Two transcription factors act redundantly to regulate the transcriptional responses to phosphate starvation at genomic level.
Abstract
When confronted with inorganic phosphate (Pi) starvation, plants activate an array of adaptive responses to sustain their growth. These responses, in a large extent, are controlled at the transcriptional level. Arabidopsis (Arabidopsis thaliana) PHOSPHATE RESPONSE1 (PHR1) and its close homolog PHR1-like 1 (PHL1) belong to a 15-member family of MYB-CC transcription factors and are regarded as the key components of the central regulatory system controlling plant transcriptional responses to Pi starvation. The knockout of PHR1 and PHL1, however, causes only a partial loss of the transcription of Pi starvation-induced genes, suggesting the existence of other key components in this regulatory system. In this work, we used the transcription of a Pi starvation-induced acid phosphatase, AtPAP10, to study the molecular mechanism underlying plant transcriptional responses to Pi starvation. We first identified a DNA sequence on the AtPAP10 promoter that is critical for the transcription of AtPAP10. We then demonstrated that PHL2 and PHL3, two other members of the MYB-CC family, specifically bind to this DNA sequence and activate the transcription of AtPAP10. Unlike PHR1 and PHL1, the transcription and protein accumulation of PHL2 and PHL3 are upregulated by Pi starvation. RNA-sequencing analyses indicated that the transcription of most Pi starvation-induced genes is impaired in the phl2 mutant, indicating that PHL2 is also a key component of the central regulatory system. Finally, we showed that PHL2, and perhaps also PHL3, acts redundantly with PHR1 to regulate plant transcriptional response to Pi starvation.
Phosphorus (P) is an essential macronutrient for plant growth, development, and metabolism. Plants uptake P from soil in the form of inorganic phosphate (Pi) (Raghothama, 2000; Nussaume et al., 2011). In most soils, however, Pi is one of the least available nutrients due to its low diffusion rate, its fixation with metals, and its conversion to organophosphates by microorganisms (Bieleski, 1973). To sustain their growth under Pi deficiency, plants activate a suite of biochemical, physiological, and developmental responses (Vance et al., 2003). These responses are controlled by a sophisticated gene regulatory network through both local and systemic signaling (Chiou and Lin, 2011). The studies of the major responses to Pi starvation, such as the increased activity of high-affinity Pi transporters, the induction of acid phosphatases (APases), and accumulation of anthocyanins indicate that transcriptional regulation is a crucial step in controlling these responses (Franco-Zorrilla et al., 2004; Jain et al., 2012). Transcriptomic analyses in several plant species has revealed a dramatic change in gene expression profiles in Pi-starved plants (Hammond et al., 2003; Misson et al., 2005; Wu et al., 2003; Hernández et al., 2007; Morcuende et al., 2007; Calderon-Vazquez et al., 2008; O’Rourke et al., 2013; Secco et al., 2013), reinforcing the importance of transcriptional regulation in plant Pi responses.
PHOSPHATE RESPONSE1 (PHR1) encodes a protein that belongs to a 15-member family of transcription factors that contain an MYB domain and a coiled-coiled (CC) domain (Rubio et al., 2001). The loss-of-function mutation of PHR1 causes the reduction in the expression of phosphate starvation-induced (PSI) genes, a decrease in cellular Pi content and shoot-to-root ratio, and the impairment of anthocyanin accumulation. In contrast, the overexpression of PHR1 results in an increased accumulation of cellular Pi and plant tolerance to Pi deprivation (Nilsson et al., 2007; Matsui et al., 2013). PHR1 is also involved in the remodeling of membrane lipids (Pant et al., 2015a), the change of primary and secondary metabolisms (Pant et al., 2015b), and the adaptation to high light (Nilsson et al., 2012) in Pi-deficient plants. In addition, PHR1 serves as the convergent point for the cross talk between Pi and other essential nutrients, such as sulfate (Rouached et al., 2011), zinc (Khan et al., 2014), and iron (Bournier et al., 2013). Orthologs of PHR1 have been found in other plant species, including Brassica napus (Ren et al., 2012), rice (Oryza sativa; Zhou et al., 2008), and wheat (Triticum aestivum; Wang et al., 2013a), and they function similarly to PHR1 in plant responses to Pi starvation.
PHR1 binds to an imperfect palindromic motif (5′-GNATATNC-3′), termed the P1BS element (PHR1 biding site), that is prevalent in the promoters of PSI genes. OsPHR2 is the functional equivalent of PHR1 in rice and binds to the same P1BS element (Zhou et al., 2008; Ruan et al., 2015). The transcription and protein accumulation of PHR1 and OsPHR2 are not responsive to changes in Pi availability (Rubio et al., 2001; Zhou et al., 2008). Instead, their activities in regulating PSI gene transcription are modulated by the SPX domain-containing proteins, which either control the translocation of PHR1 from cytoplasm to the nucleus (Lv et al., 2014) or compete with the P1BS element for binding to PHR1 (Wang et al., 2014b; Puga et al., 2014). In the MYB-CC family, PHR1-like 1 (PHL1) is a close relative to PHR1 (Bustos et al., 2010). The functional disruption of PHL1 has little effect on the expression of PSI genes; in the phr1phl1 double mutant, however, the expression of the PSI genes is further reduced relative to that in the phr1 single mutant. Using microarray analysis, Bustos et al. (2010) found that PHR1 could directly bind to the promoter of 340 genes and that the expression of most PSI genes was more or less impaired in phr1 and phr1phl1 mutants. Thus, PHR1 and PHL1 are regarded as the key components of a central regulatory system controlling plant transcriptional responses to Pi starvation. Notably, the double mutation in PHR1 and PHL1 only partially affects the transcription of Pi-responsive genes (Bustos et al., 2010), suggesting a functional redundancy between PHR1/PHL1 and other transcription factors.
In this work, we used the transcription of AtPAP10 mRNA as a readout to investigate the molecular mechanism underlying plant transcriptional responses to Pi starvation. AtPAP10 (Arabidopsis purple acid phosphatase 10) is a major Pi starvation-induced secreted APase (Wang et al., 2011, 2014a; Zhang et al., 2014). Both the transcription and protein accumulation of AtPAP10 are upregulated by Pi starvation. Its gene expression patterns, biochemical properties, and function in plant acclimation to Pi starvation have been well characterized (Wang et al., 2011). In addition, the transcription of AtPAP10 mRNA can be easily monitored by using quantitative PCR (qPCR) or by using a AtPAP10:GUS marker line. The AtPAP10 activity on the root surface of Arabidopsis (Arabidopsis thaliana) seedlings can also be easily detected by histochemical staining and can be quantified with a spectrophotometer (Wang et al., 2011). Through the study of the regulatory mechanism of AtPAP10 transcription, we show that the other members of the MYB-CC family, PHL2 and perhaps also PHL3, acts redundantly with PHR1 as the key components of the central regulatory system controlling plant transcriptional responses to Pi starvation.
RESULTS
PHR1 Is Partially Involved in the Regulation of AtPAP10 Transcription
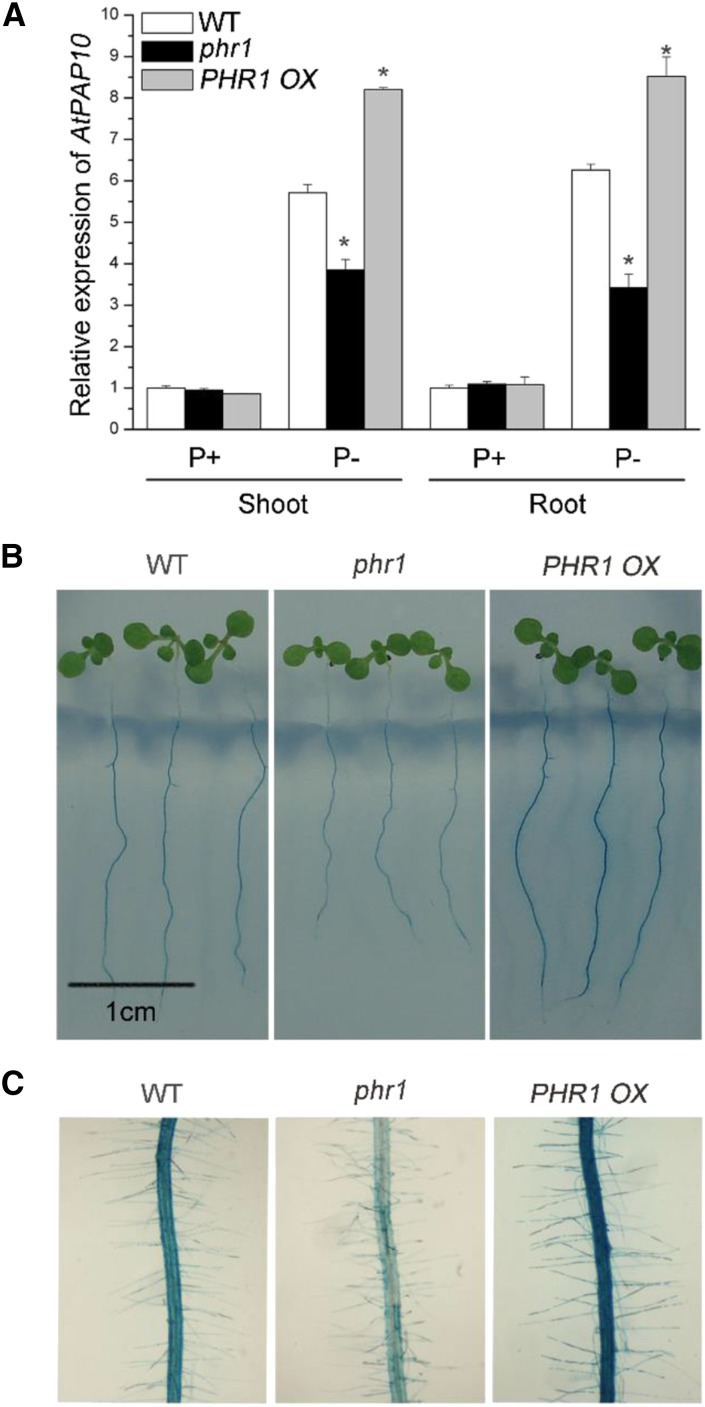
Using microarray analysis, Bustos et al. (2010) previously showed that PHR1 could directly bind to the promoter of AtPAP10 and is a positive regulator of AtPAP10 transcription. To confirm this, we examined the expression of AtPAP10 mRNA in the phr1 mutant (SALK_067629; Nilsson et al., 2007) and in PHR1-overexpressing (PHR1 OX) lines by qPCR. Our results showed that the expression of AtPAP10 was up-regulated in both shoots and roots of Pi-starved wild-type seedlings (Fig. 1A), consistent with what was previously reported by Wang et al. (2011). The level of AtPAP10 mRNA in phr1 grown on Pi-deficient (P-) medium was about 70% of that of the wild type. In contrast, the level of AtPAP10 mRNA in three PHR1 OX lines was significantly higher than that of the wild type (the result from one representative line is shown).
Figure 1.
Expression of AtPAP10 and root-associated APase activity in wild-type, phr1, and PHR1 OX seedlings. A, qPCR analysis of AtPAP10 transcript level in shoots and roots of 8-d-old wild-type, phr1, and PHR1OX seedlings grown under P+ and P− conditions. Values are means ± se of three replicates. Asterisks indicate a significant difference from the wild type under the same growth conditions (Student’s t test, P < 0.05). B, BCIP staining of root-associated APase activities of 8-d-old wild-type, phr1, and PHR1 OX seedlings grown under Pi deficiency. C, A close-up view of the BCIP staining in the roots of the plants shown in B.
