Two homologous kinase inhibitor proteins play differential roles in regulating plant cell cycle and defense.
Abstract
Precise cell-cycle control is critical for plant development and responses to pathogen invasion. Two homologous cyclin-dependent kinase inhibitor genes, SIAMESE (SIM) and SIM-RELATED 1 (SMR1), were recently shown to regulate Arabidopsis (Arabidopsis thaliana) defense based on phenotypes conferred by a sim smr1 double mutant. However, whether these two genes play differential roles in cell-cycle and defense control is unknown. In this report, we show that while acting synergistically to promote endoreplication, SIM and SMR1 play different roles in affecting the ploidy of trichome and leaf cells, respectively. In addition, we found that the smr1-1 mutant, but not sim-1, was more susceptible to a virulent Pseudomonas syringae strain, and this susceptibility could be rescued by activating salicylic acid (SA)-mediated defense. Consistent with these results, smr1-1 partially suppressed the dwarfism, high SA levels, and cell death phenotypes in acd6-1, a mutant used to gauge the change of defense levels. Thus, SMR1 functions partly through SA in defense control. The differential roles of SIM and SMR1 are due to differences in temporal and spatial expression of these two genes in Arabidopsis tissues and in response to P. syringae infection. In addition, flow-cytometry analysis of plants with altered SA signaling revealed that SA is necessary, but not sufficient, to change cell-cycle progression. We further found that a mutant with three CYCD3 genes disrupted also compromised disease resistance to P. syringae. Together, this study reveals differential roles of two homologous cyclin-dependent kinase inhibitors in regulating cell-cycle progression and innate immunity in Arabidopsis and provides insights into the importance of cell-cycle control during host-pathogen interactions.
Properly controlled cell-cycle progression is critical for plant growth and development (Inzé and De Veylder, 2006; De Veylder et al., 2011; Polyn et al., 2015). The cell-cycle machinery has recently been shown to be important for plant defense (Bao et al., 2013; Chandran et al., 2014; Wang et al., 2014b; Bao and Hua, 2015). However, how the host cell-cycle machinery is modulated during host-pathogen interactions has not been completely understood.
Mitotic cell cycle is typically divided into five major phases: quiescent phase (G0), postmitotic interphase (G1), DNA synthesis phase (S), postsynthetic interphase (G2), and mitosis (M). Transitions between phases in the cell cycle are tightly regulated at checkpoints in plants, animals, and yeast (Harashima et al., 2013). Heterodimeric protein kinase complexes, which consist of catalytic subunits (cyclin-dependent kinases, or CDKs) and regulatory subunits (cyclins, or CYCs), are the main gatekeepers of the checkpoints. Activities of CYC/CDK complexes can be regulated at multiple levels (Potuschak and Doerner, 2001; Berckmans and De Veylder, 2009; Harashima et al., 2013). For instance, CYCs are prone to degradation via ubiquitin-mediated pathways involving at least two types of ubiquitin E3 ligases, the anaphase-promoting complex and the Skp1/Cullin/F-box related complex (Vodermaier, 2004; Heyman and De Veylder, 2012). Activities of CYC/CDK complexes can also be regulated by CDK-activating kinases and CDK inhibitors (CKIs; Sherr and Roberts, 1999; Umeda et al., 2005; De Clercq and Inzé, 2006). Arabidopsis (Arabidopsis thaliana) has two classes of CKIs, the Kip-related proteins (KRPs), which are similar to the mammalian Kip/Cip proteins (De Veylder et al., 2001), and SIAMESE/SIAMESE-RELATED (SIM/SMR) proteins, which are more distantly related to KRPs and are plant specific (Churchman et al., 2006; Peres et al., 2007).
Active CYC/CDK complexes can target downstream cell-cycle core components to trigger specific cell-cycle events. For instance, a CDKA/CYCD complex can phosphorylate the retinoblastoma-related protein (RBR), and such a modification releases the binding of RBR to the transcription factors E2F, subsequently activating E2F and promoting expression of genes necessary for the transition from G1 to S phase, while CYCA- and CYCB-containing CDK complexes trigger entry into mitosis. However, when activities of mitotic CYC/CDK complexes are suppressed, the cell cycle is reprogrammed to enter endoreplication in which multiple rounds of DNA replication occur without subsequent mitosis and cytokinesis (De Veylder et al., 2011). Endoreplication, also called endoreduplication, endocycling, or endoploidization, is a common variant of the cell cycle of many cell types during plant development (Bramsiepe et al., 2010; De Veylder et al., 2011).
Many studies have also linked cell-cycle control with plant responses to pathogen invasions. Pathogens, such as the bacteria Pseudomonas syringae and Xanthomonas citri, the actinomycete Rhodococcus fascians, the fungal pathogen powdery mildew, and nematodes, can induce endoreplication in some cells at or near the infected loci of host tissue (Swarup et al., 1991; Chandran et al., 2010; Wildermuth, 2010; de Almeida Engler and Gheysen, 2013; Hamdoun et al., 2013). Consistent with pathogen-induced endoreplication in the host, some pathogen effectors are known to interact with host cell-cycle components, either through direct physical protein-protein interaction and/or through modulating expression of genes critical for cell-cycle progression (Kay et al., 2007; Mukhtar et al., 2011). Expression of several core cell-cycle genes of plants was also known to be induced or suppressed during infections (Niebel et al., 1996; de Almeida Engler et al., 1999; Favery et al., 2002; Ascencio-Ibáñez et al., 2008; Chandran et al., 2010). In addition, several core cell-cycle genes were shown to be important for plant defense against pathogens (Favery et al., 2002; Depuydt et al., 2009a, 2009b; Bao et al., 2013; Chandran et al., 2014; Wang et al., 2014b). Thus, cell-cycle control is intimately interconnected with plant defense responses.
Among the defense-related core cell-cycle genes, SIM and SMR1 belong to a plant-specific CKI family with 17 members, functions of which have not been well understood (Yi et al., 2014). SIM is the founding member of the family and was previously studied for its role in trichome development and endoreplication in trichome cells (Walker et al., 2000; Churchman et al., 2006). Leaves of wild-type plants have unicellular trichomes, and each has three to four branches and a single nucleus containing 16 to 32 C-value of DNA. In contrast, most trichomes of sim loss-of-function mutants are multicellular and consist of up to 15 cells with significantly reduced nuclear DNA content (Walker et al., 2000; Churchman et al., 2006). Thus, SIM was proposed to be a positive regulator of endoreplication in trichomes. SMR1, also called LOSS OF GIANT CELLS FROM ORGANS, is the closest homolog of SIM in the SMR family (Roeder et al., 2010). SMR1 affects the formation of giant endoreplicated pavement cells on Arabidopsis sepals (Roeder et al., 2010). However, the role of SMR1 in trichome development has not been well studied. Recently, a sim smr1 double mutant was shown to have compromised responses to P. syringae and the oomycete pathogen Hyaloperonospera arabidopsidis (Wang et al., 2014b). However, the individual single mutants were not analyzed in this study. It is likely that the observed defense phenotypes in the sim smr1 double mutant are due to the synergistic effect of the two genes. It is also possible that these two genes could play differential roles in affecting Arabidopsis defense. Therefore, it is important to further elucidate the roles of SIM and SMR1 in regulating defense and cell-cycle progression in order to gain better understanding of functions of these two genes.
In this report, we analyzed mutants impaired in SIM, SMR1, or both genes for cell-cycle and defense phenotypes. We found that SIM promotes endoreplication predominately in the trichome while SMR1 in nontrichome leaf cells. The two genes also act synergistically to affect endoreplication in leaf cells. In addition, we found that that SMR1 plays a greater role than SIM in regulating resistance to P. syringae. The defense function of SMR1 is at least partly through signaling mediated by salicylic acid (SA). Such differential roles of SIM and SMR1 are most likely due to differential expression of these genes in Arabidopsis tissues and in response to pathogen infection. On the other hand, cell ploidy analyses of SA mutants and plants treated with an SA analog indicate that SA signaling is necessary, but not sufficient, to disrupt cell-cycle progression. Interestingly, similar to sim smr1 that has reduced endoreplication, a mutant with three CYCD3 genes disrupted that has increased endoreplication also showed compromised disease resistance to P. syringae. In addition, we found that expression of SMR1 and several cell-cycle genes were suppressed under defense conditions, suggesting a negative feedback regulation between cell-cycle progression and defense activation. Together, our study reveals differential roles of two homologous CKIs in regulating cell-cycle progression and innate immunity in Arabidopsis and provides important insight into the role of cell cycle in affecting host-pathogen interactions.
