Chloroplast chaperone Hsp93 associates with the inner envelope membrane via interaction with the Tic110 translocon and likely functions in quality control but not in import propulsion of translocating preproteins.
Abstract
The Hsp100-type chaperone Hsp93/ClpC has crucial roles in chloroplast biogenesis. In addition to its role in proteolysis in the stroma, biochemical and genetic evidence led to the hypothesis that this chaperone collaborates with the inner envelope TIC complex to power preprotein import. Recently, it was suggested that Hsp93, working together with the Clp proteolytic core, can confer a protein quality control mechanism at the envelope. Thus, the role of envelope-localized Hsp93, and the mechanism by which it participates in protein import, remain unclear. To analyze the function of Hsp93 in protein import independently of its ClpP association, we created a mutant of Hsp93 affecting its ClpP-binding motif (PBM) (Hsp93[P-]), which is essential for the chaperone’s interaction with the Clp proteolytic core. The Hsp93[P-] construct was ineffective at complementing the pale-yellow phenotype of hsp93 Arabidopsis (Arabidopsis thaliana) mutants, indicating that the PBM is essential for Hsp93 function. As expected, the PBM mutation negatively affected the degradation activity of the stromal Clp protease. The mutation also disrupted association of Hsp93 with the Clp proteolytic core at the envelope, without affecting the envelope localization of Hsp93 itself or its association with the TIC machinery, which we demonstrate to be mediated by a direct interaction with Tic110. Nonetheless, Hsp93[P-] expression did not detectably improve the protein import efficiency of hsp93 mutant chloroplasts. Thus, our results do not support the proposed function of Hsp93 in protein import propulsion, but are more consistent with the notion of Hsp93 performing a quality control role at the point of import.
Chloroplasts are essential organelles in plant cells as they are responsible for performing a variety of functions (Jarvis and López-Juez, 2013). Although chloroplasts have their own genome (encoding approximately 100 proteins), the majority of the proteins found in these organelles are nucleus-encoded (approximately 3,000) (Leister, 2003), synthesized in the cytosol, and imported into the chloroplast as precursor proteins (preproteins), each one with a cleavable N-terminal extension or transit peptide (Shi and Theg, 2013a; Paila et al., 2015). The preprotein import mechanism is initiated by the interaction of the transit peptide with the translocon at the outer envelope membrane of chloroplasts (TOC) complex and subsequently involves transport through the translocon at the inner envelope membrane of chloroplasts (TIC) machinery in an energy-dependent process (Theg et al., 1989; Shi and Theg, 2013b). The Tic110 and Tic40 components have long been described as central TIC components, but these proteins were absent from a recently described 1-MD TIC complex (consisting of Tic20, Tic56, Tic100, and Tic214; Kovács-Bogdan et al., 2010; Nakai, 2015). One possible explanation is that two TIC complexes act sequentially during protein import (e.g. a Tic110-containing complex may act downstream of the 1-MD complex). A TIC complex associated import motor is proposed to exist at the stromal side of the inner envelope, and several stromal chaperones, including Hsp93/ClpC and Hsp70, have been proposed to act as motors to drive protein translocation into the stroma (for review, see Flores-Pérez and Jarvis, 2013).
Hsp93 is closely related to bacterial ClpC and is a member of the Class I subfamily of Hsp100 chaperones, which themselves belong to the wider AAA+ (ATPases associated with various cellular activities) superfamily (Hanson and Whiteheart, 2005; Flores-Pérez and Jarvis, 2013). AAA+ enzymes are involved in a variety of cellular processes, such as protein folding, unfolding for proteolysis, and disassembly of protein aggregates or protein complexes. Although AAA+ chaperones are well characterized in bacteria, they are found in all kingdoms (Hanson and Whiteheart, 2005). Such proteins possess one or two nucleotide binding domains, both of which contain conserved Walker A and B motifs. These chaperones may also contain a conserved ClpP-binding motif (PBM), or P-loop, which is essential for interaction with the unrelated, proteolytic ClpP subunit (Weibezahn et al., 2004; Hanson and Whiteheart, 2005).
In the chloroplast, Hsp93/ClpC partitions between the inner envelope membrane and the chloroplast stroma. Most Hsp93/ClpC protein is located in the stroma. Nonetheless, a large proportion of the total chloroplast Hsp93/ClpC pool (30%) associates with the envelope (Sjögren et al., 2014). Hsp93 has frequently been copurified with TIC and TOC complex components, which led to the hypothesis that it provides the driving force for preprotein import (Akita et al., 1997; Nielsen et al., 1997). Also, Hsp93 was found to specifically coimmunoprecipitate with preproteins under limiting ATP conditions and to stably bind to transit peptides in vitro (Nielsen et al., 1997; Rosano et al., 2011). Genetic and molecular studies have suggested that it functions in close association with Tic110 and Tic40 (Chou et al., 2003; Kovacheva et al., 2005; Chou et al., 2006). More recently, it was shown that the N-terminal domain of Hsp93 is important for its membrane association (Chu and Li, 2012). Despite all this evidence, the nature of the interaction between Hsp93 and the TIC apparatus has not been fully characterized.
Analysis of mutants also supported the involvement of the Hsp93 chaperone in protein import. In Arabidopsis (Arabidopsis thaliana), two homologous genes, atHSP93-V (CLPC1) and atHSP93-III (CLPC2), code for Hsp93/ClpC, and the resulting protein isoforms share 91% amino acid sequence identity (Kovacheva et al., 2007). The Hsp93-V protein is the most abundant isoform, and mutations in the atHSP93-V gene lead to a pale-green plant phenotype with protein import defective chloroplasts. In contrast, atHSP93-III knockout plants are indistinguishable from the wild type, most likely due to the compensatory presence of functionally redundant and abundant atHsp93-V (Kovacheva et al., 2005, 2007). Complete loss of both proteins in Arabidopsis is lethal during embryo development, whereas double mutants lacking Hsp93-V but retaining partial Hsp93-III activity are viable but exhibit severe chlorosis and protein import defects (Kovacheva et al., 2007).
More typically, as expected by its close relationship to bacterial orthologs, Hsp93/ClpC is a functional component of the caseinolytic protease (Clp) in the chloroplast stroma, where it recognizes and unfolds substrates for degradation (Shanklin et al., 1995). Significantly, the Clp proteolytic core is also bound to the envelope membranes, in quantities which are sufficient to bind to all of the similarly localized Hsp93/ClpC (Sjögren et al., 2014). This recent finding suggested a role for the Clp protease in protein quality control at the envelope. The structure of the Clp protease complex comprises a cylinder-like protease core and an AAA+ chaperone ring complex, and it is generally conserved throughout evolution (Nishimura and van Wijk, 2015). In Arabidopsis, the plastid Clp proteolytic core contains two distinct heptameric rings (the P-ring consisting of ClpP3-P6 and the R-ring consisting of ClpP1 and ClpR1-R4; Sjögren et al., 2006), and attached to this are accessory ClpT proteins involved in core assembly (Sjögren and Clarke, 2011). Several studies have shown that deficiency of the proteolytic subunits of the core complex leads to sick plant phenotypes (Sjögren et al., 2004; Rudella et al., 2006; Sjögren et al., 2006), highlighting the essential nature of Clp proteolytic activity to chloroplast function and plant viability.