The root surface-associated APase activity in Arabidopsis can be detected by overlaying an agar solution containing the substrate of APase, BCIP (5-bromo-4-chloro-3-indolyl phosphate), on the roots of the seedlings (Wang et al., 2011). The cleavage of the substrate by APases produces a blue precipitate. Accordingly, under Pi deficiency, phr1 root surfaces showed a reduced level of BCIP staining, whereas PHR1 OX root surfaces showed an enhanced level of BCIP staining (Fig. 1, B and C). However, the incomplete loss of AtPAP10 transcription and root-associated APase activity in phr1 indicated that other transcription factors also participate in the transcription of AtPAP10.
Identification of a Key DNA Sequence Required for AtPAP10 Transcription
Using the promoter analysis program PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), we found several cis-acting elements within the 1-kb sequence upstream of the AtPAP10 start codon. These cis-elements include those responsive to light, methyl jasmonate, gibberellin, drought, salicylic acid, defense, and stress (Supplemental Fig. S1). Among them, there is one copy of the P1BS element (GAATATTC, −115 to −123) and three copies of P1BS-like elements, namely, P1BS-like I (ACATATTC, −257 to −264), P1BS-like II (AAATATCC, −277 to −284), and P1BS-like III (ACATATTC, −780 to −787) (Fig. 2A). Among the three P1BS-like elements, P1BS-like I and III contain the same sequence.
Figure 2.
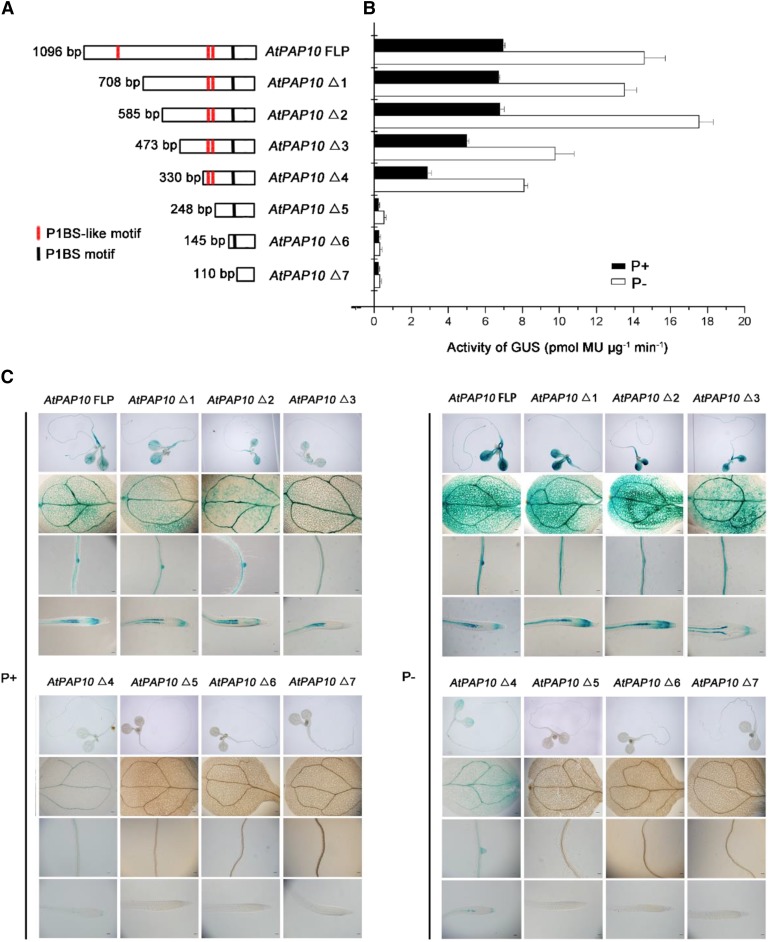
Deletion analysis of the AtPAP10 promoter. A, A diagram showing eight deletion constructs of the AtPAP10 promoter. The length of the promoter (from its 5′ end to the start codon) for each construct is indicated on the left, and the name of the construct is indicated on the right. The location of the P1BS element and P1BS-like elements is indicated by a black line and red lines, respectively. B, GUS activity of a representative line for each AtPAP10:GUS construct. Values are means ± se of three biological replicates. C, Expression patterns of eight AtPAP10:GUS constructs in 8-d-old seedlings grown under P+ and P− conditions. The names of the constructs are indicated at the top. For each construct, the GUS expression patterns in whole seedling, cotyledon, root maturation zone, and root tip are shown (from top to bottom). Bars = 20 μm.
To identify the cis-elements required for AtPAP10 transcription, we carried out a promoter-deletion analysis in transgenic plants. A set of eight AtPAP10:GUS constructs were generated with different lengths of the AtPAP10 promoter sequences fused to a GUS reporter gene. The 5′ starting positions of these promoter fragments relative to the start codon of AtPAP10 were as follows: −1096 (designated as the full-length promoter [FLP]), −708 (Δ1), −585 (Δ2), −473 (Δ3), −330 (Δ4), −248 (Δ5), −145 (Δ6), and −110 (Δ7) (Fig. 2A). These constructs were transformed into wild-type Arabidopsis plants, and at least 20 independent transgenic lines were obtained for each construct. In these transgenic lines, the GUS activity was visually rated as strong, weak, and none based on staining intensity. The results of GUS staining are summarized in Supplemental Figure S2. The GUS expression pattern of a representative line for each construct is shown in Figure 2C. For the plants carrying the constructs from AtPAP10 FLP to AtPAP10 Δ4, the GUS gene was mainly expressed in vascular tissues of cotyledons and roots and in root tips under Pi sufficiency, whereas the GUS gene expression was enhanced and extended to intervine regions in the cotyledons under Pi deficiency. Furthermore, when the deletion went from position −330 to −248, the transgenic plants carrying the construct AtPAP10 Δ5 not only lost the response to low Pi but also lost the basal expression level of the GUS gene, although this construct contained one copy of P1BS (Fig. 2, B and C). These results indicated that the DNA sequence from −330 to −248 (designated as the P sequence) contained some cis-elements that are critical for the expression of the AtPAP10 gene.
Two PHR1-Like Proteins, PHL2 and PHL3, Bind to the P Sequence
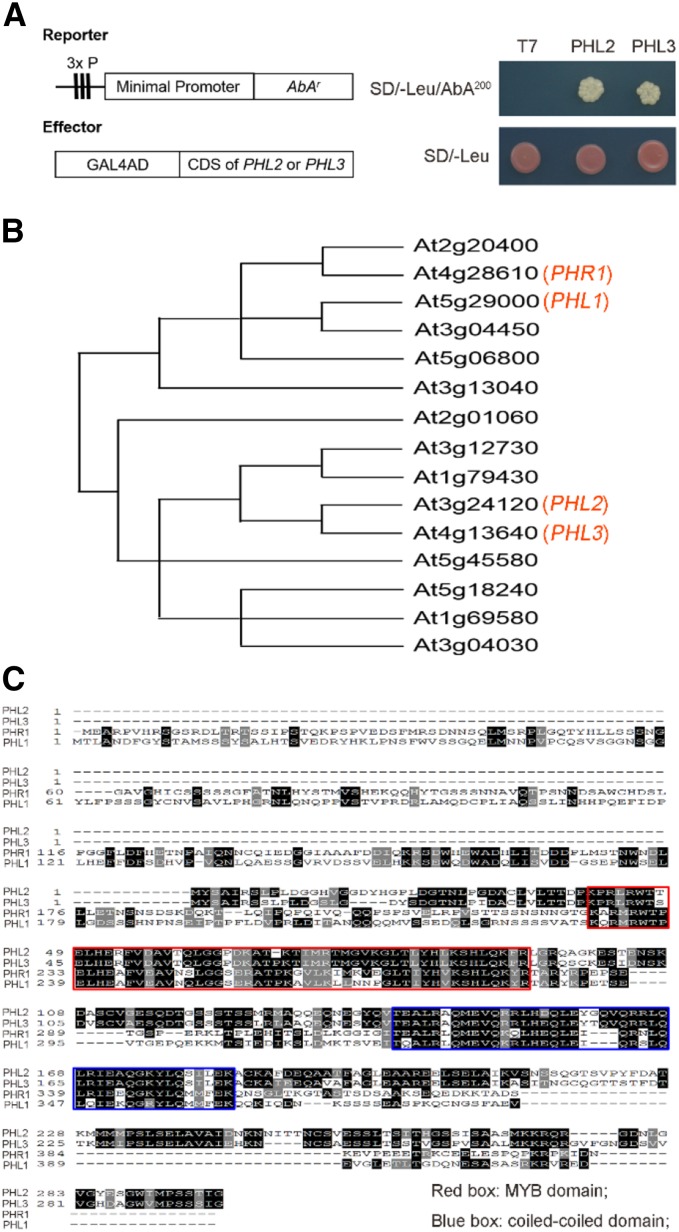
To identify the transcription factor(s) that bind to the P sequence, we performed a yeast one-hybrid (Y1H) screen against a cDNA library made from the Pi-starved Arabidopsis seedlings using the P sequence as a bait. Among the several dozens of positive clones that we obtained, there were only two genes, AT3G24120 and AT4G13640, that encode putative transcription factors (Fig. 3A). These two proteins belong to the MYB-CC family that contains PHR1 and PHL1 and share 92% sequence identity at the protein level (Fig. 3, B and C). Thus, we renamed these two proteins as PHL2 and PHL3, respectively. Compared to PHR1 and PHL1, both PHL2 and PHL3 lack an N-terminal extension of 187 amino acids but have two extra segments of 42 and 17 amino acids at their C terminus (Fig. 3C). PHL2 and PHL3 are highly conserved in higher plants (Supplemental Fig. S3).
Figure 3.
Identification of PHL2 and PHL3 as the P sequence-binding proteins by Y1H assay. A, Yeast cells harboring the 3×P:AbAr reporter gene were transformed with the expression vector carrying a fusion gene of the GAL4 activation domain and the CDS of PHL2 or PHL3. The transformants were grown on control (SD/-Leu) and selection medium (SD/-Leu/200 nm aureobasidin A). B, The relative position of PHL2 and PHL3 in the MYB-CC family. The diagram showing the phylogenetic relationship of MYB-CC proteins is modified from Bustos et al. (2010). C, Alignment of protein sequences of PHL2, PHL3, PHR1, and PHL1. The two conserved regions corresponding to the MYB domain and coiled-coiled domain are indicated in red and blue boxes, respectively.
To confirm that PHL2 and PHL3 can directly bind to the P sequence, we performed electrophoretic mobility shift assays (EMSAs). The full-length recombinant PHL2 and PHL3 proteins were produced in Escherichia coli cells and purified to near homogeneity. Each of the recombinant PHL2 and PHL3 proteins contains an MBP (maltose-binding protein) tag in its N terminus and a 6X His tag in its C terminus. The EMSA results indicated that the recombinant PHL2 proteins could cause a mobility shift of the biotin-labeled P sequence, whereas MBP alone could not (Fig. 4B). The degree of the shifted band was correlated with the quantity of PHL2 proteins added to the reaction mixture. Furthermore, the unlabeled P sequence (cold probe) could specifically compete with labeled P sequence in binding to PHL2, indicating that the binding is sequence-specific. Similar results were obtained for PHL3 proteins (Supplemental Fig. S4).
Figure 4.