RESULTS
The SIM and SMR1 Genes Act Synergistically to Affect Trichome Development
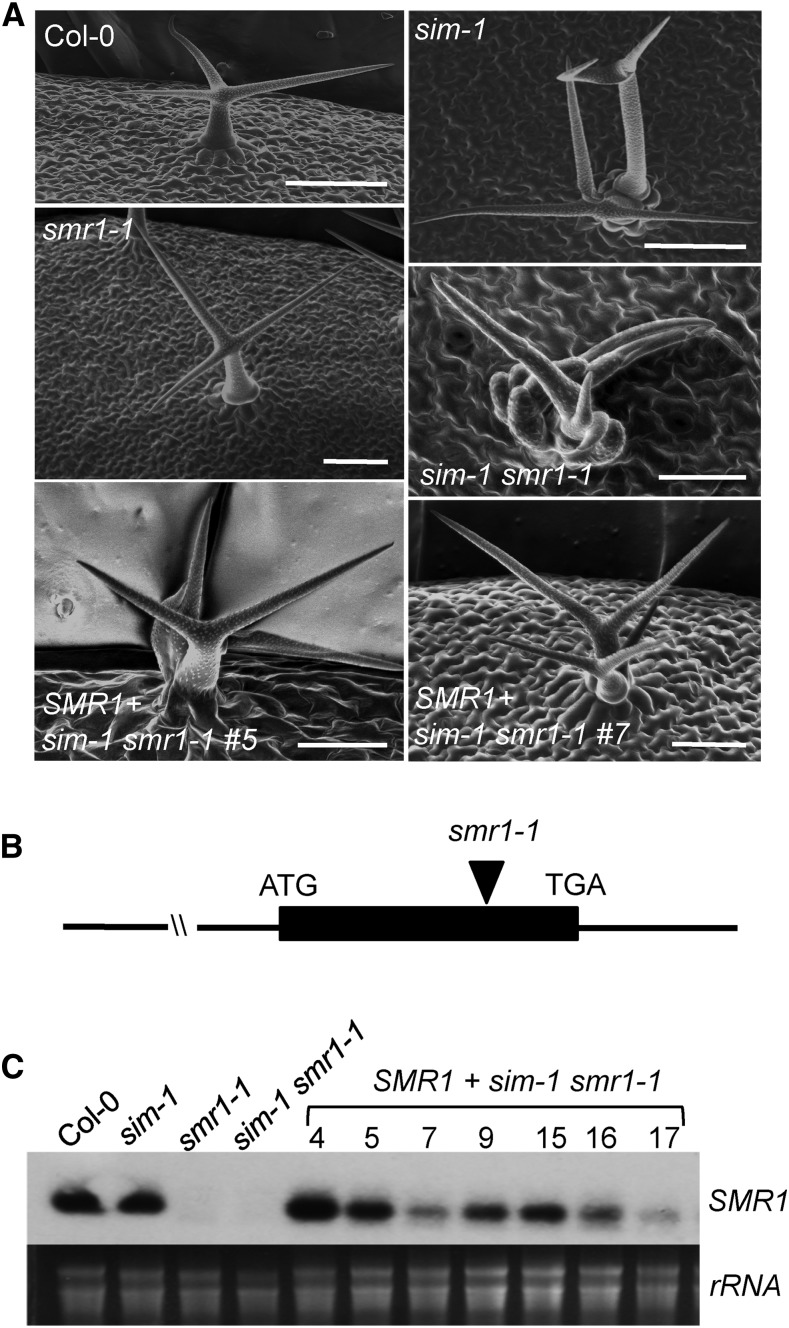
Wild-type Columbia-0 (Col-0) plants form single trichomes, each consisting of a single cell with multiple branches. In contrast, most trichomes of the sim-1 mutant are multicellular, with about 2.5 cells per trichome (Fig. 1A; Supplemental Figs. S1 and S2; Table I; Walker et al., 2000; Churchman et al., 2006). Trichomes of sim-1 have reduced DNA content than those of Col-0, leading to the conclusion that SIM acts as a positive regulator of endoreplication in trichomes (Walker et al., 2000; Churchman et al., 2006). Among the SMR family members, SMR1 exhibits the highest homology to SIM with 62% identity at the amino acid level. However, a null mutation caused by a T-DNA insertion in the SMR1 gene, smr1-1, did not affect trichome morphology. Like Col-0, the trichomes of smr1-1 were unicellular (Fig. 1; Supplemental Figs. S1 and S2; Table I). On the other hand, the double mutant sim-1 smr1-1 produced much smaller and deformed trichomes than sim-1 along. A closer inspection revealed that trichomes of sim-1 smr1-1 branched from the bases of the trichome initiation sites and contained more than twice the number of cells per initiation site as in sim-1. On the other hand, smr1-1 and Col-0 had a similar number of cells per trichome initiation site (Fig. 1A; Table I).
Figure 1.
Rescue sim-1 smr1-1-conferred phenotypes by a SMR1 genomic fragment. A, Scanning electron micrographs. Two-week-old plants were observed with a scanning electron microscope. Bars in Col-0, sim-1, and smr1-1 are 100 μm. Bars in sim-1 smr1-1 and the lines 5 and 7 are 50 μm. B, The SMR1 construct used for the rescue experiment. The position of the smr1-1 mutation was indicated. C, Expression of SMR1 in transgenic sim-1 smr1-1 plants. A construct containing a 2367 bp SMR1 genomic fragment was used to transform the sim-1 smr1-1 mutant, and independently transformed homozygous lines (4, 5, 7, 9, 15, 16, and 17) were obtained. Total RNA was extracted and analyzed by northern blotting. These experiments were repeated twice with similar results.
Table I. SMR1 rescued the multicellular phenotype of the sim-1 smr1-1 double mutant.
Nuclei/TIS, Nuclei per trichome initiation site, a measure of the number of cells per trichome initiation site. Statistical analysis was performed with one-way ANOVA. Letters indicate significant difference among the samples (P < 0.05).
| Genotype | Nuclei/TIS | No. of TIS |
|---|---|---|
| Col-0 | 1.0 ± 0 a | 50 |
| sim-1 | 2.5 ± 1.1 b | 50 |
| smr1-1 | 1.0 ± 0 a | 50 |
| sim-1 smr1-1 | 5.9 ± 2.1 c | 50 |
| SMR1+sim-1 smr1-1 #5 | 2.2 ± 1.1 b | 50 |
| SMR1+sim-1 smr1-1 #7 | 2.8 ± 1.3 b | 50 |
We were able to rescue the sim-1 smr1-1 mutant with a SMR1 genomic fragment, including sequences of 1424 bp 5′ end upstream and 637 bp 3′ end downstream of the SMR1 coding region (Fig. 1B). The transgenic sim-1 smr1-1 plants expressed variable levels of the SMR1 transgene and were rescued for the severe trichome phenotype, as shown in the two representative lines 5 and 7 (Fig. 1, A and C; Supplemental Fig. S2; Table I). These transgenic lines still showed the weaker multicellular trichome phenotype of the sim-1 mutant, suggesting that increased SMR1 expression did not affect the sim-1 phenotype. Thus, SIM and SMR1 may function differently in regulating trichome development. Together these data suggest that SIM plays a major role in regulating endoreplication of trichome cells and trichome development. While SMR1 by itself may play a lesser role in regulating these processes, it could contribute synergistically to such function of SIM.
SIM and SMR1 Act Synergistically to Affect Cell Ploidy in the Leaf
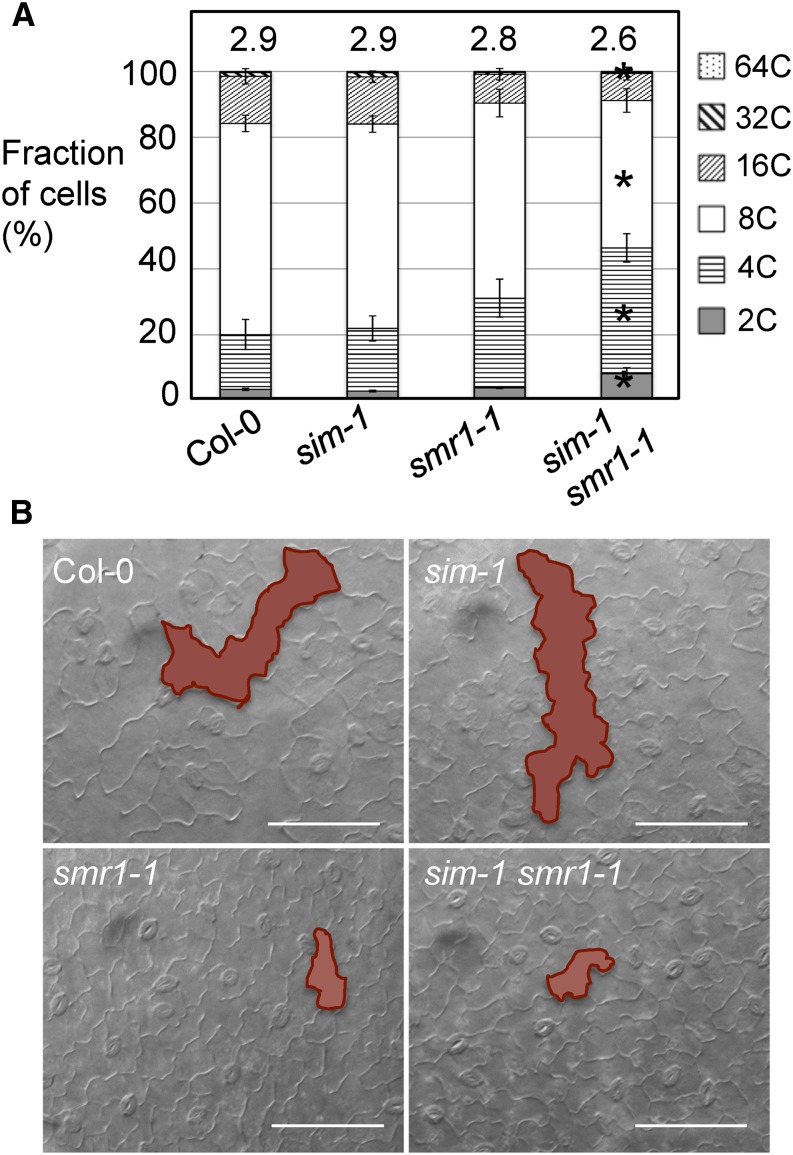
To further determine if SIM and SMR1 affect cell-cycle progression in other cell types besides the trichome, we measured the ploidy of leaf cells using flow cytometry. We found that although showing strong trichome defects, the sim-1 mutant did not affect overall cell ploidy pattern in the leaf, compared with Col-0 (Fig. 2A). The smr1-1 mutant repeatedly showed lower cell ploidy, having increased 4C but decreased 16C cells. However, the difference between Col-0 and smr1-1 was not statistically significant in our analysis, although it was reported so in a previous study (Roeder et al., 2010). The sim-1 smr1-1 mutant, on the other hand, showed a significant increase in 2C and 4C cells but a decrease in 8C and 64C cells. sim-1 smr1-1 also had significantly reduced ploidy index (2.6 ± 0.2), compared with those of sim-1 (2.9 ± 0.1), smr1-1 (2.8 ± 0.2), and Col-0 (2.9 ± 0.2).