As described above, the putative interacting partners of Hsp93 at the envelope are Tic110 and Tic40. Tic110 is a highly abundant protein and is essential for plastid biogenesis (Inaba et al., 2005; Kovacheva et al., 2007). It has two N-terminal transmembrane α-helices, and it projects a large C-terminal hydrophilic domain into the stroma (Jackson et al., 1998; Inaba et al., 2003). A stromal region proximal to the second transmembrane helix selectively associates with transit peptides, serving as a docking site for preproteins as they emerge from the TIC channel (Inaba et al., 2003). The hydrophilic domain of algal Tic110 possesses a rod-shaped helix-repeat structure similar to HEAT-repeat domains (and plant Tic110 proteins are predicted to be similar), and these typically function as scaffolds for protein-protein interactions (Tsai et al., 2013). Tic40 is topologically similar to Tic110 and is proposed to act as a cochaperone in the preprotein import motor (Chou et al., 2003). In the corresponding model, a transit peptide emerging from the TIC channel binds to the stromal domain of Tic110; this binding causes a conformational change of Tic110 to recruit Tic40, which in turn triggers transit peptide release to enable association of the preprotein with Hsp93 (Inaba et al., 2003; Chou et al., 2006). Finally, Tic40 is proposed to stimulate ATP hydrolysis by Hsp93 so that the chaperone pulls the preprotein into the stroma (Chou et al., 2006).
Although there is good evidence that Hsp93 is involved in protein import, the ability of Hsp93 to associate with the Clp protease core means that, in principle, any aspect of the hsp93 mutant phenotype could be due to disruption of the ClpP-linked functions of the protein. Bearing this in mind, we aimed to further characterize the role of Hsp93 at the inner envelope membrane. First, we analyzed the putative interactions of Hsp93 with the TIC components, Tic110 and Tic40, in a complementary set of in vitro and in vivo studies. Second, we evaluated the proposed role of Hsp93 in protein import independently of its role in proteolysis by creating a PBM mutant of the major Hsp93 isoform, atHsp93-V, and studying its activity in planta.
RESULTS
Hsp93 Association with the Envelope Involves a Direct Interaction with Tic110
It has long been known that Hsp93 associates with the stromal face of the TIC apparatus, but the means by which it does so remain ill-defined (Akita et al., 1997; Nielsen et al., 1997; Chou et al., 2006). To characterize this critical Hsp93-TIC interaction, we conducted a set of complementary in vitro and in vivo interaction studies, as detailed below.
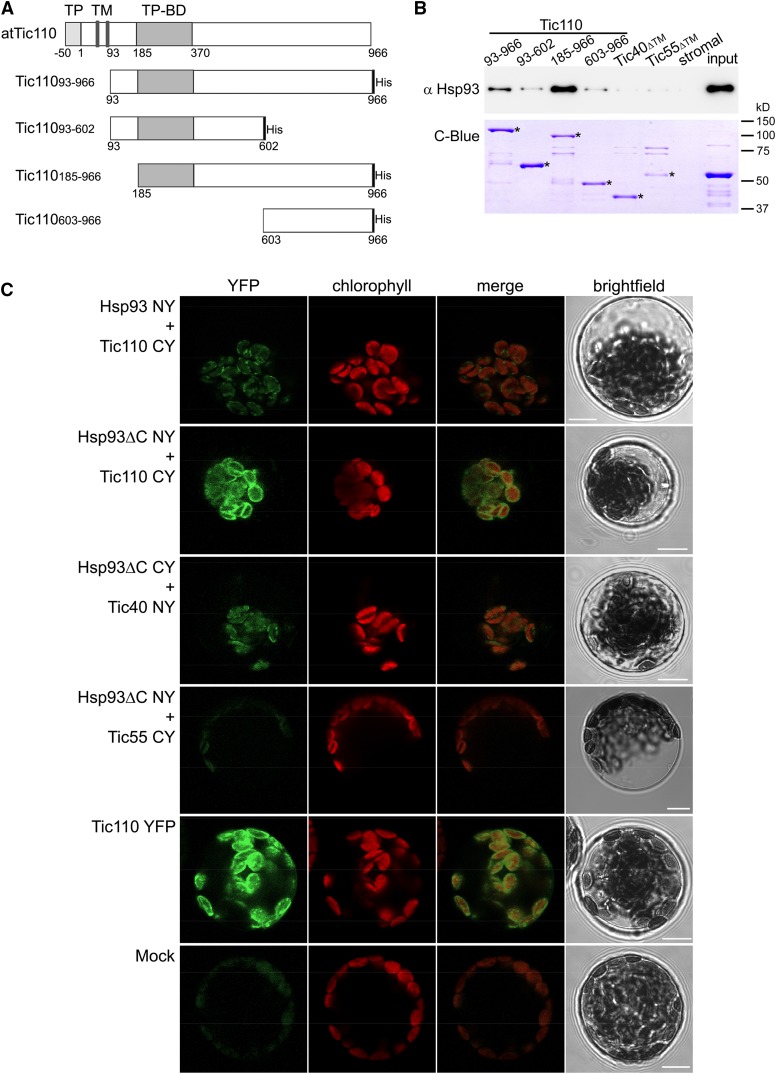
The Tic110 protein is reported to have a large domain protruding into the stroma that has been proposed to coordinate the activities of stromal chaperones (Jackson et al., 1998; Inaba et al., 2003). To investigate the possibility of a direct interaction between Hsp93 and Tic110, we first performed in vitro pull-down assays. To this end, we generated and purified various recombinant atTic110 domains, as shown in Figure 1A. All of the Tic110 domain constructs were made in pET vectors that add a 6×His-tag at the C terminus. The different constructs encode Tic11093-966 (complete stromal domain; Inaba et al., 2003), Tic11093-602 (lacks the extreme C terminus of the stromal domain), Tic110185-966 (lacks the extreme N terminus of the stromal domain; Inaba et al., 2003), and Tic110603-966 (comprises the C terminus of the stromal domain). The different domains were expressed in Escherichia coli and purified efficiently using nickel-nitrilotriacetic acid agarose (Ni-NTA) resin. All four proteins, together with Tic40ΔTM and Tic55ΔTM (ΔTM, lacking the transmembrane domain) controls that had been expressed and purified in the same manner, were tested for their interaction with Hsp93 by incubation with a soluble stromal extract followed by sample analysis using immunoblotting with Hsp93 antiserum. It is important to note that the Clp protease is not involved in the turnover of any these envelope subunits (i.e. Tic110, Tic40, and Tic55), so we would not expect to observe normal chaperone substrate-type interactions in these assays (Sjögren et al., 2014).
