A metabolic quantitative trait loci study combined with a regulatory network unraveled the genetic architecture of the natural variation of 155 metabolites in mature maize kernels.
Abstract
Metabolic quantitative trait locus (QTL) studies have allowed us to better understand the genetic architecture underlying naturally occurring plant metabolic variance. Here, we use two recombinant inbred line (RIL) populations to dissect the genetic architecture of natural variation of 155 metabolites measured in the mature maize (Zea mays) kernel. Overall, linkage mapping identified 882 metabolic QTLs in both RIL populations across two environments, with an average of 2.1 QTLs per metabolite. A large number of metabolic QTLs (more than 65%) were identified with moderate effects (r2 = 2.1%–10%), while a small portion (less than 35%) showed major effects (r2 > 10%). Epistatic interactions between these identified loci were detected for more than 30% of metabolites (with the proportion of phenotypic variance ranging from 1.6% to 37.8%), implying that genetic epistasis is not negligible in determining metabolic variation. In total, 57 QTLs were validated by our previous genome-wide association study on the same metabolites that provided clues for exploring the underlying genes. A gene regulatory network associated with the flavonoid metabolic pathway was constructed based on the transcriptional variations of 28,769 genes in kernels (15 d after pollination) of 368 maize inbred lines. A large number of genes (34 of 58) in this network overlapped with previously defined genes controlled by maize PERICARP COLOR1, while three of them were identified here within QTL intervals for multiple flavonoids. The deeply characterized RIL populations, elucidation of metabolic phenotypes, and identification of candidate genes lay the foundation for maize quality improvement.
Knowledge concerning plant metabolism is important for crop improvement as well as in exploring the potential for enhancing the accumulation of high-value products by metabolic engineering strategies. Recent studies that utilized a wide range of techniques, including biochemistry, informatics, genetics, and genomics, have boosted our understanding of the genetics of plant metabolism (Luo, 2015). Metabolic quantitative trait locus (QTL) studies that combined different techniques have aided us to better understand the genetic architecture underlying naturally occurring phenotypic variance and promise to better facilitate future plant-breeding strategies (Fernie and Schauer, 2009).
Maize (Zea mays) kernels make a very large contribution to the diets of humans and animals. The chemical composition and impact of genetic variation on the metabolic diversity of maize kernels have been widely studied (Chander et al., 2008c; Yan et al., 2010; Li et al., 2013; Wen et al., 2014, 2015). The most important storage chemical components in mature maize kernels are starch (70%–75% of dry matter), protein (8%–10% of dry matter), and oil (4%–5% of dry matter), and the underlying genes and related pathways of these have been well studied during the past two decades (Moose et al., 2004; Fernie and Schauer, 2009; Li et al., 2013; Luo, 2015). Although only accumulating to low levels in the maize kernel, many secondary metabolites, such as carotenoids, tocopherols, and flavonoids, also hold great economic and biological importance. Thus, studying the chemical composition and uncovering the genetic contribution to the natural variation of metabolite abundance will enhance efforts in breeding highly nutritious maize. Recently, we conducted a metabolome-based genome-wide association study (GWAS) to identify genes and genomic regions that control the metabolite level in mature maize kernels (Wen et al., 2014). Despite the high resolution and precise mapping results obtained via GWAS, linkage analysis can provide higher statistical power as well as complementary evidence to GWAS in terms of validating and identifying causal polymorphisms. Additionally, it is possible to detect the interaction between QTLs in linkage populations, which was demonstrated previously to be important in determining genetic and phenotypic variation in Arabidopsis (Arabidopsis thaliana; Rowe et al., 2008) and maize (Wen et al., 2015) metabolomes. The identification of metabolic QTLs using experimental populations such as recombinant inbred line (RILs) and introgression lines is a powerful tool that has been utilized in several plant species (Lisec et al., 2008; Fernie and Klee, 2011; Gong et al., 2013; Alseekh et al., 2015).
With the rapid development of next-generation sequencing technology and high-density single-nucleotide polymorphism (SNP) arrays, it is feasible to easily construct ultra-high-density linkage maps and thereby narrow QTLs down to smaller regions (Pan et al., 2012). Here, we genotyped two RIL populations (B73/By804 [BB; n = 197] and Zong3/Yu87-1 [ZY; n = 197]) with the commercial maize SNP50 array (Ganal et al., 2011) and constructed a high-density bin map for each population. Natural variations of 155 metabolites measured from liquid chromatography-tandem mass spectrometry (MS/MS)-based metabolite profiling in mature maize kernels of the two RIL populations were dissected, and QTL mapping was performed for each metabolite. Moreover, a network-based analysis of gene expression associated with the flavonoid pathway was carried out in order to discover novel genes and afford further cross-validation. The obtained results are discussed in the context both of contemporary approaches to enhance genetic resolution in quantitative trait analyses and of our current understanding of the genetic architecture of metabolite accumulation in maize.
RESULTS
High-Density Linkage Maps of Two RIL Populations
Two previously developed RIL populations (i.e. BB and ZY; Ma et al., 2007; Chander et al., 2008a) were used in this study. These populations were subjected to maize SNP50 array analyses (Ganal et al., 2011) as detailed in “Materials and Methods,” and high-density bin maps were subsequently constructed based on the 15,285 and 13,759 polymorphic SNP markers for the BB and ZY populations, respectively. In brief, for the BB and ZY populations, 2,496 and 3,071 recombinant bins (a region in which no recombination was genotyped) were distributed throughout the genome, and the genetic linkage maps were 1,790.2 and 2,716 centimorgan (cM) in length, respectively. The linkage map information of the two RILs was subsequently used for QTL mapping and is available at http://github.com/panqingchun/linkage_map.