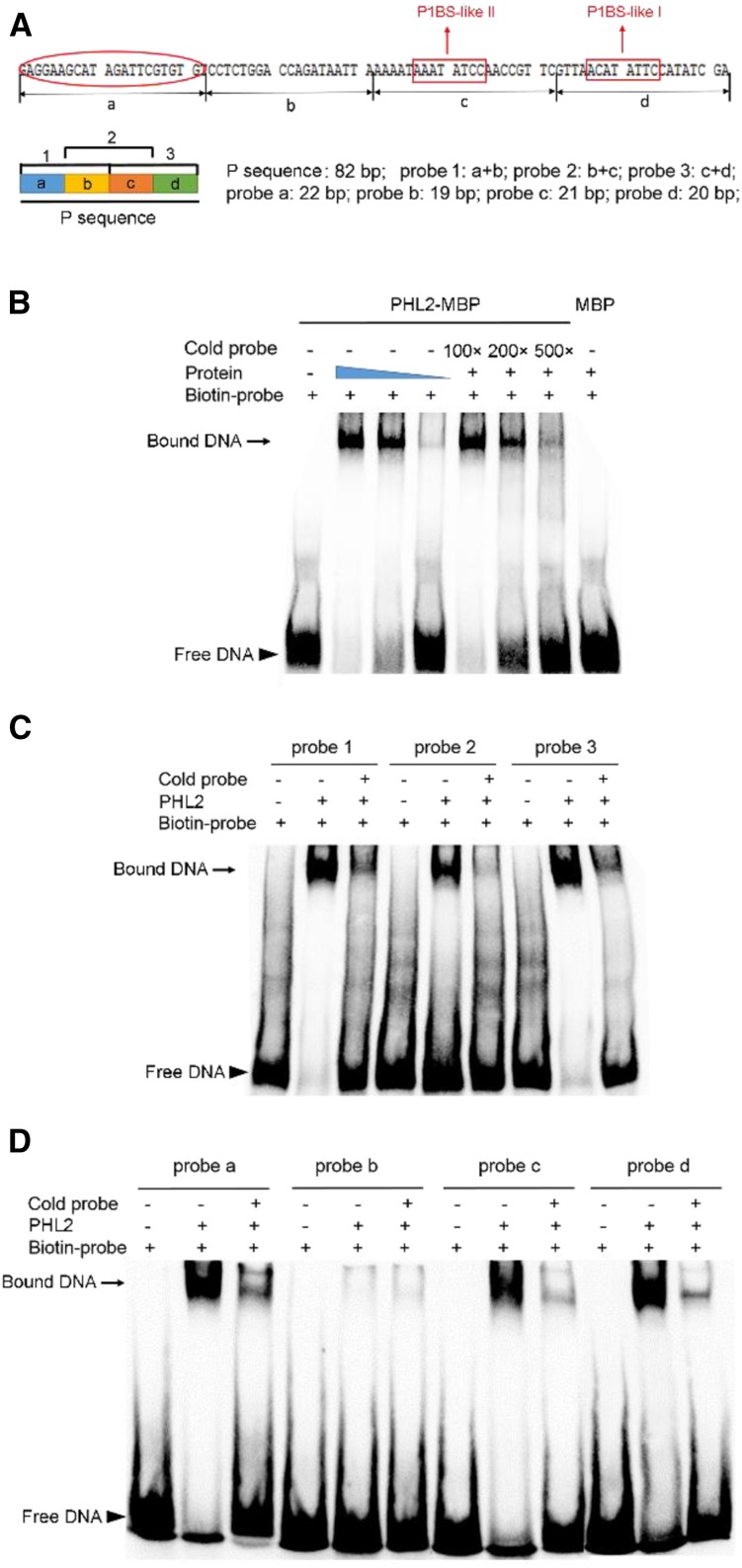
EMSA assays showing the binding of PHL2 to the P sequence. A, Top: Diagram showing the DNA sequence of the P sequence. Two P1BS-like elements are indicated by red rectangular boxes. The fragment a, which contains a novel DNA-binding site, is indicated by a red oval. Bottom: Diagram showing the different fragments used in EMSA assays. The length of each fragment is indicated on the right. B, EMSA assay showing that PHL2-MBP fusion proteins specifically bind to the P sequence. The experiment was performed using 0.1, 0.05, and 0.01 μg of PHL2-MBP proteins. The filled blue triangle indicates the decrease in protein concentration. The biotin-labeled probe contained 10 fmol of the P sequence. The unlabeled probe of the P sequence was used as a competitor at an excess molar ratio of 100:1, 200:1, and 500:1 to labeled probe. C, EMSA assay performed with three overlapping fragments in the P sequence. This experiment was performed using 0.05 μg PHL2-MBP fusion protein. The ratio of unlabeled probe to labeled probe was 500:1. D, EMSA assay performed with four fragments of the P sequence as indicated at the top. Experimental conditions were the same as in C.
To further define the binding sequence for PHL2, we divided the 82-bp P sequence into three overlapping fragments, which were termed fragments (probes) 1, 2, and 3 (Fig. 4A). Each fragment was duplicated in tandem and was used for EMSA assays. As shown in Figure 4C, PHL2 was able to bind to all three probes and had high affinity to probes 1 and 3 and weak affinity to the probe 2. We then divided the P sequence into four equal parts (a, b, c, and d; Fig. 4A) and individually tested the ability of these fragments (each fragment was duplicated in tandem) to bind to PHL2. The results showed that PHL2 bound strongly to probes a and d, bound weakly to probe c, and did not bind to probe b (Fig. 4D). Probes c and d each contained one copy of the P1BS-like element (Fig. 4A). However, no annotated cis-element in the public database was found in probe a. Thus, we identified a novel protein binding site within the P sequence; however, the exact binding sequence remained to be further defined.
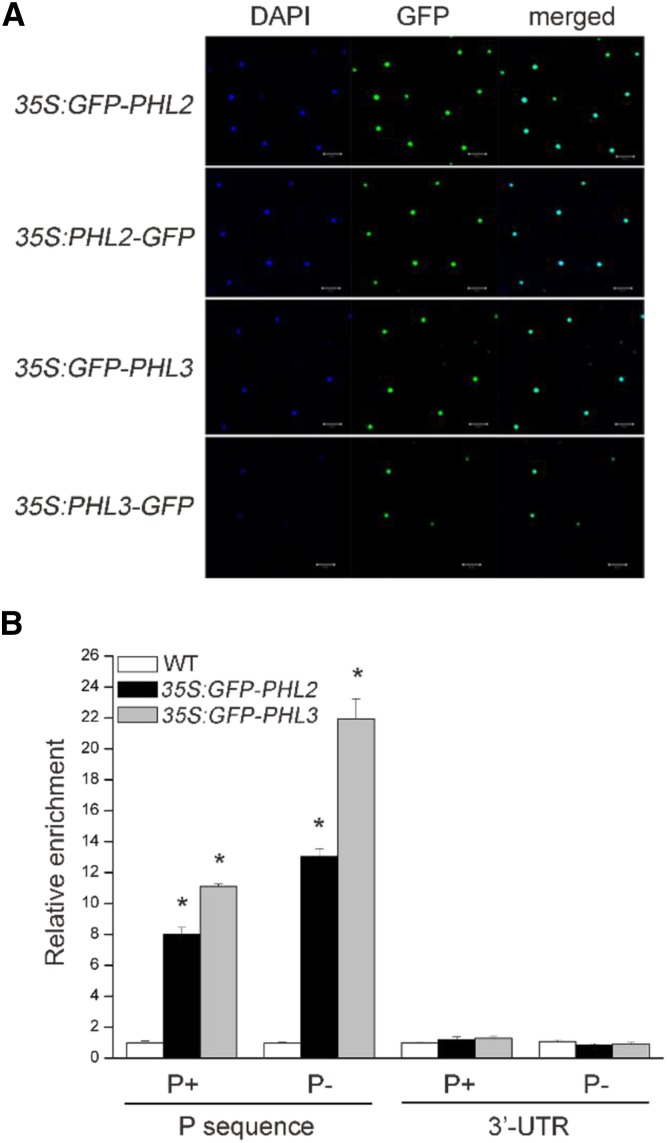
Next, we examined the subcellular localization of PHL2 and PHL3. The full-length coding sequence (CDS) of PHL2 and PHL3 was individually fused to the N terminus or C terminus of green florescence protein (GFP), and the resulting constructs were transiently expressed in the leaves of Nicotiana benthamiana under the control of the Cauliflower mosaic virus (CaMV) 35S promoter. Both PHL2-GFP and PHL3-GFP fusion proteins (fused to either the N terminus or C terminus of GFP) were exclusively colocalized with the site of 4′,6-diamino-phenylindole (DAPI; a nucleus-specific dye) staining, indicating that they are nuclear proteins (Fig. 5A). Finally, we confirmed the binding of PHL2 and PHL3 to the P sequence in vivo using chromatin immunoprecipitation (ChIP) assays. Transgenic Arabidopsis plants expressing PHL2-GFP or PHL3-GFP driven by the 35S CaMV promoter (in the wild-type background) were generated. The chromatins were isolated from the whole seedlings of these transgenic plants and precipitated by anti-GFP antibodies. The DNA fragments that precipitated with PHL2-GFP or PHL3-GFP proteins were analyzed by qPCR using the primers flanking the P sequence or the 3′ untranslated region (UTR) of the AtPAP10 gene. The results showed that the PCR-amplified P sequence, but not the 3′ UTR, was enriched in the immunoprecipitated chromatins from the GFP-PHL2 or GFP-PHL3 transgenic plants compared to the wild type (Fig. 5B). Moreover, the enrichment of the P sequence in the precipitated chromatins was higher for Pi-starved GFP-PHL2 or GFP-PHL3 plants than for plants grown under normal conditions (Fig. 5B). Taken together, our results demonstrated that PHL2 and PHL3 could directly bind to the P sequence in vitro and in vivo and that such binding was enhanced by Pi starvation.
Figure 5.
PHL2 and PHL3 are localized in the nucleus and bind to the P sequence in vivo. A, Subcellular localization of PHL2 and PHL3. The expression constructs as indicated on the left were infiltrated into the leaves N. benthamiana and GFP fluorescence was observed 2 d after infiltration. The nuclei were stained by DAPI. B, ChIP-PCR analysis of the in vivo binding of the GFP-PHL2 or GFP-PHL3 to the P sequence. Chromatins from the wild type and the transgenic plants expressing GFP-PHL2 or GFP-PHL3 were immunoprecipitated with GFP-Trap. The levels of enrichment of the precipitated DNA fragments were quantified by qPCR assay. Values are means ± se of three replicates. Asterisks indicate a significant difference from the wild type under the same growth conditions (Student’s t test, P < 0.05).
PHR1 Also Binds to the P Sequence
Because PHR1 shares high homology with PHL2 and PHL3, we wondered whether PHR1 could also bind to the P sequence. To test this hypothesis, we performed EMSA assays using the purified PHR1 proteins produced in E. coli cells. Like PHL2 and PHL3, PHR1 could specifically bind to the biotin-labeled P sequence (Supplemental Fig. S5A). Further analyses indicated that PHR1 binds to the same sites within the P sequence as PHL2 and PHL3 (Supplemental Fig. S5B). Reciprocally, PHL2 and PHL3 could specifically bind to the P1BS element (Supplemental Fig. S5C). To compare the relative binding affinity of PHR1 to P1BS and the P sequence, we first incubated PHR1 with the biotin-labeled P1BS element and then added different amounts of unlabeled probes of P1BS or the P sequence for competition assays. As shown in Supplemental Figure S5D, the unlabeled P1BS competed more efficiently than the unlabeled P sequence, indicating that PHR1 had higher binding affinity for the P1BS element than for the P sequence.
