allen for providing the code for fitting voxel t values of individual components to the Normal‐Gamma‐Gamma mixture model. Financial Disclosures: Authors have no financial conflicts of interest.
REFERENCES
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD (2011): A baseline for the multivariate comparison of resting‐state networks. Front Syst Neurosci 5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Li Y, Wang Y, Wu J, Gao S, Bukhari L, Mathews VP, Kalnin A, Lowe MJ (2005): Activity and connectivity of brain mood regulating circuit in depression: A functional magnetic resonance study. Biol Psychiatry 57:1079–1088. [DOI] [PubMed] [Google Scholar]
- Antonini A, Stryker MP (1993): Rapid remodeling of axonal arbors in the visual cortex. Science 260:1819–1821. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ (1999): Nonlinear spatial normalization using basis functions. Hum Brain Mapp 7:254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM (2005): Investigations into resting‐state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci 360:1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS (1995): Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magn Reson Med 34:537–541. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, Dogonowski AM, Ernst M, Fair D, Hampson M, Hoptman MJ, Hyde JS, Kiviniemi VJ, Kotter R, Li SJ, Lin CP, Lowe MJ, Mackay C, Madden DJ, Madsen KH, Margulies DS, Mayberg HS, McMahon K, Monk CS, Mostofsky SH, Nagel BJ, Pekar JJ, Peltier SJ, Petersen SE, Riedl V, Rombouts SA, Rypma B, Schlaggar BL, Schmidt S, Seidler RD, Siegle GJ, Sorg C, Teng GJ, Veijola J, Villringer A, Walter M, Wang L, Weng XC, Whitfield‐Gabrieli S, Williamson P, Windischberger C, Zang YF, Zhang HY, Castellanos FX, Milham MP (2010): Toward discovery science of human brain function. Proc Natl Acad Sci USA 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokland GA, McMahon KL, Thompson PM, Martin NG, de Zubicaray GI, Wright MJ (2011): Heritability of working memory brain activation. J Neurosci 31:10882–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boker S, Neale M, Maes H, Wilde M, Spiegel M, Brick T, Spies J, Estabrook R, Kenny S, Bates T, Mehta P, Fox J (2011): OpenMx: An open source extended structural equation modeling framework. Psychometrika 76:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briley DA, Tucker‐Drob EM (2013): Explaining the increasing heritability of cognitive ability across development: A meta‐analysis of longitudinal twin and adoption studies. Psychol Sci 24:1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broyd SJ, Demanuele C, Debener S, Helps SK, James CJ, Sonuga‐Barke EJ (2009): Default‐mode brain dysfunction in mental disorders: A systematic review. Neurosci Biobehav Rev 33:279–296. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ (2001a): Group ICA of functional MRI data: Separability, stationarity, and inference In: International Conference on ICA and BSS. San Diego, CA: pp 155–160. [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ (2001b): A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp 14:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AR, Shulman GL, Corbetta M (2012): Why use a connectivity‐based approach to study stroke and recovery of function? Neuroimage 62:2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li X (2014): Genetic and environmental etiologies of adolescent dysfunctional attitudes: A twin study. Twin Res Hum Genet 17:16–22. [DOI] [PubMed] [Google Scholar]
- Clemens B, Voss B, Pawliczek C, Mingoia G, Weyer D, Repple J, Eggermann T, Zerres K, Reetz K, Habel U (2014): Effect of MAOA genotype on resting‐state networks in healthy participants. Cereb Cortex 25:1771–1781. [DOI] [PubMed] [Google Scholar]
- Cocchi L, Bramati IE, Zalesky A, Furukawa E, Fontenelle LF, Moll J, Tripp G, Mattos P (2012): Altered functional brain connectivity in a non‐clinical sample of young adults with attention‐deficit/hyperactivity disorder. J Neurosci 32:17753–17761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvy‐Duchesne B, Blokland GA, Hickie IB, Thompson PM, Martin NG, de Zubicaray GI, McMahon KL, Wright MJ (2014): Heritability of head motion during resting state functional MRI in 462 healthy twins. Neuroimage 102(Pt 2):424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF (2006): Consistent resting‐state networks across healthy subjects. Proc Natl Acad Sci USA 103:13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM (2006): fMRI resting state networks define distinct modes of long‐distance interactions in the human brain. Neuroimage 29:1359–1367. [DOI] [PubMed] [Google Scholar]