Phenotypic Variation and Heritability of the Measured Metabolic Traits
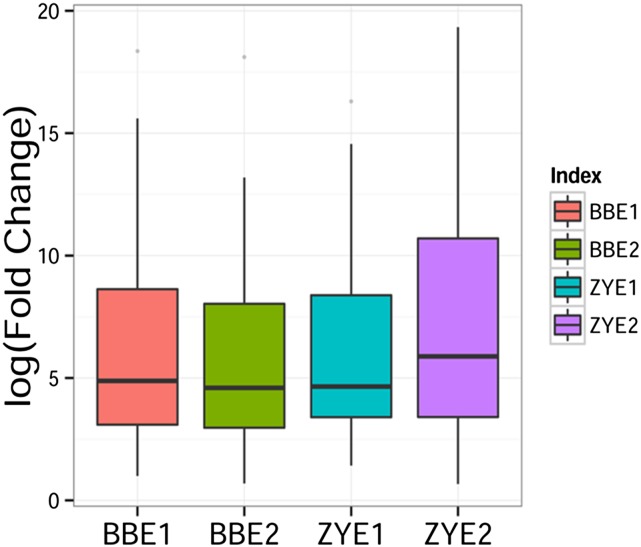
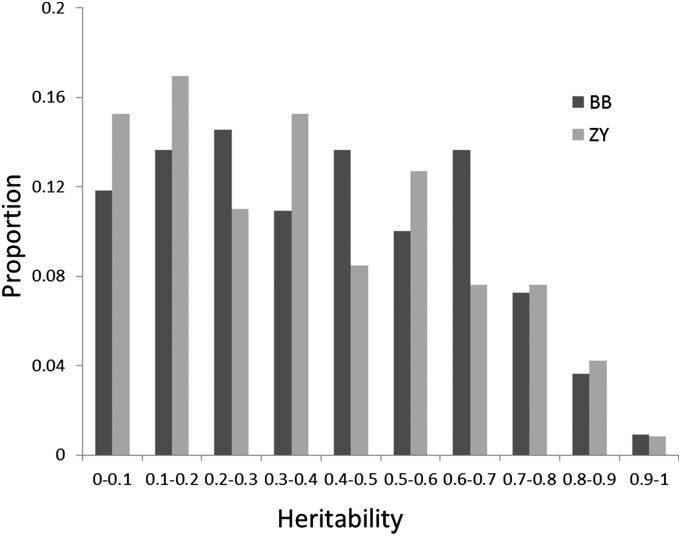
Both BB and ZY RIL populations were planted in two independent environments (simply called experiments 1 and 2 here but described in detail in “Materials and Methods”), and the kernel samples were harvested from these four field experiments for metabolite profiling. While 983 metabolic features were found in previous association mapping studies (Wen et al., 2014), only 184 of these were chemically annotated. Of these 184 compounds, only 155 metabolites were found in the mature kernels of both RIL populations. These 155 metabolites can be broadly classified into alkaloid, amino acid, fatty acid, flavonoid, lysophosphatide, phenolamide, and vitamin classes. Detailed information of each metabolite is provided following recent metabolite reporting recommendations (Fernie et al., 2011) in Supplemental Table S1. Both RIL populations harbored great diversity in terms of metabolite level, as indicated by the distribution of the log value of fold changes among the recombinant lines (Fig. 1; Supplemental Table S2). In both RIL populations, more than half of these metabolites have broad-sense heritability (H2) greater than 0.3 and over 30% of metabolites have H2 greater than 0.5 (Fig. 2). Among the metabolites with H2 greater than 0.5, flavonoids, phenolamides, and amino acids were dominant.
Figure 1.
Distribution of log-fold changes of metabolic traits measured in two RIL populations. Box plots show the log-fold changes of all metabolites among the BB and ZY RILs. Data from different environments (experiments) for each RIL population are shown.
Figure 2.
Distribution of the heritability of metabolic traits. The vertical bars show the proportion of metabolic traits that were detected in both experiments of each RIL population.
The Genetic Basis Underlying Metabolic Variation in the Mature Maize Kernel
In the two experiments using the BB population, 211 and 219 QTLs were identified for 107 and 106 metabolites in experiments 1 and 2, respectively. Each QTL could explain phenotypic variation ranging from 2.1% to 76.8%, with a mean of 9.7%. Of the 86 metabolites for which we detected QTLs in both experiments, a total of 316 QTLs were detected, 38 of which were conserved in both experiments. In the ZY population, 223 QTLs were mapped for 105 metabolites in the first experiment, while in the second experiment, 229 QTLs were detected for 103 metabolites (Table I). Forty-eight QTLs for 34 metabolites were conserved in both experiments. Each QTL could explain between 2.8% and 51.6% of phenotypic variation, with an average variation of 9.8%. The identified QTLs are evenly distributed across the maize genome, and detailed information for the QTL results, including confidence interval, logarithm of odds (LOD) value, and explained phenotypic variation (r2) of each QTL, is indicated in Supplemental Table S3. Linkage mapping using both RIL populations indicated that a large number of metabolic QTLs were identified with moderate effects (72.4% of the mapped QTLs from the BB population and 66.3% of the mapped QTLs from the ZY population, with r2 ranging between 2.1% and 10%), while a relatively small portion showed major effects (17.6% of the mapped QTLs from BB and 33.7% of the mapped QTLs from ZY, with r2 greater than 10%).
Table I. Summary of QTL mapping for metabolic traits measured in two RIL populations.
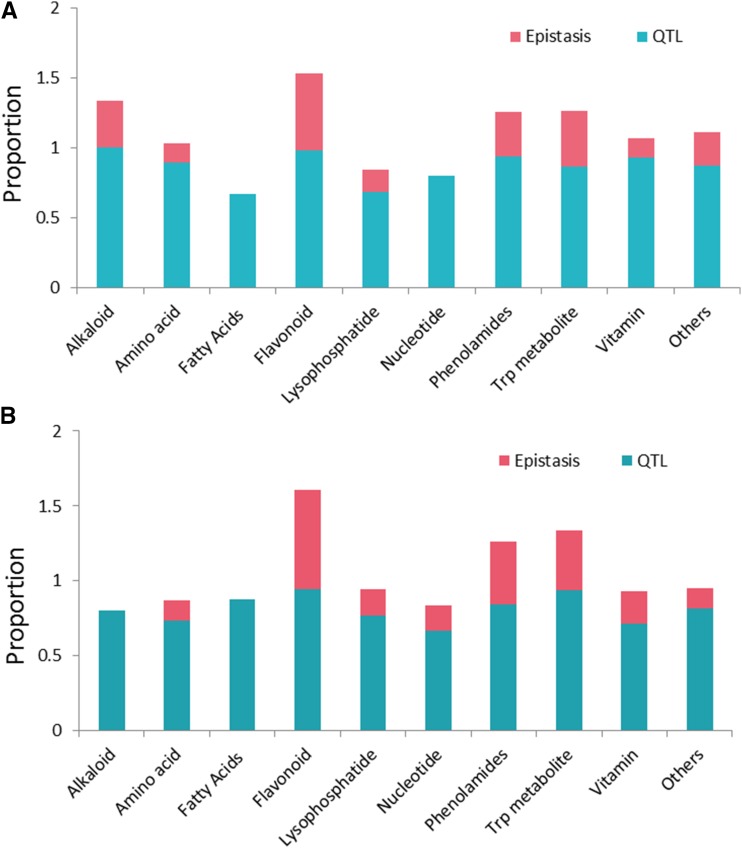
We further investigated the pairwise epistatic interactions between these identified QTLs for each metabolic trait measured in each experiment. In the BB population, epistatic interaction between QTLs was found for 26.1% (31 of 119) and 28.7% (35 of 122) of all the metabolic traits detected in experiments 1 and 2, respectively. In the two experiments in the ZY population, genetic epistasis could partly explain the phenotypic variation for 29.1% (37 of 127) and 30.1% (37 of 123) of all the identified metabolic traits, respectively. The fraction of the phenotypic variation explained by epistasis for each metabolic trait ranged from 1.6% to 37.8% and from 1.6% to 29.3% across the two experiments of the BB and ZY populations, respectively. Significant pairwise epistatic interactions (P < 0.05) between QTLs and their explained phenotypic variances are listed in Supplemental Table S4. A summary of QTL identification and epistasis investigation for different classes of metabolites is given in Figure 3. In each class, more than one QTL was identified for the majority of metabolites (greater than 65%) in both populations. Interestingly, epistasis between the QTL for fatty acids was not found in either population; however, epistatic effects explained a considerable portion of the phenotypic variation of other metabolites, especially flavonoids (Fig. 3).
Figure 3.
Schematic representation of QTL identification for metabolic traits measured in BB (A) and ZY (B) RIL populations across different environments. The blue bars represent the proportion of metabolic traits that have at least one QTL identified; the red bars represent the proportion of metabolic traits for which epistatic effects were identified.