PHL2 and PHL3 Physically Interact with Each Other But Not with PHR1
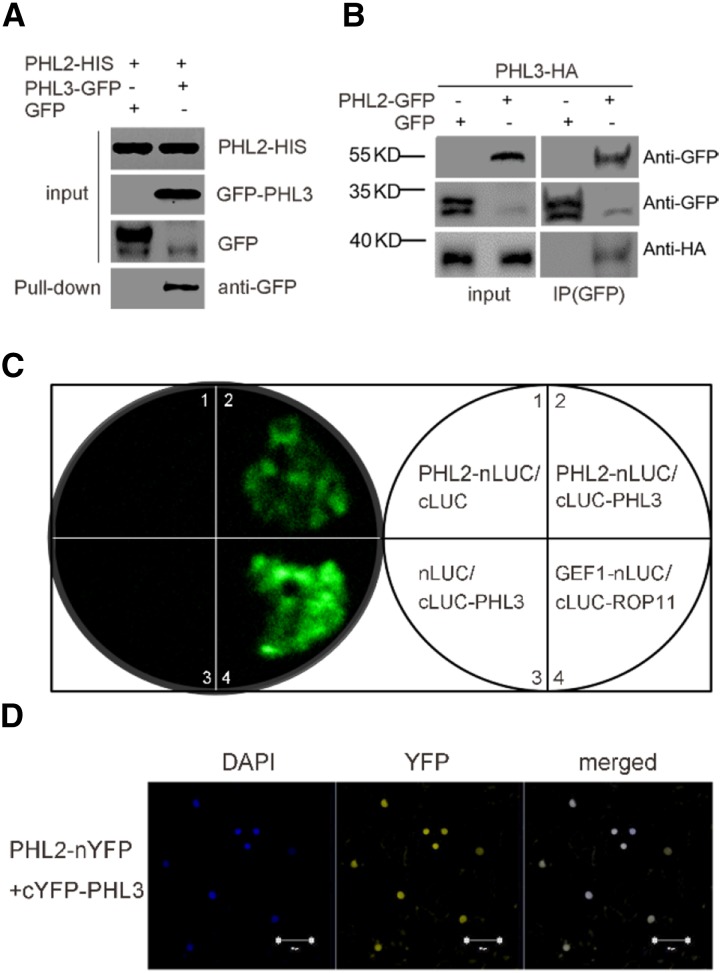
Because PHL2 and PHL3 share high sequence homology at the protein level, we wanted to know whether these two proteins could interact. In pull-down assays, the GFP-tagged PHL3 but not GFP alone expressed in the leaves of N. benthamiana could be pulled down by the His-tagged PHL2 produced from E. coli cells (Fig. 6A). To confirm their interactions in vivo, we performed a coimmunoprecipitation experiment. The GFP-tagged PHL2 or GFP alone was transiently coexpressed with HA-tagged PHL3 in the leaves of N. benthamiana. The expressed proteins were immunoprecipitated with anti-GFP antibodies. The HA-PHL3 proteins could be detected by anti-HA antibodies in the precipitated protein fractions from GFP-PHL2-expressing leaves but not from GFP-expressing leaves (Fig. 6B). The in vivo interactions between PHL2 and PHL3 were also tested using luciferase complementation imaging (LCI) assays and bimolecular fluorescence complementation (BiFC) assays in the leaves of N. benthamiana. For the LCI assay, the full-length CDS of PHL2 and PHL3 was individually fused to the C-terminal domain (PHL2-nLUC) and N-terminal domain (cLUC-PHL3) of the luciferase (LUC) gene. A strong florescence signal resulting from the reconstitution of a functional luciferase was observed when these two constructs were coexpressed in the leaves of N. benthamiana (Fig. 6C). In contrast, no florescence was detected when PHL2-nLUC or PHL3-cLUC was expressed with the empty vector cLUC or nLUC. The BiFC assays further confirmed the interaction between PHL2 and PHL3 (Fig. 6D). The interaction between PHL2 and PHL3 was also observed in yeast two-hybrid assays (Supplemental Fig. S6A). Self-interactions for PHL2 and PHL3 were demonstrated in the yeast two-hybrid assays and BiFC assays (Supplemental Fig. S6, A and B). Taken together, these results suggested that PHL2 and PHL3 might form homodimers or heterodimers as PHR1 and PHL1 (Bustos et al., 2010). However, the interaction between PHL2 and PHR1 or between PHL3 and PHR1 was not observed in LCI assays (Supplemental Fig. S7) or in yeast two-hybrid assays.
Figure 6.
PHL2 physically interacts with PHL3. A, Pull-down assay for the interaction of PHL2 and PHL3. PHL3-GFP or GFP was transiently expressed in the leaves of N. benthamiana. The expressed PHL3-GFP or GFP was pulled down by E. coli-produced PHL2-His proteins using anti-His antibodies. PHL3-GFP proteins in the pull-downed fraction were detected by immunoblot using anti-GFP antibody. B, Co-IP assay for the interaction between PHL2 and PHL3. Total proteins extracted from the leaves of N. benthamiana coexpressing 35S:PHL3-HA and 35S:PHL2-GFP or 35S:GFP were immunoprecipitated with anti-GFP antibodies. The PHL3-HA proteins in the precipitated fraction were detected by immunoblot using anti-HA antibody. C, LCI assays for the interaction of PHL2 and PHL3. The various pairs of the nLUC and cLUC constructs were coexpressed in the leaves of N. benthamiana, and LUC activity was observed 2 d after Agrobacterium tumefaciens infiltration. Two previously reported interacting proteins, GEF1 and ROP11, were used as the positive control (Li and Liu, 2012). D, BiFC assays for the interaction between PHL2 and PHL3. The constructs PHL2-nYFP and cYFP-PHL3 were coinfiltrated into the leaves of N. benthamiana. YFP fluorescence was detected 2 d after infiltration. The nuclei were revealed by DAPI staining. Bars = 50 μm.
Transcription and Protein Accumulation of PHL2 and PHL3 Are Up-Regulated by Low Pi
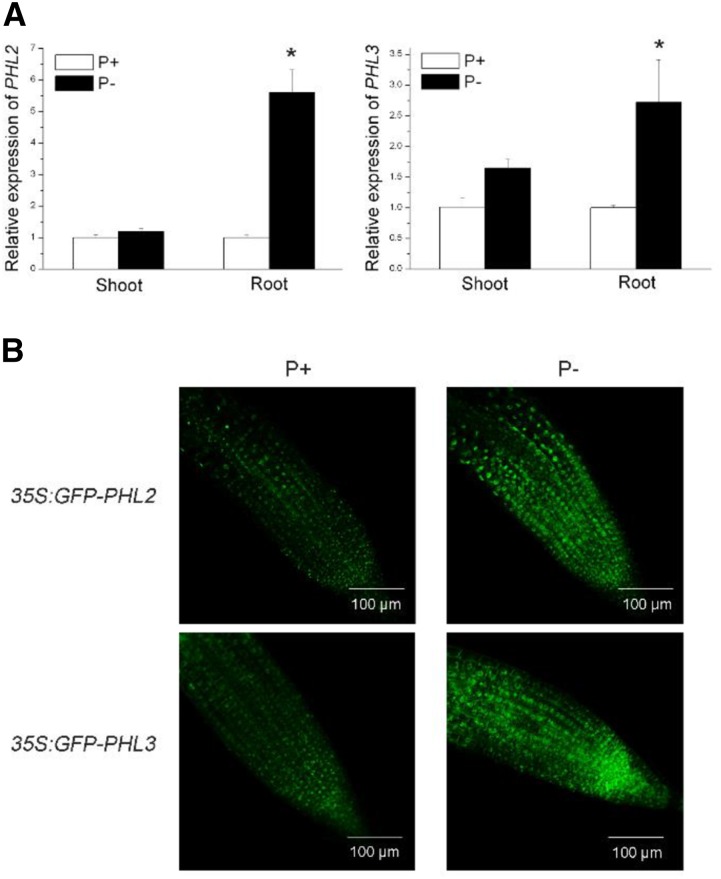
To determine whether the transcription of PHL2 and PHL3 is induced by Pi starvation, we extracted total RNAs from 8-d-old Arabidopsis seedlings grown on P-sufficient (P+) and P-deficient (P−) media and then performed qPCR analysis. The results showed that the levels of both PHL2 and PHL3 mRNA were increased in roots but not in shoots under Pi deficiency (Fig. 7A). The effects of low Pi on protein accumulation of PHL2 and PHL3 were also examined. The Arabidopsis seedlings carrying the 35S:GFP-PHL2 or 35S:GFP-PHL3 constructs were grown under Pi-sufficient and -deficient conditions. As shown in Figure 7B, the green fluorescence signals of both GFP-PHL2 and GFP-PHL3 proteins were greatly enhanced in the nucleus of root cells under Pi deficiency.
Figure 7.
The transcription and protein accumulation of PHL2 and PHL3 under P+ and P− conditions. A, qPCR analysis of the relative expression of PHL2 and PHL3 in shoots and roots of 8-d-old wild-type seedlings under P+ and P− conditions. Values are means ± se of three replicates. Asterisks indicate a significant difference from the wild type under the same growth conditions (Student’s t test, P < 0.05). B, GFP fluorescence signals in the root cells of the 8-d-old transgenic plants expressing 35S:GFP-PHL2 or 35S:GFP-PHL3 under P+ and P− conditions.
To determine whether the enhanced accumulation of GFP-PHL2 and GFP-PHL3 proteins under Pi deficiency was due to the increased expression of GFP-PHL2 and GFP-PHL3 mRNAs, we measured the expression levels of these two transgenes under both P+ and P− conditions by qPCR. Although the 35S promoter is a strong constitutive promoter, we found that the expression levels of GFP-PHL2 and GFP-PHL3 were enhanced under the P− condition (Supplemental Fig. S8), indicating that Pi deficiency might increase the stability of PHL2 and PHL3 mRNAs. Thus, at this point, we could not exclude the possibility that the enhanced accumulation of PHL2 and PHL3 proteins under Pi deficiency was due to the increased expression of PHL2 and PHL3 mRNAs.
PHL2 and PHL3 Are Transcriptional Activators of AtPAP10 Transcription
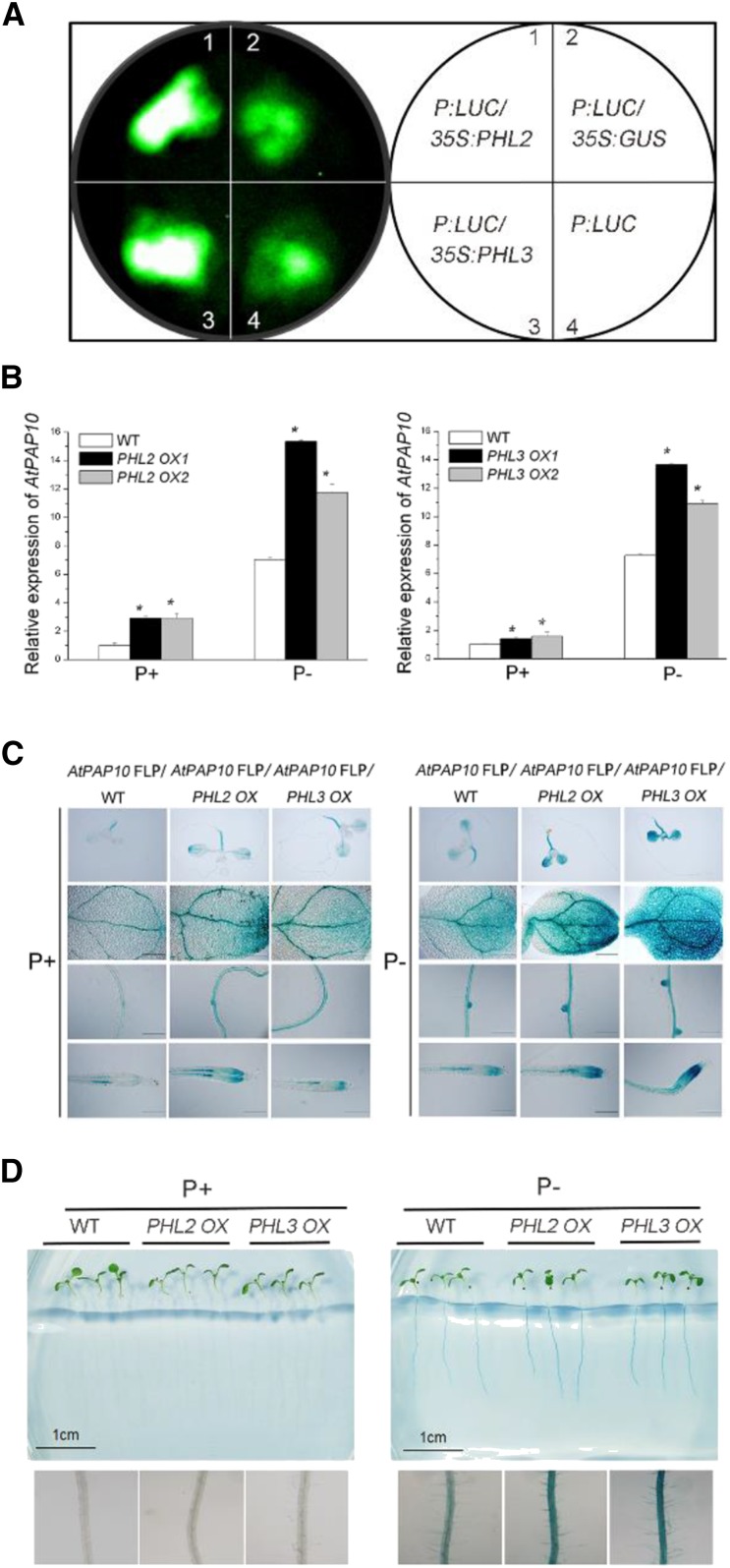
To determine whether PHL2 and PHL3 function in the regulation of the transcription of AtPAP10, we fused the P sequence with a 35S minimal promoter and placed it in the front of the LUC reporter gene. The resulting construct P: LUC was infiltrated alone or coinfiltrated with the construct 35S:PHL2 or 35S:PHL3 into the leaves of N. benthamiana. Two days after infiltration, a basal level of LUC activity was observed for the P:LUC construct (Fig. 8A). LUC activity was greatly enhanced in the leaves coexpressing 35S:PHL2 or 35S:PHL3, but not in the leaves coexpressing 35S:GUS. These results indicated that PHL2 and PHL3 bound to the P sequence and activated the expression of the reporter gene in vivo.