- Dominicus A, Skrondal A, Gjessing HK, Pedersen NL, Palmgren J (2006): Likelihood ratio tests in behavioral genetics: Problems and solutions. Behav Genet 36:331–340. [DOI] [PubMed] [Google Scholar]
- Fair DA, Cohen AL, Dosenbach NU, Church JA, Miezin FM, Barch DM, Raichle ME, Petersen SE, Schlaggar BL (2008): The maturing architecture of the brain's default network. Proc Natl Acad Sci USA 105:4028–4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bassett DS, Meunier D, Ellison‐Wright I, Yucel M, Wood SJ, Shaw K, O'Connor J, Nertney D, Mowry BJ, Pantelis C, Bullmore ET (2011): Genetic influences on cost‐efficient organization of human cortical functional networks. J Neurosci 31:3261–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME (2007): Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci 8:700–711. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T (2002): Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 15:870–878. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J, Gottesman II, Almasy L (2014): Arguments for the sake of endophenotypes: Examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet 165B:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM, Beckmann CF, Fox PT, Blangero J (2010): Genetic control over the resting brain. Proc Natl Acad Sci USA 107:1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD (2003): The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry 160:636–645. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Flores BH, Menon V, Glover GH, Solvason HB, Kenna H, Reiss AL, Schatzberg AF (2007): Resting‐state functional connectivity in major depression: Abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol Psychiatry 62:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V (2004): Default‐mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA 101:4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JV, Wilde EA, Tong KA, Holshouser BA (2012): Emerging imaging tools for use with traumatic brain injury research. J Neurotrauma 29:654–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy DP, Redcay E, Courchesne E (2006): Failing to deactivate: Resting functional abnormalities in autism. Proc Natl Acad Sci USA 103:8275–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar MS, Ram K, Williams LM, Gatt JM, Grieve SM (2014): Establishing the resting state default mode network derived from functional magnetic resonance imaging tasks as an endophenotype: A twins study. Hum Brain Mapp 35:3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N (2011): Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J Neurosci 31:3776–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N (2012a): Anticorrelated resting‐state functional connectivity in awake rat brain. Neuroimage 59:1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N (2012b): Intrinsic organization of the anesthetized brain. J Neurosci 32:10183–10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N (2014): Neuroplasticity to a single‐episode traumatic stress revealed by resting‐state fMRI in awake rats. Neuroimage 103:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Li T, King J, Zhang N (2013): Mapping thalamocortical networks in rat brain using resting‐state functional connectivity. Neuroimage 83:237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Liu X, Zhang N (2015): Dynamic resting state functional connectivity in awake and anesthetized rodents. Neuroimage 104:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe MJ, Phillips MD, Lurito JT, Mattson D, Dzemidzic M, Mathews VP (2002): Multiple sclerosis: Low‐frequency temporal blood oxygen level‐dependent fluctuations indicate reduced functional connectivity initial results. Radiology 224:184–192. [DOI] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y (2012): Rat brains also have a default mode network. Proc Natl Acad Sci USA 109:3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig C, Snyder AZ, Bhakta M, O'Brien KC, McAvoy M, Raichle ME, Morris JC, Buckner RL (2003): Functional deactivations: Change with age and dementia of the Alzheimer type. Proc Natl Acad Sci USA 100:14504–14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Wang B, Chen X, Xiong J (2007): Detecting functional connectivity in the resting brain: A comparison between ICA and CCA. Magn Reson Imaging 25:47–56. [DOI] [PubMed] [Google Scholar]
