Immunity triggered by high doses a and hormetic growth enhancement triggered by low doses of a synthetic elicitor doses are associated with distinct transcriptional profiles.
Abstract
Synthetic elicitors are drug-like compounds that induce plant immune responses but are structurally distinct from natural defense elicitors. Using high-throughput screening, we previously identified 114 synthetic elicitors that activate the expression of a pathogen-responsive reporter gene in Arabidopsis (Arabidopsis thaliana). Here, we report on the characterization of one of these compounds, 2-(5-bromo-2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid (BHTC). BHTC induces disease resistance of plants against bacterial, oomycete, and fungal pathogens and has a unique mode of action and structure. Surprisingly, we found that low doses of BHTC enhanced root growth in Arabidopsis, while high doses of this compound inhibited root growth, besides inducing defense. These effects are reminiscent of the hormetic response, which is characterized by low-dose stimulatory effects of a wide range of agents that are toxic or inhibitory at higher doses. Like its effects on defense, BHTC-induced hormesis in Arabidopsis roots is partially dependent on the WRKY70 transcription factor. Interestingly, BHTC-induced root hormesis is also affected in the auxin-response mutants axr1-3 and slr-1. By messenger RNA sequencing, we uncovered a dramatic difference between transcriptional profiles triggered by low and high doses of BHTC. Only high levels of BHTC induce typical defense-related transcriptional changes. Instead, low BHTC levels trigger a coordinated intercompartmental transcriptional response manifested in the suppression of photosynthesis- and respiration-related genes in the nucleus, chloroplasts, and mitochondria as well as the induction of development-related nuclear genes. Taken together, our functional characterization of BHTC links defense regulation to hormesis and provides a hypothetical transcriptional scenario for the induction of hormetic root growth.
Plant innate immunity against pathogens depends on a network of functionally interconnected genes involved in the regulation and execution of defense reactions (Glazebrook et al., 2003; Tsuda et al., 2009; Sato et al., 2010). A fundamental form of innate immunity in plants involves conserved molecular signatures common to many pathogens termed microbe-associated molecular patterns (MAMPs), which are recognized by pattern recognition receptors on the surface of plant cells (Jones and Dangl, 2006; Hein et al., 2009; Zipfel, 2014). MAMP recognition activates a comprehensive set of defense reactions collectively referred to as pattern-triggered immunity (PTI). Adapted pathogens have acquired the ability to attenuate PTI through the secretion of effector molecules, suppressing defense and, thus, enabling infection (effector-triggered susceptibility; Chisholm et al., 2006). In this case, the pathogen is virulent and the host is susceptible. During such compatible interactions, plants can still mount a weakened immune response, called basal defense, which limits pathogen spread but is typically not capable of fully preventing disease (Glazebrook, 2001; Ahmad et al., 2011). As a countermeasure to effector-triggered susceptibility, plants can recognize effectors by highly specific plant RESISTANCE proteins, which mediate effector-triggered immunity (ETI), resulting in incompatible interactions and leaving pathogens avirulent (Jones and Dangl, 2006). Numerous studies have shown that ETI, basal defense, and PTI utilize a common set of signaling components including multiple messenger substances, such as reactive oxygen species, Ca2+, salicylic acid (SA), ethylene, and jasmonic acid (Nimchuk et al., 2003). While basal defense seems to be a weakened form of PTI, ETI has been proposed to result from boosted basal defense- or PTI-associated responses (Tao et al., 2003).
The plant immune network can be subdivided into various defined sectors that can interact with each other (Tsuda et al., 2009; Sato et al., 2010). For example, distinct defense signaling sectors dependent on early MAMP-activated mitogen-activated protein kinases or the messenger molecules SA or jasmonic acid have been described for Arabidopsis (Arabidopsis thaliana).
Plant diseases cause dramatic losses in crop production. Global agriculture depends heavily on the use of pesticides to control such crop diseases. Pesticides typically rely on direct toxic, antipathogenic activity, which leads to undesirable ecological side effects (Casida, 2009). The disquiet over the dangers of pesticides has spawned considerable interest in alternative methods of disease control (Pimentel and Pimentel, 2008). Plant defense-inducing chemicals (plant activators and synthetic elicitors), which protect plants from diseases by activating their innate immune responses without the need of being toxic to pathogens, offer an attractive alternative for disease-control regimes that can be environmentally friendly (Bektas and Eulgem, 2014). Examples of such compounds include 2,6-dichloroisonicotinic acid (INA) and acibenzolar-S-methyl benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester (BTH; Métraux et al., 1991; Ward et al., 1991; Uknes et al., 1992). Interactions of these compounds with the plant immune system have been well characterized, and both INA and BTH are known to trigger a profile of defense-associated responses related to those triggered by SA-dependent signaling mechanisms (Ward et al., 1991; Görlach et al., 1996; Lawton et al., 1996; Bektas and Eulgem, 2014).
We have initiated a chemical genomics-based approach to identify, characterize, and utilize new types of synthetic elicitors for the dissection of the plant immune system and the development of novel types of environmentally safe pesticide alternatives (Knoth et al., 2009). By high-throughput chemical screening, we identified 114 drug-like organic compounds that induce the pathogen-responsive CaBP22−333::GUS reporter gene in transgenic Arabidopsis. One of them, 3,5-dicholoroanthranilic acid (DCA), triggered fast, strong, and transient disease resistance against the pathogenic oomycete Hyaloperonospora arabidopsidis (Hpa) and the bacterial pathogen Pseudomonas syringae (Knoth et al., 2009). Experiments addressing the defense-inducing activity of DCA in various Arabidopsis defense mutants showed that this synthetic elicitor activates a signaling route partially dependent on the WRKY70 transcription factor. In contrast to INA- and BTH-mediated immunity, which is fully dependent on the transcriptional cofactor and SA coreceptor NPR1 (Cao et al., 1994; Dong, 2004), DCA-mediated immunity is only weakly NPR1 dependent (Knoth et al., 2009). In addition, immunity mediated by BTH and INA is long lasting, while DCA acts transiently (Knoth et al., 2009). Thus, the mode of action utilized by DCA in defense induction is distinct from that of INA and BTH.
Here, we report on another representative of the 114 novel synthetic elicitors we identified, 2-(5-bromo-2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid (BHTC). Like DCA, BHTC also induces disease resistance quickly and transiently. However, its mode of action is distinct from that of DCA, as it strongly depends on NPR1. In addition, we found that low doses of BHTC enhanced the elongation of Arabidopsis roots, while high concentrations inhibited root elongation. These effects are reminiscent of the phenomenon of hormesis, which has been described in various biological systems and which is characterized by enhanced biological performance in response to low doses of a wide range of stimuli that are toxic or otherwise detrimental at higher doses (Calabrese and Baldwin, 2002; Mattson and Calabrese, 2010; Calabrese and Blain, 2011; Calabrese, 2013). At least in some cases, hormesis may constitute an adaptive evolutionary response of organisms to detrimental or otherwise unfavorable biological conditions (Mattson, 2010). Hormetic responses have been proposed to be generally based on compensatory processes following an initial disruption in homeostasis (Calabrese, 2010). Although such phenomena have been described for a wide variety of organismal types and stimuli, their mechanistic basis has only been established in several nonplant systems (Mattson, 2008, 2010; Mattson and Calabrese, 2010; Son et al., 2010), and it is unclear if distinct forms of hormesis share common regulatory processes.
Interestingly, we found the transcriptional profiles triggered in Arabidopsis by low and high BHTC doses to be very different. In addition, wrky70 mutants, which exhibit reduced BHTC-mediated immunity, as well as the auxin-response mutants axr1-3 and slr-1, are compromised in BHTC-triggered root hormesis. Taken together, our results link plant defense signaling to hormetic developmental responses and provide a genetic and transcriptional framework for future studies on the mechanistic basis of plant hormesis.
RESULTS
BHTC, a Small Molecule Elicitor of CaBP22−333::GUS Expression and Transient Resistance of Arabidopsis to Hpa
We previously identified 114 compounds that induce the expression of the pathogen-responsive CaBP22−333::GUS reporter gene in Arabidopsis (Knoth et al., 2009). One of them, BHTC, has not been reported as a synthetic elicitor and has a chemical structure distinct from DCA or any other plant defense inducers described so far (Schreiber and Desveaux, 2008; Knoth et al., 2009; Bektas and Eulgem, 2014). BHTC activated reporter gene expression in 1-week-old CaBP22−333::GUS Arabidopsis seedlings submerged in liquid growth medium at a concentration as low as 1 µm (Supplemental Fig. S1A). To examine if BHTC induces phytotoxicity, we stained CaBP22−333::GUS seedlings after BHTC treatment with Trypan Blue. We observed dark blue staining indicating cell death in 100% of the seedlings treated for 24 h with 500 µm BHTC (Supplemental Fig. S1B). No cell death was observed at lower concentrations (1–100 µm), which resulted in CaBP22−333::GUS activation, indicating that BHTC-induced phytotoxicity is not responsible for its effect on the expression of this pathogen-responsive reporter gene.
We further examined if BHTC, like DCA, has the ability to induce pathogen resistance in soil-grown plants. Single foliar spray application of 100 µm BHTC 1 h to 1 d prior to infection with the virulent Hpa isolate Noco2 strongly reduced numbers of Hpa spores by up to 73% (Fig. 1). Maximal levels of immunity against Hpa Noco2 were observed with 50 to 100 μm BHTC (Fig. 1B). We estimated the median effective concentration (EC50) of BHTC regarding its ability to protect Col-0 from Hpa Noco2 as 5.5 μm (Supplemental Fig. S2A). EC50 values represent the concentration of a bioactive compound at which half-maximal biological activity is observed and reflect its potency regarding uptake and/or ability to interact with its target(s). Compounds with lower EC50 values are likely more efficiently taken up by biological systems and/or have a higher affinity for their targets than compounds with higher EC50 values. While DCA triggered higher levels of immunity, suppressing Hpa Noco2 formation in Col-0 by nearly 100% (Knoth et al., 2009), its estimated EC50 value of 6.5 μm (Supplemental Fig. S2B) regarding this response is slightly higher than that of BHTC.