Figure 1.
Interaction of Hsp93 with the envelope protein Tic110. A, The depicted Tic110 domains were generated for in vitro interaction studies. TP, Transit peptide; TM, transmembrane domain; TP-BD, TP binding domain. The respective recombinant proteins, carrying a His-tag at the C terminus, were expressed in a bacterial system and purified, as were recombinant Tic40 and Tic55 (with no TM, ΔTM) control proteins. B, In vitro pull-down analysis to evaluate the interaction of Hsp93 with the Tic110, Tic40, and Tic55 recombinant proteins. Equimolar amounts (175 pmol) of immobilized His-tagged recombinant proteins were incubated (3 h at 4°C) with a stromal protein fraction (input) from isolated wild-type chloroplasts (approximately 200 μg). A control assay (stromal) conducted in the absence of His-tagged protein enabled evaluation of potential unspecific affinity between the stromal proteins and the Ni-NTA beads. The figure shows immunoblot analysis of 30% of the eluted proteins using an antibody against Hsp93. A fraction (20%) of the pulled-down protein was also analyzed by SDS-PAGE and Coomassie Brilliant Blue staining (C-Blue) to visualize the recombinant proteins; asterisks indicate the correct band for each recombinant protein. A sample (approximately 2 μg) of the stromal fraction input was similarly analyzed. C, In vivo analysis of the protein interactions by BiFC. Wild-type protoplasts were (co)transfected with the indicated constructs and analyzed by confocal microscopy for YFP fluorescence and chlorophyll autofluorescence (merged images of these two are also presented), and by bright-field illumination. Interaction of Tic110 or Tic40 with Hsp93 or with Hsp93ΔC was indicated by the reconstitution of YFP fluorescence, as can be seen in the representative images shown. No YFP signal was detected in assays for a potential Hsp93-Tic55 interaction (this served as a negative control). The Tic110 YFP panel illustrates a typical distribution of fluorescence at the envelope of the chloroplast. The Mock panel illustrates typical background levels of fluorescence seen in a nontransfected protoplast using imaging settings equivalent to those employed in the other panels. Bars = 10 μm.
Figure 1B shows a representative immunoblot identifying the extent of Hsp93 pull-down by the various Tic110 domains, Tic40ΔTM and Tic55ΔTM in these experiments. Because Tic110 possesses a transit peptide (TP) binding site near its N terminus, we had expected the C-terminal part to be responsible for binding Hsp93. However, the results (Fig. 1B) indicated that Hsp93 interacted most strongly with the Tic110185-966 form. It appears that when the putative TP binding site is exposed at the N terminus and the full-length C-terminal part of Tic110 is present, then the association of Hsp93 is enhanced. Other Tic110 domains displayed a weak affinity for Hsp93. No interaction of Hsp93 with either Tic40ΔTM or Tic55ΔTM was observed. These results were consistently observed in repeated experiments (each construct was tested up to four times), with no significant differences. The specificity of our results was further confirmed by the fact that we did not detect pull-down of other stromal proteins, including two different chaperones (Hsp70 and Cpn60), with any of the recombinant TIC proteins (Supplemental Fig. S1A).
To provide in vivo corroboration of the pull-down data, we assessed the Hsp93-Tic110 interaction by bimolecular fluorescence complementation (BiFC). For this purpose, yellow fluorescent protein (YFP) fragments (N-terminal YFP and C-terminal YFP; NY and CY, respectively) were fused to the C termini of the full-length atTic110 and atHsp93-V proteins (Bédard et al., 2007). As controls, we also analyzed the in vivo interaction of atHsp93 with atTic40 and atTic55. We additionally fused the full-length YFP to all the proteins tested for use as positive controls in the transfection experiments (Supplemental Fig. S1B).
Because BiFC analysis using the full-length Hsp93 construct presented certain technical difficulties (i.e. very low efficiency of transfection and low-intensity YFP fluorescence; data not shown and Supplemental Fig. S1B), we also created constructs encoding just the N-terminal domain of atHsp93-V (Hsp93ΔC), comprising its transit peptide (preprotein residues 1–93) and its N-terminal domain (preprotein residues 94–237; Fig. 2A); this domain of Hsp93 has been shown to be important for its membrane association (Chu and Li, 2012). We reasoned that this truncated construct might perform better in the BiFC assays.
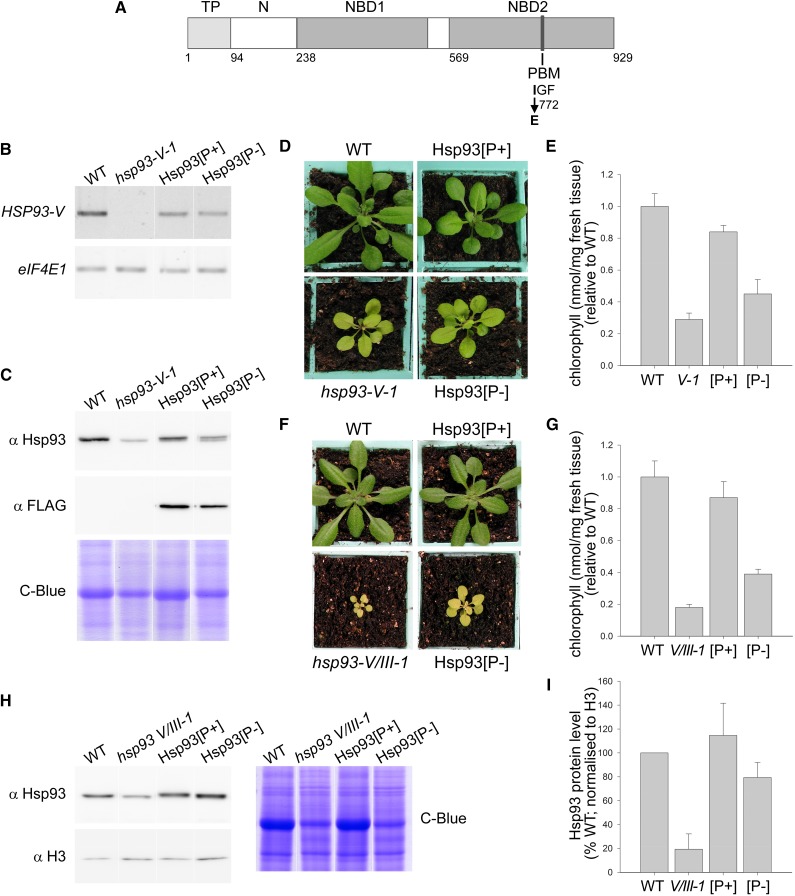
Figure 2.