Figure 2.
SMR1 plays a stronger role than SIM in regulating ploidy of leaf cells. A, Analysis of leaf cell ploidy by flow cytometry. The fourth to sixth leaves of 25-d-old plants were collected for nuclei isolation, followed by flow cytometry analysis. Data represent the average of five experiments ± se of mean. Statistical analysis was performed with Mann-Whitney test (http://vassarstats.net/). Asterisks indicate significant difference between samples of different genotypes in the same ploidy group (P < 0.05). Ploidy indices are shown above the bars. B, Epidermal cell morphology. The fourth to sixth leaves of 25-d-old plants were cleared overnight with ethanol and photographed using a dissecting microscope connected to a CCD camera. Highlighted shapes indicate typical pavement cells in each genotype. Note giant pavement cells in Col-0 and sim-1, but not in smr1-1 and sim-1 smr1-1. Bars, 100 μm.
Consistent with the change of cell ploidy detected by flow cytometry, we found that while both sim-1 and Col-0 showed large pavement cells on the leaf surface, smr1-1 and sim-1 smr1-1 mutants lacked these giant cells (Fig. 2B). Together, these results suggest that SMR1 plays a stronger role than SIM in regulating endoreplication of nontrichome leaf cells in Arabidopsis.
SMR1 Plays a Greater Role in Disease Resistance to P. syringae
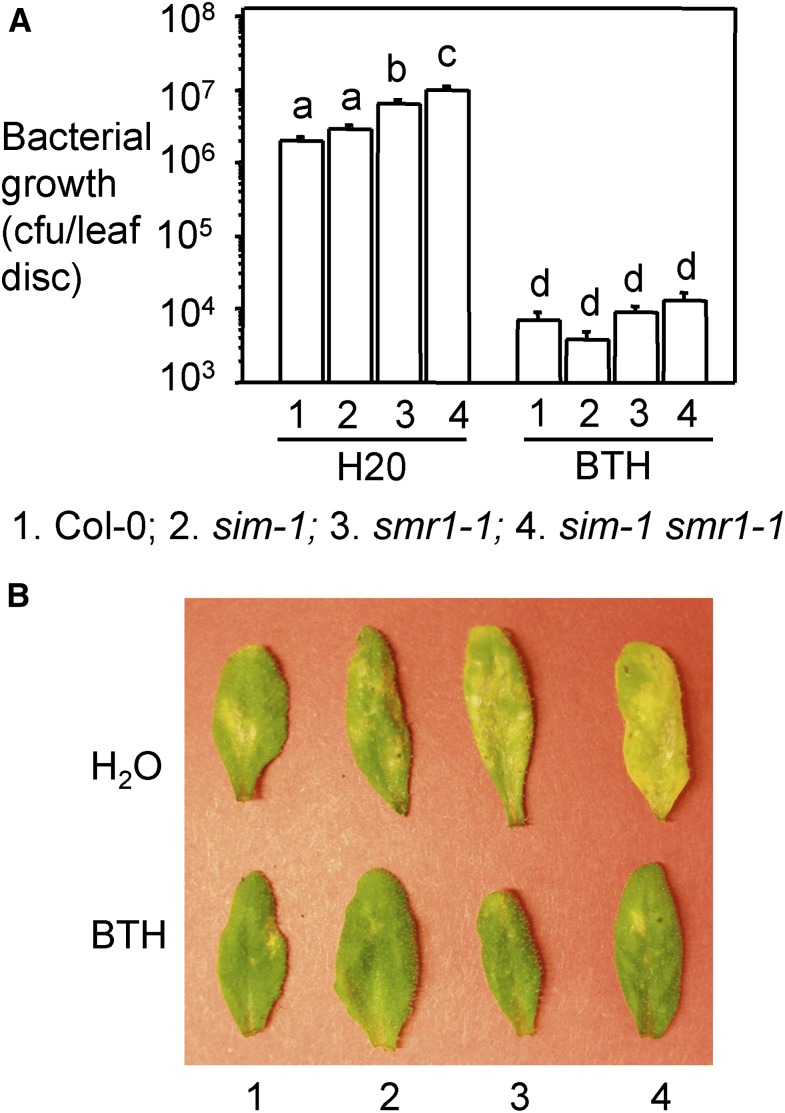
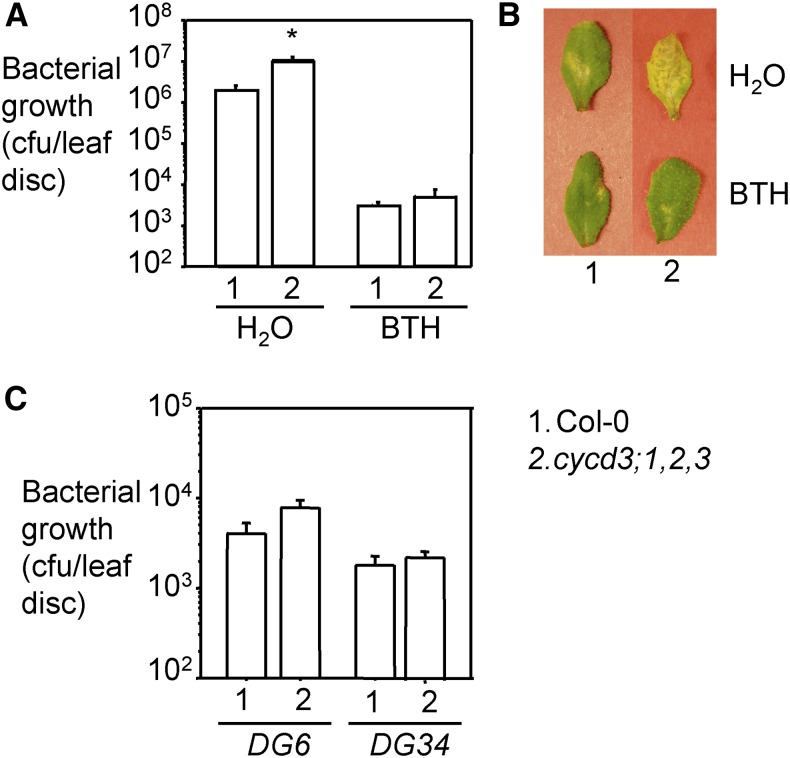
Recently SIM and SMR1 were shown to affect effector triggered immunity in response to P. syringae and H. arabidopsidis (Wang et al., 2014b). However, in this study only a sim smr1 double mutant, but not the individual single mutants, were tested. Our data showing differential roles of SIM and SMR1 in cell-cycle control prompted us to ask whether these two genes also play differential roles in defense control. To address this question, we infected sim-1 and smr1-1 mutants with the virulent strain P. syringae pv maculicola ES4326 DG3 (PmaDG3). We found that while sim-1 had a similar level of bacterial growth as Col-0, smr1-1 was more susceptible to P. syringae than Col-0 (Fig. 3). The sim-1 smr1-1 mutant was even more susceptible than smr1-1. We were able to rescue the enhanced susceptibility of smr1-1 and sim-1 smr1-1 with the SMR1 genomic construct (Supplemental Fig. S3). In addition, treating smr1-1 and sim-1 smr1-1 plants with an SA analog, 300 μm benzo (1, 2, 3) thiadiazol-7-carothioic acid (BTH) that induces similar defense responses as SA (Lawton et al., 1996), rescued the susceptible phenotype of smr1-1 and sim-1 smr1-1 (Fig. 3). These results suggest that SMR1 plays a greater role than SIM in defense against P. syringae in Arabidopsis and SMR1 likely acts upstream of the SA pathway.
Figure 3.
Enhanced disease susceptibility of smr1-1 and sim-1 smr1-1 mutants was rescued by BTH treatment. A, Bacterial growth assay. Plants were sprayed with 300 μm BTH or water for 24 h, and the fifth to seventh leaves of the treated plants were infiltrated with the virulent strain PmaDG3 (OD600 = 0.0001). Leaf discs were taken 3 d post infection for the measurement of bacterial growth. Statistical analysis was performed with Student’s t test (StatView 5.0.1). Different letters indicate significant difference among the samples (n = 6; P < 0.01). B, Disease symptoms. Pictures of the infected leaves were taken at 4 d post infection. These experiments were repeated twice with similar results.