- Mayer AR, Mannell MV, Ling J, Gasparovic C, Yeo RA (2011): Functional connectivity in mild traumatic brain injury. Hum Brain Mapp 32:1825–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Gill A, Stevens MC, Lorenzoni RP, Glahn DC, Calhoun VD, Sweeney JA, Tamminga CA, Keshavan MS, Thaker G, Pearlson GD (2012): Differences in resting‐state functional magnetic resonance imaging functional network connectivity between schizophrenia and psychotic bipolar probands and their unaffected first‐degree relatives. Biol Psychiatry 71:881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A (2009): Neural connectivity as an intermediate phenotype: Brain networks under genetic control. Hum Brain Mapp 30:1938–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Cardon LR (1992): Methodology for Genetic Studies of Twins and Families. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
- Pawela CP, Biswal BB, Cho YR, Kao DS, Li R, Jones SR, Schulte ML, Matloub HS, Hudetz AG, Hyde JS (2008): Resting‐state functional connectivity of the rat brain. Magn Reson Med 59:1021–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, DeFries JC; Knopik VS, Neiderhiser JM (2013): Behavioral Genetics. New York: Worth Publishers; 519 p. [Google Scholar]
- Sforazzini F, Schwarz AJ, Galbusera A, Bifone A, Gozzi A (2014): Distributed BOLD and CBV‐weighted resting‐state networks in the mouse brain. Neuroimage 87:403–415. [DOI] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP (2009): The resting brain: Unconstrained yet reliable. Cereb Cortex 19:2209–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver M, Montana G, Nichols TE, Alzheimer's Disease Neuroimaging Initiative (2011): False positives in neuroimaging genetics using voxel‐based morphometry data. Neuroimage 54:992–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF (2009): Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci USA 106:13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song XW, Dong ZY, Long XY, Li SF, Zuo XN, Zhu CZ, He Y, Yan CG, Zang YF (2011): REST: A toolkit for resting‐state functional magnetic resonance imaging data processing. PLoS One 6:e25031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Ge T, Glahn DC, Jahanshad N, Nichols TE (2013): Genetics of the connectome. Neuroimage 80:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Jiang T, Wang Y, Zang Y, He Y, Liang M, Sui M, Cao Q, Hu S, Peng M, Zhuo Y (2006): Altered resting‐state functional connectivity patterns of anterior cingulate cortex in adolescents with attention deficit hyperactivity disorder. Neurosci Lett 400:39–43. [DOI] [PubMed] [Google Scholar]
- Trachtenberg AJ, Filippini N, Ebmeier KP, Smith SM, Karpe F, Mackay CE (2012): The effects of APOE on the functional architecture of the resting brain. Neuroimage 59:565–572. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, van Soelen IL, Stam CJ, Kahn RS, Boomsma DI, Hulshoff Pol HE (2013): Genetic control of functional brain network efficiency in children. Eur Neuropsychopharmacol 23:19–23. [DOI] [PubMed] [Google Scholar]
- van Meer MP, Otte WM, van der Marel K, Nijboer CH, Kavelaars A, van der Sprenkel JW, Viergever MA, Dijkhuizen RM (2012): Extent of bilateral neuronal network reorganization and functional recovery in relation to stroke severity. J Neurosci 32:4495–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese D, Wray NR, Scott JG, Williams GM, Najman JM, McGrath JJ (2013): The heritability of delusional‐like experiences. Acta Psychiatr Scand 127:48–52. [DOI] [PubMed] [Google Scholar]
- Whitfield‐Gabrieli S, Thermenos HW, Milanovic S, Tsuang MT, Faraone SV, McCarley RW, Shenton ME, Green AI, Nieto‐Castanon A, LaViolette P, Wojcik J, Gabrieli JD, Seidman LJ (2009): Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first‐degree relatives of persons with schizophrenia. Proc Natl Acad Sci USA 106:1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH (1963): Effects of visual deprivation on morphology and physiology of cells in the cats lateral geniculate body. J Neurophysiol 26:978–993. [DOI] [PubMed] [Google Scholar]
- Yan C, Zhang Y (2010): DPARSF: A MATLAB toolbox for "Pipeline" data analysis of resting‐state fMRI. Front Syst Neurosci 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MJ, Tzeng CH, Tseng JY, Huang CY (2006): Determination of twin zygosity using a commercially available STR analysis of 15 unlinked loci and the gender‐determining marker amelogenin–a preliminary report. Hum Reprod 21:2175–2179. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Egan GF, Pantelis C, Bullmore ET (2012): The relationship between regional and inter‐regional functional connectivity deficits in schizophrenia. Hum Brain Mapp 33:2535–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Rane P, Huang W, Liang Z, Kennedy D, Frazier JA, King J (2010): Mapping resting‐state brain networks in conscious animals. J Neurosci Methods 189:186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP (2010): Reliable intrinsic connectivity networks: Test‐retest evaluation using ICA and dual regression approach. Neuroimage 49:2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