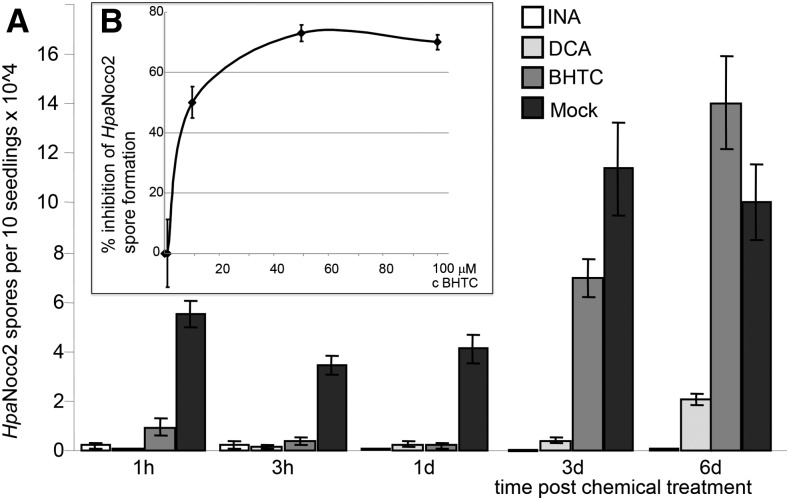
Figure 1.
Kinetic and dose-response analyses of BHTC-induced immunity of Arabidopsis against Hpa. A, Kinetic analysis of chemically induced disease resistance. Three-week-old soil-grown Columbia-0 (Col-0) seedlings were sprayed with 100 µm BHTC, DCA, INA, or mock solution (1% [v/v] dimethyl sulfoxide [DMSO]) at the indicated times prior to infection with 2 × 104 mL−1 Hpa Noco2 spores (2 mL per pot). Hpa spores were counted 7 d post infection (dpi). Mean and se values were calculated from a minimum of three biological replicates, and the average of those is shown. Student’s t test (P < 0.05) showed significant differences for all of the synthetic elicitor treatments relative to the mock-treated control, except for 6 d after treatment with BHTC. B, Dose-response curve for BHTC-mediated immunity of Arabidopsis against Hpa Noco2. Plotted is the relative inhibition of Hpa spore formation versus the concentration of BHTC used in single foliar spray applications.
We further compared the kinetics of defense induction in Col-0 seedlings sprayed once with 100 µm BHTC, DCA, or INA at various time points prior to pathogen challenge (Fig. 1A). Mock treatment itself diminished spore growth when time points between pathogen challenge and elicitor pretreatment were less than 1 d apart. This effect may be due to residual liquid coating Arabidopsis seedlings before being sprayed with the Hpa spore suspension. Already at 1 to 3 h post treatment, all three tested synthetic elicitors strongly suppressed Hpa spore production. However, at 3 d post treatment, BHTC-triggered immunity to Hpa Noco2 was reduced, and no effect of this compound on immunity was detectable at 6 d post treatment. As reported previously, DCA also induces plant defense transiently (Knoth et al., 2009), while the activity of INA is long lasting (Métraux et al., 1991; Görlach et al., 1996; Bowling et al., 1997). Based on our data, the defense-inducing activity of BHTC is even more transient than that of DCA. Taken together, BHTC, like DCA, is a fast, potent, but reversible inducer of Arabidopsis immunity against Hpa Noco2.
Structure-Activity Analysis with BHTC Derivatives
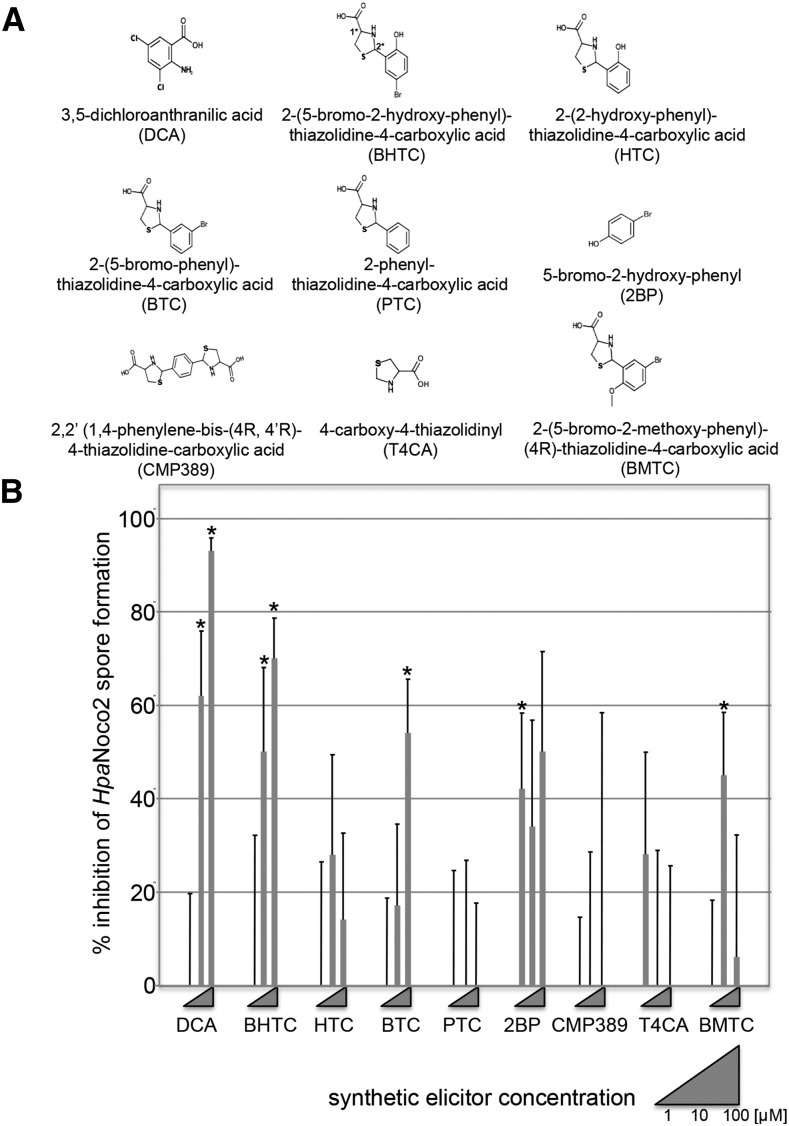
To determine which substituents or moieties of the BHTC molecule are critical for its defense-inducing activity, seven commercially available BHTC derivatives were analyzed that differed only minimally from the original synthetic elicitor structure (Fig. 2A). We tested the ability of these compounds, next to DCA and BHTC, to inhibit Hpa Noco2 spore development in Col-0 plants after a single foliar spray application (Fig. 2B). DCA and BHTC provided the highest protection against Hpa Noco2 infection, clearly suppressing Hpa spore formation at 10 and 100 μm and reaching levels of over 70% protection. 2-(5-Bromo-phenyl)-thiazolidine-4-carboxylic acid (BTC) and 2-(5-bromo-2-methoxy-phenyl)-thiazolidine-4-carboxylic acid (BMTC) mediated at one of the tested concentrations significant levels of intermediate spore reduction. Compared with BHTC, BTC lacks the hydroxy group of the phenyl moiety, while this substituent is replaced by a methoxy group in BMTC. 2-(2-Hydroxy-phenyl)-thiazolidine-4-carboxylic acid (HTC), which lacks the bromine of the phenyl moiety, did not suppress spore formation at 10 and 100 μm. However, this compound induced CaBP22−333::GUS expression, and we observed high levels of spore suppression with this compound at a concentration of 200 μm (Supplemental Fig. S3). 2-Phenyl-thiazolidine-4-carboxylic acid (PTC) did not mediate any protection against Hpa Noco2. Thus, substitution of the phenyl moiety of phenylthiazolidine-4-carboxylic acid derivatives seems to be critical for their ability to induce plant immune responses.
Figure 2.
Structure-activity analysis of BHTC analogs. A, Chemical structures of DCA, BHTC, and tested BHTC derivatives. Chiral centers of the BHTC skeleton are indicated by 1* and 2* in BHTC. B, Hpa Noco2 spore inhibition assay. Three-week-old soil-grown Col-0 seedlings were spray infected 24 h after treating with varying concentrations of each synthetic elicitor and then assayed at 7 dpi for spore growth; 100% inhibition = zero spores. The assay was repeated three times with similar results. The average of those three replicates is shown. Significant differences of compound-treated compared with mock-treated seedlings determined by Student’s t test (P < 0.05) are marked by asterisks. T4CA, 4-Carboxy-4-thiazolidinyl.
The isolated thiazolidine-4-carboxylic acid moiety of BHTC, 4-carboxy-4-thiazolidinyl, as well as CMP389, which consists of a phenyl moiety with two 4-carboxy-4-thiazolidinyl substituents, also did not mediate protection against Hpa Noco2. Interestingly, 5-bromo-2-hydroxy-phenyl (2BP), which consists only of the substituted phenyl moiety of BHTC, was sufficient to trigger some protection against Hpa Noco2. However, significant levels of immunity were only observed at one of the tested concentrations (1 μm), and levels of Hpa spore suppression did not exceed 50%. Except for 2BP, all other tested BHTC derivatives that induced significant protection of Col-0 against Hpa Noco2 also triggered GUS expression in our CaBP22−333::GUS reporter gene assays at 100 μm or lower concentrations (data not shown). Thus, 2BP seems to be a weaker and less reliable plant defense inducer than BHTC. Compared with its tested derivatives, BHTC seems to be the most robust and efficient synthetic elicitor. Therefore, we used this compound as a representative for the new class of PTC synthetic elicitors for all further experiments in this study.