SMR1 Contributes to SA-Mediated Defense
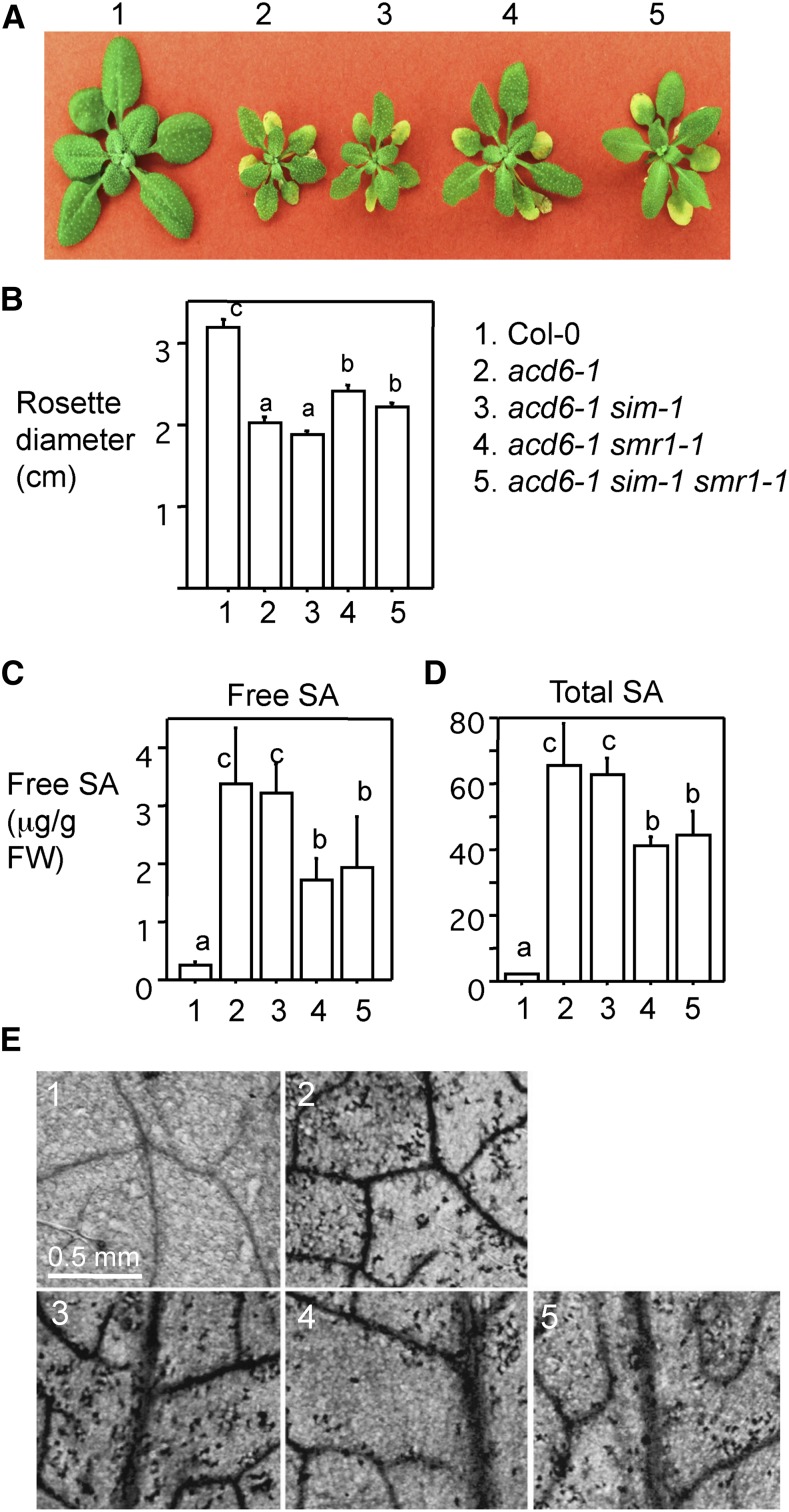
To further test whether SMR1 regulates SA-mediated defense, we crossed sim-1 and smr1-1 mutants into the acd6-1 background. acd6-1 is caused by a point mutation in the ACD6 gene, encoding an ankyrin repeat protein with a transmembrane domain (Rate et al., 1999; Lu et al., 2003). Although its biochemical function is still unclear, ACD6 was shown as a positive regulator of plant defense. The acd6-1 mutant is gain-of-function in nature and it shows dwarfism, constitutive defense, high levels of SA, and spontaneous cell death phenotypes. Interestingly, the small size of acd6-1 was shown largely in an inverse correlation with defense levels of the plant. Thus, the change of acd6-1 dwarfism induced by second-site mutations could be conveniently used as a visual readout to provide a rapid assessment of the effect of these second-site mutations in some genes on defense responses. acd6-1 has been successfully used in genetic analyses to reveal epistatic relationships between defense mutants and in a genetic screen to identify, to our knowledge, novel defense genes (Song et al., 2004; Lu et al., 2009; Ng et al., 2011; Wang et al., 2011a, 2011b, 2014a). We found that while acd6-1 sim-1 resembled acd6-1 in plant morphology, acd6-1 smr1-1 and acd6-1 sim-1 smr1-1 were significantly larger than acd6-1 plants (Fig. 4, A and B). Associating with increased plant size, acd6-1 smr1-1 and acd6-1 sim-1 smr1-1 plants accumulated lower levels of SA and exhibited less cell death, compared with acd6-1 (Fig. 4, C–E). No significant difference was detected between acd6-1 smr1-1 and acd6-1 sim-1 smr1-1 in plant size, SA accumulation, and cell death. Together, these data indicate that the role of SMR1 in regulating Arabidopsis defense is through influencing SA accumulation.
Figure 4.
smr1-1 suppresses dwarfism, SA accumulation, and cell death in acd6-1. A, Pictures of 25-d-old plants. Plant genotypes are as follows: 1, Col-0; 2, acd6-1; 3, acd6-1 sim-1; 4, acd6-1 smr1-1; 5, acd6-1 sim-1 smr1-1. B, Plant size comparison. Rosette diameters of plants (n > 15) were measured to determine plant size. C, Quantification of free SA level. D, Quantification of total SA level. Free and total SA were extracted from 25-d-old plants and analyzed with an HPLC instrument. E, Images of cell death. The fourth to sixth leaves of each genotype were stained with trypan blue and photographed. Statistical analysis was performed with Student’s t test (StatView 5.0.1). Different letters indicate significant difference among the samples (P < 0.05). These experiments were repeated twice with similar results.
Expression Analysis of SIM and SMR1
Recently Kumar et al. (2015) showed that the multicellular trichome phenotype conferred by sim-1 could be rescued by a number of SMR genes, including SMR1, when these genes were artificially expressed under the control of a trichome-specific promoter. Thus, the differential roles of SIM and SMR1 in cell-cycle and defense control are unlikely due to the difference in biochemical function of the gene products, but rather due to transcriptional and/or other posttranscriptional regulations. Indeed we found that expression of SMR1 was much higher than SIM in the leaf tissue (Fig. 5; Supplemental Fig. S4). On the other hand, a microarray study showed that expression of SIM is 28-fold enriched in trichomes, compared to leaves with trichomes removed. SMR1 did not show up as a significantly trichome-enriched gene in this study (Jakoby et al., 2008). In addition, another microarray study showed that SIM expression was found to be highest in first leaves at 9 d post germination, when leaves are about 200 microns long and trichome initiation is the greatest, and decline thereafter, while SMR1 expression increases approximately 5-fold from 9 to 22 d post germination (Beemster et al., 2005; Supplemental Fig. S4A). Thus, spatial and temporal expression patterns of SIM and SMR1 are important in determining the biological processes regulated by these genes.
Figure 5.
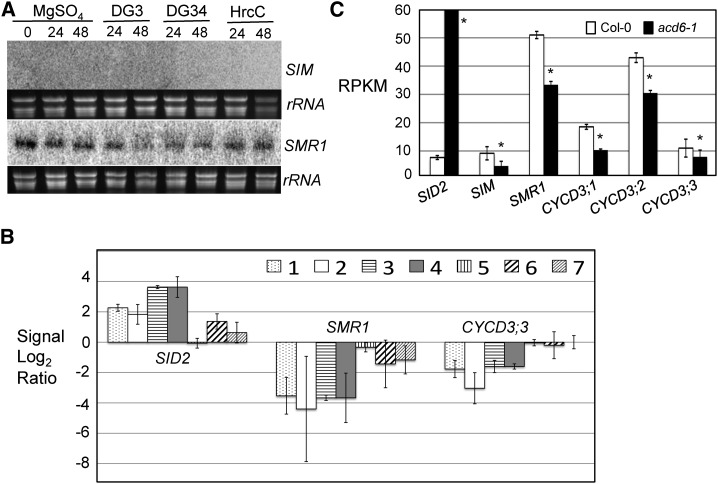
Expression analysis of cell-cycle genes. A, Northern blotting. Total RNA was extracted from 25-d-old plants infected with P. syringae strains at the indicated time points. These experiments were repeated twice with similar results. B, Gene expression analysis based on published microarray data. The comparisons are as follows: 1, Pst DC3000 versus mock, 1 × 106 bacteria/mL 24 h post infection (plants were infected with Pst DC3000 at 1 × 106 bacteria/mL or mock-treated and harvested 24 h post infection); 2, Pst DC3000 versus mock, 1 × 108 bacteria/mL 7 h post infection; 3, Pst COR− versus mock, 1 × 106 bacteria/mL 24 h post infection; 4, Pst COR− versus mock, 5 × 107 bacteria/mL 10 h post infection; 5, Pst COR−hrpS versus mock, 1 × 106 bacteria/mL 24 h post infection; 6, Pst COR−hrpS versus mock, 5 × 107 bacteria/mL 10 h post infection; and 7, Pst hrpA versus mock, 1 × 108 bacteria/mL 7 h post infection. The signal log2 ratio for each pathogen infection per gene was average of three samples ± SD. Details for the experiments were described previously (Thilmony et al., 2006). The original values for signal log2 ratio are listed in Supplemental Table S1 in the article. Only genes whose transcript levels showed 2-fold or more changes are listed in the table. Expression of SID2 was included as a control. C, Quantification of gene expression based on RNAseq. Total RNA extracted from Col-0 and acd6-1 was used for making cDNA libraries, followed by high-throughput sequencing using Illumina HiSEquation 2000 platform. RPKM were average of three samples ± SD. The RPKM value for SID2 is 174 and is over the scale limit, which was set to show the lower expression of other genes. Asterisks indicate that expression of a gene in acd6-1 is significantly different from its expression in Col-0 (P < 0.05).