Figure 8.
PHL2 and PHL3 function as transcriptional activators. A, The P:LUC construct was infiltrated alone or was coinfiltrated with 35S:PHL2, 35S:PHL3, or 35S:GUS into the leaves of N. benthamiana. LUC activity was detected 2 d after infiltration. B, The relative expression of AtPAP10 in 8-d-old transgenic plants overexpressing PHL2 or PHL3 as determined by qPCR. Values are means ± se of three replicates. Asterisks indicate a significant difference from the wild type under the same growth conditions (Student’s t test, P < 0.05). C, The transgenic plants overexpressing PHL2 or PHL3 were crossed with the AtPAP10 FLP:GUS transgenic line. The GUS expression patterns are shown for the whole seedling, cotyledon, root maturation zone, and root tip of F1 plants. Bars = 100 μm. D, The root-associated APase activity in the 8-d-old wild-type, PHL2 OX, and PHL3 OX seedlings grown under P+ and P− conditions. Bottom: A close-up view of the BCIP staining of the roots of the plants shown at the top of D.
To further demonstrate that PHL2 and PHL3 function as transcriptional activators, we generated transgenic plants overexpressing the PHL2 gene (PHL2 OX) or the PHL3 gene (PHL3 OX) driven by the 35S promoter (Supplemental Fig. S9). The expression of AtPAP10 was greater in these transgenic plants than in wild-type plants under both P+ and P− conditions (Fig. 8B). We then crossed the PHL2 OX and PHL3 OX plants with the transgenic line carrying the AtPAP10 FLP:GUS construct. The F1 plants derived from this cross exhibited greater GUS activity than AtPAP10 FLP:GUS plants in wild-type background (Fig. 8C). Accordingly, the PHL2 OX and PHL3 OX lines showed increased APase activity on the root surface under Pi deficiency (Fig. 8D). In addition, we crossed PHL2 OX to phr1 and selected phr1 plants overexpressing PHL2 in the F2 population. In these plants, the level of AtPAP10 mRNA and root-associated APase were restored to that of the wild type (Supplemental Fig. S10, A and B). These results indicated that PHL2 and PHL3 function as transcriptional activators of AtPAP10 and that the overexpression of PHL2 can compensate the loss of function of PHR1.
To provide additional evidence that PHL2 and PHL3 are involved in the control of AtPAP10 transcription, we tried to analyze AtPAP10 expression in PHL2 and PHL3 knockout mutants. The SALK_114420C line contains a T-DNA insertion in the first intron of PHL2 (Supplemental Fig. S11A). In this line, the transcript of PHL2 was barely detected, and it therefore could be regarded as a null mutant (Supplemental Fig. S11B). qPCR analysis indicated that the expression level of AtPAP10 in the mutant was about 75% of that in the wild type under Pi deficiency (Fig. 10). However, the BCIP staining of the root-associated APase activity in phl2, was not significantly reduced (Supplemental Fig. S12). For PHL3, it was not possible to analyze AtPAP10 expression in its knockout mutant because a previous report has shown that the insertion of a Ds transposon into the PHL3 gene results in an embryo-lethal phenotype (Pagnussat et al., 2005).
Figure 10.
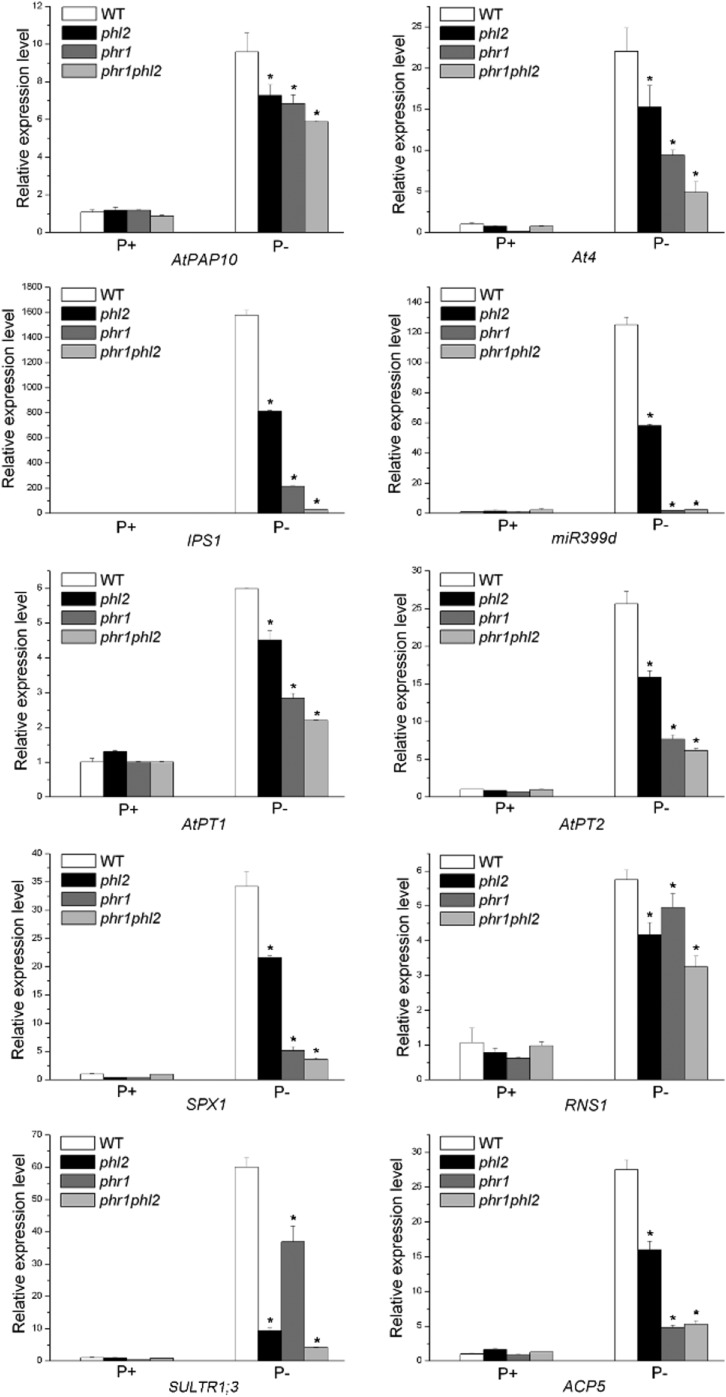
Relative expression of ten PSI genes in the wild type, phl2, phr1, and phr1phl2. Total RNAs were extracted from 8-d-old seedlings grown under P+ and P− conditions. Values are means ± se of three replicates. Asterisks indicate a significant difference from the wild type under the same growth conditions (Student’s t test, P < 0.05). The names of the genes examined are indicated at the bottom of each panel.
PHL2 Is a Key Component Controlling Plant Transcriptional Response to Pi Starvation
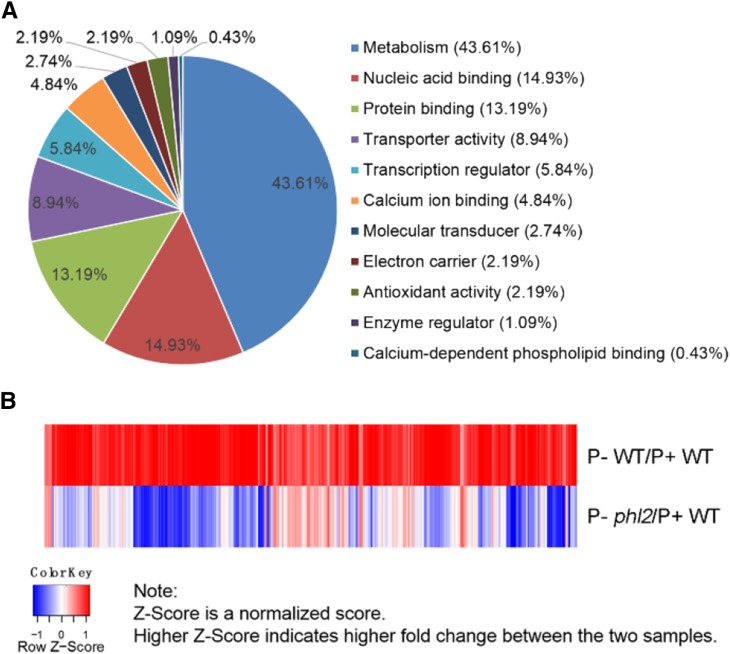
To investigate how widely PHL2 regulates the expression of PSI genes at the genomic level, we performed a RNA-sequencing (RNA-seq) analysis on wild-type and phl2 plants. Total RNAs were extracted from the roots of P+ and P− wild-type seedlings (i.e. seedlings grown on P-sufficient and P-deficient media) and of P− phl2 seedlings. The extracted RNAs were subjected to RNA deep sequencing. We first compared the transcriptomes of the roots of wild-type seedlings grown under P+ and P− conditions. Using a 2-fold change and a false discovery rate (FDR) of <0.05 as the cutoff for selecting the differentially expressed transcripts, we found that the expression of 581 genes was induced and that the expression of 120 genes was repressed by Pi starvation (Supplemental Tables S1 and S2). The induced genes included the typical PSI marker genes IPS1, At4, ACP5, and AtPT2, indicating that the experimental conditions were proper. The accuracy of the RNA-seq results was validated by the qPCR analysis of another six PSI genes, Pht1;5, OCT1, MGD3, PAP22, SPX2, and STP12 (Supplemental Fig. S13). The Gene Ontology (GO) analysis indicated that these 581 PSI genes fell into the category of metabolism (43.61%), followed by nucleic acid binding (14.93%), protein binding (13.19%), transporter activity (8.94%), transcription regulator (5.85%), and others (Fig. 9A). In this study, we focused on the genes that were up-regulated by Pi starvation. We then compared the expression levels of these 581 PSI genes in the wild type and phl2. The results showed that the average expression level of these genes was 51% lower in phl2 relative to the wild type. Among the 581 genes, only 2.4% showed higher expression in P− phl2 than in P− wild-type seedlings (Fig. 9B). Moreover, the expression levels for 4.6% of these 581 PSI genes were even lower in P− phl2 than in P+ wild-type seedlings. These results indicated that for some PSI genes, the mutation of PHL2 not only impairs their Pi inducibility, but also reduces their basal expression level. If we arbitrarily defined a 30% reduction in the expression level of PSI genes as “a significant reduction,” the expression of 87.6% of PSI genes was significantly reduced in P− phl2 relative to P− wild-type seedlings (Fig. 9B; Supplemental Table S3). GO analysis of these affected genes showed the similar patterns as that for all PSI genes, indicating that the affected genes are involved in almost all aspects of plant responses to Pi starvation (Supplemental Fig. S14). Supplemental Table S4 lists 30 PSI genes whose expression was most reduced in phl2. Because the induction of most PSI genes was impaired in phl2, we conclude that PHL2 is a key component of the central regulatory system controlling transcriptional responses to Pi starvation.