BHTC Is Functionally Distinct from DCA
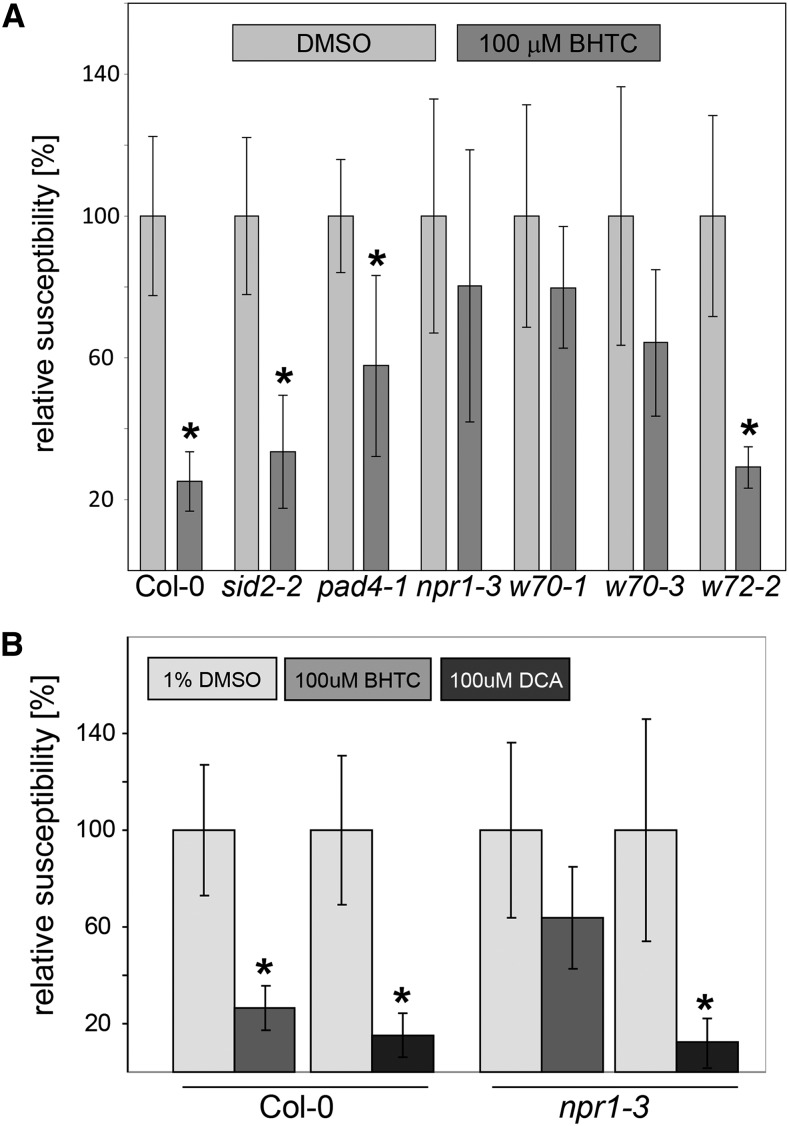
DCA and BHTC differ regarding the timing of their defense-inducing activity. While both synthetic elicitors trigger transient protection of Col-0 against Hpa Noco2, immunity mediated by BHTC is of even shorter duration than that triggered by DCA (Fig. 1A). To genetically establish whether the mode of action of BHTC differs from that of DCA, we tested the defense-inducing activity of this new synthetic elicitor in various Arabidopsis mutants after a single foliar spray application. We previously reported that full immunity mediated by DCA requires both NPR1 and the WRKY70 transcription factor (Knoth et al., 2007, 2009). However, the dependency of DCA on WRKY70 is more pronounced than that on NPR1. While BHTC triggered significant levels of immunity against Hpa Noco2 in Col-0 plants as well as the sid2-2, pad4-1, and wrky72-2 mutants, no significant protection of the npr1-3, wrky70-1, and wrky70-3 mutants against this pathogen was observed (Fig. 3A). The sid2-2 and pad4-1 mutants are compromised in the defense-associated accumulation of SA (Feys et al., 2001; Wildermuth et al., 2001). We previously reported the wrky72-2 mutant to be deficient in signaling processes that seem independent of SA (Bhattarai et al., 2010). Based on this, BHTC, like DCA (Knoth et al., 2009), appears to interfere with signaling processes operating downstream from SA (or independent from this defense hormone) and to require NPR1 as well as WRKY70 for defense induction. A critical difference between BHTC and DCA, however, seems to be their level of NPR1 dependency. In contrast to DCA, which can trigger significant levels of immunity against Hpa Noco2 in the npr1-3 mutant (Knoth et al., 2009), BHTC is unable to provide significant protection against this pathogen in npr1-3 plants (Fig. 3B).
Figure 3.
Analysis of BHTC activity in Arabidopsis Col-0 or Col-0 defense mutants. Experiments were conducted with 3-week-old soil-grown seedlings sprayed with 100 µm BHTC, 100 µm DCA, or mock solution (1% [v/v] DMSO) 24 h prior to infection with 3 × 104 virulent Hpa Noco2 spores mL−1 (2 mL per pot). Spores were counted at 7 dpi. Shown are relative numbers of spores per seedling compared with values obtained with the respective mock-treated controls. Error bars represent se based on at least four independent biological replicates. For each replicate, at least three pots, each containing more than 20 seedlings, were examined. Relative spore numbers that are significantly reduced based on Student’s t test (P < 0.05) are marked by asterisks. w, wrky.
BHTC Can Provide Disease Protection in a Variety of Plant-Pathogen Interactions
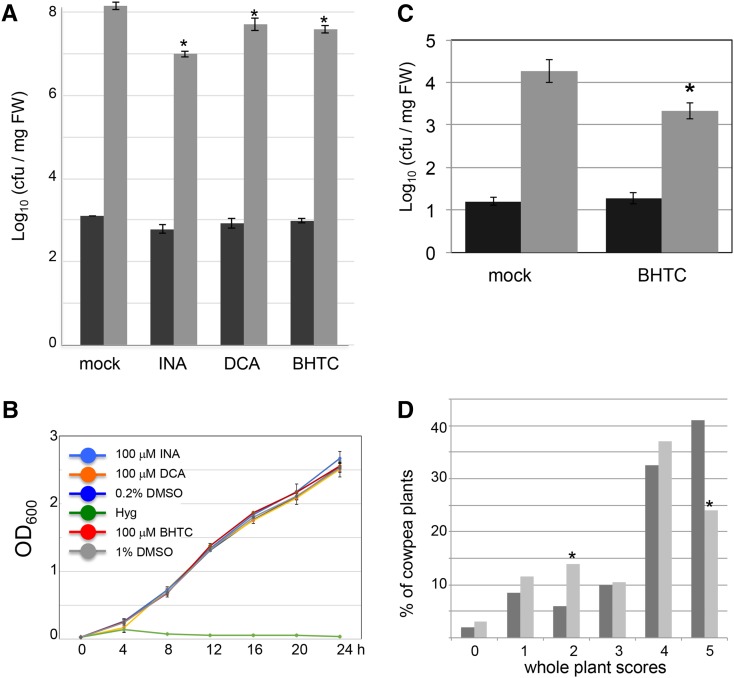
We further tested if BHTC can mediate disease protection in additional plant-pathogen interactions. Like DCA and INA, BHTC reduced the growth of the virulent bacterial pathogen Pseudomonas syringae pv tomato (Pst) strain DC3000 growth in Arabidopsis after a single foliar spray application at a concentration of 100 μm (Fig. 4A). To determine the direct antibacterial activity of BHTC, we monitored the growth of Pst in liquid medium containing 100 μm BHTC, DCA, INA, or the antibiotic hygromycin (Fig. 4B). None of the tested synthetic elicitors reduced bacterial growth, while hygromycin completely eliminated the growth of the bacteria. Taken together, these data show that BHTC can protect Arabidopsis against Pst by inducing plant defense reactions and not by direct toxicity against these bacteria.
Figure 4.
BHTC induces defense reactions in multiple plant species against diverse pathogens. A, Quantification of Pst DC3000 growth on 2-week-old Arabidopsis Col-0 plants by colony-forming units (cfu). Col-0 seedlings were pretreated with 100 μm of the indicated chemical or mock solution (solvent only) 24 h prior to dip inoculation with virulent Pst DC3000 (optical density at 600 nm [OD600] = 0.005). Bacterial titer was evaluated at day 0 (black bars) or day 3 (gray bars). Significant differences were determined using Student’s t test (P < 0.05). The data shown represent a typical example of five nearly identical biological replicates. FW, Fresh weight. Error bars represent se based on individual measurements of each of three pots, each with more than 20 plants. B, Pst DC3000 grown in liquid culture with 100 μm of the indicated chemicals or 100 μg mL−1 hygromycin (Hyg). The OD600 was measured at the indicated times (hours) after inoculation. Error bars represent se based on at least three independent replicates. C, Tomato plants (cv Moneymaker) root drenched with 200 µm BHTC display lower levels of Pst growth in leaves relative to mock-treated (solvent only) plants 3 dpi (n = 4; Student’s t test, P = 0.027). The data shown are representative of at least three independent experiments. D, Cowpea plants sprayed with BHTC exhibit reduced severity of Fot3-induced disease symptoms. Whole-plant scores were rated on a scale of 0 to 5, based on the percentage of the plant that displayed Fot3-induced symptoms (including chlorosis, wilting, vascular discoloration, and tissue necrosis): 0 = no disease symptoms; 1 = 10%; 2 = 25%; 3 = 50%; 4 = 75%; and 5 = 100%. χ2 tests of independence showed significant differences between BHTC- and mock-treated plants for scores of 2 (P < 0.012) and 5 (P < 0.003), indicated with asterisks. The data shown are combinations of two independent experiments including a total of 200 individual plants for each type of treatment.
We further tested the effects of BHTC on the compatible interaction of tomato (Solanum lycopersicum ‘Moneymaker’) with Pst. At 3 dpi, tomato plants treated with 200 μm BHTC exhibited clearly reduced numbers of colony-forming units of Pst compared with mock-treated control plants (Fig. 4C). Similarly, BHTC mildly, but significantly, delayed the development of disease symptoms in the legume cowpea (Vigna unguiculata ‘California Blackeye 5’) after infection with the fungal pathogen Fusarium oxysporum f. sp. tracheiphilum race 3 (Fot3). As shown in Figure 4D, compared with mock-treated plants, a larger number of BHTC-treated plants exhibited an intermediate level of disease symptoms (category 2) while a reduced number of BHTC-treated plants showed severe disease symptoms (category 5).
BHTC Induces Hormesis-Like Responses in Arabidopsis Roots
Surprisingly, we found BHTC at concentrations below 1 µm to enhance the root length of Arabidopsis plants grown on BHTC-laced one-half-strength Murashige and Skoog (MS) agar plates, while higher doses of BHTC resulted in reduced root length (Fig. 5). In contrast to our Hpa defense assays (see above), plants were continuously exposed to BHTC in our root growth assays. The observation that low doses of BHTC stimulate root elongation while high doses of this compound reduce root length is reminiscent of the known phenomenon of hormesis, which has been described for a large variety of physical and chemical stimuli in numerous organisms, including humans (Calabrese and Baldwin, 2002; Mattson and Calabrese, 2010; Calabrese and Blain, 2011). Hormesis is generally characterized as low-dose stimulation and high-dose inhibition of biological responses.
Figure 5.