A QTL located on the short arm of chromosome 1 was identified for about 40% of the flavonoids identified in this study. This locus colocalized with the p (pericarp color) locus on chromosome 1. The p locus is a complex of duplicated MYB-homologous genes p1 and p2 (Zhang et al., 2000). P1 encodes an R2R3-MYB transcription factor in maize, and its main function is to regulate flavonoid biosynthesis (flavonols and 3-deoxyflavonoids) in a pathway that potentially competes with the formation of anthocyanins. Another QTL on chromosome 4 was also frequently identified for flavonoids (mapped for 37.1% flavonoids), which colocated with the known gene c2. C2 encodes the enzyme chalcone synthase, which is responsible for the first dedicated step in the pathway (Wienand et al., 1986). P1 was only detected by QTL mapping using the BB population, while c2 was only detected by QTL mapping using the ZY population. The cob and pericarp color can serve as visible markers for the p1 locus, since B73 (p1-wr) has white pericarp and red cob color while By804 (p1-ww) has white pericarp and cob color. The two parents of the ZY population (i.e. Zong3 and Yu87-1) have an identical phenotype of red cob and white pericarp. These facts also explain the mapping results of p1 from two RIL populations. Other known genes involved in flavonoid biosynthesis were also found within the QTL intervals for flavonoids. For instance, a2 was identified to be associated with naringenin, apigenin C-pentoside, and apigenin C-pentosyl-C-pentoside; b1 and c1 were found within the QTL for apigenin C-pentoside; pr1 was within the QTL for chrysoeriol di-C-hexoside; and r1 was colocated with the QTL for 4-coumaric acid. Significant epistatic interactions between QTLs for flavonoids were frequently identified (Supplemental Tables S3 and S4). Interestingly, significant epistatic interactions between p1 and other QTLs were often observed. The fact that p1 has been identified as a major regulator for a set of genes involved in flavonoid biosynthesis could be one of the reasons, as the transcriptional factor and enzyme interaction can generate this observed genetic epistasis (Grotewold et al., 1998; Kliebenstein, 2009; Morohashi et al., 2012). QTLs containing another R2R3-MYB transcription factor, c1, were also involved in significant epistatic interactions with other QTLs for flavonoids. For example, three QTLs that colocated with a2, b1, and c1 were identified as associated with apigenin C-pentoside, and the QTL containing c1 was in significant epistatic interaction with both of the other two QTLs (Supplemental Tables S3 and S4). Although c1 is not expected to directly control flavones (e.g. apigenin derivatives), the compounds known to be controlled by c1 might compete for naringenin and other pathway intermediates for the formation of flavones. These significant epistatic interactions between QTLs may also be due to the known gene interactions, since extensive genetic and molecular studies have demonstrated that the c1 gene requires a member of the basic helix-loop-helix-containing R or B gene family to activate the transcription of anthocyanin biosynthetic genes such as a2 (Hernandez et al., 2004).
Cross-Validation of Metabolic QTLs and Identification of Candidate Genes
The high-density linkage map generated and utilized in this study helped narrow most of the identified QTLs down to small regions (Supplemental Table S3). Between-RIL population cross-validation of the QTLs facilitates candidate gene selection and will aid in the identification of functional genetic variants. Thirty-two QTLs for 32 metabolites were consistently identified using BB and ZY populations (Supplemental Table S3). Significant SNP metabolic trait associations identified in our previous GWAS on maize kernels offered another resource for cross-validating the present mapping results. In total, 57 QTLs overlapped with loci significantly associated with the same metabolic traits detected in our previous GWAS (Wen et al., 2014; Supplemental Table S3). QTLs that were either consistently identified in both experiments for the same metabolite or cross-validated in different mapping populations or by different mapping approaches will be preferentially selected for future in-depth analysis.
For each QTL identified in this study, candidate genes within the peak bin were annotated and are listed in Supplemental Table S5, providing a database for further study on specific metabolites of interest. Taking advantage of the mapping results, information on genome annotation, and prior knowledge of related metabolic pathways to first distill the most likely genes from the candidate list could be feasible and helpful for revealing the casual genes of the identified QTLs for the metabolites of interest (Wen et al., 2015). Here, we summarize a list of candidate genes for metabolic QTLs that were repeatedly identified or cross-validated using different populations and approaches in Table II. The 35 genes listed in Table II are located within 27 QTLs for 24 metabolites, which are regarded as candidates with high possibility to underlie the identified QTLs. For each gene, the corresponding metabolic trait, physical position in the maize genome, annotation of gene function, and information regarding cross-validation are indicated (Table II; Supplemental Table S5).
Table II. Summary of candidate genes of metabolic QTLs repeatedly identified or cross-validated using different populations and approaches.
| Metabolic Trait | Candidate Gene | Gene Interval | Annotation | Cross-Validation |
|---|---|---|---|---|
| bp | ||||
| l-Pipecolate | GRMZM2G139852 | chr1:24225117-24229054 | Unknown | ZYE1, ZYE2, GWAS |
| l-Met | GRMZM2G450498 | chr1:223161919-223165331 | Met γ-lyase | BBE1, BBE2, ZYE1 |
| 4-Coumaric acid | r1 | chr10:138462252-138471072 | Basic helix-loop-helix DNA-binding | BBE1, ZYE1, ZYE2 |
| Dicoumaroylputrescine | GRMZM2G311059 | chr10:139838952-139839963 | R2R3-MYB domain protein | BBE1, BBE2, ZYE2 |
| LPC(1-acyl 16:1)-2 | GRMZM2G121783 | chr8:171204852-171206144 | RNase III | ZYE1, GWAS |
| Phenolamides | GRMZM2G011655 | chr6:55648330-55654264 | Terpenoid synthase | ZYE2, GWAS |
| LPC(1-acyl 18:1) | GRMZM5G864771 | chr6:166954568-166958991 | Triglyceride lipase | BBE1, ZYE2 |
| GRMZM2G156620 | chr6:165800173-165802545 | β-Ketoacyl-acyl-carrier protein synthaseI | BBE1, ZYE2 | |
| Triornicin | GRMZM2G154687 | chr6:165626650-165628898 | Triacylglycerol lipase, class 3 | BBE1, ZYE2, GWAS |
| Tryptamine | GRMZM2G021277 | chr10:82848693-82850823 | Aromatic-l-amino acid decarboxylase | BBE2, GWAS |
| GRMZM2G021388 | chr10:82837506-82839784 | Aromatic-l-amino acid decarboxylase | BBE2, GWAS | |
| l-Carnitine | GRMZM5G808017 | chr2:193648937-193649473 | Palmitoyl-CoA hydrolase, acyl-CoA hydrolysis | BBE1, BBE2, ZYE1, ZYE2 |