Hsp93 PBM mutant complementation analysis. A, Diagram of the atHsp93-V protein showing functional domains and the I772E mutation. TP, Transit peptide; N, N-terminal domain; NBD, nucleotide binding domain; PBM, ClpP-binding tripeptide motif. B, Analysis of the expression of the transgenes by RT-PCR using gene-specific primers for HSP93-V and for translation initiation factor eIF4E1 as a control. Analyzed RNA samples were isolated from 10-d-old seedlings grown in vitro. C, Immunoblot analysis of the expression levels of the Hsp93[P+] and Hsp93[P-] proteins. The Hsp93-specific antibody detects total Hsp93 protein (i.e. it binds to Hsp93-V and Hsp93-III). Analyzed samples (10 μg per lane or 20 μg for Coomassie staining) were total protein extracts from 14-d-old plants grown in vitro. Coomassie Brilliant Blue (C-Blue) staining served as an equal loading control. D, Visible phenotypes of typical transgenic plants in the hsp93-V single-mutant background (T2 generation), after 27 d growing on soil. Wild-type and hsp93-V mutant plants were grown together as controls. E, Relative chlorophyll concentrations in the plant genotypes described in D. Analyzed leaves were from 35-d-old plants grown on soil. F, Visible phenotypes of typical transgenic plants in the hsp93-V hsp93-III-1 double-mutant background (V/III-1; hsp93-III-1 is a knockdown allele) (F3 homozygotes), after 25 d growing on soil. Wild-type and hsp93-V/III-1 double-mutant plants were grown together as controls. G, Relative chlorophyll concentrations in the plant genotypes described in F. Analyzed leaves were from 52-d-old plants grown on soil. H, Immunoblot analysis of Hsp93 protein levels in transgenic lines in the double-mutant background. Analyzed samples were total protein extracts from 14-d-old seedlings grown in vitro. Coomassie Brilliant Blue staining served as an equal loading control. I, Quantification of the immunoblots shown in H, together with additional similar blots. Data for Hsp93 protein levels were normalized using equivalent data for Histone 3 (H3). All values shown are means, and error bars indicate sd (n = 8, 8 and 3 for E, G and I, respectively).
As expected, YFP fluorescence in protoplasts transfected with the full-length YFP fusions was exclusively localized at the chloroplast envelope (for Tic110-YFP, Tic40-YFP, and Tic55-YFP; Fig. 1C; Supplemental Fig. S1B) or in the stroma (for Hsp93-YFP and Hsp93ΔC-YFP; Supplemental Fig. S1B), confirming the suitability of our constructs for the BiFC assay.
When we analyzed the double-transfected protoplasts expressing different NY+CY combinations, the results validated our earlier in vitro observations. Hsp93 and Tic110 were seen to interact at the chloroplast envelope, as revealed by the reconstitution of the YFP fluorescence in protoplasts coexpressing the Hsp93-NY and Tic110-CY constructs (Fig. 1C). Moreover, the results showed that the N-terminal domain of Hsp93 (Hsp93ΔC) successfully associates with Tic110 at the envelope, indicating that this domain is sufficient for the interaction (Hsp93ΔC -NY + Tic110-CY; Fig. 1C). The truncated Hsp93 protein (Hsp93ΔC) produced a stronger YFP signal than the full-length Hsp93 construct, both in the context of the BiFC interaction assay and when fused to intact YFP (Supplemental Fig. S1B). The frequency of the transformed (fluorescent) protoplasts, relative to untransformed (nonfluorescent) protoplasts, observed for Hsp93ΔC-Tic110 heterodimers, was comparable to that seen for the full-length Tic110-YFP construct, suggesting that this interaction is highly efficient in vivo (Supplemental Fig. S1C).
Previous biochemical and genetic data indicated that Hsp93 likely cooperates functionally with both Tic110 and Tic40 (Chou et al., 2003; Kovacheva et al., 2005). Accordingly, in our BiFC experiments, we found that Hsp93 can also interact with Tic40 (Hsp93ΔC -NY + Tic40-CY; Fig. 1C). However, we consistently found that the fluorescent signal arising from this interaction was considerably weaker than that resulting from the corresponding Tic110 interaction (Fig. 1C). Moreover, protoplasts showing such an Hsp93ΔC-Tic40 interaction occurred at low efficiency (Supplemental Fig. S1C). We interpret these observations to indicate that the Hsp93-Tic40 interaction is intrinsically weak or transient in nature. The fact that this interaction could be detected by BiFC but not in the context of our in vitro pull-down experiments (Fig. 1B) may reflect its stabilization by the reconstitution of the YFP structure in the BiFC assays (Hu et al., 2002). Alternatively, it may reflect an indirect interaction of Hsp93 and Tic40, mediated via the close proximity of both proteins to Tic110, which would not be detectable in the in vitro system (Fig. 1B; Ohad et al., 2007). As expected, we did not detect any interaction between Hsp93ΔC and Tic55 (Fig. 1C), confirming the specificity of the results.
Generating Transgenic Plants Expressing PMB Mutant Hsp93
In all tested Hsp100 chaperones (e.g. E. coli ClpA and ClpX, Helicobacter pylori ClpX, Synechococcus ClpC, and Arabidopsis ClpC1, ClpC2, and ClpD), the PBM is a strictly conserved IG(F/L) motif, and in atHsp93-V/ClpC1 it is at position 772–774 of the preprotein (Fig. 2A; Kim et al., 2001; Kim and Kim, 2003; Joshi et al., 2004; Tryggvesson et al., 2012; Nishimura and van Wijk, 2015). To specifically disrupt the ClpP-dependent functions of atHsp93-V, we generated an I772E mutant by PCR mutagenesis of the atHSP93-V cDNA; corresponding mutations in several other Hsp100 proteins are well known to disrupt PBM function and ClpP binding (Kim et al., 2001; Singh et al., 2001; Joshi et al., 2004). To assess the effect of this mutation in vivo, we used the pH2GW7 vector (modified to include the atHSP93-V native promoter), which adds a C-terminal FLAG-tag, to create transgenic plant lines expressing the PBM mutant form of Hsp93, as well as control transgenic lines expressing the wild-type form of Hsp93 from the same vector. The resultant transgenic lines are denoted Hsp93[P-] and Hsp93[P+], respectively. For this complementation analysis, we used Arabidopsis hsp93 mutants (Kovacheva et al., 2005, 2007).
Assuming that Hsp93 is involved in both protein import and proteolysis, as has been proposed, the chlorosis in hsp93-V is likely to be due to a combination of both import and proteolysis defects. Thus, it was expected that Hsp93[P-] and Hsp93[P+] transgenic lines would display different results: The former, where ClpP-related defects persist, would exhibit only partial complementation, whereas the latter should display complete complementation of the hsp93 mutant phenotype.
Several transformants were obtained for each construct (Hsp93[P+] and Hsp93[P-]) in the hsp93-V-1 mutant background, and the lines were initially selected based on visible phenotype. For the Hsp93[P+] construct, we analyzed seven T1-transformants, and all of them showed a full recovery of the hsp93 mutant phenotype, as expected. Together with the visible phenotype, we used transgene segregation and mRNA and protein expression data to select a suitable homozygous Hsp93[P+] line for further analysis; during the course of the selection process, we also took into consideration corresponding data on the Hsp93[P-] lines. For the Hsp93[P-] construct, the results were quite different: None of the lines displayed full complementation, but six T1-transformants did display partial complementation of the hsp93 phenotype.