Consistent with the major role of SMR1 in defense control, we found that expression of SMR1 was suppressed by infection of both virulent and avirulent P. syringae strains, PmaDG3 and PmaDG34 avrRpm1, respectively (Fig. 5A; Supplemental Fig. S4B). This observation is further supported by in silico analysis of a microarray dataset involving RNA samples isolated from Arabidopsis leaves infected with different P. syringae strains (Thilmony et al., 2006). P. syringae pv tomato DC3000 (Pst DC3000) is a virulent strain producing the phytotoxin coronatine (COR), while Pst COR− lacks COR production (Mittal and Davis, 1995; Bender et al., 1999). HrpS is a regulatory gene of P. syringae that could affect the formation of type 3 secretion system and COR production (Roine et al., 1997). HrpA encodes the main structural protein for the of type 3 secretion system pilus (Wei et al., 2000). Both P. syringae hrpS and hrpA mutant strains are unable to deliver bacterial effector proteins into the host cells, thus only inducing PAMPs-triggered immunity, but not effector triggered immunity in the host. Like PmaDG3, Pst DC3000 infection at both doses (1 × 106 bacteria/mL and 1 × 108 bacteria/mL) suppressed expression of SMR1 (Fig. 5B). The lack of COR production by the Pst DC3000 did not affect such suppression, suggesting Pst DC3000-induced suppression of SMR1 is COR independent. Infection with Pst COR− hrpS and Pst hrpA also suppressed SMR1 expression, albeit at a lower level. These results suggest that bacterial effector(s) have stronger influence than PAMP molecules in suppressing SMR1 expression.
Since our data showed that SMR1 affects SA-mediated defense (Fig. 4), we sought to further examine whether expression of SMR1 is reciprocally affected by SA. We first analyzed the microarray dataset that has triplicate samples harvested two hours after mock or 1 mm SA treatment of Arabidopsis leaves (Thibaud-Nissen et al., 2006; data accessible at NCBI GEO database, accession no. GSE3984). We found that there was a small suppression of SMR1 expression in the SA-treated sample (7.89 ± 0.27 counts), compared with the control (9.72 ± 0.60 counts), suggesting that SA negatively regulates SMR1 expression. To further validate SA suppression of SMR1 expression, we performed a high-throughput RNAseq experiment, using the SA-hyperaccumulating mutant acd6-1 and Col-0 as a control. Analysis of the RNAseq data revealed that expression of SMR1 was significantly lower in acd6-1 than in Col-0 (Fig. 5C). SIM expression was low in Col-0 and was further reduced in acd6-1. On the other hand, expression of SID2 (a positive control) was highly induced in acd6-1. Together these data suggest that while SIM and SMR1 genes can positively affect defense, defense activation can also feedback to inhibit expression of these genes. Such a negative feedback loop between the cell cycle and immune responses of Arabidopsis could be important for a tight control of these two biological processes during plant growth and development and biotic stress conditions.
SA Signaling Affects Cell-Cycle Progression
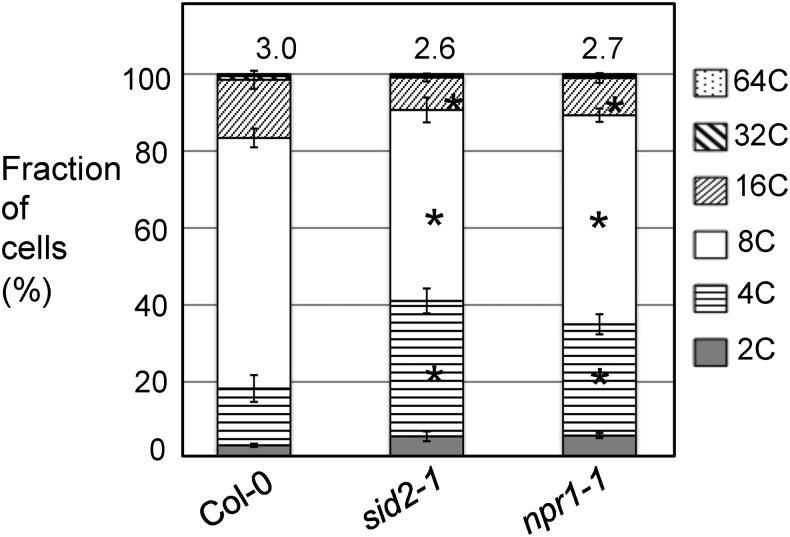
Besides SMR1, two other cell-cycle regulators, CPR5 and the noncanonical E2F gene DEL1, were shown to affect SA-mediated defense (Chandran et al., 2014; Wang et al., 2014b). These results prompted us to ask whether SA could affect cell-cycle progression as part of downstream signaling of some cell-cycle genes. To test this, we analyzed cell ploidy in the major SA biosynthetic mutant sid2-1 (Wildermuth et al., 2001; Ng et al., 2011) and the SA signaling mutant npr1-1 (Dong, 2004; Lu, 2009; Fu and Dong, 2013). Our results show that both sid2-1 and npr1-1 had significantly increased numbers of 4C cells but reduced numbers of 8C and 16C cells (Fig. 6). The ploidy indices of sid2-1 and npr1-1 were 2.6 ± 0.2 and 2.7 ± 0.1, respectively, significantly lower than that of Col-0 (3.0 ± 0.2). Thus, these data suggest that reducing SA levels and blocking SA signaling could lead to reduced endoreplication in Arabidopsis. However, when we treated Col-0 plants with the SA activator, 300 μm BTH, we did not detect any changes in cell ploidy pattern 24 and 72 h post BTH treatment (Supplemental Fig. S5). Longer BTH treatment up to 12 d with plants being BTH-sprayed every 4 d did not reveal significant difference in the cell ploidy pattern (data not shown). Thus, these results suggest that SA is necessary, but not sufficient, to affect cell-cycle progression.
Figure 6.
Altered cell ploidy in sid2-1 and npr1-1 mutants. The fourth to sixth leaves of 25-d-old plants were collected for nuclei isolation, followed by flow cytometry analysis. Data represent the average of five experiments ± se of mean. Statistical analysis was performed with Mann-Whitney test (http://vassarstats.net/). Asterisks indicate significant difference between Col-0 and sid2-1 or npr1-1 in the same ploidy group (P < 0.05). Ploidy indices were shown above the bars.
The CYCD3 Genes Contribute to Defense
Our results showed that decreased cell ploidy in smr1-1, sim-1 smr1-1, and the SA mutants is associated with increased disease susceptibility to P. syringae. To further test how an increase in cell ploidy could affect Arabidopsis defense, we examined defense responses of a triple mutant with three CYCD3 genes disrupted, cycd3;1,2,3, which was shown to have a higher ploidy level than wild type (Dewitte et al., 2007). We chose to work on this mutant is also because CYCD3 proteins are potential targets of CKIs and are known to play a key role in transitions from G1-S and from cell proliferation to endoreplication (Riou-Khamlichi et al., 1999; Dewitte et al., 2003; Churchman et al., 2006; Van Leene et al., 2010).
While SIM and SMR1 are positive regulators of endoreplication, the CYCD3 genes are considered negative regulators. Interestingly, when infected with PmaDG3, the cycd3;1,2,3 mutant showed increased disease susceptibility associated with more severe disease symptoms (Fig. 7, A and B). This susceptibility of cycd3;1,2,3 could be rescued with exogenous BTH treatment. However, cycd3;1,2,3 was not susceptible to the avirulent strains PmaDG6 (expressing the avirulence effector avrRpt2) and PmaDG34 (expressing the avirulence effector avrRpm1; Hamdoun et al., 2013; Fig. 7C). Neither did we detect a compromised hypersensitive response in this mutant to the infection of the two avirulent strains (Supplemental Fig. S6). These results suggest that CYCD3;1,2,3 genes are involved in affecting basal defense in Arabidopsis.
Figure 7.
Enhanced disease susceptibility of the cycd3;1,2,3 mutant was rescued by BTH treatment. A, Bacterial growth assay with PmaDG3. Plants were sprayed with 300 μm BTH or water for 24 h, and the fifth to seventh leaves of the treated plants were infiltrated with the virulent strain PmaDG3 (105 bacteria/mL). Leaf discs were taken 3 d post infection for the measurement of bacterial growth. Statistical analysis was performed with Student’s t test (StatView 5.0.1). The asterisk indicates significant difference between Col-0 and cycd3;1,2,3 with water treatment (n = 6; P < 0.05). B, Disease symptoms. Pictures of the infected leaves were taken at 4 d post infection. C, Bacterial growth assay with PmaDG6 or PmaDG34 infection. These experiments were repeated twice with similar results.
Similar to SMR1, expression of a CYCD3 gene, CYCD3;3, was suppressed by Pst DC3000 infection independent of bacterial COR production (Fig. 5B). In addition, the transcript levels of CYCD3;1, CYCD3;2, and CYCD3;3 were significantly lower in the constitutive defense mutant acd6-1 than in Col-0 (Fig. 5C). These results further support a negative feedback regulatory mechanism in controlling expression of some cell-cycle genes and immune activation in Arabidopsis.
DISCUSSION
In this report we presented data to show that two homologous CKI genes, SIM and SMR1, play differential roles in regulating cell ploidy and disease resistance to the bacterial pathogen P. syringae. We provide a mechanistic explanation of such differential roles of the two genes, which is mainly due to transcriptional regulation of these two genes during development, in different tissue, and in response to pathogens. Our data further reveal a negative feedback regulation between SMR1-mediated defense and cell-cycle control. While supporting that cell-cycle progression is intimately related to plant defense, our data also raise an interesting question regarding relationship between the ploidy level and defense.