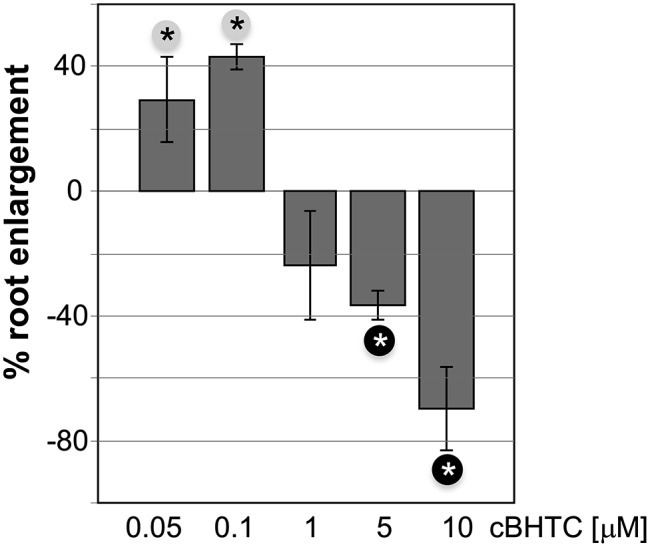
Relative root length of Col-0 plants grown on BHTC. Col-0 seedlings were grown for 14 d on one-half-strength MS medium containing the indicated concentrations of BHTC (cBHTC) or the respective solvent only. Plotted is the enhancement in root length (%) of BHTC-treated plants relative to the root length of mock (solvent only)-treated plants at day 14. Significant differences between BHTC- and mock-treated plants were determined by Student’s t test (P < 0.05) and are marked by asterisks. Black asterisks (on gray background) and white asterisks (on black background) mark the root elongation of BHTC-treated plants compared with mock controls that is significantly enhanced and reduced, respectively. Error bars represent the combined se of the respective BHTC-treated samples and their mock controls. The numbers of tested plants (n) for the respective treatments are as follows: n (0.05 µm BHTC) = 16, n (0.1 µm BHTC) = 154, n (1 µm BHTC) = 16, n (5 µm BHTC) = 39, and n (10 µm BHTC) = 16.
Dose Dependency of BHTC-Triggered Transcriptome Changes
In order to uncover transcriptional patterns associated with defense activation or hormesis induction, we profiled by mRNA sequencing (mRNA-seq) the responses triggered in 14-d-old plate-grown Arabidopsis seedlings by continuous exposure to a high dose (hd) of 5 μm BHTC or a low dose (ld) of 0.1 μm BHTC. These conditions were chosen because continuous exposure to 5 μm BHTC resulted in strong activation of the CaBP22−333::GUS reporter gene and suppression of root elongation in Arabidopsis, while the same kind of application of 0.1 μm BHTC did not induce the expression of this pathogen-responsive reporter and stimulated enhanced root elongation (Fig. 5; Supplemental Fig. S1). As controls, we used mock treatment (solvent only). For each treatment type, root and shoot tissues were analyzed separately. We performed two independent biological replicates for each experimental condition and sequenced the respective complementary DNA libraries using the Illumina HiSeq2500 platform. Differentially expressed genes (DEGs) were identified by comparing read counts from BHTC-treated samples versus their respective mock controls using a Bonferroni-corrected false discovery rate cutoff of 0.05 (Table I; Supplemental Table S1).
Table I. Set of Arabidopsis genes significantly differentially expressed in response to low-dose or high-dose BHTC treatment in plate-grown Col-0 seedlings.
| Gene Set | No. of Genes in Set | Enriched GO Termsa (with P Values) |
|---|---|---|
| hd-BHTC-shoots-up | 445 | Response to stress (P = 3.562e-73) |
| Response to abiotic or biotic stimulus (P = 1.187e-55) | ||
| Signal transduction (P = 6.848e-48) | ||
| Other biological processes (P = 1.133e-38) | ||
| Transport (P = 2.520e-21) | ||
| hd-BHTC-shoots-down | 54 | Response to abiotic or biotic stimulus (P = 6.084e-04) |
| Response to stress (P = 4.145e-03) | ||
| Protein metabolism (P = 4.965e-03) | ||
| Unknown biological processes (P = 9.490e-03) | ||
| Cell organization and biogenesis (P = 9.584e-03) | ||
| hd-BHTC-roots-up | 26 | Signal transduction (P = 6.546e-03) |
| Other cellular processes (P = 0.026) | ||
| hd-BHTC-roots-down | 10 | Other metabolic processes (P = 0.029) |
| ld-BHTC-shoots-up | 34 | Developmental processes (P = 4.062e-05) |
| Other cellular processes (P = 8.555e-04) | ||
| Other biological processes (P = 1.359e-03) | ||
| Response to stress (P = 4.527e-03) | ||
| Transport (P = 4.699e-03) | ||
| Cell organization and biogenesis (P = 0.025) | ||
| ld-BHTC-shoots-down | 132 | Electron transport or energy pathways (P = 2.718e-80) |
| DNA-dependent transcription (P = 2.054e-21) | ||
| Other metabolic processes (P = 5.610e-14) | ||
| Other cellular processes (P = 1.218e-07) | ||
| ld-BHTC-roots-up | 1 | – |
| ld-BHTC-roots-down | 51 | Electron transport or energy pathways (P = 1.656e-18) |
| Other metabolic processes (P = 1.901e-06) | ||
| DNA-dependent transcription (P = 7.534e-05) | ||
| Unknown biological processes (P = 1.549e-04) | ||
| Other cellular processes (P = 9.358e-04) | ||
| Developmental processes (P = 7.095e-03) |
Listed are all significantly enriched GO terms regarding the biological function based on the Botany Array Resource classification super viewer (http://bar.utoronto.ca/welcome.htm).
A total of 499 genes exhibited significantly altered transcript levels in shoots after high-dose BHTC treatment, with 445 of these DEGs up-regulated (hd-BHTC-shoots-up) and 54 down-regulated (hd-BHTC-shoots-down). In roots, the number of DEGs was substantially lower (35 DEGs), with 25 up-regulated genes (hd-BHTC-roots-up) and 10 down-regulated genes (hd-BHTC-roots-down). The high-dose BHTC treatment in shoots resulted in a typical defense-associated transcript profile, including transcript up-regulation of standard defense marker genes, such as PR1, PR5, CaBP22, and LURP1, as well as numerous genes encoding WRKY transcription factors and disease resistance protein family members (Supplemental Table S1). Highly significantly enriched Gene Ontology (GO) terms in the hd-BHTC-shoots-up set calculated by the Botany Array Resource classification super viewer (http://bar.utoronto.ca/welcome.htm; Toufighi et al., 2005) suggested collective roles of these genes in responses to stress and abiotic/biotic stimuli as well as signal transduction (Table I). Consistent with a role in defense, 1,000-bp upstream sequences of the hd-BHTC-shoots-up gene set are highly enriched for known defense-associated promoter motifs. According to The Arabidopsis Information Resource motif analysis tool (https://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp), the hexameric motifs TTGACT (P = 4.25e-19) and TTGACC (P = 2.51e-06), which match the WRKY-binding W-box element (TTGACC/T; Eulgem et al., 2000), as well as the TGACGT hexamer (P = 6.23e-08) containing the TGA box core motif (TGACG; Eulgem, 2005), are significantly overrepresented in these promoter regions.
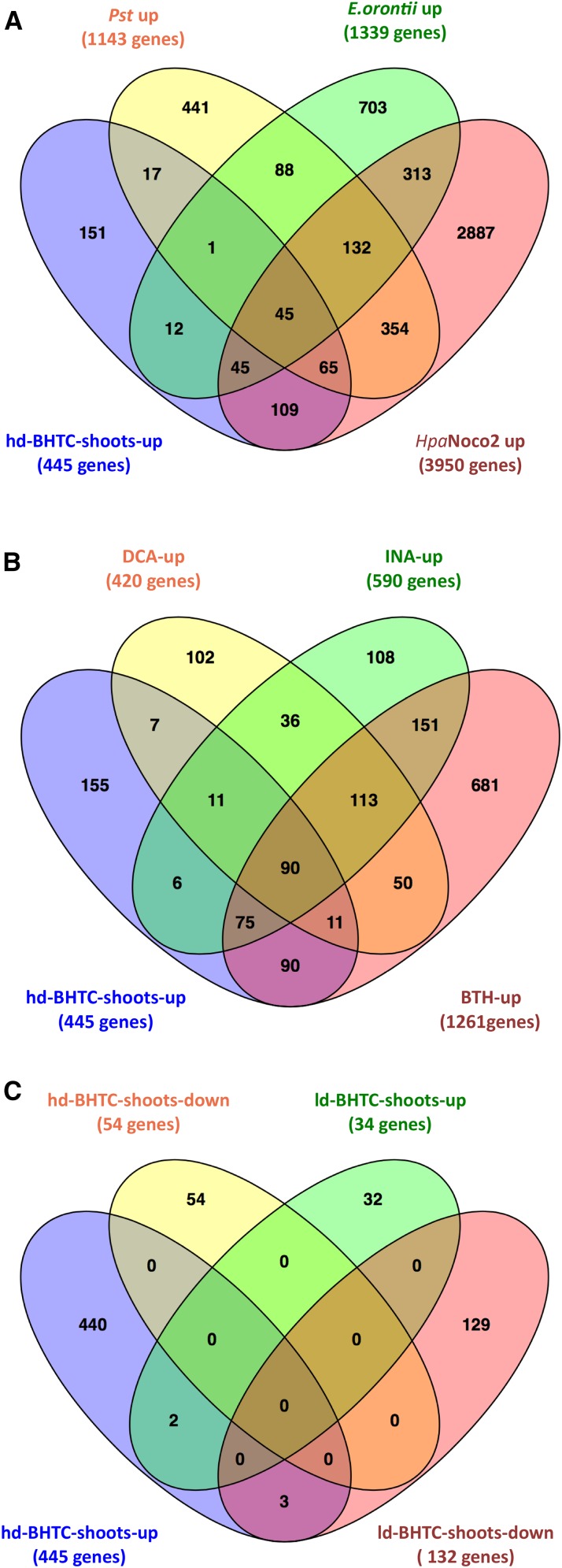
Furthermore, many genes of the hd-BHTC-shoots-up set are responsive to pathogen infection, with 66% (294 of all 445 genes) being up-regulated in Arabidopsis after infections with the oomycete Hpa (Bhattarai et al., 2010), the bacterium P. syringae (Thilmony et al., 2006), and/or the powdery mildew fungus Erysiphae orontii (Pandey et al., 2010; Fig. 6A). In addition, 65% (290) of all 445 hd-BHTC-shoots-up members are inducible by the SA analogs DCA, INA, and/or BTH (Wang et al., 2006; Knoth et al., 2009; Fig. 6B), suggesting that BHTC also mimics some SA functions and acts as a partial agonist of this defense hormone.
Figure 6.
Arabidopsis gene sets responsive to low-dose and high-dose BHTC treatments differ profoundly. Four-way Venn diagram analysis highlights the commonalities and differences between the gene set induced by a high dose of BHTC and various pathogens (A), other SA analogs (B), and low-dose BHTC treatment (C). High-dose BHTC treatment triggers typical defense-associated transcriptome changes that are qualitatively different from the responses triggered by low-dose BHTC treatment. A, Sets of genes up-regulated by Pst, E. orontii, and Hpa are from Thilmony et al. (2006), Pandey et al. (2010), and Bhattarai et al. (2010), respectively. B, Sets of genes up-regulated by DCA and INA or BTH are from Knoth et al. (2009) and Wang et al. (2006), respectively. C, For details about the gene sets shown, see Supplemental Table S1.