In five of the six partially complementing Hsp93[P-] lines, the selectable marker associated with the T-DNA insertion showed a resistance:sensitivity ratio of 3:1 when the T2 generation seeds were plated on selective medium, indicating the presence of a single T-DNA insertion site in each one. Upon analysis of the lines in the T3 generation, we were able to identify homozygous lines for only two of the five single-locus Hsp93[P-] lines. Therefore, we selected these two homozygous Hsp93[P-] lines (#4 and #7) for preliminary analysis and characterized them based on visible phenotype, mRNA, and protein expression (data not shown; see below). Based on the data, line Hsp93[P-] #4 was employed for all subsequent, detailed analyses. This line will henceforth be called Hsp93[P-].
To confirm that each of the lines was expressing the relevant construct, we isolated total RNA from seedlings grown in vitro and analyzed transgene expression by RT-PCR using HSP93-V gene-specific primers. The results indicated that the Hsp93[P+] and Hsp93[P-] transgenes were both expressed in the selected lines (Fig. 2B). Because the selected Hsp93[P-] line (#4) expressed HSP93-V at a lower level than the wild type, we selected an Hsp93[P+] line (line “B”) with a similarly low expression level, in order to enable more accurate comparisons between the two constructs. This low mRNA expression level was seen to produce a sufficient amount of Hsp93[P+] protein to complement the mutant phenotype (see below), so we reasoned that the levels of Hsp93[P-] protein would also not be limiting. Semiquantitative RT-PCR analysis revealed that the Hsp93[P+] and Hsp93[P-] constructs differed in expression level by <20% (data not shown). Moreover, immunoblot analysis confirmed that both transgenes drive expression of Hsp93 (Fig. 2C). As expected, the amount of Hsp93 protein produced in the transgenic lines was low compared to that seen in the wild type (Fig. 2C); note that the Hsp93 antibody used binds to both Hsp93-V and Hsp93-III, so that in the immunoblot it was possible to detect Hsp93-III in the hsp93-V mutant and the transgenic lines. Immunodetection of the C-terminal FLAG-tag also showed that the Hsp93[P+] and Hsp93[P-] proteins were expressed and that they accumulated at broadly equivalent levels (Fig. 2C).
The FLAG-tag Does Not Interfere with the Functionality of Hsp93 in the Transgenic Lines
We expected that the eight amino acids of the FLAG tag would not interfere with Hsp93 function. However, as a control to formally eliminate this possibility, we identified and selected, using similar procedures and criteria, Hsp93[P+] and Hsp93[P-] lines using nontagged constructs, and we distinguished them as Hsp93[P+]* and Hsp93[P-]* (Supplemental Fig. S2). We analyzed 7 T1-transformants for the Hsp93[P+]* construct, and all of them showed a full recovery of the hsp93 mutant phenotype, as expected. For the Hsp93[P-]* construct, we analyzed 12 T1 transformants that displayed partial complementation of the hsp93 phenotype. In seven of the 12 partially complemented lines, the selectable marker associated with the T-DNA insertion showed a resistance:sensitivity ratio of 3:1 in the T2 generation, and these lines were all carefully analyzed. Using the same criteria as used for the FLAG-tagged lines, we selected Hsp93[P-]* line #3 and Hsp93[P+]* line #8, which displayed similar levels of transgene expression enabling better comparisons between the constructs. Semiquantitative RT-PCR analysis showed that the expression level of Hsp93[P+]* and Hsp93[P-]* constructs differed by <2% (data not shown). The selected nontagged lines showed similar results to the tagged transformants in terms of visible complementation of the hsp93-V phenotype (i.e. growth and chlorophyll content; Supplemental Fig. S2, A and B) and in terms of the level of expression (i.e. mRNA and protein expression; Supplemental Fig. S2, C and D). Therefore, we concluded that the FLAG-tag does not interfere with the results.
In general, during the screening for Hsp93[P-] and Hsp93[P-]* T1-transformants, we observed that both constructs had a negative effect on plant vigor in high-level expressing lines (Supplemental Fig. S3). We previously demonstrated that Hsp93 is essential for plant viability, as the double knockout mutant genotype (affecting both Arabidopsis genes) causes lethality (Kovacheva et al., 2007). We speculate that Hsp93[P-] interferes with the functionality of residual, native Hsp93-III when expression levels are high, such that most of the Hsp93 hexamers formed are unable to associate with the Clp core complex, leading to the observed worsened phenotypes (Supplemental Fig. S3). However, when the expression of Hsp93[P-] is low, it may partially complement the mutant phenotype by enabling the formation of more Hsp93 hexamers; these hexamers would be hetero-oligomeric, comprising Hsp93[P-] and native Hsp93-III, and so able to associate with the core complex (see below).
The PBM of Hsp93 Is Essential for Normal Functionality of the Protein
The visible phenotypes of the Hsp93[P+] and Hsp93[P-] plants were observed under in vitro growth conditions (data not shown) and following growth on soil (Fig. 2D). The hsp93-V mutant displays a clear pale, chlorotic phenotype compared to the wild type, but when carrying the Hsp93[P+] construct, it developed to nearly full, wild-type size and accumulated almost wild-type levels of chlorophyll (Fig. 2, D and E). However, the effect of the Hsp93[P-] construct was quite different, as it only partially complemented the hsp93-V-1 phenotype in relation to both the size of the plants and chlorophyll accumulation (Fig. 2, D and E).
In order to assess the properties of the two Hsp93 constructs with less potentially complicating effects of Hsp93-III, the selected Hsp93[P+] and Hsp93[P-] lines were transferred into the hsp93-V hsp93-III double-mutant background. The hsp93-III-2 mutation is a complete knockout allele, while hsp93-III-1 is a knockdown allele (approximately 50% wild-type expression level; Kovacheva et al., 2007), and both corresponding double mutants exhibit more severe phenotypes than the single hsp93-V mutant (Kovacheva et al., 2007). We tried to identify complemented double homozygotes using the hsp93-III-2 knockout allele but, we were unable to obtain any (data not shown). However, when the constructs were transferred into the hsp93-V hsp93-III-1 (V/III-1) background, we observed that, as expected, the double-mutant V/III-1 phenotype was fully complemented by Hsp93[P+] and only slightly rescued by Hsp93[P-], as judged by both visible phenotype and chlorophyll content analyses (Fig. 2, F and G). These lines were also assessed for Hsp93 content by immunoblotting and were found to contain roughly the same amount of protein as the wild type after normalizing with histone H3 (Fig. 2, H and I). Our failure to identify double homozygotes using the knockout hsp93-III allele may indicate that plants need a background pool of Hsp93 with an intact PBM to be viable. All together, these results are entirely consistent with those obtained in the hsp93-V single mutant background and indicate normal functionality of the Hsp93[P+] construct and seriously impaired functionality of Hsp93[P-]. The data clearly indicate the essential role of the PBM for Hsp93 function.