Figure 9.
RNA-seq analysis of PSI genes in the wild type and phl2. A, GO analysis of 581 PSI genes. B, A heat map of the RNA-seq analysis showing the differentially expressed PSI genes in the wild type and phl2. The map shows the expression of 581 upregulated genes in the wild type with a cut-off value = 2-fold (FDR < 0.05) and expression levels of the corresponding genes in phl2. The formula for calculating the z-score was z = (x − μ)/σ, where x is the fold-change value, μ is the mean, and σ is the sd.
Besides, we investigated whether the mutation of PHL2 affects the expression of non-PSI genes. RNA-seq results indicated that the expression of 4178 genes was not significantly altered by Pi deficiency (the changes was below 2-fold). Therefore, these genes were regarded as non-PSI genes. We then compared the expression levels of these non-PSI genes between the P− wild type and P− phl2 mutant (this was because the sample of P+ phl2 was not included in our RNA-seq experiment). In this comparison, we found that the expression of 575 genes was significantly reduced in phl2 using 50% reduction as a cutoff (Supplemental Table S5). To validate the effect of PHL2 mutation on the expression of these non-PSI gens, we analyzed the expression of eight genes by qPCR analysis for the wild-type and phl2 seedlings grown under normal condition. These eight genes were selected from different functional categories, including indole-3-acetic acid biosynthesis (NIT2), jasmonate signaling (MYC3), chloride transport (CLCA), lignin catabolism (LAC7), etc. (Supplemental Table S6). The results showed that the expression of these nine genes were significantly reduced in phl2 (Supplemental Figure S15), indicating that the mutation of PHL2 might also affected other biological processes under normal growth condition.
PHL2 Acts Redundantly with PHR1 in Regulating PSI Gene Expression
The incomplete loss of AtPAP10 transcription in phl2 indicated that PHL2 functions redundantly with other transcription factors, perhaps PHR1 and PHL1, in regulating the transcription of PSI genes. To test this hypothesis, we examined the expression of 10 PSI genes in the wild type, phl2 and phr1 single mutants, and the phr1phl2 double mutant. These PSI genes included two high-affinity phosphate transporters, AtPT1 (Pht1;1) and AtPT2 (Phtl;4) (Muchhal et al.,1996); two noncoding transcripts, IPS1 and At4 (Burleigh and Harrison, 1999); two acid phosphatase, AtPAP10 and ACP5 (AtPAP17; del Pozo et al.,1999); an RNase, RNS1 (Bariola et al., 1994); miR399d (Fujii et al., 2005); an SPX domain-containing protein, SPX1 (Duan et al., 2008); and a sulfate transporter, SULTR1;3 (Rouached et al., 2011). As shown in Figure 10, the expression of all of these PSI genes was impaired to some degree in phl2 relative to the wild type. Similarly, the expression of all of these PSI genes was reduced in phr1, which was consistent with previous reports (Rubio et al., 2001; Bustos et al., 2010). Among these 10 PSI genes, the reduction of expression for seven genes was greater in phr1 than in phl2. The average reduction of the expression of these 10 PSI genes was 61.4% in phr1 and 41.1% in phl2, indicating that PSI gene expression was affected more by the mutation in PHR1 than in PHL2. Furthermore, the expression of these 10 PSI genes in the phr1phl2 double mutant was lower (reduced 75.9% of the wild type) than that in either phr1 or phl2, indicating a functional redundancy between PHR1 and PHL2.
We then examined the root-associated APase activity in the phr1phl2 double mutant. Although the expression level of AtPAP10 was further reduced in phr1phl2 compared to that in phr1 or phl2, the root-associated APase activity in phr1phl2 was similar to that in phr1 (Supplemental Fig. S11). Considering phr1 and phl2 had similar reduction of AtPAP10 transcription compared to the wild type, but phl2 did not show an obvious decrease of root-associated APase activity as phr1, it indicated that PHR1 and PHL2 might have different effects on the posttranscriptional regulation involved in the induction of root-associated APase activity.
DISCUSSION
The responses of plants to Pi starvation are controlled at multiple levels and involve transcriptional, posttranscriptional, and posttranslational regulation. It is now well known that plants undergo a dramatic change in gene expression when the external Pi level decreases. The functional characterization of the Arabidopsis transcription factor PHR1 provided a framework of a central regulatory system controlling plant transcriptional responses to Pi starvation. However, the experimental evidence accumulated so far, including the partial impairment of AtPAP10 transcription in phr1 (Bustos et al., 2010; this work, Fig. 1) indicated that PHR1 is not the only master regulator of this system. Thus, to further understand how this regulatory system functions, it is important to identify other key transcription factors that globally regulate plant transcriptional responses to changes in Pi availability.
To search for other transcriptional regulators of AtPAP10, we first dissected the sequence architecture of the AtPAP10 promoter. In silico analysis indicated that the AtPAP10 promoter contains one copy of the P1BS element and three copies of P1BS-like elements (Fig. 2). The importance of the P1BS element has been experimentally demonstrated for the transcription of several PSI genes in Arabidopsis and rice (Schünmann et al., 2004; Karthikeyan et al., 2009; Bustos et al., 2010; Oropeza-Aburto et al., 2012; Wu et al., 2013; Ruan et al., 2015); however, how this element functions seems quite complicated. The copy number of the P1BS element in the promoters of PSI genes varies from gene to gene, and the function of the same P1BS element can differ significantly depending on its position. The promoter of the Arabidopsis RNS1 gene contains one copy of P1BS, and mutation of this element completely abolished Pi starvation-induced gene expression (Bustos et al., 2010). Also, the fusing of four tandem copies of this P1BS element to a 35S minimal promoter was sufficient to confer Pi inducibility to the reporter gene. Bustos et al. (2010) also found that the Arabidopsis IPS1 promoter contains two P1BS elements and that one is completely dispensable, indicating that only one P1BS elements is required for Pi starvation-induced expression. Besides, the sequences that flanked the functional P1BS element in the IPS1 promoter are critical for the function of the P1BS element, although these flanking sequences themselves are not sufficient to confer gene expression. The analyses of the promoter of Arabidopsis PLDZ2 (phospholipase D2) gene led to similar conclusions, i.e. a P1BS element is important for the Pi starvation-induced transcription, but not all the P1BS elements on a given promoter is indispensable, and sequences that flank the P1BS element are also critical for maintaining the function of this element (Oropeza-Aburto et al., 2012).
In the case of the AtPAP10 promoter, our results showed that just one P1BS element proximal to the transcription start site is not sufficient to confer expression of a reporter gene and the distal P1BS-like element is completely dispensable (Fig. 2). Instead, the P sequence (−330 to −248), which contains two P1BS-like elements, is critical for not only Pi inducibility but also for the basal level of expression of a reporter gene (Fig. 2). The EMSA assays confirmed that PHR1 can specifically bind to these two elements (Supplemental Fig. S5). This is not surprising because these two elements closely resemble P1BS. Besides, we identified a novel binding site for PHR1 that is located within the first 20 nucleotides of the P sequence, although the exact binding sequence remains to be determined (Fig. 4). Interestingly, these two P1BS-like elements and the novel protein binding site can also bind to PHL2 and PHL3. At this point, it is not clear whether all these three protein binding sites (two P1BS-like elements and the novel protein binding site) are required for gene transcription. Currently, we are generating stable transgenic plants carrying the various deletions or mutations of these two P1BS-like elements and the novel protein binding site to elucidate the function of these elements. Once the role for each of these three elements in the controlling of AtPAP10 transcription is elucidated, next, it will be important to determine the stoichiometry of the DNA-protein complex formed between the cis-elements and PHR1(-like) proteins to better understand how these transcription factors operate in regulating PSI gene expression.
Using a variety approaches (Y1H assay, EMSA assays, and CHIP assays), we identified two proteins that directly bind to the P sequence (Fig. 3). These two proteins also belong to the family of MYB-CC transcription factors and are the most closely related among all members of this family. The high conservation of PHL2 and PHL3 sequences in diverse plant species emphasizes the importance of these two proteins in regulating gene transcription (Supplemental Fig. S3). Compared to PHR1 and PHL1, PHL2 and PHL3 lack an N-terminal extension and have two extra segments in their C terminus (Fig. 3). Although the sequences containing the MYB domain and coil-coil domain among these four proteins are highly conserved, there are still some small variations between PHR1/PHL1 and PHL2/PHL3. The similarity and differences in protein sequences between PHR1/PHL1 and PHL2/PHL3 suggests that they may bind to similar DNA sequences but differ somewhat in their biochemical functions. For example, PHR1 and PHL2/PHL3 can bind to both the P sequence and P1BS element, but PHR1 has a higher binding affinity for the P1BS element than for the P sequence (Supplemental Fig. S5). Also, PHL2 and PHL3 can physically interact with each other and with themselves (Fig. 6; Supplemental Fig. S7), but they cannot interact with PHR1. These results suggest that PHL2 and PHL3 may form homodimers and heterodimers just as PHR1 and PHL1 do (Bustos et al., 2010) but that they will not be able to form a protein complex with PHR1 or PHL1 through the direct interaction. In addition, the accumulation of PHL2 and PHL3 proteins in roots is enhanced by Pi starvation (Fig. 7), whereas that of PHR1 and PHL1 is not, although at this point we cannot be sure whether the enhanced protein accumulation is due to the increased transcription of their corresponding genes under Pi deficiency. Such differences in the protein properties and expression patterns between PHL2/PHL3 and PHR1/PHL1 may reflect different mechanisms that these proteins regulate the transcription of PSI genes.
Several lines of evidence indicate that PHL2 and PHL3 act as transcriptional activators of AtPAP10 transcription (Figs. 8 and 10). Furthermore, our RNA-seq analyses showed that among the 581 PSI genes, the expression of 87% is significantly reduced in phl2 (Fig. 9), demonstrating that PHL2 positively regulates the transcription of a large number of PSI genes at the genomic level, although the number of genes that are the direct targets for PHL2 remains to be determined. These results demonstrated that, like PHR1 and PHL1, PHL2 is also a key component of the central regulatory system controlling the transcriptional responses to Pi starvation. For PHL3, currently, it is not feasible to perform an RNA-seq analysis in its knockout mutant because phl3 is embryo-lethal. However, its high sequence homology with PHL2, its Pi deficiency-induced expression, its binding ability to the P sequence, and its ability to activate transcription of AtPAP10 strongly suggest that PHL3 may behave like PHL2 in regulating PSI gene expression. Interestingly, we found that PHL2 is also involved in the control of the transcription of many non-PSI genes, indicating that it also participates in the regulation of other types of biological processes under normal growth condition. Taken together, these results implicated that the gene duplication and divergence events in this MYB-CC transcription factor family are not only used to build a central regulatory system controlling Pi transcriptional responses, but can also play roles in other regulatory networks of distinct biological processes.