The set of hd-BHTC-roots-up genes is substantially smaller (only 25 genes) and features no strongly enriched GO terms. However, like hd-BHTC-shoots-up, this set also contains several canonical defense genes, including CaBP22 and WRKY genes, and promoters of this set are enriched for the two W-box derivatives TTGACT (P = 1.99e-02) and TTGACC (P = 1.13e-01) as well as the TGA-box core-containing hexamer TGACGT (P = 1.00e-02). Thus, a collective role of hd-BHTC-roots-up members in defense is likely. Genes down-regulated by high-dose BHTC treatment in shoots or roots are not strongly enriched for any informative GO terms, and common biological roles of either of these two gene sets are unclear (Table I).
Responses triggered by the low-dose BHTC treatment in shoots and roots were in stark contrast to those triggered by the high BHTC dose. A set of 166 genes was found to be differentially expressed after low-dose BHTC treatment in shoots. This set can be subdivided into 34 genes that exhibit transcriptional up-regulation after 0.1 μm BHTC (ld-BHTC-shoots-up) and 132 genes that are transcriptionally down-regulated by this treatment (ld-BHTC-shoots-down; Supplemental Table S1). The most strongly enriched GO term of the ld-BHTC-shoots-up set is developmental processes (P = 4.062e-05). While all other BHTC-responsive gene sets we identified exclusively feature nuclear genes, genes down-regulated by the low-dose BHTC treatment in shoots and roots consist of nuclear, chloroplast-resident, and mitochondrial genes (Supplemental Table S1). The set of 132 ld-BHTC-shoots-down genes can be nearly evenly subdivided into 33 nuclear, 53 chloroplast-resident, and 44 mitochondrial genes (Supplemental Table S1). All three of these subsets are strongly enriched for genes encoding proteins involved in electron transport or energy pathways, DNA-dependent transcription, other metabolic processes, or other cellular processes (Table I). Particularly strongly represented are genes involved in photosynthetic or respiratory energy production, such as components of the photosynthetic and respiratory electron transport chains, ATPases, Rubisco, or components of the photosynthetic reaction centers (Supplemental Table S1).
As for high-dose BHTC treatment, low-dose BHTC treatment resulted in roots with a smaller set of transcriptional changes, with only one gene significantly up-regulated (AT3G15450) and 51 genes significantly down-regulated (ld-BHTC-roots-down; Table I; Supplemental Table S1). The response to low-dose BHTC treatment qualitatively resembles very much the response triggered by a low dose of this compound in shoots. The set of ld-BHTC-roots-down genes also features nuclear, chloroplast-resident, and mitochondrial genes. Furthermore, as in the case of ld-BHTC-shoots-down genes, genes involved in photosynthetic and respiratory energy production are strongly represented among ld-BHTC-roots-down genes, and significantly enriched GO terms of this set are electron transport or energy pathways, DNA-dependent transcription, other metabolic processes, and other cellular processes. Of the 51 ld-BHTC-roots-down genes, 20 (39%) are also present in the ld-BHTC-shoots-down set.
Taken together, two clearly recognizable trends of BHTC-induced transcriptional changes in both shoots and roots are (1) the up-regulation of typical defense genes by the high-dose treatment with this compound and (2) a coordinated intercompartmental response triggered by low-dose BHTC treatment manifested in the suppression of photosynthesis- and respiration-related genes in the nucleus, chloroplasts, and mitochondria. While it is unclear how the collective differential expression of low-dose BHTC-responsive genes may contribute to hormesis mediated by low doses of this compound, it is striking that transcriptional responses triggered by a low dose of BHTC are qualitatively entirely distinct from the responses we observed after treatment with a high BHTC dose (Fig. 6C).
BHTC-Mediated Root Hormesis Partially Depends on the Defense Regulator WRKY70 as Well as the Auxin-Related Signaling Components AXR1 and SLR
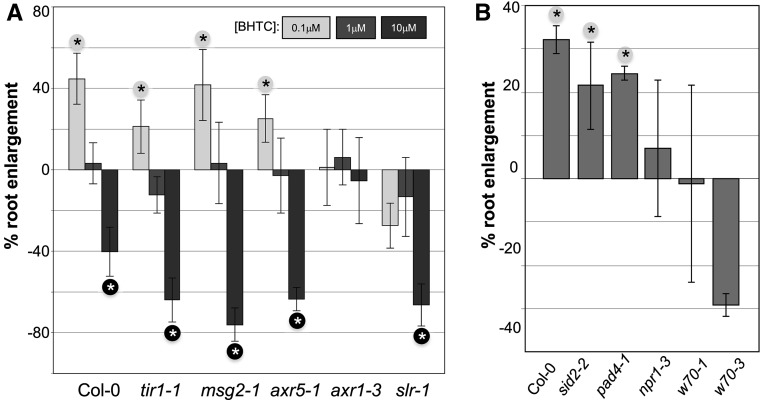
Auxin is known to trigger hormetic growth effects in roots, and the Arabidopsis axr1-3 auxin-response mutant has been reported previously to exhibit a reduction in enhanced root growth induced by low doses of the auxin indole-3-acetic acid (IAA; Evans et al., 1994). Therefore, we tested several Arabidopsis auxin-related mutants for BHTC-triggered hormetic effects. Continuous exposure to 0.1 μm BHTC triggered enhanced elongation of roots in plate-grown Col-0, while an intermediate BHTC dose of 1 μm had no effect and a high dose of 10 μm triggered a severe suppression of root elongation (Fig. 7A). In the tir1-1, msg2-1, and axr5-1 auxin-response mutants, this profile was largely unchanged. However, the axr1-3 and slr-1 mutants exhibited a significantly altered BHTC-response profile. In both cases, no positive growth response to 0.1 μm BHTC could be detected. In addition, axr1-3 plants exhibited no clear reduction of root growth after high-dose BHTC treatment. We further tested several Arabidopsis defense mutants for BHTC-induced root hormesis. BHTC-induced growth enhancement of roots was unaffected in the tested mutants when this compound was applied at a concentration of 0.1 μm (shown for wrky70 mutants in Supplemental Fig. S4). However, the wrky70-1 and wrky70-3 mutants did not respond with enhanced root elongation when exposed to 0.05 μm (Fig. 7B). While a similar trend was also observed with npr1-3 plants, this effect seems to be less pronounced in this mutant. Strikingly, both wrky70 mutants and npr1-3 plants are also compromised in BHTC-mediated immunity against Hpa (Fig. 3).
Figure 7.
The Arabidopsis axr1-3, slr-1, and wrky70 mutants are compromised in hormetic root elongation by low BHTC doses. Relative root enlargement is shown for Arabidopsis seedlings of the indicated genotypes grown for 14 d on 0.1, 1, or 10 µm BHTC (A) or 0.05 µm BHTC (B). Relative root enlargement was determined as in the experiments shown in Figure 5. w70, wrky70. Significantly enlarged or shortened roots in BHTC-treated plants compared with mock-treated plants were determined by Student's t test (P < 0.05) and are marked by black asterisks (on gray background) or white asterisks (on black background), respectively. Error bars represent the combined se of the respective BHTC-treated samples and their mock controls. The numbers of tested plants (n) for the respective treatments are as follows: n (0.1 µm BHTC/Col-0) = 45, n (1 µm BHTC/Col-0) = 50, n (10 µm BHTC/Col-0) = 47, n (0.1 µm BHTC/tir1) = 16, n (1 µm BHTC/tir1) = 17, n (10 µm BHTC/tir1) = 17, n (0.1 µm BHTC/msg2) = 16, n (1 µm BHTC/msg2) = 19, n (10 µm BHTC/msg2) = 12, n (0.1 µm BHTC/axr5) = 16, n (1 µm BHTC/axr5) = 18, n (10 µm BHTC/axr5) = 18, n (0.1 µm BHTC/axr1) = 15, n (1 µm BHTC/axr1) = 16, n (10 µm BHTC/axr1) = 13, n (0.1 µm BHTC/slr) = 18, n (1 µm BHTC/slr) = 16, n (10 µm BHTC/slr) = 17, n (0.05 µm BHTC/Col-0) = 16, n (0.05 µm BHTC/sid2) = 23, n (0.05 µm BHTC/pad4) = 18, n (0.05 µm BHTC/npr1) = 11, n (0.05 µm BHTC/wrky70-1) = 24, and n (0.05 µm BHTC/wrky70-3) = 16.
Although axr1-3 and slr-1 plants were compromised in the BHTC-induced hormetic response, we did not observe any reduction in BHTC-mediated resistance to Hpa in these auxin-response mutants (Supplemental Fig. S5A). However, we unexpectedly found basal defense to this pathogen to be reduced in axr1-3 and slr-1 plants (Supplemental Fig. S5B).
DISCUSSION
Besides the benzoic acid derivative DCA, our chemical screen for inducers of CaBP22−333::GUS in Arabidopsis (Knoth et al., 2009) led to the identification of the PTC derivative BHTC as a new synthetic elicitor. To our knowledge, compounds of this class have not been described as plant defense inducers. While plant-based studies on PTCs seem not to be available, studies in other biological systems have shown some of these compounds to have anticancer, antioxidant, or antimicrobial activities (Włodek et al., 1996; Ferrández et al., 1999; Alhamadsheh et al., 2006; Sriharsha et al., 2007). None of these studies, however, examined the effect of PTCs on plant pathogens. The diversity of biological activities of PTCs suggests that these compounds are highly suitable for interactions with a wide range of different cellular targets. Although some PTCs were shown to have antimicrobial activities, BHTC clearly provided disease protection in the tested interactions with Pst by inducing plant immune responses and not by having direct biocidal effects against this pathogen. Hpa is a strict biotroph and cannot be grown in vitro. Thus, it was not possible to test the direct effects of BHTC against this pathogen. However, suppression of Hpa growth in Arabidopsis required the plant immune system to be intact. In addition, application of BHTC induced a typical defense-associated transcriptional profile. Hence, BHTC can protect plants against microbial diseases by stimulating natural plant immunity.
Both major moieties of BHTC are necessary for strong elicitor activity, as neither the 4-carboxy-4-thiazolidinyl portion nor the 5-bromo-2-hydroxy-phenyl portion robustly induced immunity in our assays. While changes of the substituents of the phenyl group resulted in reduced elicitor activity, the PTC derivatives HTC, BTC, and BMTC, which carry at the phenyl group at least one substituent distinct from the thiazolidine group, still significantly suppressed the formation of Hpa Noco2 spores in Arabidopsis. Thus, phenyl-substituted PTCs can be considered a novel class of synthetic elicitors. Of those PTCs tested in our study, BHTC is the most potent and robust plant defense inducer.