The PBM Mutation Negatively Affects Proteolytic Activity of the Stromal ClpP Core
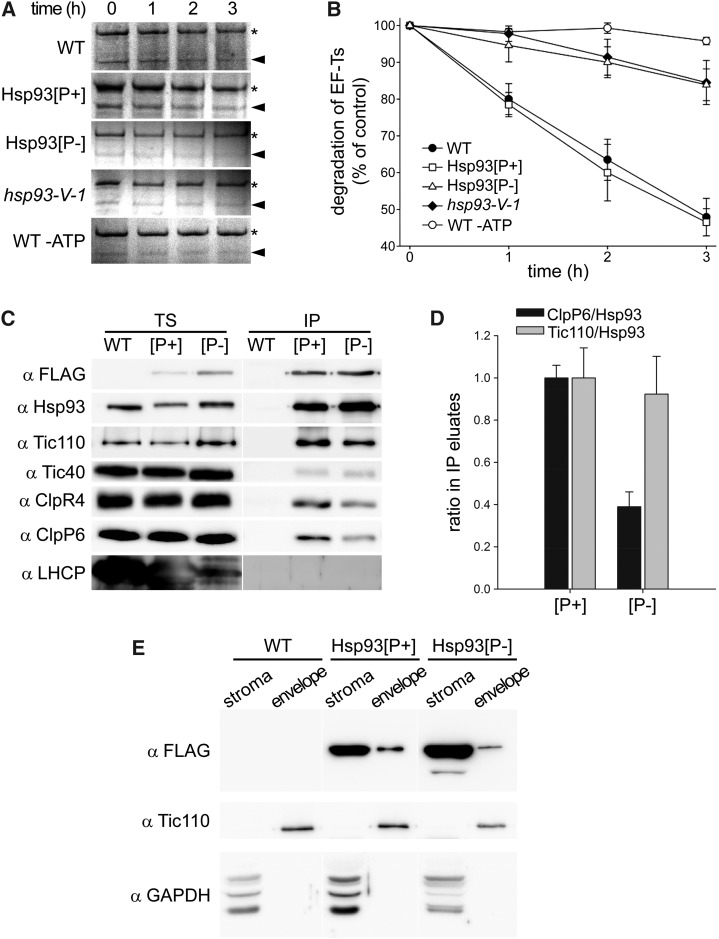
We next examined if the failure of the Hsp93[P-] protein to complement the hsp93 mutants was related to its presumed inability to associate with the Clp proteolytic core. To test this, we used an in organello degradation assay that had previously identified almost 30 putative substrates for the stromal Clp protease in wild-type Arabidopsis (Sjögren et al., 2006; Stanne et al., 2009). Intact chloroplasts were isolated from the Hsp93[P-] and Hsp93[P+] lines as well as from the hsp93-V mutant and wild-type Arabidopsis. The Clp proteolytic activity in these chloroplasts was measured using one of the previously identified degradation targets (elongation factor-Ts [EF-Ts]) as a model substrate. As shown in Figure 3A, the Hsp93[P+] line exhibited the same degradation rates of EF-Ts as the wild type, confirming that the Hsp93[P+] protein restores Hsp93-V function to the hsp93-V mutant. In contrast, the Hsp93[P-] protein failed to increase EF-Ts degradation beyond that observed in the hsp93-V mutant, implying that mutation of the PBM site in the Hsp93[P-] protein had indeed abolished its ability to bind to the Clp proteolytic core. It should be noted that the low degradation rates of EF-Ts in hsp93-V and the Hsp93[P-] line are almost certainly due to the Hsp93-III protein that remains in these plants, the activity of which can be inhibited by excluding ATP in the assay (Fig. 3B).
Figure 3.
The Hsp93 PBM mutation compromises Clp proteolytic activity but does not prevent the TIC complex association or envelope localization of Hsp93. A and B, Degradation of stromal EF-Ts in wild-type Arabidopsis (WT), hsp93-V single mutant (hsp93-V-1), and the Hsp93[P+] and Hsp93[P-] lines in the single-mutant background. Equal amounts of intact chloroplasts were isolated from each line and incubated for 0 to 3 h in the presence of light and ATP, with the exception of the wild-type control reaction, which was incubated in the dark in the absence of ATP (WT -ATP). Fractionated stroma from different time points was separated by SDS-PAGE and proteins visualized by Coomassie Brilliant Blue staining. The previously identified substrate protein EF-Ts (indicated by arrowheads) was quantified during the time course, with a representative replicate shown in A and the quantification shown in B. Asterisks in A indicate an unrelated band of unknown identity employed as an internal point of reference. Values shown in the graph are means, and error bars indicate se (n = 3); the amount of EF-Ts in the 0 h sample was set to 100% and other values were normalized accordingly. C, Analysis of Hsp93 interactions by anti-FLAG immunoprecipitation of Hsp93[P+] and Hsp93[P-] FLAG-tagged proteins from cross-linked (0.25 mm DSP) isolated chloroplasts. Analyzed samples were from 14-d-old plants grown in vitro; the Hsp93[P+] and Hsp93[P-] transgenic lines were in the double-mutant background for this analysis. TS, Total soluble input; IP, immunoprecipitates. Fractions of each TS and IP (approximately 3% and 30%, respectively) were analyzed by immunoblotting for the presence of transgene-encoded Hsp93 (αFLAG; αHsp93 additionally detects residual levels of native Hsp93-III) and its interaction partners (αTic110, αTic40, αClpR4, and αClpP6); analysis for the presence of LHCP served as a negative control. Representative figures for each immunoblot are shown. D, Calculation of the ClpP6/Hsp93 and Tic110/Hsp93 ratios in the IP eluates in C and in two further similar experiments. Ratio values were calculated relative to the respective transgenic Hsp93 protein ([P+] or [P-]) by employing the anti-FLAG data. Values shown in the graph are means, and error bars indicate se (n = 3). E, Analysis of Hsp93 localization by immunoblot analysis of purified fractions (stroma and envelope) from isolated chloroplasts of the wild type and the Hsp93[P+] and Hsp93[P-] transgenic lines (single-mutant background). Plants grown for 14 d in vitro were used to isolate chloroplasts for fractionation (150 million per genotype), and 5% of each purified fraction was analyzed by immunoblotting as shown. An anti-FLAG antibody was used to detect transgene-encoded Hsp93. Tic110 and GAPDH served as envelope and stromal protein controls, respectively.