The SIM and SMR1 genes belong to a family of plant-specific CKIs. Although sharing over 60% identity in their encoded amino acids, SIM and SMR1 show both distinct and overlapping functions in cell-cycle and defense regulation. We presented in this report several pieces of evidence to support this notion: (1) SIM plays a stronger role in affecting endoreplication in trichomes, while SMR1 affects leaf cell ploidy more strongly; (2) the smr1-1 mutant, but not sim-1, was more susceptible to P. syringae infection and showed suppression of dwarfism, cell death, and SA accumulation in acd6-1; (3) a SMR1 genomic fragment rescued severe multicellular trichome phenotype of sim-1 smr1-1 but did not rescue trichome defects of sim-1; and (4) knockout of both genes resulted in more severe ploidy defects in trichomes and leaf cells and further enhanced susceptibility to P. syringae infection. Since SIM and SMR1 were shown to bear similar biochemical activities when being overexpressed in trichome cells (Kumar et al., 2015), the differential functions displayed by the two homologous genes are likely due to their differential expression patterns. Indeed, we found that SIM was enriched in the trichome while SMR1 was enriched in nontrichome leaf cells. In addition, expression of SMR1 was suppressed by P. syringae infection and in the constitutive defense mutant acd6-1, consistent with a stronger role of SMR1 in defense. Other cell-cycle genes, such as SIM and CYCD3, were also suppressed during defense activation (Fig. 5). The initial high levels of SMR1 (possibly also other cell-cycle genes) could preemptively guard against pathogens. Suppression of these genes after defense activation could reflect an adjustment in balancing development and the costly defense responses in Arabidopsis.
Besides the SIM and SMR1 genes, other SMR family members have not been demonstrated to have roles in plant defense, although several studies have implicated some SMR genes in stress responses. For instance, expression of several Arabidopsis SMR genes, as well as a rice homolog of SIM, is altered in response to biotic and/or abiotic stress conditions (Peres et al., 2007; Chandran et al., 2009; Yi et al., 2014). Two SIM homologs, SMR5 and SMR7, were shown to be important for plant response to DNA damage (Yi et al., 2014). In addition, several SMR genes, including SMR1, -2, -3, -4, -5, -7, -11, and -13, complemented the multicellular trichome defects of sim-1, when these genes were expressed in the trichome using a trichome-specific promoter (Kumar et al., 2015). Thus, SMR proteins share high conservation as SIM, and it is possible that additional SMR family members are important for plant innate immunity. It would be interesting to identify and characterize such SMR genes and determine their functions in cell-cycle and defense control.
SA is a small signaling molecule important for biotic responses of plants. Our data presented here and in earlier studies (Vanacker et al., 2001; Hamdoun et al., 2013) suggest SA-mediated pathway is an integral part of cell-cycle signaling. The lack of SA biosynthesis and signaling in sid2-1 and npr1-1, respectively, had reduced overall ploidy pattern of leaf cells when assayed by flow cytometry (Fig. 6). Interestingly, our previous study using nuclear quantification of mesophyll cells from fixed sections near the middle vein of npr1-1 showed increased ploidy, suggesting a possibility that npr1-1 leaves harbor cells with different ploidy. Both sid2-1 and npr1-1 mutants were also compromised in P. syringae-induced large cell growth (Hamdoun et al., 2013). Since activation of SA signaling with exogenous application of an SA analog did not change cell ploidy and the formation of tumor-like growth in the leaf (Supplemental Fig. S5; Hamdoun et al., 2013), it is possible that SA signaling requires the activation of additional pathways to regulate cell-cycle progression.
Our data show that SMR1 acts partly through SA in defense regulation. We found that the susceptibility of smr1-1 was rescued by exogenous activation of SA signaling and smr1-1 suppressed SA accumulation in acd6-1 (Figs. 3 and 4). Consistent with our data, Wang et al. showed that the sim smr1 mutant suppressed high SA levels of the cpr5 mutant and exhibited a delay in SA accumulation with the infection of the avirulent P. syringae strain expressing avrRpt2 (Wang et al., 2014b). CPR5 encodes a nuclear membrane protein that interacts with SIM and SMR1. Mutations in the CPR5 gene confer constitutive defense and higher SA accumulation (Bowling et al., 1997; Clarke et al., 2001; Jirage et al., 2001; Brininstool et al., 2008). Thus, SIM/SMR1 and CPR5 are positive and negative regulators of the SA pathway, respectively.
On the other hand, like SIM and SMR1, studies suggest that CPR5 promotes endoreplication, possibly through the canonical cell-cycle signaling pathway involving RBR-E2F (Kirik et al., 2001; Wang et al., 2014b). The SA pathway influenced by CPR5-SIM/SMR1 could be convergent with the canonical RBR-E2F pathway; for instance, CPR5-SIM/SMR1 act through RBR-E2F signaling to regulate SA accumulation. However, it is currently unknown if RBR-E2F signaling could affect SA-mediated defense besides its role in cell-cycle control. Another possible downstream cell-cycle component of CPR5-SIM/SMR1 could be the noncanonical E2F gene, DEL1, whose protein product was shown to bind directly to the main SA regulatory gene EDS5 (Nawrath and Métraux, 1999; Nawrath et al., 2002; Chandran et al., 2014). It is also possible that CPR5-SIM/SMR1 may act through a pathway independent of cell-cycle signaling in regulating SA-mediated defense.
One interesting observation that we had during the course of the study is that there appears to be no direct correlation between the ploidy level and defense. sim-1, smr1-1, sid2-1, and npr1-1 mutants had lower cell ploidy and reduced disease resistance, suggesting a positive association of the two processes. However, results with the cycd3;1,2,3 mutant that has increased cell ploidy and decreased resistance to P. syringae would challenge such a statement. Indeed, analysis of additional cell-cycle mutants recently reported to have defense defects showed no direct association between the ploidy level and defense response. For instance, cpr5-1 had a lower ploidy but was highly resistant (Bowling et al., 1997; Clarke et al., 2001; Jirage et al., 2001; Kirik et al., 2001). The loss-of-function and gain-of-function mutations in the Omission of the Second Division 1 gene, encoding a negative regulator of anaphase-promoting complex/cyclosome, resulted in higher and lower cell ploidy, respectively. Despite of their opposite effects on the cell cycle, both types of plants were resistant to P. syringae infection (Bao et al., 2013; Bao and Hua, 2014). Thus, it is not the level of cell ploidy that affects host defense. Rather, it is the disruption of the normal cell-cycle progression that results in altered host response to pathogen invasion.
How does one explain such a dissociation of ploidy and defense levels? One possibility is that there are different modes of action used by cell-cycle genes in cell-cycle and defense control. For instance, CPR5, SIM, and SMR1 are positive regulators to promote endoreplication, and these proteins may execute this function by physical interaction with each other (Wang et al., 2014b). The increased phosphorylation in the RBR protein in the sim smr1 and cpr5 mutants is consistent with the roles of CPR5-SIM/SMR1 proteins in inhibiting activities of CYC/CDK complexes and promoting endoreplication. On the other hand, sim smr1 and cpr5 mutants exhibited opposing defense phenotypes, suggesting that CPR5 antagonizes SMR1 (and/or SIM) in defense control. In this regard, it is possible that SMR1 (and/or SIM) act downstream of CPR5 in regulating disease resistance based on the suppression of defense phenotypes of a cpr5 mutant by sim and smr1 mutations (Wang et al., 2014b). In CPR5-SIM/SMR-mediated defense pathway, there might be additional genes that could differentially interact with CPR5 and SIM/SMR1 to positively or negatively regulate plant defense.
SIM and SMR1 are CKIs that target CYC/CDK complexes to promote endoreplication. One of such targets could be CYCD3 proteins. Indeed, SIM was shown to interact with the CYCD3;2 protein when both proteins transiently expressed in Arabidopsis leaf cells (Churchman et al., 2006). SIM was also shown to interact with CDKA;1, which is activated by CYCD3;1, and inhibits activities of CYCD3;1/CDKA;1 and several other CYC/CDK complexes (Kumar et al., 2015). The decreased ploidy in sim-1 smr1-1 and increased ploidy in cycd3;1,2,3 is consistent with SIM and SMR1 being inhibitor of CYCD3/CDKA complexes and that these two genes act in opposing ways to regulate endoreplication. However, SIM, SMR1, and CYCD3 genes appear to be positive regulators of plant defense because mutations in the corresponding genes conferred enhanced disease susceptibility to P. syringae. In this case, SIM and SMR1 could positively affect CYCD3 function in defense control. Such positive and negative roles of SIM and SMR1 have already been reported for other cell-cycle genes (Sherr and Roberts, 1999). Alternatively, SIM/SMR1 could act in separate pathways from the CYCD3 genes to affect the cell cycle and defense in Arabidopsis. To support this latter speculation, CYCD3;2 was found to copurify with two other SMR family members, SMR4 and SMR6 (Van Leene et al., 2010), implicating that CYCD3;2 could interact with different partners in executing its functions in control of cell-cycle progression and defense.
The molecular mechanism underlying the crosstalk between cell-cycle progression and innate immune control is far from being understood. It is critically important to elucidate the mechanisms of action of core cell-cycle genes in defense control. Further identification and characterization of additional genes that are involved in cell-cycle and defense control should also help to gain a better understanding of processes involved in cell-cycle control and plant defense.