Our studies of the PSI gene expression in the phr1 and phl2 single mutant and phr1phl2 double mutant indicate a functional redundancy between PHL2 and other transcription factors, such as PHR1 and PHL1. Furthermore, when PHL2 was overexpressed in the phr1 background, it restored the transcription of AtPAP10 and the root-associated APase activity to the levels of the wild type, suggesting that the overexpression of PHL2 could compensate for the loss of PHR1 activity in regulating AtPAP10 transcription. This provides additional evidence that PHL2 and PHR1 act redundantly in regulating PSI gene expression. The functional redundancy among the transcription factors of the same family has also been observed for plant responses to deficiency of other nutrients, such as iron (Wang et al., 2013b) and nitrate (Konishi and Yanagisawa, 2013). In Arabidopsis, the bHLH transcription factor FIT is the key regulator of the expression of iron deficiency-responsive genes (Yuan et al., 2008). In addition, other four AtbHLH proteins interact with FIT to regulate the expression of iron deficiency-responsive genes by forming heterodimers with FIT (Wang et al., 2013b). Under iron deficiency, the single knockout mutant of these four AtbHLH genes did not exhibit obvious defects in the responses to iron deficiency, while the double and triple knockout mutants displayed the progressively aggravated mutant phenotypes. Through the analysis of the mutants with various combinations of the AtbHLH genes, the significance of these four bHLH genes in regulating plant responses to iron deficiency was determined. In the case of the MYB-CC transcription factor family, PHR1 may play a major role in regulating Pi-responsive genes, and its functions may be fine-tuned by PHL1, PHL2, and PHL3. Thus, it will also be important to analyze the transcriptional responses as well as the developmental and physiological phenotypes of the single, double, and triple mutants of PHR1, PHL1, and PHL2. Because PHL2 and PHL3 do not form heterodimer with PHR1, they may interact with PHR1 in a different way from that observed for bHLH transcription factors in regulating iron deficiency responses. Furthermore, by identifying the common and distinct targets for PHR1, PHL1, and PHL2, we will have a better understanding of the different roles of these three transcription factors in the central regulatory system controlling plant transcriptional responses to Pi starvation.
MATERIALS AND METHODS
Plant Material and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Colombia-0 ecotype background. The T-DNA insertional lines SALK_067629 (phr1) and SALK_114420C (phl2) were obtained from the Arabidopsis Biological Resource Center. The SALK_067629 line has been previously identified as a null mutant of PHR1 (Nilsson et al., 2007). The primers used to confirm the insertion of the T-DNA in SALK_114420C are listed in Supplemental Table S7. The phr1phl2 double mutant was generated through genetic crossing. Seeds were surface sterilized with 20% bleach for 15 min and washed three times with sterile-distilled water. The seeds were then sown on agar plates containing a Pi-sufficient (P+) medium or a Pi-deficient (P−) medium. The P+ medium used in this study contained half-strength Murashige and Skoog salt, 1.0% (w/v) Suc, 0.5% MES, and 1.2% (w/v) agar (Sigma-Aldrich). In the Pi− medium, the 1.25 mm KH2PO4 in the P+ medium was replaced with 0.65 mm K2SO4. After seeds were stratified for 2 d at 4°C, the agar plates were placed vertically in a growth room with a photoperiod of 16 h light:8 h dark at 22 to 24°C.The light intensity was 100 µmol m−2 s−1. The Nicotiana benthamiana plants were grown in soil at 26°C under the same lighting conditions.
Analysis of Root-Associated APase Activity
For histochemical staining of APase activity on the root surface of Arabidopsis seedlings, an agar solution (0.5%, w/v) containing 0.01% (w/v) BCIP was evenly overlaid on the roots growing on agar plates (Wang et al., 2011). After 12 h of color development, the roots were photographed with a camera attached to a stereomicroscope (Olympus SZ61).
qPCR Analysis
Total RNAs of 8-d-old seedlings were extracted using the Tiangen RNAeasy kit. DNase I-digested RNA (1 μg) was reverse transcribed to cDNA using M-MLV reverse transcriptase (Takara). qPCR analyses were carried out using SYBR Fast qPCR Master Mix (KAPA) on a Bio-Rad CFX96 real-time PCR detection system according to the manufacturer’s instructions. The ACTIN gene was used as an internal control. The primers used for qPCR analysis are listed in Supplemental Table S7.
Vector Construction and Plant Transformation
For the overexpression of the AtPAP10 gene, the genomic sequence of PHR1 was PCR amplified from the genomic DNA isolated from Arabidopsis seedlings. The amplified PHR1 gene was used to replace the GUS gene between the CaMV 35S promoter and the NOS terminator on the plant expression vector pBI121 using SmaI and SacI restriction enzymes. The resulting construct carried a kanamycin-resistant gene as a selectable marker for plant transformation. For the overexpression of PHL2 and PHL3 genes, their corresponding genomic sequences were PCR amplified from the genomic DNA and were used to replace the LUC gene between the CaMV 35S promoter and the NOS terminator on the plant expression vector pZH01 using the BamHI and SacI restriction enzymes. The resulting constructs carried a hygromycin-resistant gene as the selectable marker for plant transformation.
For the deletion analysis of the AtPAP10 promoter, a set of eight AtPAP10 promoter sequences with different lengths was generated by PCR from a 2-kb sequence upstream of the start codon of AtPAP10 (Wang et al., 2011). During amplification, HindIII and BamHI restriction enzyme sites were added to the 5′ and 3′ ends of the PCR products, respectively. The amplified fragments were cloned into HindIII and BamHI sites between the CaMV 35S promoter and the GUS reporter gene in the plant transformation vector pBI101. The resulting construct carried a kanamycin-resistant gene as a selectable marker for plant transformation. For testing the function of the P sequence (−330 to −248), the P sequence was PCR amplified and fused to a 35S minimal promoter sequence (−46 to +8). The fused sequence was used to replace the 35S promoter in front of the LUC gene on the plant expression vector pZH01.
For subcellular localization analysis, the full-length CDS of PHL2 and PHL3 was PCR amplified from plant cDNA and fused to the 5′ terminus of GFP in the vector pJG186 or to the 3′ terminus of GFP in the vector pJG053 (both vectors were modified from the vector CAMBIA 1300-35S-GFP). For LCI assays, the full-length CDSs of PHL2 and PHL3 were inserted into the vectors pCAMBIA-nLUC and pCAMBIA-cLUC (Chen et al., 2008), respectively, to generate PHL2-nLUC and cLUC-PHL3 constructs. For BiFC assays, the full-length CDSs of PHL2 and PHL3 were individually inserted into the vector nYFP or cYFP through the Gateway cloning method.
The primers used for vector construction are listed in Supplemental Table S4. All constructs were mobilized into the Agrobacterium tumefaciens strain GV3101 and transformed into Arabidopsis plants via the Agrobacterium-mediated flower dip method (Clough and Bent, 1998) or were used for transient expression in the leaves of N. benthamiana as described by Batoko et al. (2000). The stable transgenic lines were selected using antibiotic-containing media.
LCI and BiFC Assays
The Agrobacterium strains harboring different constructs were grown to a cell density of OD600 = 0.5 and were harvested and resuspended in infiltration buffer (10 mm MES, 0.2 mm acetosyringone, and 10 mm MgCl2) to the same concentration. The Agrobacterium strains were infiltrated into the leaves of 4-week-old N. benthamiana plants. For LCI assays, the leaves were sprayed uniformly with luciferin (100 mM) dissolved in 0.1% Triton X-100 2 d after infiltration. After a 5-min exposure in the dark, the luminescence images were captured with an Andor iXon CCD camera (Andor Technology). For BiFC assays, the leaves of N. benthamiana plants were coinfiltrated with the various combination of nYFP and cYFP constructs. The DAPI solution (5 μg/mL) was infiltrated 2 d after first infiltration. Two hours later after second infiltration, the fluorescence signals of YFP and DAPI staining were examined with a Zeiss confocal microscope (LSM710).
Analysis of GUS Activity
The histochemical and quantitative analyses of GUS activity were performed as described by Jefferson (1989).
Y1H Assays
The Y1H assays were carried out using the MatchMaker One-Hybrid System (Clontech) according to the manufacturer’s instructions. The P sequence was triplicated in tandem and cloned into the vector pAbAi to create a bait construct. The yeast bait/reporter strain was generated by integrating the pBait-AbAi plasmid into the URA3-52 locus of the Y1HGold yeast genome. SD/-Ura medium was used to select for the integration of the bait-reporter construct into the genome of Y1HGold. The Y1H cDNA library was generated from 8-d-old Pi-starved Arabidopsis seedlings using SMART cDNA synthesis technology (Takara). The cDNA library and the linearized pGADT7-Rec vector were cotransformed into the Y1HGold bait strain. The transformed cells were plated on SD/-Ura/AbA medium for selection of aureobasidin A (AbA) resistance, which was activated by the prey proteins that specifically bound to the bait sequence.
The full-length CDS of the positive clones obtained from the Y1H screening was cloned into the EcoRI and BamHI sites of the yeast expression vector pGADT7. The resulting constructs were transformed into the yeast strain Y1HGold, which carried the reporter construct. The transformed cells were plated on both SD/-Leu and SD/-Leu/AbA media and were grown for 2 to 4 d. The cells that could grow on both media indicated an interaction between the prey protein and the bait sequence.
EMSAs
The full-length CDSs of PHL2, PHL3, and PHR1 were cloned in-frame between the 3′ terminus of an MBP tag and 5′ terminus of a 6X HIS tag sequence in the vector pMAL-c5x (NEB) using NotI and SalI restriction sites. The MBP-PHL2-His, MBP-PHL3-His, and MBP-PHR1-His constructs were separately transformed into E. coli strain BL21 (DE3). The recombinant proteins were purified using Ni-NTA agarose columns. The biotin-labeled probes of the P sequences and its derivatives were generated by annealing the biotin-labeled complementary oligonucleotides. The sequences of the oligonucleotides used for generating various probes are listed in Supplemental Table S4. The labeled probes were incubated with the recombinant proteins in a buffer solution (25 mm HEPES, 40 mm KCl, 5 mm MgCl2, 1 mm DTT, 1 mm EDTA, and 8% glycerol, pH 8.0) for 20 min at room temperature. The DNA-protein reaction mixtures were then loaded onto a 4.5% nonreducing gel (4.5% acrylamide, 0.5× Tris-borate/EDTA, 0.1% ammonium persulfate, and 0.1% N,N,N′,N′-tetramethylethylenediamine) and were subjected to electrophoresis for 1 to 2 h. The free and protein-bound DNAs were detected using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific) according to the manufacturer’s instructions.
Yeast Two-Hybrid Assays
The yeast two-hybrid assays were performed using the Matchmaker GAL4 Two-Hybrid System (Clontech) according to the manufacturer’s instructions. Full-length CDSs of PHL2 and PHL3 were cloned into either pGADT7 or pGBKT7 vectors. The various combinations of the two constructs were cotransformed into the yeast strain AH109. Transformants were selected on SD/-Trp/-Leu/-His medium supplemented with 2.5 mm 3-amino-1,2,4-triazole.
Pull-Down Assays
The GFP-PHL3 fusion protein and GFP protein were transiently and separately expressed in the leaves of N. benthamiana. Two days after infiltration, the total proteins were extracted from the infiltrated leaves using ice-cold buffer (50 mm NaCl, 50 mm Tris-HCl, 0.1% Tween 20, 10% glycerol, 0.143% β-mercaptoethanol, 25 mm imidazole, and 1× cocktail protease inhibitor [Roche], pH 7.8). The protein extracts were incubated with the recombinant PHL2-His fusion proteins, which had been immobilized to Ni-NTA agarose beads, for 3 h at 4°C in RB buffer. After they were washed three times with RB buffer, the beads were boiled for 10 min in 30 μL of 2× protein loading buffer. The proteins released from the beads were separated by SDS-PAGE and were subjected to immunoblot analysis using anti-GFP antibodies.