The PBM Mutation Disrupts Association to the ClpP Proteolytic Core But Not to the TIC Complex
To assess the effect of the PBM mutation on the association of Hsp93[P-] with interacting partners, we employed anti-FLAG immunoprecipitation of Hsp93 using detergent dodecyl maltoside (DDM)-solubilized, isolated chloroplasts treated with dithiobis succinimidylpropionate (DSP) cross-linker. Analysis of the precipitates by immunoblotting using specific antibodies against the proteolytic core subunits ClpR4 and ClpP6 revealed substantially reduced ClpRP binding to the Hsp93[P-] protein (Fig. 3, C and D). In this analysis, the work was conducted in the double-mutant V/III-1 background to minimize the complicating effect of the Hsp93-III isoform. Amounts of ClpP6 in the Hsp93[P-] immunoprecipitation samples were significantly reduced, such that the ClpP6/Hsp93 ratio in the Hsp93[P-] eluate was reduced to approximately one-third of that seen in the Hsp93[P+] eluate (Fig. 3, C and D). We attribute the residual association of the ClpRP proteolytic core with Hsp93[P-] to the formation of mixed complexes (stabilized by DSP) containing native atHsp93-III (the minor isoform), which retains a normal PBM. On the other hand, the interaction of Hsp93[P-] with the TIC apparatus component Tic110 was not compromised by the PMB mutation (Fig. 3, C and D), which is consistent with results described below indicating normal envelope association of the Hsp93[P-] protein. In accordance with the BiFC data (Fig. 1C; Supplemental Fig. S1C), this analysis also showed a weak interaction between the Hsp93[P+] and Hsp93[P-] proteins and Tic40. In this analysis, a control protein (light-harvesting chlorophyll-binding protein [LHCP]) was not detected in the precipitates, indicating the specificity of the experiment (Fig. 3C).
To evaluate the effect of the I772E PBM mutation on the localization of Hsp93[P-], we purified stromal and envelope fractions from isolated chloroplasts of our selected lines (in the hsp93-V single-mutant background) and analyzed for the presence of Hsp93 by immunoblotting (Fig. 3E). We used antibody against the FLAG-tag to detect the products of the transgenes and antibodies against Tic110 and GAPDH as markers for the envelope and stroma, respectively. The analysis showed that both Hsp93[P+] and Hsp93[P-] are appropriately abundant in the stroma and present to a lesser extent in the envelope, in line with a previous report (Sjögren et al., 2014). Thus, the PBM mutation does not prevent the envelope localization of the Hsp93[P-] protein.
Effect of the PBM Mutation on the Association of the ClpP Proteolytic Core at the Envelope
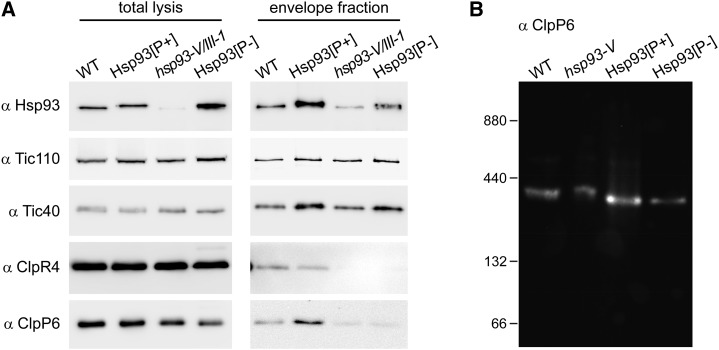
As described above, the PBM mutation disrupts the interaction of Hsp93 with its ClpP6 interacting partner (Fig. 3, C and D). To specifically evaluate the effect of the PBM mutation on Hsp93 at the envelope, and on association of the Clp proteolytic core with the envelope, we isolated envelopes from the Hsp93[P+] and Hsp93[P-] lines (in the hsp93-V/III-1 double-mutant background) and analyzed them for the presence of Hsp93 and other subunits of the Clp protease complex (i.e. ClpR4 and ClpP6) by immunoblotting, as well as for TIC apparatus components as envelope markers (Fig. 4A). The results were consistent with those shown in Figure 3, C and D, which were generated using total chloroplast protein samples. Specifically, the analysis in Figure 4A demonstrated that the PBM mutation interferes with the association of the proteolytic core with the envelope: Levels of ClpP6 and ClpR4 were normal in Hsp93[P+] envelope fractions, but very low in Hsp93[P-] envelope fractions (comparable to those detected in hsp93-V/III-1 envelopes). This indicates that the disruptive effect of the PBM mutation on Clp proteolytic core binding also occurs with the envelope-associated fraction of Hsp93 and implies that association of the core with the envelope is mediated by its interaction with Hsp93 (which in turn is mediated by interaction between the Hsp93 N terminus and Tic110; this study; Chu and Li, 2012). These conclusions were supported by a blue native PAGE analysis of envelope fractions isolated from the selected lines (in the hsp93-V single-mutant background), which confirmed that levels of the Clp proteolytic core (approximately 325 kD in size) (Sjögren and Clarke, 2011) attached to the envelope membrane are restored in the Hsp93[P+] line, but not in the Hsp93[P-] line (Fig. 4B).
Figure 4.
Effect of the Hsp93 PBM mutation on the association of the Clp proteolytic core to the envelope membranes. A, Analysis of the envelope association of the Clp proteolytic core by envelope fractionation and immunoblotting. Representative immunoblot analyses of whole isolated chloroplasts (total lysis) (isolated from 18 d old seedlings grown in vitro) and of purified envelope fractions, from the Hsp93[P+] and Hsp93[P-] transgenic lines (double-mutant background) and from the indicated controls, are shown. Amounts equivalent to approximately 1% of each total lysate and approximately 10% of each envelope fraction were analyzed using the indicated antibodies. Envelope samples did not show any stromal contamination when analyzed for the presence of a stromal marker protein, GAPDH, as in Figure 3E (data not shown). B, Analysis of the Clp proteolytic core in envelope fractions by blue native PAGE. Envelope membrane samples (equivalent to 15 µg of protein) isolated from leaves of the Hsp93[P+] and Hsp93[P-] transgenic lines (single-mutant background), and from the indicated controls, were separated by blue native PAGE. The Clp proteolytic core was detected using antibodies specific for the ClpP6 subunit. Sizes of the molecular mass markers (ferritin dimer [880 kD] and monomer [440 kD], BSA dimer [132 kD] and monomer [66 kD]) are shown on the left.