| GRMZM5G860226 | chr2:193644136-193645990 | Palmitoyl-CoA hydrolase, acyl-CoA hydrolysis | BBE1, BBE2, ZYE1, ZYE2 | |
| GRMZM2G395244 | chr2:217644444-217647139 | EF-Hand1, calcium-binding site | ZYE1, ZYE2, GWAS | |
| N-Acetyldopamine | bx1 | chr4:3256234-3258478 | Indole-3-glycerol phosphate lyase | ZYE1, ZYE2 |
| DIMBOA | bx1 | chr4:3256234-3258478 | Indole-3-glycerol phosphate lyase | ZYE1, ZYE2 |
| Acetylintermedine | GRMZM2G032962 | chr1:277141292-277151152 | Chaperone DnaJ domain superfamily protein | BBE1, BBE2, ZYE2 |
| GRMZM2G031138 | chr1:278022167-278026951 | UDP-glucuronosyl/UDP-glucosyltransferase | BBE1, BBE2, ZYE2 | |
| GRMZM2G174570 | chr8:21445191-21449321 | Glutathione metabolism-like domain | ZYE1, ZYE2, GWAS | |
| GRMZM2G371276 | chr8:21295629-21296981 | Calmodulin-dependent protein kinases | ZYE1, ZYE2, GWAS | |
| Trp metabolite | GRMZM2G021277 | chr10:82848693-82850823 | Aromatic-l-amino acid decarboxylase | BBE2, GWAS |
| GRMZM2G021388 | chr10:82837506-82839784 | Aromatic-l-amino acid decarboxylase | BBE2, GWAS | |
| Caffeic acid derivative | GRMZM2G177412 | chr10:144614306-144619343 | Shikimate dehydrogenase | BBE1, ZYE1 |
| Thiamin | GRMZM2G112956 | chr4:156756845-156760798 | NADH dehydrogenase (ubiquinone) | BBE1, ZYE1 |
| N-Coumaroyl-spermidine derivative | GRMZM2G066049 | chr10:1087065-1088682 | Agmatine coumaroyltransferase | BBE2, GWAS |
| GRMZM2G066142 | chr10:1084103-1085734 | Agmatine coumaroyltransferase | BBE2, GWAS | |
| Vitexin | GRMZM2G043295 | chr9:130524722-130526606 | Flavanol-3-O-glucosyltransferase | ZYE1, GWAS |
| Apigenin C-pentosyl-C-pentoside | GRMZM2G059590 | chr8:153858072-153864534 | Unknown | BBE1, GWAS |
| Tricin derivative | GRMZM2G097841 | chr10:143746242-143748475 | Anthocyanidin reductase | BBE1, BBE2, GWAS |
| GRMZM2G097854 | chr10:143738314-143740641 | Leucoanthocyanidin reductase | BBE1, BBE2, GWAS | |
| GRMZM2G431504 | chr10:143752078-143754011 | Leucoanthocyanidin reductase | BBE1, BBE2, GWAS | |
| Chrysoeriol O-rhamnosyl-O-hexoside | p1 | chr1:48117497-48128047 | A-type R2R3-MYB protein | BBE1, BBE2, GWAS |
| N-Feruloylagmatine | GRMZM5G836567 | chr10:137286273-137294014 | Spc97/Spc98 family of spindle pole body component | BBE1, ZYE2, GWAS |
| Methoxylated flavonoid 3-O-hexoside | GRMZM2G104710 | chr10:1919386-1920869 | O-Methyltransferase family protein | BBE1, BBE2, GWAS |
| Methoxylated flavonoid di-O-hexoside | GRMZM2G104710 | chr10:1919386-1920869 | O-Methyltransferase family protein | BBE1, BBE2, GWAS |
Among these 35 genes, several are well characterized in maize. For example, bx1 (benzoxazinless1), which encodes an indole-3-glycerol phosphate lyase, was within a QTL for the level of 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA), and this QTL was identified in both experiments in the ZY population (Table II). DIMBOA is the main benzoxazinoid in maize that confers resistance to herbivores and microbes, and bx1 is a signature gene in the DIMBOA biosynthetic pathway (Meihls et al., 2013; Zheng et al., 2015).
Except for the genes of known function, most candidate genes listed in Table II have a putative function that connects directly to the corresponding metabolite or pathway. For example, a candidate gene annotated as Met γ-lyase was associated with the level of Met, as indicated by linkage mapping using both BB and ZY populations; genes annotated as aromatic-l-amino acid decarboxylase were candidates for the content of tryptamine and another Trp metabolite in maize kernels based on linkage mapping in the BB population and the GWAS result. We also found a couple of candidate genes for the level of multiple flavonoids, although a large number of genes have already been identified for the flavonoid biosynthetic pathway. For instance, a gene encoding a putative O-methyltransferase (OMT) in maize was found by both linkage mapping in the BB population and GWAS for methoxylated flavonoid-3-O-hexoside and methoxylated flavonoid di-O-hexoside (Table II). The gene is an ortholog of AtOMT1 in Arabidopsis, which encodes a flavonol 3′-O-methyltransferease, as indicated by Muzac et al. (2000). Products of other candidate genes for the identified flavonoids here include a putative flavonol-3-O-glucosyltransferase for vitexin content and a putative anthocyanidin or leucoanthocyanidin reductase for the level of tricin derivative (Table II).
Two other genes with unknown function are listed in Table II, which were identified by both GWAS and linkage mapping. The gene corresponding to the peak of the GWAS signal was usually selected if there was no legitimate candidate within that locus. Functional annotation of the gene may ultimately be updated in this case; however, this will necessitate further molecular and biochemical validation, as suggested by Wen et al. (2014).
Regulatory Network of Genes Associated with the Flavonoid Metabolic Pathway
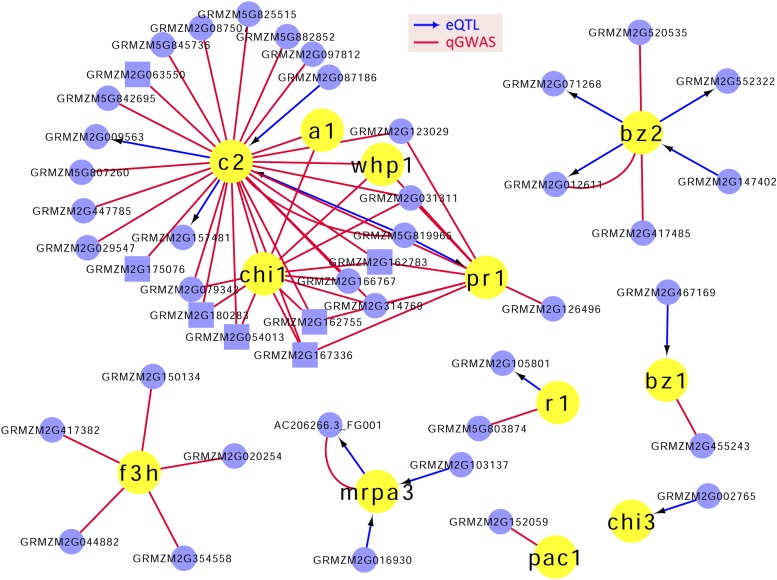
Here, we employed an expression quantitative trait locus (eQTL)- and quantitative genome-wide association study (qGWAS)-based network analysis (see “Materials and Methods”) to search for additional genes that may underlie the identified natural variation of metabolic traits. Flavonoids are plant-specialized metabolites that play important roles in terms of plant growth and development, such as male fertility, UV light protection, and insect pest resistance (Grotewold, 2006; Falcone Ferreyra et al., 2012; Saito et al., 2013; Nakabayashi et al., 2014). Many genes in the flavonol and anthocyanin biosynthetic pathway have been identified and characterized in maize based on genetic and molecular studies of a wide spectrum of mutants (Dooner et al., 1991; Koes et al., 2005). These genes include enzymes, regulatory genes, and transporters (Grotewold et al., 1994; Koes et al., 2005). Expression of 15 of these genes (a1, a2, b1, bz1, bz2, c1, c2, chi1, chi3, f3h, pr1, pac1, mrpa3, r1, and whp1) with known functions was detected in maize kernels at 15 d after pollination of 368 inbred lines according to a previous RNA sequencing study (Fu et al., 2013; Supplemental Table S6). A primary network between all the expressed genes and the 15 genes based on a relationship of transcriptional regulation (revealed by eQTL analysis) or coexpression (revealed by qGWAS) was constructed that contains a total of 58 genes (Fig. 4). Twelve of these 15 genes were present in the primary network, and additionally, five of them formed a subnetwork consisting of a1 (GRMZM2G026930), c2 (GRMZM2G422750), chi1 (GRMZM2G155329), pr1 (GRMZM2G025832), and whp1 (GRMZM2G151227). Two genes (GRMZM2G119186 and GRMZM2G058292) had connectivity with only one node (gene). C2 (GRMZM2G422750) was connected with more nodes than the other 11 genes, indicating its involvement in a larger regulatory network (Fig. 4).