Coimmunoprecipitation Assays
The plant constructs of 35S:GFP-PHL2 and 35S:HA-PHL3 were cotransformed into the leaves of N. benthamiana. After 2 d, the total proteins were extracted from the infiltrated leaves with ice-cold immunoprecipitation buffer (150 mm NaCl, 50 mm Tris-HCl, 10% glycerol, 5 mm DTT, 1× cocktail protease inhibitor [Roche], and 0.5% Nonidet P-40, pH 8.0). Protein extracts were incubated with GFP-Trap A beads (Chromo Tek) for 3 h at 4°C. The beads were then washed three times with ice-cold immunoprecipitation buffer to remove the unbound proteins. Finally, the proteins bound to the beads were released by boiling for 10 min, and the released proteins were then subjected to immunoblot analysis using anti-HA antibody.
ChIP-qPCR Analysis
ChIP-qPCR assays were performed essentially as described by Saleh et al. (2008). Briefly, the chromatins were isolated from the transgenic plants overexpressing PHL2 or PHL3. The isolated chromatins were cross-linked with 1% formaldehyde, sonicated, and precipitated by GFP-Trap A beads. The precipitated DNA fragments were released by 200 μm NaCl and subjected to qPCR analysis.
RNA-Seq Analysis
The total RNAs were extracted from the roots of 8-d-old wild-type and phl2 seedlings grown on P+ or P− medium using the RNeasy plant mini kit (Qiagen). The RNA-seq analyses were performed at Bionova Company. The RNA-seq libraries were constructed through adaptor ligation and were subjected to single-ended sequencing with a 50-nucleotide reading length. FastQC software was used to assess the quality of raw sequencing reads. The adapter and the low-quality reads were filtered before data analysis. The remaining reads were aligned to the Arabidopsis TAIR 10.0 reference genome using TopHat2. After the sequences of rRNA or tRNA were removed, TopHat’s read alignments were assembled by Cufflinks to produce a transcriptome annotation of the genome. The expression levels for all transcripts were normalized to per million mapped reads. The differentially expressed genes were identified using DESeq2 (Love et al., 2014). The cutoff value for differentially expressed transcripts was a >2-fold change in expression with an FDR ≤ 0.05.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome initiative or GenBank/EMBL databases under the following accession numbers: PHR1 (AT4G28610), PHL1 (AT5G29000), PHL2 (AT3G24120), PHL3 (AT4G13640), AtPAP10 (AT2G16430), AtPAP22 (AT3G52820), IPS1 (AT3G09922), At4 (AT5G03545), miR399d (AT2G34202), SPX1 (AT5G20150), SPX2 (AT2G26660), AtPT1 (AT5G43350), AtPT2 (AT2G38940), Pht1;5 (AT2G32830), ACP5 (AT3G17790), RNS1 (AT2G02990), SULTR1;3 (AT1G22150), STP12 (AT4G21480), OST1 (AT4G33950), and MGD3 (AT2G11810). The RNA-seq data generated in this study have been deposited in the Gene Expression Omnibus database under accession number GSE74972.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A diagram showing the cis-elements in the AtPAP10 gene.
Supplemental Figure S2. Summary of the GUS activity for the transgenic plants carrying various deletion constructs of the AtPAP10 promoter.
Supplemental Figure S3. Alignment of protein sequences of PHL2 and its orthologs in other plant species.
Supplemental Figure S4. EMSA assays showing the binding of recombinant PHL2-MBP fusion protein to the P sequence.
Supplemental Figure S5. EMSA assays showing the binding of PHL2 and PHL3 to the P1BS element and the binding of PHR1 to the P sequence.
Supplemental Figure S6. Interactions between PHL2 and PHL3 and with themselves as indicated by yeast two-hybrid and BiFC assays.
Supplemental Figure S7. The LCI assays showing no interaction between PHR1 and PHL2 or PHL3.
Supplemental Figure S8. The relative expression of GFP-PHL2 and GFP-PHL3 under P+ and P− conditions.
Supplemental Figure S9. The relative expression of PHL2 and PHL3 genes in the wild type and in the PHL2- or PHL3-overexpressing lines.
Supplemental Figure S10. Relative expression of AtPAP10 and root-associated APase activity the PHL2 OX/phr1 line.
Supplemental Figure S11. Identification of the phl2 knockout mutant.
Supplemental Figure S12. BCIP staining of root-associated APase activity in 8-d-old wild-type, phr1, phl2, and phr1phl2 seedlings grown under P− condition.
Supplemental Figure S13. Validation of the RNA-seq results by qPCR analysis.
Supplemental Figure S14. GO analysis of PSI genes whose expression is significantly reduced in phl2.
Supplemental Figure S15. Expression analysis of nine non-PSI genes under normal growth condition.
Supplemental Table S1. The genes whose expression is upregulated by Pi starvation in the wild type.
Supplemental Table S2. The genes whose expression is downregulated by Pi starvation in the wild type.
Supplemental Table S3. The PSI genes whose expression is significantly reduced in phl2.
Supplemental Table S4. The 30 PSI genes whose expression is most reduced in phl2.
Supplemental Table S5. The non-PSI genes whose expression is down-regulated in phl2.
Supplemental Table S6. The non-PSI genes whose expression was validated by qPCR analysis in P+ phl2 and P+ wild type.
Supplemental Table S7. The sequences of the primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Javier Paz-Ares for insightful discussions during this research. We also thank the Arabidopsis Biological Resource Center for providing seeds of phr1 and phl2 T-DNA lines.
Glossary
- APase
acid phosphatase
- PSI
phosphate starvation-induced
- Pi
inorganic phosphate
- qPCR
quantitative PCR
- BCIP
5-bromo-4-chloro-3-indolyl phosphate
- Y1H
yeast one-hybrid
- EMSA
electrophoretic mobility shift assay
- CDS
coding sequence
- CaMV
Cauliflower mosaic virus
- DAPI
4′,6-diamino-phenylindole
- ChIP
chromatin immunoprecipitation
- UTR
untranslated region
- LCI
luciferase complementation imaging
- BiFC
bimolecular fluorescence complementation
- RNA-seq
RNA-sequencing
- FDR
false discovery rate
- GO
Gene Ontology
Footnotes
This work was supported by funds from the Natural Science Foundation of China (grant no. 31370290) and the Ministry of Agriculture of China (grant no. 2014ZX0800932B).
References
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ (1994) The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J 6: 673–685 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL. (1973) Phosphate pools, phosphate transport, and phosphate availability. Annu Rev Plant Physiol 24: 225–252 [Google Scholar]
- Bournier M, Tissot N, Mari S, Boucherez J, Lacombe E, Briat JF, Gaymard F (2013) Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J Biol Chem 288: 22670–22680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ (1999) The down-regulation of Mt4-like genes by phosphate fertilization occurs systemically and involves phosphate translocation to the shoots. Plant Physiol 119: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon-Vazquez C, Ibarra-Laclette E, Caballero-Perez J, Herrera-Estrella L (2008) Transcript profiling of Zea mays roots reveals gene responses to phosphate deficiency at the plant- and species-specific levels. J Exp Bot 59: 2479–2497 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P (2008) Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54: 965–975 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, González E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol 15: 2038–2043 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Ramírez M, Valdés-López O, Tesfaye M, Graham MA, Czechowski T, Schlereth A, Wandrey M, Erban A, Cheung F, et al. (2007) Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol 144: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Nagarajan VK, Raghothama KG (2012) Transcriptional regulation of phosphate acquisition by higher plants. Cell Mol Life Sci 69: 3207–3224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA. (1989) The GUS reporter gene system. Nature 342: 837–838 [DOI] [PubMed] [Google Scholar]
- Karthikeyan AS, Ballachanda DN, Raghothama KG (2009) Promoter deletion analysis elucidates the role of cis elements and 5'UTR intron in spatiotemporal regulation of AtPht1;4 expression in Arabidopsis. Physiol Plant 136: 10–18 [DOI] [PubMed] [Google Scholar]
- Khan GA, Bouraine S, Wege S, Li Y, de Carbonnel M, Berthomieu P, Poirier Y, Rouached H (2014) Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J Exp Bot 65: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S (2013) Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat Commun 4: 1617. [DOI] [PubMed] [Google Scholar]
- Li Z, Liu D (2012) ROPGEF1 and ROPGEF4 are functional regulators of ROP11 GTPase in ABA-mediated stomatal closure in Arabidopsis. FEBS Lett 586: 1253–1258 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, Wu P (2014) SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell 26: 1586–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Togami J, Mason JG, Chandler SF, Tanaka Y (2013) Enhancement of phosphate absorption by garden plants by genetic engineering: a new tool for phytoremediation. BioMed Res Int 2013: 182032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. (2005) A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci USA 102: 11934–11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, Scheible WR (2007) Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ 30: 85–112 [DOI] [PubMed] [Google Scholar]
- Muchhal US, Pardo JM, Raghothama KG (1996) Phosphate transporters from the higher plant Arabidopsis thaliana. Proc Natl Acad Sci USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Lundmark M, Jensen PE, Nielsen TH (2012) The Arabidopsis transcription factor PHR1 is essential for adaptation to high light and retaining functional photosynthesis during phosphate starvation. Physiol Plant 144: 35–47 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH (2007) Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ 30: 1499–1512 [DOI] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oropeza-Aburto A, Cruz-Ramírez A, Acevedo-Hernández GJ, Pérez-Torres CA, Caballero-Pérez J, Herrera-Estrella L (2012) Functional analysis of the Arabidopsis PLDZ2 promoter reveals an evolutionarily conserved low-Pi-responsive transcriptional enhancer element. J Exp Bot 63: 2189–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JA, Yang SS, Miller SS, Bucciarelli B, Liu J, Rydeen A, Bozsoki Z, Uhde-Stone C, Tu ZJ, Allan D, Gronwald JW, Vance CP (2013) An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol 161: 705–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Yu HJ, Ngo QA, Rajani S, Mayalagu S, Johnson CS, Capron A, Xie LF, Ye D, Sundaresan V (2005) Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132: 603–614 [DOI] [PubMed] [Google Scholar]
- Pant BD, Burgos A, Pant P, Cuadros-Inostroza A, Willmitzer L, Scheible WR (2015a) The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J Exp Bot 66: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Pant P, Erban A, Huhman D, Kopka J, Scheible WR (2015b) Identification of primary and secondary metabolites with phosphorus status-dependent abundance in Arabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ 38: 172–187 [DOI] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111: 14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. (2000) Phosphate transport and signaling. Curr Opin Plant Biol 3: 182–187 [PubMed] [Google Scholar]
- Ren F, Guo QQ, Chang LL, Chen L, Zhao CZ, Zhong H, Li XB (2012) Brassica napus PHR1 gene encoding a MYB-like protein functions in response to phosphate starvation. PLoS One 7: e44005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H, Secco D, Arpat B, Poirier Y (2011) The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Guo M, Cai L, Hu H, Li C, Liu Y, Wu Z, Mao C, Yi K, Wu P, Mo X (2015) Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol Biol 87: 429–440 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Alvarez-Venegas R, Avramova Z (2008) An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Schünmann PHD, Richardson AE, Vickers CE, Delhaize E (2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136: 4205–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol 157: 423–447 [DOI] [PubMed] [Google Scholar]
- Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, Tong Y, Li Z (2013a) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot (Lond) 111: 1139–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Z, Qian W, Guo W, Gao X, Huang L, Wang H, Zhu H, Wu JW, Wang D, Liu D (2011) The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol 157: 1283–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D (2014a) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56: 299–314 [DOI] [PubMed] [Google Scholar]
- Wang N, Cui Y, Liu Y, Fan H, Du J, Huang Z, Yuan Y, Wu H, Ling HQ (2013b) Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant 6: 503–513 [DOI] [PubMed] [Google Scholar]
- Wang Z, Ruan W, Shi J, Zhang L, Xiang D, Yang C, Li C, Wu Z, Liu Y, Yu Y, et al. (2014b) Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc Natl Acad Sci USA 111: 14953–14958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16: 205–212 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Lu S, Liu D (2014) A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J Exp Bot 65: 6577–6588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.