MATERIALS AND METHODS
Plant Materials
All plants used in this report were in Col-0 background and were grown in growth chambers with a 12 h light/12 h dark cycle, light intensity at 200 µmol m−2 s−1, 60% humidity, and 22°C. The mutants acd6-1, sim-1, cycd3;1,2,3, npr1-1, and sid2-1 were previously described (Walker et al., 2000; Lu et al., 2003; Churchman et al., 2006; Dewitte et al., 2007; Wang et al., 2011a). The smr1-1 (SALK_33950) was obtained from the Arabidopsis Biological Research Center. sim-1 smr1-1, acd6-1 sim-1, acd6-1 smr1-1, and acd6-1 sim-1 smr1-1 mutants were generated by genetic crosses. All mutations were confirmed by genotyping with specific primers (Supplemental Table S1; Lu et al., 2003).
Generate Transgenic Plants
A 2367 bp SMR1 genomic fragment including 1424 bp promoter and 637 bp 3′ downstream of the SMR1 gene was amplified by PCR with the primers genomicSMR1-For1 and genomicSMR1-R1 (Supplemental Table S1) and cloned into the binary vector pGreen0229 (Hellens et al., 2000) for transformation of smr1-1 or sim-1 smr1-1 plants. At least 10 independent transformants from each background were obtained for further selection of homozygous transgenic plants, followed by analyses for gene expression and disease resistance to P. syringae.
Pseudomonas syringae Infection
The virulent strain P. syringae pv maculicola ES4326 DG3 (PmaDG3) and the avirulent strains PmaDG6 (expressing the avirulence effector avrRpt2) and PmaDG34 (expressing the avirulence effector avrRpm1; Hamdoun et al., 2013) were used to infect plants. Bacterial culture, preparation, inoculation by infiltration, and bacterial growth assay were conducted as described (Lu et al., 2013). For BTH treatment, plants were sprayed with 300 μm BTH (a kind gift from Robert Dietrich [Syngenta]) or water for 24 h before being infected with PmaDG3. For the hypersensitive response, a half area of a leaf at the fourth to sixth position of each genotype was infiltrated with a P. syringae strain (OD600 = 0.1) and examined for leaf wilting and collapse 18 h post infection. At least 15 leaves from at least six plants were used for each treatment group.
RNA Analysis
For P. syringae-challenged plants, the infected leaves were harvested for RNA extraction. For noninfected plants, all leaves were used for RNA preparation. The SIM and SMR1 probes were made by PCR with gene specific primers (Supplemental Table S1) and labeled with [32P]dCTP, using a corresponding antisense primer.
For quantitative reverse transcription PCR, 2 μg of total RNA was reversed-transcribed into cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. cDNA was diluted 20 times for subsequent quantitative PCR reactions, which were set up using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Scientific). Three milliliters of diluted cDNA template was used in a 20 mL PCR reaction. Primers for detection of transcripts of SIM, SMR1, and PP2AA3 (as a control) are listed in Supplemental Table S1. The quantitative PCR reactions were run in a CFX96 Touch Real-Time PCR System (Biorad) with one step at 95°C for 5 min, 40 cycles at 95°C for 10 s, 52°C for 15 s, and 72°C for 15 s. PCR products were checked using melting curve analysis to ensure amplification of a single product and the absence of primer-dimers. For relative gene expression at different time points, the 2−ΔCT method was applied as described (Schmittgen and Livak, 2008), using the cycle threshold values of PP2AA3 for normalization.
RNAseq Library Preparation, Sequencing, and Analysis
Total RNA was extracted from 25-d-old Col-0 and acd6-1 plants using Qiagen RNeasy plant mini kit (catalog no. 74903). On-column digestion was performed using Qiagen RNase free DNase kit (catalog no. 79254) to remove genomic DNA contamination. Each sample had three biological replicates, and 0.5 μg RNA per replicate was used to generate cDNA libraries using Illumina TruSeq RNA sample preparation kit (catalog no. RS-122-2001) for deep sequencing in Genomics Resources Core Facility at Weill Cornell Medical College. The samples were multiplexed and sequenced with the standard run of 51-cycle and single-read. At least 150 million reads per lane were obtained.
The raw reads in FASTQ format were imported into CLC Genomics Workbench 8 (Qiagen) and mapped to the TAIR 10 Arabidopsis reference genome annotated with genes and transcripts. Gene Expression files of each sample were generated based on the total number of reads mapped to individual genes. Gene Expression files from different samples were compared in order to identify differentially expressed genes using the following parameters: the absolute value of fold change is > 2; the P value of a false discovery rate is < 0.05, and the value of reads per kilobase of exon per million reads mapped (RPKM) is > 0.3. For the comparison of expression of individual genes between different samples, the original RPKM was used. Statistic analysis was performed using ANOVA post hoc Fisher’s PLSD test.
SA Measurement
Free and total (glucosylated) SA were extracted from plants and quantified with an HPLC instrument as previously described (Ng et al., 2011; Wang et al., 2011a).
Cell Death Staining
The fourth to sixth leaves of 25-d-old plants were collected, boiled in lactophenol containing 0.01% trypan blue for 2 min, cleared off by boiling in alcoholic lactophenol, and rinsed with 50% ethanol. The stained leaves were visualized with a dissecting microscope (Leica M80, Leica Microsystems) and photographed with a CCD camera (Leica IC80 HD) connected to the microscope. At least four leaves from four plants of each treatment were stained and examined for cell death.
Epidermal Cell Morphology
The fourth to sixth leaves of 25-d-old plants were harvested, incubated with ethanol to clear off chlorophyll, and examined for epidermal cell morphology. At least three leaves from three plants of each genotype were photographed with a CCD camera (Cool Snap HQ2, Photometrics) connected to a dissecting microscope (Leica M205 FA, Leica Microsystems).
Scanning Electron Microscopy
First leaves of 2-week-old Arabidopsis plants were mounted on the specimen stubs using double-stick tape and were observed under high vacuum mode at 5.0 kV in a JEOL JSM 6610LV scanning electron microscope, working quickly to avoid drying and damage from the beam.
Quantification of Trichome Initiation Site
Trichome nuclei staining with 4′,6-diamidino-2-phenylindole was performed as described (Walker et al., 2000). DAPI-stained nuclei per trichome initiation site were counted at either 200× or 400× magnification using a Leica DM RXA2 microscope, and images were captured with a SensiCamQE 12-bit cooled CCD camera.
Flow Cytometry Analysis
The fourth to sixth leaves of 25-d-old plants were harvested and coarsely chopped with a single-edge razor blade into a mince in “Aru” buffer containing 97.5% MgSO4 (0.246% MgSO4 7H2O, 0.37% KCl, and 0.12% HEPES), 0.1% DTT, and 2.5% Triton X-100 (Arumuganathan and Earle, 1991) in order to release nuclei. At least 10 leaves were used for each sample. The mince was passed through a nylon mesh with pore size 40 µm to collect nuclei. Nuclei were stained with propidium iodide at the final concentration 30 µg/mL and were subjected to flow analysis with a CyAn ADP flow cytometer (Beckman Coulter). Data were analyzed using Summit software (v4.3.02). Propidium-iodide-stained nuclei were detected using a blue laser (emission 488 nm and excitation 613/620 nm) and 605 voltages power output. At least 20,000 events were recorded at the flow rate of 150 events/second for each sample. Triplicate samples per treatment and/or per genotype were included. Ploidy index was calculated as follows: ploidy index = (% 2C nuclei × 1) + (% 4C nuclei × 2) + (% 8C nuclei × 3) + (% 16C nuclei × 4) + (% 32C nuclei × 5) + (% 64C nuclei × 6).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers At5g04470 for SIM and At3g10525 for SMR1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Trichome morphology.
Supplemental Figure S2. Rescue trichome phenotype of sim-1 smr1-1 to that of sim-1 by a SMR1 genomic fragment.
Supplemental Figure S3. Complementation of enhanced disease susceptibility of sim-1 smr1-1 and smr1-1 by a SMR1 genomic fragment.
Supplemental Figure S4. Expression analysis of SIM and SMR1.
Supplemental Figure S5. BTH treatment does not affect cell ploidy in Arabidopsis.
Supplemental Figure S6. The cycd3;1,2,3 mutant is not compromised to the hypersensitive response induced by PmaDG6 or PmaDG34.
Supplemental Table S1. Primers used in this paper.
Supplementary Material
Acknowledgments
We thank Dr. Jean Greenberg for her support of this research and members in the Lu laboratory for assistance with this work. We also thank Dr. Sue Ostrand-Rosenberg and Ms. Virginia Clements for sharing the flow cytometry instrument and for flow cytometry technical support. We thank Dr. Xinnian Dong for helpful discussions about this research.
Glossary
- SA
salicylic acid
- Col-0
Columbia-0
- BTH
benzo (1, 2, 3) thiadiazol-7-carothioic acid
- COR
coronatine
- RPKM
reads per kilobase of exon per million reads mapped
Footnotes
This work was partially supported by a grant from the National Science Foundation (NSF RIG–0818651) to H.L. Work by J.C.L. and N.K. was supported by a National Science Foundation grant (no. IOS1146620) to J.C.L.
Articles can be viewed without a subscription.