Figure 4.
Primary regulatory network of genes associated with the flavonoid biosynthetic pathway. Yellow circles represent the 12 genes of known function associated with flavonoids. Blue nodes represent newly identified genes, including genes involved in flavonoid biosynthesis (squares) based on functional annotation. Relationships between genes are indicated by blue arrows (revealed by eQTL analysis) or red lines (revealed by qGWAS).
Gene Ontology (GO) term analysis revealed that genes related to lipid biosynthetic and metabolic process and oxidation reduction process were significantly enriched among the 58 genes (Supplemental Table S7). Similar enrichment results were observed when these 12 known flavonoid-related genes were removed in the GO term analysis (Supplemental Table S8). This result coupled with findings from several previous studies may suggest cross talk between the lipid and flavonoid metabolism potentially driven by the fact that acetyl-CoA can act as a substrate for both pathways (Okazaki et al., 2009; Morohashi et al., 2012).
We additionally constructed a secondary network based on the primary network using the same eQTL- and qGWAS-based method. A total of 190 genes were present in the secondary network; the new genes found on top of the genes from the primary network are demarcated as gray nodes in Supplemental Figure S1. The eight subnetworks were expanded but without links between them. Likewise, GO term enrichment analysis was performed on these new genes. However, no significant enrichment was identified in specific biological process, cellular organization, or molecular function (Supplemental Fig. S2).
The primary network provided us a relatively short list of candidate genes for the flavonoid biosynthetic pathway. Notably, a large number of genes identified in this primary network overlapped with the genes found previously by Morohashi et al. (2012). Specifically, the expression of 34 of these 58 genes was modulated by P1, as indicated by RNA sequencing of P1-rr and P1-ww developing pericarps and silks. Moreover, 13 of the 34 genes were identified as putative direct targets of P1 by chromatin immunoprecipitation sequencing analysis, which included four known flavonoid pathway genes (a1, c2, whp1, and mrpa3). Of the remaining nine genes, two putative UDP-glycosyltransferases (GRMZM2G162755 and GRMZM2G063550) and a putative Rha synthase (GRMZM2G031311) were highlighted by Morohashi et al. (2012) for their possible involvement in the flavonoid biosynthetic pathway. Except for the 12 genes of known function, seven genes in the primary network were regarded to have a direct or strong relevance to the flavonoid pathway based on their functional annotations (Fig. 4). Recent studies have examined and updated the functions of two (GRMZM2G167336 and GRMZM2G162783) of these seven genes (Morohashi et al., 2012; Falcone Ferreyra et al., 2013). By transforming WAT11 yeast (Saccharomyces cerevisiae) cells using empty vector and vectors harboring the open reading frame of GRMZM2G167336, Morohashi et al. (2012) identified that this gene product has F2H activity (i.e. is capable of converting naringenin or eriodictyol into the corresponding 2-hydroxyflavanones). The UDP-dependent glycosyltransferase (UGT708A6) encoded by GRMZM2G162783 is a bifunctional enzyme with the ability to form both C-glycoside and O-glycoside derivatives, as confirmed by both in vivo bioconversion assays and in vitro assays (Falcone Ferreyra et al., 2013). Among the remaining five genes, three (GRMZM2G180283, GRMZM2G162755, and GRMZM2G063550) belong to the UDP-glycosyltransferase1 family (Supplemental Fig. S3). These three genes along with GRMZM2G162783 (UGT708A6) were highly up-regulated in P1-rr compared with P1-ww pericarps, and all of them contain the plant secondary product glycosyltransferase motif with 10 conserved amino acids proposed to be involved in the interaction with the UDP-sugar molecule (Morohashi et al., 2012; Falcone Ferreyra et al., 2013). GRMZM2G162755 is orthologous to the rice (Oryza sativa) gene flavonoid-C-6-glucosyltransferase (Os06g18010), which was characterized by Brazier-Hicks et al. (2009; Supplemental Fig. S3).
The candidate glycosyltransferases in the flavonoid biosynthesis identified in this study are in line with previous findings, which together provided a strong foundation to support further investigation of the next step in the pathway. One of the remaining two genes (GRMZM2G175076; annotated as a putative chalcone-flavanone isomerase, orthologous to Arabidopsis CHI-like; Tohge et al., 2013) is homologous to the maize gene chi1 (chalcone-flavonone isomerase; with identity of 34%), which converts chalcone to naringenin (Dong et al., 2001). In addition, the expression of this putative chi (GRMZM2G175076) in pericarp at 25 d after pollination was controlled by P1, as indicated by Morohashi et al. (2012). The other gene (GRMZM2G054013) is orthologous to a well-characterized gene in Arabidopsis (4cl3; with identity of 56%), which encodes 4-coumarate-CoA-ligase3. 4CL3 is involved in the last step of the general phenylpropanoid pathway and transcriptionally regulated by a flavonoid MYB transcription factor in Arabidopsis and tomato (Solanum lycopersicum). Expression of this putative 4cl3 (GRMZM2G054013) in silk was also controlled by P1 (Morohashi et al., 2012). Notably, three of these seven genes were also identified within the QTL intervals of flavonoids identified in this study. For instance, GRMZM2G054013 (annotated as 4-coumarate-CoA-ligase3) was found in the peak bin of a QTL for tricin O-rhamnosyl-O-hexoside. Both GRMZM2G162755 and GRMZM2G063550 belong to UDP-glycosyltransferase gene family 1, and they were identified in the peak bin of a QTL interval for C-pentosyl-apigenin O-caffeoylhexoside and apigenin di-C-hexoside, respectively (Supplemental Table S5). Genetic and/or transcriptional network analyses in this study identified various candidate genes and provided certain evidence for their possible function in the pathway, which can lead to a more complete understanding of flavonoid biosynthesis. Further molecular and biochemical studies are required to fully understand how these newly identified genes are involved in the flavonoid pathway.