All synthetic elicitors identified by our previous chemical screen should induce a common set of defense reactions, which include transcriptional activation of the LURP gene cluster, including CaBP22, and other responses known to be dependent on SA (Knoth et al., 2009). Nonetheless, we found DCA and BHTC to employ different modes of action, as their defense-inducing activities differ in the Arabidopsis npr1-3 mutant. While DCA-mediated immunity is only weakly NPR1 dependent, no significant levels of disease resistance could be observed in npr1-3 plants after BHTC application. In contrast to other NPR1-dependent synthetic elicitors/plant activators (e.g. INA or BTH), which mediate long-lasting defense induction after a single application, BHTC treatment resulted only in transient immunity under these conditions. Thus, the mode of action of BHTC seems to differ from those of INA or BTH as well. A comparison of the transcriptional profiles induced by DCA, INA, BTH, or BHTC suggested that all four compounds act as SA analogs and induce related, yet partly distinct, subsets of transcriptional changes. Thus, with DCA, BHTC, INA, or BTH, a set of synthetic elicitors is available that can be used to study distinct aspects of immune responses and regulatory processes associated with the defense hormone SA.
A major strategy of disease control in agriculture and horticulture has been the use of pesticides. Chemical pesticides typically rely on direct antibiotic or biocidal activity, which often leads to undesirable toxic environmental side effects (Kessmann et al., 1994; Gilliom et al., 2007). Synthetic elicitors, however, can protect plants by inducing their natural immune responses, do not rely on toxic effects, and therefore are attractive alternatives to conventional pesticides (Bektas and Eulgem, 2014). A possible disadvantage of the use of synthetic elicitors for crop protection is that permanent defense activation often results in fitness costs, due to the phytotoxicity of some defensive plant products and resource allocation away from growth or reproduction. For example, as a result of its long-term activity, INA was insufficiently tolerated by some crop plants to warrant practical use as a plant protection compound (Ryals et al., 1996). However, we found DCA and BHTC to be promising in this respect, as their defense-inducing activity is only transient and weakens within several days after application (Fig. 1; Knoth et al., 2009).
Both DCA and BHTC have similar EC50 values regarding their ability to protect Arabidopsis against Hpa (6.5 and 5.5 μm, respectively). This suggests that both compounds are equally potent with respect to their uptake, stability in planta, and/or affinity to their cellular targets (Katzung, 2007). However, we observed maximal inhibition of Hpa growth in Arabidopsis to be 100% with DCA, while BHTC can only reduce the growth of this pathogen by 73%. Thus, defense reactions induced by DCA appear to be more efficient than those induced by BHTC.
Despite its clear ability to protect Arabidopsis against diseases, the efficiency of BHTC in the tested crop systems seems weak. While we detected a significant reduction of pathogen growth or disease symptoms in tomato or cowpea, respectively, these effects were quantitatively mild, and higher BHTC doses compared with Arabidopsis were necessary. Possibly the uptake, compound stability, and/or target affinity of BHTC may be weaker in these crop species. Testing of additional phenyl-substituted PTC derivatives in tomato or cowpea may lead to the identification of synthetic elicitors better suited for crop protection than BHTC. Nonetheless, BHTC seems to induce defense reactions in multiple plant species that are effective against phylogenetically distinct types of pathogens. The oomycete Hpa and the bacterium Pst typically infect and reproduce in shoot tissues, whereas Fot3, a soil-borne fungus, invades plants through the roots, entering the shoot through vascular tissue, causing disease symptoms (Pietro et al., 2003; Slusarenko and Schlaich, 2003; Zeng et al., 2011). Accordingly, synthetic elicitors that promote broad-spectrum disease resistance in crop plants can have a potential application that is far more efficient than the use of pesticides that target one type of pathogen.
Unexpectedly, we found continuous BHTC exposure to trigger hormetic effects in Arabidopsis. While high doses of BHTC activated defense gene expression and strongly reduced root length, low doses of this compound stimulated root elongation. We also found several other synthetic elicitors as well as SA to trigger similar hormetic effects (data not shown). However, of those we tested, BHTC is the most efficient compound in this respect. Hormesis (Greek for excite) is characterized by low-dose stimulation and high-dose inhibition of biological performance parameters, such as growth, metabolic rate, or stress tolerance, often resulting in inverse U-shaped dose-response curves instead of the sigmoid curves predicted by standard pharmacological threshold models (Calabrese and Baldwin, 2002, 2003). Hormetic phenomena have been described for a wide variety of physical or chemical stimuli in various types of organisms, including humans and plants. For example, radioactive radiation, which is a powerful mutagen, metabolic inhibitors, toxic heavy metals, and carcinogenic chemicals, such as dioxins, are known to trigger hormetic effects (Calabrese and Baldwin, 2001; Kaiser, 2003; Calabrese and Blain, 2005).
Hormesis seems to be as common among plants as it is among animals (Calabrese and Blain, 2009). In particular, herbicides, natural phytotoxins, and radioactivity were found to be potent stimuli of plant hormesis. In the vast majority of cases, growth and metabolic rate were found to be end points stimulated by low doses of hormetic agents in plants. Despite the potential significance of hormetic performance enhancement for commercial crop production, the genetic and biochemical basis of hormesis in plants is completely unclear. Surprisingly, no systematic studies on hormesis seem to have been performed using the versatile molecular genetic plant model system Arabidopsis (Calabrese and Blain, 2009). Thus, our results on BHTC-induced root hormesis in Arabidopsis can serve as a starting point for more extended studies on the mechanistic basis underlying this and related phenomena in plants.
Low- and high-dose BHTC treatments elicited profoundly distinct transcriptional profiles. In both shoots and roots, only high levels of BHTC induced typical defense-related transcriptional changes, while low BHTC levels triggered a coordinated intercompartmental transcriptional response manifested by the suppression of photosynthesis- and respiration-related genes in the nucleus, chloroplasts, and mitochondria. In shoots, low-dose BHTC treatment also up-regulated transcript levels of a set of 34 genes associated with developmental processes. Inspection of publicly available Arabidopsis microarray data in the Botany Array Resource (http://bar.utoronto.ca/welcome.htm) showed that nearly all representatives of this ld-BHTC-shoots-up set are specifically up-regulated in various root cell types and only weakly expressed or not expressed in other tissues. In our own mRNA-seq data set, this general trend is also very obvious (Supplemental Table S1). Compared with mock-treated seedlings, all ld-BHTC-shoots-up members exhibit very high transcript levels in mock-treated roots. Thus, a plausible assumption is that these genes play important roles in root development or growth. Possibly, low-dose BHTC treatment triggers a transition to root development-specific gene expression patterns in the entire seedling. As transcript levels of ld-BHTC-shoots-up members are already extremely high in untreated roots but low in untreated shoots, the triggered change may only be clearly detectable in the latter tissues. Consistent with this assumption, we also observed a trend of ld-BHTC-shoots-up members to be weakly up-regulated in low-dose BHTC-treated roots (average fold change of low-dose BHTC-treated roots versus mock-treated roots = 1.25). While this observation was not statistically significant in our data set, additional mRNA-seq replicates, or use of a more sensitive method, may confirm this trend. We observed the opposite trend for these genes in a comparison between high-dose BHTC- and mock-treated roots (average fold change of high-dose BHTC-treated roots versus mock-treated roots = 0.79), and in a direct comparison, 26% of all ld-BHTC-shoots-up members exhibited significantly elevated transcript levels in low-dose BHTC-treated roots versus high-dose BHTC-treated roots (Supplemental Table S1). Thus, differential expression of root-specific genes may contribute to the dramatic growth differences we observed between low-dose and high-dose BHTC-treated roots.
It is unclear, however, how the collective down-regulation of ld-BHTC-roots-down genes may contribute to enhanced root growth. In any case, the highly distinct nature of transcriptional responses triggered by low and high doses of BHTC is striking. Both responses do not differ much in a quantitative manner (e.g. in the amplitude of expression responses of a common set of genes) but are profoundly distinct regarding the identity and predicted roles of the gene sets they affect. This observation may suggest that different BHTC response processes are triggered by dose-sensitive recognition mechanisms. Recently, a dose-dependent perception mechanism for SA that involves NPR1 as well as the NPR1-related NPR3 and NPR4 proteins has been proposed in Arabidopsis (Fu et al., 2012; Fu and Dong, 2013). Future studies will have to address if related mechanisms are responsible for the dose-dependent perception of BHTC.
Our tests in Arabidopsis defense and auxin-response mutants provided further insight into processes mediating BHTC-triggered root hormesis. The WRKY70 transcription factor seems to be involved in both BHTC-mediated immunity and hormetic root elongation, while components of the auxin response pathway appear to be required for BHTC-mediated root hormesis but not defense triggered by this compound. The phytohormone auxin is involved in a wide variety of developmental processes. One well-known function of auxin is the suppression of root elongation when applied at relatively high doses. Interestingly, at low doses, auxin can enhance root elongation in Arabidopsis (Evans et al., 1994). Some perception mechanisms of this hormone are well understood and seem to generally involve ARF transcription factors that can be repressed by auxin/IAA proteins. Auxin-response processes are initiated by the auxin-induced ubiquitylation of auxin/IAA proteins by SCFAFB E3 ubiquitin ligase complexes. Several AFB (auxin signaling F box) proteins, including TIR1, have been identified that mediate specific interactions of auxin-responsive SCF complexes with their respective auxin/IAA targets (Dharmasiri et al., 2005a, 2005b; Kepinski and Leyser, 2005). The accumulation of auxin beyond certain threshold levels can trigger SCFAFB-mediated ubiquitylation of defined auxin/IAA members followed by the targeted proteasome-dependent degradation of these transcriptional repressors. This results in the derepression of certain ARFs that induce the transcription of auxin-response genes upon binding to auxin-responsive promoter elements.
While some auxin-signaling mutants we tested did not show any clear reduction of BHTC-mediated hormesis, the axr1-3 and slr-1 mutants were clearly compromised in this response. AXR1 encodes an E1 enzyme subunit that plays a central role in the perception of auxin by transferring the ubiquitin-related peptide RUB to SCFAFB complexes and, thereby, activating them (Leyser et al., 1993; Quint and Gray, 2006). Mutants of AXR1 are known to comprehensively block multiple aspects of auxin responses (Gray and Estelle, 2000). Described phenotypes of axr1 mutants include reduced auxin sensitivity in roots as well as several abnormalities or defects in leaf, inflorescence, and flower morphology (Estelle and Somerville, 1987). Most importantly, the axr1-3 mutant has been shown to exhibit a reduction in auxin-mediated root hormesis (Evans et al., 1994). Thus, an AXR1-dependent mechanism may be common to auxin and BHTC-mediated root hormesis.