References
- Arumuganathan K, Earle E (1991) Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol Biol Rep 9: 229–233 [Google Scholar]
- Ascencio-Ibáñez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, Hanley-Bowdoin L (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Hua J (2014) Interaction of CPR5 with cell cycle regulators UVI4 and OSD1 in Arabidopsis. PLoS One 9: e100347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Hua J (2015) Linking the cell cycle with innate immunity in Arabidopsis. Mol Plant 8: 980–982 [DOI] [PubMed] [Google Scholar]
- Bao Z, Yang H, Hua J (2013) Perturbation of cell cycle regulation triggers plant immune response via activation of disease resistance genes. Proc Natl Acad Sci USA 110: 2407–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster GT, De Veylder L, Vercruysse S, West G, Rombaut D, Van Hummelen P, Galichet A, Gruissem W, Inzé D, Vuylsteke M (2005) Genome-wide analysis of gene expression profiles associated with cell cycle transitions in growing organs of Arabidopsis. Plant Physiol 138: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CL, Alarcón-Chaidez F, Gross DC (1999) Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol Mol Biol Rev 63: 266–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berckmans B, De Veylder L (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hülskamp M, Schnittger A (2010) Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brininstool G, Kasili R, Simmons LA, Kirik V, Hülskamp M, Larkin JC (2008) Constitutive Expressor Of Pathogenesis-related Genes5 affects cell wall biogenesis and trichome development. BMC Plant Biol 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Inada N, Hather G, Kleindt CK, Wildermuth MC (2010) Laser microdissection of Arabidopsis cells at the powdery mildew infection site reveals site-specific processes and regulators. Proc Natl Acad Sci USA 107: 460–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran D, Rickert J, Huang Y, Steinwand MA, Marr SK, Wildermuth MC (2014) Atypical E2F transcriptional repressor DEL1 acts at the intersection of plant growth and immunity by controlling the hormone salicylic acid. Cell Host Microbe 15: 506–513 [DOI] [PubMed] [Google Scholar]
- Chandran D, Tai YC, Hather G, Dewdney J, Denoux C, Burgess DG, Ausubel FM, Speed TP, Wildermuth MC (2009) Temporal global expression data reveal known and novel salicylate-impacted processes and regulators mediating powdery mildew growth and reproduction on Arabidopsis. Plant Physiol 149: 1435–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, Kirik V, Hülskamp M, Inzé D, De Veylder L, Walker JD, Zheng Z, Oppenheimer DG, Gwin T, Churchman J, et al. (2006) SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell 18: 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE (2001) Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J 26: 409–420 [DOI] [PubMed] [Google Scholar]
- de Almeida Engler J, De Vleesschauwer V, Burssens S, Celenza JL Jr, Inzé D, Van Montagu M, Engler G, Gheysen G (1999) Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11: 793–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Engler J, Gheysen G (2013) Nematode-induced endoreduplication in plant host cells: why and how? Mol Plant Microbe Interact 26: 17–24 [DOI] [PubMed] [Google Scholar]
- De Clercq A, Inzé D (2006) Cyclin-dependent kinase inhibitors in yeast, animals, and plants: a functional comparison. Crit Rev Biochem Mol Biol 41: 293–313 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D (2001) Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13: 1653–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Depuydt S, De Veylder L, Holsters M, Vereecke D (2009a) Eternal youth, the fate of developing Arabidopsis leaves upon Rhodococcus fascians infection. Plant Physiol 149: 1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S, Trenkamp S, Fernie AR, Elftieh S, Renou JP, Vuylsteke M, Holsters M, Vereecke D (2009b) An integrated genomics approach to define niche establishment by Rhodococcus fascians. Plant Physiol 149: 1366–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X. (2004) NPR1, all things considered. Curr Opin Plant Biol 7: 547–552 [DOI] [PubMed] [Google Scholar]
- Favery B, Complainville A, Vinardell JM, Lecomte P, Vaubert D, Mergaert P, Kondorosi A, Kondorosi E, Crespi M, Abad P (2002) The endosymbiosis-induced genes ENOD40 and CCS52a are involved in endoparasitic-nematode interactions in Medicago truncatula. Mol Plant Microbe Interact 15: 1008–1013 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Hamdoun S, Liu Z, Gill M, Yao N, Lu H (2013) Dynamics of defense responses and cell fate change during Arabidopsis-Pseudomonas syringae interactions. PLoS One 8: e83219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Heyman J, De Veylder L (2012) The anaphase-promoting complex/cyclosome in control of plant development. Mol Plant 5: 1182–1194 [DOI] [PubMed] [Google Scholar]
- Inzé D, De Veylder L (2006) Cell cycle regulation in plant development. Annu Rev Genet 40: 77–105 [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148: 1583–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Zhou N, Cooper B, Clarke JD, Dong X, Glazebrook J (2001) Constitutive salicylic acid-dependent signaling in cpr1 and cpr6 mutants requires PAD4. Plant J 26: 395–407 [DOI] [PubMed] [Google Scholar]
- Kay S, Hahn S, Marois E, Hause G, Bonas U (2007) A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318: 648–651 [DOI] [PubMed] [Google Scholar]
- Kirik V, Bouyer D, Schöbinger U, Bechtold N, Herzog M, Bonneville JM, Hülskamp M (2001) CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol 11: 1891–1895 [DOI] [PubMed] [Google Scholar]
- Kumar N, Harashima H, Kalve S, Bramsiepe J, Wang K, Sizani B, Bertrand LL, Johnson MC, Faulk C, Dale R, Simmons LA, Churchman ML, et al. (November 6, 2015) Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell http://dx.doi.org/10.1105/tpc.15.00489 [DOI] [PMC free article] [PubMed]
- Lawton KA, Friedrich L, Hunt M, Weymann K, Delaney T, Kessmann H, Staub T, Ryals J (1996) Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J 10: 71–82 [DOI] [PubMed] [Google Scholar]
- Lu H. (2009) Dissection of salicylic acid-mediated defense signaling networks. Plant Signal Behav 4: 713–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Rate DN, Song JT, Greenberg JT (2003) ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zhang C, Albrecht U, Shimizu R, Wang G, Bowman KD (2013) Overexpression of a citrus NDR1 ortholog increases disease resistance in Arabidopsis. Front Plant Sci 4: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Salimian S, Gamelin E, Wang G, Fedorowski J, LaCourse W, Greenberg JT (2009) Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis. Plant J 58: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, Pevzner SJ, Donovan SE, et al. ; European Union Effectoromics Consortium (2011) Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333: 596–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Métraux JP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng G, Seabolt S, Zhang C, Salimian S, Watkins TA, Lu H (2011) Genetic dissection of salicylic acid-mediated defense signaling networks in Arabidopsis. Genetics 189: 851–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebel A, de Almeida Engler J, Hemerly A, Ferreira P, Inzé D, Van Montagu M, Gheysen G (1996) Induction of cdc2a and cyc1At expression in Arabidopsis thaliana during early phases of nematode-induced feeding cell formation. Plant J 10: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Van Der Schueren E, Beemster GT, Frankard V, Larkin JC, et al. (2007) Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem 282: 25588–25596 [DOI] [PubMed] [Google Scholar]
- Polyn S, Willems A, De Veylder L (2015) Cell cycle entry, maintenance, and exit during plant development. Curr Opin Plant Biol 23: 1–7 [DOI] [PubMed] [Google Scholar]
- Potuschak T, Doerner P (2001) Cell cycle controls: genome-wide analysis in Arabidopsis. Curr Opin Plant Biol 4: 501–506 [DOI] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Roeder AH, Chickarmane V, Cunha A, Obara B, Manjunath BS, Meyerowitz EM (2010) Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol 8: e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine E, Wei W, Yuan J, Nurmiaho-Lassila EL, Kalkkinen N, Romantschuk M, He SY (1997) Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA 94: 3459–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Song JT, Lu H, McDowell JM, Greenberg JT (2004) A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J 40: 200–212 [DOI] [PubMed] [Google Scholar]
- Swarup S, de Feyter R, Brlansky RH, Gabriel DW (1991) A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of X. campestris to elicit cankerlike lesions on citrus. Phytopathology 81: 802–809 [Google Scholar]
- Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD (2006) Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J 47: 152–162 [DOI] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Umeda M, Shimotohno A, Yamaguchi M (2005) Control of cell division and transcription by cyclin-dependent kinase-activating kinases in plants. Plant Cell Physiol 46: 1437–1442 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, De Bodt S, Maere S, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Lu H, Rate DN, Greenberg JT (2001) A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis. Plant J 28: 209–216 [DOI] [PubMed] [Google Scholar]
- Vodermaier HC. (2004) APC/C and SCF: controlling each other and the cell cycle. Curr Biol 14: R787–R796 [DOI] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC (2000) SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127: 3931–3940 [DOI] [PubMed] [Google Scholar]
- Wang G, Zhang C, Battle S, Lu H (2014a) The phosphate transporter PHT4;1 is a salicylic acid regulator likely controlled by the circadian clock protein CCA1. Front Plant Sci 5: 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GF, Seabolt S, Hamdoun S, Ng G, Park J, Lu H (2011a) Multiple roles of WIN3 in regulating disease resistance, cell death, and flowering time in Arabidopsis. Plant Physiol 156: 1508–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GY, Shi JL, Ng G, Battle SL, Zhang C, Lu H (2011b) Circadian clock-regulated phosphate transporter PHT4;1 plays an important role in Arabidopsis defense. Mol Plant 4: 516–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Gu Y, Zebell SG, Anderson LK, Wang W, Mohan R, Dong X (2014b) A noncanonical role for the CKI-RB-E2F cell-cycle signaling pathway in plant effector-triggered immunity. Cell Host Microbe 16: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Plovanich-Jones A, Deng WL, Jin QL, Collmer A, Huang HC, He SY (2000) The gene coding for the Hrp pilus structural protein is required for type III secretion of Hrp and Avr proteins in Pseudomonas syringae pv. tomato. Proc Natl Acad Sci USA 97: 2247–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC. (2010) Modulation of host nuclear ploidy: a common plant biotroph mechanism. Curr Opin Plant Biol 13: 449–458 [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, Conklin P, Britt A, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.