DISCUSSION
There are more and more forward genetic studies on a broad range of metabolic traits in plants, including major crops such as maize and rice (Keurentjes et al., 2006; Lisec et al., 2008; Rowe et al., 2008; Riedelsheimer et al., 2012; Gong et al., 2013; Sauvage et al., 2014; Alseekh et al., 2015; Zhang et al., 2015). In most cases, the effects of these QTLs are modest. In this study, several hundred metabolic QTLs were identified and the effects of both single locus and pairwise epistatic interactions on the metabolic variation were assessed. The effect size of each individual QTL was generally modest (r2 < 10%); however, a reasonable metabolite QTL (17.6% for BB and 33.7% for ZY) was of major effect (r2 > 10%). Epistasis was identified for more than 30% of metabolites here, implying that it might also play broad and important roles in determining metabolic variation. The influence of epistasis in shaping the metabolic variation, however, was already highlighted in previous studies (Rowe et al., 2008; Gong et al., 2013; Wen et al., 2015).
Maize is a widely grown cereal crop mainly for food and feed; thus, increasing its yield while providing added nutritional value is becoming ever more important. Many metabolites detected in this study have important roles as nutrients. For instance, flavones have potent antiinflammatory and anticarcinogenic activities (Falcone Ferreyra et al., 2013; Casas et al., 2014). Obtaining the genetic architecture and regulatory network information for the target nutritional metabolites here is a crucial step toward a sustainable strategy of maize grain quality improvement (Martin et al., 2011; Fitzpatrick et al., 2012). Furthermore, the deeply characterized RIL populations used here are attractive resources not only for powerful and high-resolution genetic analysis of maize traits but also for providing valuable plant materials and genes for better nutritional maize breeding. Moreover, major QTLs or genes affecting carotenoids, tocopherols, and fatty acids have already been mapped and cloned in the BB population, and some have been used directly for breeding practices (Chander et al., 2008a, 2008b; Yan et al., 2010; Yang et al., 2010; Chai et al., 2012; Li et al., 2013). For example, the gene crtRB1 (β-carotene hydroxylase 1), which was a major QTL in the BB population, explaining about 16% of variation of the provitamin A content in mature maize kernel, has been widely used for maize provitamin A biofortification in order to take up the global challenge of vitamin A deficiency (Yan et al., 2010). Another major QTL in the BB population, DGAT1 (Diacylglycerol O-acyltransferase 1), could increase about 20% of the relative total oil content in a regular maize line by marker-assisted backcross selection (Chai et al., 2012).
Our QTL analysis demonstrated that the p locus as a regulator explained a great proportion of phenotypic variation for a large amount of flavonoids. In addition, significant epistatic interactions between p1 and other QTLs were observed frequently. Epistatic interactions between p and structural genes within the flavonoid pathway (e.g. a1 and c2) were also found previously by Szalma et al. (2005). These findings suggest that either further functional investigation of the flavonoid structural genes or pyramiding favorable alleles in maize improvement will require careful genetic interaction analysis and examination of the p genotype. The molecular mechanisms that underlie genetic interactions remain an open question. However, there is a long way to go from QTL-QTL interaction to gene-gene interaction, although our results here provide some useful information for further elaboration of the genetic and molecular mechanisms. Causal gene identification is ultimately the key step in this process. The maize flavonoid pathway provides a wonderful system for studying gene interaction and regulatory mechanisms, owing to its extensive characterization at the genetic, biochemical, and molecular levels (Koes et al., 2005). We found several QTL-QTL interactions in this study that could be explained by the known physical interactions between the underlying genes. Regulatory or enzymatic interactions can generate genetic epistasis through functional epistasis, as indicated by previous studies (Byrne et al., 1998; McMullen et al., 1998; Kliebenstein, 2009). However, it remains challenging to determine and validate the biologically important interactions that are indicated by statistical analysis. For large-scale metabolomics studies, the evidence of genetic interaction can also be used to aid in network generation and extension, as demonstrated by Rowe et al. (2008).
The biochemical nature of metabolic traits and the relatively large amount of prior knowledge facilitate the dissection of the molecular basis of metabolic variation. Improving the resolution of QTL mapping can definitely enhance the efficiency of causal gene identification, which also benefits from combining different forward genetic approaches such as combining linkage with genome-wide association mapping, as illustrated in this study. The eQTL- and qGWAS-based gene expression network analysis identified novel genes associated with the flavonoid pathway, whose role could be rationalized with regard to their functional annotation. Compared with the more routine coexpression analysis, the eQTL and qGWAS method is considerably more stringent and, as such, can highly reduce false-positive findings, especially in the primary network. The secondary network could also be considered, but the effect of the newly identified genes may not be as large as that of the genes in the primary network. This result of the network analysis, which is based on transcriptional data, offered another source for cross-validation. Combing quantitative genetics approaches and transcriptional network analysis, as shown in this study, represents an alternative route to dissect the genetic basis of metabolic traits and, as such, an effective strategy to prioritize candidate genes prior to embarking on laborious transgenic validations. The deeply characterized RIL populations, elucidation of metabolic phenotypes, and identification of QTLs as well as novel candidate genes lay the foundation for maize quality improvement.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Two RIL populations of maize (Zea mays), BB (Chander et al., 2008a) and ZY (Ma et al., 2007), were used for linkage analysis in this study. The 197 BB RIL lines were planted in Hainan (Sanya; E 109°51′, N 18°25′) in 2010 (experiment 1 BBE1) and Henan (Zhengzhou; E 113°42′, N 34°44′) in 2011 (experiment 2 BBE2). The 197 RILs from the ZY cross were planted in Yunnan (Kunming; E 102°30′, N 24°25′; experiment 1 ZYE1) and Henan (Zhengzhou; E 113°42′, N 34°44′; experiment 2 ZYE2) in 2011. All lines were self-pollinated, and ears of each plot were hand harvested after maturity, air dried, and shelled. For each line, ears from five plants were harvested at the same maturity and bulked.
Sample Preparation and Metabolite Profiling
We carried out metabolic profiling on mature maize kernels from lines of the two RIL populations. For each line, 12 well-grown kernels were randomly selected from five plants and bulked for grinding. The kernels were ground using a mixer mill (MM 400; Retsch) with zirconia beads for 2 min at 30 Hz. The powder of each genotype was partitioned into two sample sets and stored at −80°C until required for extraction. Samples were extracted following the procedures described in detail in our previous study (Wen et al., 2014) before analysis using a liquid chromatography-electrospray ionization (ESI)-MS/MS system. The metabolite annotation and quantification were performed as described previously (Wen et al., 2014). Briefly, an tandem mass spectral tag (MS2T) library containing 983 (almost) nonredundant metabolite signals was constructed for mature maize kernels. An accurate mass of 245 of Q1 was obtained on the basis of the fact that similar fragmentation patterns were obtained between ESI-quadrupole trap-MS/MS and ESI-quadrupole-quadrupole-time-of-flight-MS/MS. The MS2T library was annotated based on the fragmentation pattern (delivered by ESI-quadrupole trap-MS/MS and/or the accurate mass-to-charge ratio value delivered by ESI-quadrupole-quadrupole-time-of-flight-MS/MS) and the retention time of each metabolite. By comparing the mass-to-charge ratio values, the retention times, and the fragmentation patterns with the authentic standards, 43 metabolites were identified, including amino acids, flavonoids, lysophosphatidylcholine and fatty acids, and some phytohormones (Supplemental Table S1). For the metabolites that could not be identified by available standards, peaks in the MS2T library, especially the peaks that have similar fragmentation patterns to the metabolites identified by authentic standards, were used to query the MS/MS spectral data taken from the literature or to search the databases (MassBank, KNApSAcK, HMDB, MoTo DB, and METLIN). Best matches were then searched in the Dictionary of Natural Products and the Kyoto Encyclopedia of Genes and Genomes for possible structures. In total, 155 metabolites were identified in the samples harvested from two experiments of both populations. A scheduled multiple reaction monitoring method was used for the quantification of metabolites (Dresen et al., 2010). The metabolite intensities of each line measured from two experiments of both BB and ZY populations are shown in Supplemental Table S2. The fold change was calculated as the ratio of the maximum intensity to the minimum intensity for each metabolite across all the lines. H2 was calculated using the equation H2 = Vg/Vg + Ve + Ver (Holland et al., 2003).