Besides axr1-3, a second known Arabidopsis auxin-response mutant, slr-1, was compromised in hormetic root enlargement by BHTC. This mutant bears a dominant-negative mutation leading to a version of the auxin/IAA member IAA14 with an increased half-life (Fukaki et al., 2002). Several dominant-negative auxin/IAA mutants are known to affect a subset of auxin responses (Liscum and Reed, 2002). The slr mutation is known to completely block lateral root formation as well as to inhibit root hair formation and the gravitropic responses of roots and hypocotyls (Fukaki et al., 2002). However, dominant-negative auxin/IAA mutations are known to have pleiotropic effects that do not always accurately reflect the authentic roles of their respective genes (Liscum and Reed, 2002). While our results link auxin-related signaling processes to hormetic responses triggered by a synthetic plant defense elicitor, mechanistic details of this link are still enigmatic and will have to be resolved in future studies. Results from this study can serve as a basis for more detailed analyses of the connections between defense signaling and root development as well as fundamental processes generally underlying hormetic phenomena in plants.
MATERIALS AND METHODS
Growth Conditions, Plant Material, Pathogen Infections, and Tissue Staining
Arabidopsis (Arabidopsis thaliana) plants were grown on soil or medium under fluorescent lights (16 h of light/8 h of dark, 23°C, and 100 µE m–2 s–1) unless noted otherwise. The Arabidopsis mutants wrky70-1 and wrky70-3 (Knoth et al., 2007), pad4-1 (Glazebrook et al., 1997), wrky72-2 (Bhattarai et al., 2010), sid2-2 (Dewdney et al., 2000), npr1-3 (Cao et al., 1994, 1997), tir1-1 (Gray et al., 2003), msg2-1 (Tatematsu et al., 2004), axr5-1 (Yang et al., 2004), axr1-3 (Lincoln et al., 1990), and slr-1 (Fukaki et al., 2002) have been described. Hyaloperonospora arabidopsidis was grown and propagated as described previously (McDowell et al., 2000). Two- or 3-week-old Arabidopsis plants were spray infected with Hpa spore suspensions at 2 × 104 spores mL–1 for Hpa Noco2 with Preval sprayers (http://www.prevalspraygun.com). Plants were scored for Hpa growth 7 dpi by counting spores per seedling using a hemicytometer to determine the spore density of a suspension of 10 infected seedlings per 1 mL of water. Student’s t test was used to determine if the effects of the mutations or chemical treatments on sporulation were statistically significant.
Pathogen Infection Experiments with Tomato and Cowpea
Tomato (Solanum lycopersicum ‘Moneymaker’; Everwilde Farms) seeds were sown on autoclaved vermiculite. Plants were fertilized with Miracle-Gro Tomato Plant Food (18-18-21; Scott’s Miracle-Gro Products) biweekly and maintained at 25°C under 200 µmol m−2 s−1 light intensity for a 12-h-light photoperiod for 4 weeks. Each pot received 30 mL of 200 µm BHTC poured over the vermiculite as a root drench 24 h prior to pathogen inoculation and 2, 24, and 48 h post inoculation. Pseudomonas syringae pv tomato strain DC3000 was cultured on King’s B medium with 50 mg mL−1 rifampicin (Sigma-Aldrich) at 28°C. Aerial portions of tomato plants were submerged in a 10 mm MgCl2 solution containing Pst (OD600 = 0.005) and 0.02% (v/v) Silwet L-77 (Lehle Seeds) for 30 s. A single leaf was removed 1 h post inoculation at day 0, and all remaining leaves were used for the 3-dpi time point. Leaves were weighed, ground in 10 mm MgCl2, diluted, and plated. Colonies were counted 40 to 48 h after plating. Cowpea (Vigna unguiculata ‘California Blackeye 5’) seedlings grown in vermiculite were inoculated at 8 d after germination with Fusarium oxysporum f. sp. tracheiphilum race 3 using a root clip and dip inoculation method. Roots of cv California Blackeye 5 were rinsed free of vermiculite in water, cut to a length of 5 cm, submerged in a suspension of 104 spores mL−1 Fot3 for 30 s, and then replanted individually into pots containing UC Mix 3 soil. Pots were randomized on benches, and plants were fertilized with Miracle-Gro (14-14-14; Scott’s Miracle-Gro Products) biweekly and watered every other day. Aerial parts of plants were sprayed with either BHTC or a mock treatment containing the corresponding amount of DMSO solvent. Relatively mature plants could tolerate higher concentrations of DMSO better than younger plants, so the chemical concentrations increased over time and the amount sprayed increased for thorough coverage as plant size increased: 24 h prior to inoculation = 100 µm BHTC, 2 mL per plant; week 1 post inoculation = 100 µm BHTC, 2 mL per plant; week 2 post inoculation = 200 µm BHTC, 3 mL per plant; week 3 post inoculation = 500 µm BHTC, 4 mL per plant; week 4 post inoculation = 750 µm BHTC, 4 mL per plant; and week 5 post inoculation = 1 mm BHTC, 4 mL per plant. Plants were evaluated 5 weeks post inoculation for severity of disease symptoms (leaf chlorosis/wilting and vascular stem discoloration) relative to noninoculated plants (10 BHTC-treated, 10 mock-treated, and 10 untreated plants) using a 0 to 5 rating scale as described previously (Pottorff et al., 2012). One hundred DMSO-treated and 100 BHTC-treated plants were scored individually in each experiment, and statistical significance was determined using χ2 tests of independence. Fot3 was grown and inoculum prepared as described previously (Pottorff et al., 2012). Cowpea-Fot3 experiments were conducted in a greenhouse with day temperatures up to 35°C and night temperatures down to 16°C.
Analysis of GUS Activity and Treatment of CaBP22–333::GUS Plants with Synthetic Elicitor
Arabidopsis seedlings were grown on 96-well plates, treated with synthetic elicitors, and then stained histochemically for GUS expression as described previously (Knoth et al., 2009).
Synthetic Elicitors
BHTC, HTC, BTC, PTC, BMTC, and CMP389 were all ordered from Sigma TimTec. 4-Carboxy-4-thiazolidinyl, 2BP, and DCA were ordered from Sigma-Aldrich. BHTC can be easily synthesized following a protocol described previously (Khan et al., 2006; Song et al., 2009). A preparation of BHTC we synthesized using this protocol (s-BHTC) produced an NMR spectrum identical to that obtained with BHTC purchased from Sigma TimTec (p-BHTC; data not shown), and the efficacy of s-BHTC and p-BHTC in reducing Hpa spore development in Arabidopsis was nearly identical (data not shown).
Synthetic Elicitor Treatment before Pathogen Infection
Stock solutions of all synthetic elicitors were prepared in 100% (v/v) DMSO. Stock solutions were diluted in water and 2 mL per pot sprayed on soil-grown plants at the indicated times and concentrations with Preval sprayers. Final DMSO concentrations never exceeded 2% (v/v). To test for chemically induced disease resistance, the plants were sprayed with 2 mL per pot of chemicals at the indicated concentrations and times prior to pathogen challenge. Disease symptoms were analyzed as described above.
Arabidopsis Root Growth Assays
Col-0 seeds were surface sterilized in a 75% (v/v) ethanol and then 0.02% (v/v) Triton X, 10% (v/v) bleach, and water solution, for 10 and 15 min, respectively. Seeds were then rinsed with sterile water and plated on solid medium laced with one-half-strength MS medium, 1.5% (w/v) agar, 3% (w/v) Suc, and defined concentrations of synthetic elicitors or the equivalent concentration of DMSO (control) on square petri plates. Seeds were stratified for 2 d at 4°C and then placed vertically under fluorescent lights. Plates were scanned at 3, 5, 7, 11, and 14 d after stratification, and root lengths were measured using ImageJ (Schneider et al., 2012).
mRNA-seq Analysis with Plate-Grown Arabidopsis Seedlings
Col-0 seeds were surface sterilized in 75% (v/v) ethanol and a 0.02% (v/v) Triton X and 10% (v/v) bleach solution, for 10 and 15 min, respectively. Seeds were then rinsed with sterile water and plated on solid medium laced with one-half-strength MS medium, 1.5% (w/v) agar, 3% (w/v) Suc, and 0.1 or 5 µm BHTC or solvent only (0.1% [v/v] DMSO). Seeds were stratified for 2 d at 4°C and then placed on plates that were vertically positioned under fluorescent lights. After 14 d, seedling tissue was separated into shoot and root parts using a blade. To prevent any tissue contamination, seedlings were cut into three parts, and root-shoot intersection areas were discarded. Total RNA was isolated from shoot and root separately using TRIZOL (Invitrogen). RNA was processed, and libraries were prepared with the NEBNext Ultra RNA library prep kit (New England Biolabs) by following the manufacturer’s instructions. For each treatment type, root and shoot tissues were analyzed separately. We performed two independent biological replicates for each experimental condition and sequenced the respective libraries using the Illumina HiSeq2500 platform. Sequence reads were analyzed using TopHat for the alignment of reads to The Arabidopsis Information Resource 10 Arabidopsis genome annotation. DEGs were identified by comparing read counts from BHTC-treated samples versus their respective mock controls by EdgeR using a Bonferroni-corrected false discovery rate cutoff of 0.05. All mRNA-seq data generated for this study were deposited as sqn format files in the National Center for Biotechnology Information sequence read archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number SRP064491.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number SRP064491.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Analysis of BHTC activity under saturation treatment conditions.
Supplemental Figure S2. Quantitative characteristics of BHTC and DCA.
Supplemental Figure S3. The BHTC derivative HTC is a weak defense inducer.
Supplemental Figure S4. Hormetic root elongation by BHTC in wrky70 mutants.
Supplemental Figure S5. The Arabidopsis axr1-3 and slr-1 mutants are not compromised in BHTC-mediated immunity against Hpa Noco2.
Supplemental Table S1. List of differentially expressed BHTC-responsive genes identified by mRNA-seq in this study
Supplementary Material
Acknowledgments
We thank Gregory Barding and Dr. Cynthia Larive (both University of California at Riverside) for helpful discussions and advice as well as Abraar Khan, Natalie Williams, and Adilene Gomez (all University of California at Riverside) and Harrison Wedel (Vassar College) for excellent technical assistance.