Genotyping and Construction of a High-Density Bin Map
Both RIL populations have been genotyped using the Illumina MaizeSNP50 BeadChip, which contains 56,110 SNPs (Ganal et al., 2011). SNPs with both missing rate and heterozygosity of less than 10% for the RIL populations were used to construct the genetic linkage map. We developed an economic go-wrong method integrating the Carthagene software (de Givry et al., 2005) to construct the genetic linkage map for both RIL populations. Detailed information of the method for constructing the linkage map can be accessed at http://github.com/panqingchun/linkage_map.
QTL Mapping and Epistasis Analysis
A map containing 2,496 and 3,071 recombinant bins was constructed for BB and ZY RILs, respectively. Composite interval mapping implemented in Windows QTL Cartographer version 2.5 was used for QTL identification (Zeng et al., 1999; Wang et al., 2006). Zmap (model 6) with a 10-cM window and a walking speed of 0.5 cM was used. To determine a threshold for significant QTLs, 500 permutations (P = 0.05) were used for each metabolite identified in both RIL populations. The bins were clearly defined, and a uniform LOD value was assigned for each bin. The confidence interval for each QTL was assigned as a 2 LOD drop from the peak. Detailed information, including location, confidence interval, and r2 (explained phenotypic variance) of each QTL for each trait, is shown in Supplemental Table S2.
The pairwise additive-by-additive epistatic interactions for all identified QTLs of each metabolite were determined by two-way ANOVA (using P < 0.05 as a significant threshold; Yu et al., 1997). The proportion of variance explained by epistasis was tested by comparing the residual of the full model containing all single-locus effects and two-locus interaction effects with that of the reduced model containing just all single-locus effects but excluding two-locus interaction effects.
Candidate Gene Identification
Candidate genes associated with the corresponding metabolic trait were searched within the confidence interval for each QTL. We found annotated gene models within the confidence interval according to the 5b filtered set (ftp://ftp.gramene.org/pub/gramene/maizesequence.org/release-5b/filtered-set/).
Construction of the Regulatory Network for a Metabolic Pathway Based on eQTL and qGWAS Methodologies
Genes of known function that are associated with specific metabolic pathways were selected. Using the flavonoid pathway as an example in this study, we used 15 genes with well-known functions in this pathway as bait and identified genes with regulatory relationships with these genes. For the primary network, genes that met one of the following criteria were included: first, the expression levels of genes that were significantly associated with genetic variations in these 15 genes using eQTL analysis or vice versa; and second, expression levels of genes that were significantly associated with expression variation of these 15 genes using qGWAS or vice versa. Expression data of 28,769 genes were used based on our previous RNA sequencing analysis on maize kernels at 15 d after pollination (Fu et al., 2013). For eQTL mapping, the expression level of each gene was considered as a trait to associate with 0.56 million genome-wide SNP markers based on a natural population containing 368 maize inbred lines (Fu et al., 2013; Wen et al., 2014). The qGWAS method integrated the expression data of two genes in a regression model accounting for the population structure. The same population (i.e. natural maize population containing 368 maize inbred lines) was used for qGWAS. Based on the genes in the primary network, a secondary network was constructed using the same eQTL and qGWAS criteria. The genes associated directly with the 15 candidates are named as first rank, while second rank genes are those that were linked to the first rank. Cytoscape (Shannon et al., 2003) was used to display the network, and GO enrichment analyses were performed using the agriGO toolkit (Du et al., 2010), with a false discovery rate of 0.05 or less being applied as a threshold.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AFW85719.1 (UGT1, GRMZM2G063550), NP_001132650.2 (UGT708A6, GRMZM2G162783), NP_001147999.1 (UGT1, GRMZM2G162755), XP_008660140.1 (F2H, GRMZM2G167336), XP_008667488.1 (UGT1, GRMZM2G180283), NP_001151452.1 (CHI, GRMZM2G175076), and AFW63473.1 (4CL3, GRMZM2G054013).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Secondary regulatory network associated with flavonoid biosynthesis.
Supplemental Figure S2. Gene Ontology enrichment analysis of the newly found genes in the secondary network.
Supplemental Figure S3. Phylogenetic analysis of candidate genes belonging to the UGT1 family.
Supplemental Table S1. Detailed information of 155 metabolites detected in this study.
Supplemental Table S2. Metabolite intensities of each line in both BB and ZY populations.
Supplemental Table S3. Detailed information of all the identified QTLs.
Supplemental Table S4. Significant pair-wise epistatic interactions.
Supplemental Table S5. Candidate genes and their detailed information for each QTL.
Supplemental Table S6. Expression level of 15 known flavonoid pathway genes in the 368 inbred lines.
Supplemental Table S7. Gene Ontology enrichment analysis on all the 58 genes in the primary network.
Supplemental Table S8. Gene Ontology enrichment analysis on the newly found 46 genes in the primary network.
Supplementary Material
Glossary
- QTL
quantitative trait locus
- GWAS
genome-wide association study
- RIL
recombinant inbred line
- SNP
single-nucleotide polymorphism
- MS/MS
tandem mass spectrometry
- BB
B73/By804
- ZY
Zong3/Yu87-1
- cM
centimorgan
- H2
broad-sense heritability
- LOD
logarithm of odds
- DIMBOA
2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one
- eQTL
expression quantitative trait locus
- qGWAS
quantitative genome-wide association study
- GO
Gene Ontology
- ESI
electrospray ionization
Footnotes
This work was supported by the National Program on Key Basic Research Projects of China (grant nos. 2013CB127003 and 2014CB138202), the National Natural Science Foundation of China (grant nos. 31201220 and 31222041), and the Max Planck Institute of Molecular Plant Physiology (fellowship to W.W.).
Articles can be viewed without a subscription.
References
- Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, Kleessen S, Giavalisco P, Pleban T, Mueller-Roeber B, et al. (2015) Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne PF, McMullen MD, Wiseman BR, Snook ME, Musket TA, Theuri JM, Widstrom NW, Coe EH (1998) Maize silk maysin concentration and corn earworm antibiosis: QTLs and genetic mechanisms. Crop Sci 38: 461–471 [Google Scholar]
- Casas MI, Duarte S, Doseff AI, Grotewold E (2014) Flavone-rich maize: an opportunity to improve the nutritional value of an important commodity crop. Front Plant Sci 5: 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai YC, Hao XM, Yang XH, Allen WB, Li JM, Yan JB, Shen B, Li JS (2012) Validation of DGAT1-2 polymorphisms associated with oil content and development of functional markers for molecular breeding of high-oil maize. Mol Breed 29: 939–949 [Google Scholar]
- Chander S, Guo Y, Yang X, Yan J, Zhang Y, Song T, Li J (2008a) Genetic dissection of tocopherol content and composition in maize grain using quantitative trait loci analysis and the candidate gene approach. Mol Breed 22: 353–365 [Google Scholar]
- Chander S, Guo YQ, Yang XH, Zhang J, Lu XQ, Yan JB, Song TM, Rocheford TR, Li JS (2008b) Using molecular markers to identify two major loci controlling carotenoid contents in maize grain. Theor Appl Genet 116: 223–233 [DOI] [PubMed] [Google Scholar]
- Chander S, Meng Y, Zhang Y, Yan J, Li J (2008c) Comparison of nutritional traits variability in selected eighty-seven inbreds from Chinese maize (Zea mays L.) germplasm. J Agric Food Chem 56: 6506–6511 [DOI] [PubMed] [Google Scholar]
- de Givry S, Bouchez M, Chabrier P, Milan D, Schiex T (2005) CARHTA GENE: multipopulation integrated genetic and radiation hybrid mapping. Bioinformatics 21: 1703–1704 [DOI] [PubMed] [Google Scholar]
- Dong X, Braun EL, Grotewold E (2001) Functional conservation of plant secondary metabolic enzymes revealed by complementation of Arabidopsis flavonoid mutants with maize genes. Plant Physiol 127: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner HK, Robbins TP, Jorgensen RA (1991) Genetic and developmental control of anthocyanin biosynthesis. Annu Rev Genet 25: 173–199 [DOI] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresen S, Ferreirós N, Gnann H, Zimmermann R, Weinmann W (2010) Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal Bioanal Chem 396: 2425–2434 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius SP, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rodriguez E, Casas MI, Labadie G, Grotewold E, Casati P (2013) Identification of a bifunctional maize C- and O-glucosyltransferase. J Biol Chem 288: 31678–31688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Klee HJ (2011) The use of natural genetic diversity in the understanding of metabolic organization and regulation. Front Plant Sci 2: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Schauer N (2009) Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet 25: 39–48 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P, Carrari F, DellaPenna D, Fraser PD, Hellmann H, Osorio S, Rothan C, Valpuesta V, et al. (2012) Vitamin deficiencies in humans: can plant science help? Plant Cell 24: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z, et al. (2013) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4: 2832. [DOI] [PubMed] [Google Scholar]
- Ganal MW, Durstewitz G, Polley A, Bérard A, Buckler ES, Charcosset A, Clarke JD, Graner EM, Hansen M, Joets J, et al. (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6: e28334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J (2013) Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA 110: 20320–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57: 761–780 [DOI] [PubMed] [Google Scholar]
- Grotewold E, Chamberlin M, Snook M, Siame B, Butler L, Swenson J, Maddock S, St Clair G, Bowen B (1998) Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10: 721–740 [PMC free article] [PubMed] [Google Scholar]
- Grotewold E, Drummond BJ, Bowen B, Peterson T (1994) The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76: 543–553 [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Heine GF, Irani NG, Feller A, Kim MG, Matulnik T, Chandler VL, Grotewold E (2004) Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J Biol Chem 279: 48205–48213 [DOI] [PubMed] [Google Scholar]
- Holland JB, Nyquist WE, Cervantes-Martinez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22: 9–111 [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LH, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Kliebenstein D. (2009) Advancing genetic theory and application by metabolic quantitative trait loci analysis. Plant Cell 21: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10: 236–242 [DOI] [PubMed] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, et al. (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45: 43–50 [DOI] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. (2015) Metabolite-based genome-wide association studies in plants. Curr Opin Plant Biol 24: 31–38 [DOI] [PubMed] [Google Scholar]
- Ma XQ, Tang JH, Teng WT, Yan JB, Meng YJ, Li JS (2007) Epistatic interaction is an important genetic basis of grain yield and its components in maize. Mol Breed 20: 41–51 [Google Scholar]
- Martin C, Butelli E, Petroni K, Tonelli C (2011) How can research on plants contribute to promoting human health? Plant Cell 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullen MD, Byrne PF, Snook ME, Wiseman BR, Lee EA, Widstrom NW, Coe EH (1998) Quantitative trait loci and metabolic pathways. Proc Natl Acad Sci USA 95: 1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. (2013) Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Dudley JW, Rocheford TR (2004) Maize selection passes the century mark: a unique resource for 21st century genomics. Trends Plant Sci 9: 358–364 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, Mejía-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, Rodriguez E, et al. (2012) A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzac I, Wang J, Anzellotti D, Zhang H, Ibrahim RK (2000) Functional expression of an Arabidopsis cDNA clone encoding a flavonol 3′-O-methyltransferase and characterization of the gene product. Arch Biochem Biophys 375: 385–388 [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Shimojima M, Sawada Y, Toyooka K, Narisawa T, Mochida K, Tanaka H, Matsuda F, Hirai A, Hirai MY, et al. (2009) A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21: 892–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Ali F, Yang X, Li J, Yan J (2012) Exploring the genetic characteristics of two recombinant inbred line populations via high-density SNP markers in maize. PLoS ONE 7: e52777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA 109: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Sauvage C, Segura V, Bauchet G, Stevens R, Do PT, Nikoloski Z, Fernie AR, Causse M (2014) Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol 165: 1120–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalma SJ, Buckler ES IV, Snook ME, McMullen MD (2005) Association analysis of candidate genes for maysin and chlorogenic acid accumulation in maize silks. Theor Appl Genet 110: 1324–1333 [DOI] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol 48: 123–152 [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2006) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, et al. (2015) Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell 27: 1839–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienand U, Weydemann U, Niesbach-Klösgen U, Peterson PA, Saedler H (1986) Molecular cloning of the c2 locus of Zea mays, the gene coding for chalcone synthase. Mol Gen Genet 203: 202–207 [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, Bai L, Kim EH, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, et al. (2010) Rare genetic variation at Zea mays crtRB1 increases beta-carotene in maize grain. Nat Genet 42: 322–327 [DOI] [PubMed] [Google Scholar]
- Yang X, Guo Y, Yan J, Zhang J, Song T, Rocheford T, Li JS (2010) Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet 120: 665–678 [DOI] [PubMed] [Google Scholar]
- Yu SB, Li JX, Xu CG, Tan YF, Gao YJ, Li XH, Zhang Q, Saghai Maroof MA (1997) Importance of epistasis as the genetic basis of heterosis in an elite rice hybrid. Proc Natl Acad Sci USA 94: 9226–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZB, Kao CH, Basten CJ (1999) Estimating the genetic architecture of quantitative traits. Genet Res 74: 279–289 [DOI] [PubMed] [Google Scholar]
- Zhang N, Gibon Y, Wallace JG, Lepak N, Li P, Dedow L, Chen C, So YS, Kremling K, Bradbury PJ, et al. (2015) Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiol 168: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Chopra S, Peterson T (2000) A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12: 2311–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, McMullen MD, Bauer E, Schön CC, Gierl A, Frey M (2015) Prolonged expression of the BX1 signature enzyme is associated with a recombination hotspot in the benzoxazinoid gene cluster in Zea mays. J Exp Bot 66: 3917–3930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.