Glossary
- MAMP
microbe-associated molecular pattern
- PTI
pattern-triggered immunity
- ETI
effector-triggered immunity
- SA
salicylic acid
- INA
2,6-dichloroisonicotinic acid
- BTH
acibenzolar-S-methyl benzo(1,2,3)thiadiazole-7-carbothioic acid S-methyl ester
- DCA
3,5-dicholoroanthranilic acid
- Hpa
Hyaloperonospora arabidopsidis
- BHTC
2-(5-bromo-2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid
- EC50
median effective concentration
- Col-0
Columbia-0
- BTC
2-(5-bromo-phenyl)-thiazolidine-4-carboxylic acid
- BMTC
2-(5-bromo-2-methoxy-phenyl)-thiazolidine-4-carboxylic acid
- HTC
2-(2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid
- PTC
2-phenyl-thiazolidine-4-carboxylic acid
- 2BP
5-bromo-2-hydroxy-phenyl
- PTC
2-phenyl-thiazolidine-4-carboxylic acid
- Pst
Pseudomonas syringae pv tomato
- Fot3
Fusarium oxysporum f. sp. tracheiphilum race 3
- MS
Murashige and Skoog
- mRNA-seq
mRNA sequencing
- DEGs
differentially expressed genes
- GO
Gene Ontology
- IAA
indole-3-acetic acid
- dpi
days post infection
- OD600
optical density at 600 nm
- DMSO
dimethyl sulfoxide
Footnotes
This work was supported by the National Science Foundation (grant no. IOS–1313814 to T.E. and ChemGen IGERT program predoctoral fellowships to M.R.-S., M.S., and C.K.) and by the Turkish Republic Ministry of National Education (predoctoral fellowship to Y.B.).
References
- Ahmad S, Van Hulten M, Martin J, Pieterse CM, Van Wees SC, Ton J (2011) Genetic dissection of basal defence responsiveness in accessions of Arabidopsis thaliana. Plant Cell Environ 34: 1191–1206 [DOI] [PubMed] [Google Scholar]
- Alhamadsheh MM, Hudson RA, Viranga Tillekeratne LM (2006) Design, total synthesis, and evaluation of novel open-chain epothilone analogues. Org Lett 8: 685–688 [DOI] [PubMed] [Google Scholar]
- Bektas Y, Eulgem T (2014) Synthetic plant defense elicitors. Front Plant Sci 5: 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63: 229–240 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E. (2010) Hormesis: once marginalized, evidence now supports hormesis as the most fundamental dose response. In Mattson MP, Calabrese E, eds, Hormesis: A Revolution in Biology, Toxicology and Medicine. Springer, New York, NY, pp 15–56 [Google Scholar]
- Calabrese EJ. (2013) Hormetic mechanisms. Crit Rev Toxicol 43: 580–606 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA (2001) The frequency of U-shaped dose responses in the toxicological literature. Toxicol Sci 62: 330–338 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA (2002) Defining hormesis. Hum Exp Toxicol 21: 91–97 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Baldwin LA (2003) Hormesis: the dose-response revolution. Annu Rev Pharmacol Toxicol 43: 175–197 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Blain R (2005) The occurrence of hormetic dose responses in the toxicological literature, the hormesis database: an overview. Toxicol Appl Pharmacol 202: 289–301 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Blain RB (2009) Hormesis and plant biology. Environ Pollut 157: 42–48 [DOI] [PubMed] [Google Scholar]
- Calabrese EJ, Blain RB (2011) The hormesis database: the occurrence of hormetic dose responses in the toxicological literature. Regul Toxicol Pharmacol 61: 73–81 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Casida JE. (2009) Pest toxicology: the primary mechanisms of pesticide action. Chem Res Toxicol 22: 609–619 [DOI] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM (2000) Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J 24: 205–218 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005a) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Lechner E, Yamada M, Hobbie L, Ehrismann JS, Jürgens G, Estelle M (2005b) Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell 9: 109–119 [DOI] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Estelle M, Somerville C (1987) Auxin resistant mutants of Arabidopsis thaliana with an altered morphology. Mol Gen Genet 206: 200–206 [Google Scholar]
- Eulgem T. (2005) Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 10: 71–78 [DOI] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Evans M, Ishikawa I, Estelle M (1994) Responses of Arabidopsis roots to auxin studies with high temporal resolution: comparison of wild type and auxin-response mutants. Planta 194: 215–222 [Google Scholar]
- Ferrández MD, Correa R, Del Rio M, De la Fuente M (1999) Effects in vitro of several antioxidants on the natural killer function of aging mice. Exp Gerontol 34: 675–685 [DOI] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE (2001) Direct interaction between the Arabidopsis disease resistance proteins, EDS1 and PAD4. EMBO J 19: 5400–5411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M (2002) Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gilliom R, Barbash J, Crawford G, Hamilton P, Martin J, Nakagaki N, Wolock D (2007) The Quality of Our Nation’s Waters: Pesticides in the Nation’s Streams and Ground Water, 1992-2001. US Geological Survey, Reston, VA [Google Scholar]
- Glazebrook J. (2001) Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr Opin Plant Biol 4: 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Métraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Zook M, Mert F, Kagan I, Rogers EE, Crute IR, Holub EB, Hammerschmidt R, Ausubel FM (1997) Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics 146: 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlach J, Volrath S, Knauf-Beiter G, Hengy G, Beckhove U, Kogel KH, Oostendorp M, Staub T, Ward E, Kessmann H, et al. (1996) Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 8: 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Estelle I (2000) Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem Sci 25: 133–138 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein I, Gilroy EM, Armstrong MR, Birch PR (2009) The zig-zag-zig in oomycete-plant interactions. Mol Plant Pathol 10: 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Khan KM, Ullah Z, Lodhi MA, Ali M, Choudhary MI, Rahman AU, Haq ZU (2006) Successful computer guided planned synthesis of (4R)-thiazolidine carboxylic acid and its 2-substituted analogues as urease inhibitors. Mol Divers 10: 223–231 [DOI] [PubMed] [Google Scholar]
- Kaiser J. (2003) Hormesis: sipping from a poisoned chalice. Science 302: 376–379 [DOI] [PubMed] [Google Scholar]
- Katzung B. (2007) Basic and Clinical Pharmacology, Ed 10 McGraw-Hill, Boston [Google Scholar]
- Kepinski S, Leyser O (2005) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kessmann H, Staub T, Hofmann C, Maetzke T, Herzog J, Ward E, Uknes S, Ryals J (1994) Induction of systemic acquired disease resistance in plants by chemicals. Annu Rev Phytopathol 32: 439–459 [DOI] [PubMed] [Google Scholar]
- Knoth C, Ringler J, Dangl JL, Eulgem T (2007) Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol Plant Microbe Interact 20: 120–128 [DOI] [PubMed] [Google Scholar]
- Knoth C, Salus MS, Girke T, Eulgem T (2009) The synthetic elicitor 3,5-dichloroanthranilic acid induces NPR1-dependent and NPR1-independent mechanisms of disease resistance in Arabidopsis. Plant Physiol 150: 333–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Leyser HMO, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49: 387–400 [PubMed] [Google Scholar]
- Mattson M, Calabrese E (2010) Hormesis: what it is and why it matters. In Mattson MP, Calabrese E, eds, Hormesis: A Revolution in Biology, Toxicology and Medicine. Springer, New York, NY, pp 1–14 [Google Scholar]
- Mattson MP. (2008) Hormesis defined. Ageing Res Rev 7: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP. (2010) The fundamental role of hormesis in evolution. In Mattson MP, Calabrese E, eds, Hormesis: A Revolution in Biology, Toxicology and Medicine. Springer, New York, NY, pp 57–68 [Google Scholar]
- McDowell JM, Cuzick A, Can C, Beynon J, Dangl JL, Holub EB (2000) Downy mildew (Peronospora parasitica) resistance genes in Arabidopsis vary in functional requirements for NDR1, EDS1, NPR1 and salicylic acid accumulation. Plant J 22: 523–529 [DOI] [PubMed] [Google Scholar]
- Métraux JP, Ahl Goy P, Staub T, Speich J, Steinemann A, Ryals J, Ward E (1991) Induced resistance in cucumber in response to 2,6-dichloroisonicotinic acid and pathogens. In Hennecke H, Verma DPS, eds, Advances in Molecular Genetics of Plant-Microbe Interactions. Kluwer, Dordrecht, The Netherlands, pp 432–439 [Google Scholar]
- Nimchuk Z, Eulgem T, Holt BF III, Dangl JL (2003) Recognition and response in the plant immune system. Annu Rev Genet 37: 579–609 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Roccaro M, Schön M, Logemann E, Somssich IE (2010) Transcriptional reprogramming regulated by WRKY18 and WRKY40 facilitates powdery mildew infection of Arabidopsis. Plant J 64: 912–923 [DOI] [PubMed] [Google Scholar]
- Pietro AD, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MI (2003) Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Mol Plant Pathol 4: 315–325 [DOI] [PubMed] [Google Scholar]
- Pimentel D, Pimentel M (2008) Food, Energy, and Society. CRC Press, Boca Raton, FL [Google Scholar]
- Pottorff M, Wanamaker S, Ma YQ, Ehlers JD, Roberts PA, Close TJ (2012) Genetic and physical mapping of candidate genes for resistance to Fusarium oxysporum f.sp. tracheiphilum race 3 in cowpea [Vigna unguiculata (L.) Walp]. PLoS ONE 7: e41600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Gray WM (2006) Auxin signaling. Curr Opin Plant Biol 9: 448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8: 1809–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Tsuda K, Wang L, Coller J, Watanabe Y, Glazebrook J, Katagiri F (2010) Network modeling reveals prevalent negative regulatory relationships between signaling sectors in Arabidopsis immune signaling. PLoS Pathog 6: e1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber KJ, Desveaux D (2008) Message in a bottle: chemical biology of induced disease resistance in plants. Plant Pathol J 24: 245–268 [Google Scholar]
- Slusarenko AJ, Schlaich NL (2003) Downy mildew of Arabidopsis thaliana caused by Hyaloperonospora parasitica (formerly Peronospora parasitica). Mol Plant Pathol 4: 159–170 [DOI] [PubMed] [Google Scholar]
- Son T, Cutler R, Mattson MP (2010) Transcriptional mediators of cellular hormesis. In Mattson MP, Calabrese E, eds, Hormesis: A Revolution in Biology, Toxicology and Medicine. Springer, New York, NY, pp 69–94 [Google Scholar]
- Song ZC, Ma GY, Lv PC, Li HQ, Xiao ZP, Zhu HL (2009) Synthesis, structure and structure-activity relationship analysis of 3-tert-butoxycarbonyl-2-arylthiazolidine-4-carboxylic acid derivatives as potential antibacterial agents. Eur J Med Chem 44: 3903–3908 [DOI] [PubMed] [Google Scholar]
- Sriharsha SN, Pai KS, Shashikanth S, Chandra N, Prabhu KR (2007) Synthesis, docking and anti-tumor activity of beta-L-1,3-thiazolidine pyrimidine nucleoside analogues. Med Chem 3: 425–432 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X (2006) A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog 2: e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Metraux JP, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Włodek L, Wróbel M, Czubak J (1996) Selective effect of 2-(polyhydroxyalkyl)-thiazolidine-4-carboxylic acids on nonprotein sulfhydryl groups in tumor bearing mice. Gen Pharmacol 27: 1373–1376 [DOI] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY (2011) A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathog 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C. (2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.