The Hsp93[P-] Protein Cannot Complement the Protein Import Defect of hsp93 Mutant Chloroplasts
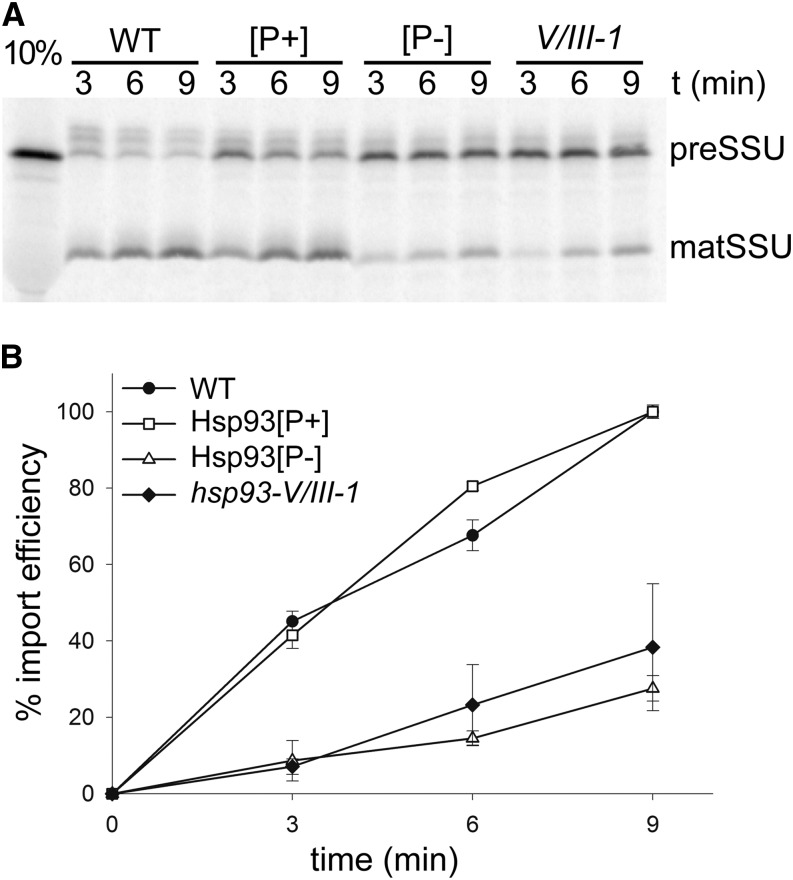
To evaluate the role of Hsp93 in the protein import mechanism at the envelope, we assessed chloroplast protein import efficiency of the Hsp93[P+] and Hsp93[P-] lines, in both hsp93-V single-mutant and double-mutant (V/III-1) backgrounds. For these experiments, we used the precursor of the small subunit of Rubisco (preSSU) as a model client to determine import efficiencies into isolated chloroplasts. Import efficiencies were determined by measuring the accumulation of imported protein (SSU, mature protein) over time, represented as a percentage of the amount of imported protein in wild-type organelles at the last point of the time course.
For the experiments assessing import in the double-mutant background, import efficiency was obviously increased in the Hsp93[P+] chloroplasts, relative control mutant plastids, as more mature SSU can be seen at each time point (Fig. 5A). However, there was no significant change in import in Hsp93[P-] chloroplasts when compared to the double mutant. After quantification of the SSU band in repeated experiments, the results indicated that the Hsp93[P+] construct fully complements the protein import deficiency of the double mutant (Fig. 5B); the amount of imported protein in Hsp93[P+] organelles was in fact identical to that in the wild type (efficiency of 100%), at the last time point of the experiment. On the other hand, quantification of the amount of SSU imported into Hsp93[P-] chloroplasts indicated that there was no complementation whatsoever of the import deficiency of the double mutant; the amount of imported protein in Hsp93[P-] chloroplasts was 27.6% (±3.4%) at the last time point, which is not more than the corresponding value for the mutant of 38.3% (±16.7%; Fig. 5B).
Figure 5.
The Hsp93 PBM mutation does not restore the protein import deficiency of the hsp93 double mutant. A, Analysis of protein import into chloroplasts isolated from the Hsp93[P+] and Hsp93[P-] transgenic lines (in the hsp93-V/III-1 double-mutant background) and from the indicated controls. Chloroplasts isolated from 14-d-old plants were incubated with in vitro translated, [35S]Met-labeled preSSU in the presence of 5 mm ATP for the indicated time periods. Samples were analyzed by SDS-PAGE and visualized using a phosphor imager, and a representative image is shown. preSSU, Precursor protein form; matSSU, mature protein form; 10%, 10% of the translation product added to each import reaction. B, The amount of protein imported into chloroplasts of each genotype in A (and in additional, similar experiments) was quantified by measuring the mature SSU band. The data are presented as a percentage of the amount of imported protein for the wild type at the last point of the time course, and expressed as “% import efficiency.” All values shown are means, and error bars indicate sd (n = 3).
Very similar results were obtained when assessing import efficiencies in the hsp93-V single-mutant background, with two different sets of constructs: Hsp93[P+] and Hsp93[P-] (data not shown) and Hsp93[P+]* and Hsp93[P-]* (Supplemental Fig. S2, E and F), providing robust support for the conclusion that the Hsp93[P-] protein is not able to significantly improve chloroplast protein import efficiency. Nonetheless, the visible phenotype and slightly enhanced chlorophyll levels (Fig. 2, D–G; Supplemental Fig. S2, A and B) of our selected Hsp93[P-] transgenic lines implied that the PBM mutant form of Hsp93 was functional in some way, improving the general condition of the chloroplasts. Taking into consideration the widely discussed hypothesis that Hsp93 has two roles (a proteolytic role in the stroma linked to ClpP and an import role at the envelope unrelated to ClpP), we expected that Hsp93[P-] chloroplasts would show improvements in protein import rates. However, our import results did not support this view, suggesting that the aforementioned hypothesis may not be wholly accurate.
DISCUSSION
In this work, we have shown that Hsp93 is associated with the TIC machinery via Tic110, a fact that has long been assumed but never clearly demonstrated. Our pull-down results showed that the Tic110185-966 domain forms the strongest or most stable interaction with Hsp93, more so even than the full-length hydrophilic domain, Tic11093-966. The Tic110185-966 was previously reported to contain a minimal transit peptide binding site (residues 185–370), displaying an in vitro precursor-binding efficiency of approximately 20% relative to that observed for full-length Tic110 (Inaba et al., 2003). Our results indicate that neither the N terminus (Tic11093-602) nor the C terminus (Tic110603-966) has high affinity for interaction in isolation, suggesting that the determinant for Hsp93 interaction spans these N-terminal and C-terminal regions. Like an algal ortholog, Tic110 of pea (Pisum sativum; Tsai et al., 2013) and Arabidopsis (this study; data not shown) are predicted to possess HEAT repeats distributed along the polypeptide chain of the stromal domain, according to the repeated-protein-motif prediction server HHrePID (Biegert and Söding, 2008). In the folded protein, such repeats are arranged about a common axis forming a superhelix or rod-like structure that is typically involved in protein interactions, and this might fail to perform properly if component sequences are lost or mutated (Andrade et al., 2001; Scheer et al., 2012).
In our in vitro system, the small N-terminal region comprising residues 93 to 185 apparently has a negative effect on the ability of Tic110 to interact with its protein partner Hsp93. This might indicate a regulatory role of the N-terminal region in coordinating interactions between Tic110 and its functional partners (e.g. in response to transit peptide arrival). Alternatively, the negative effect of the N terminus on binding could be an artificial consequence of the experimental system. For example, it might be a flexible domain which, when no longer anchored in the chloroplast envelope, can interfere with Hsp93 binding. Alternatively, the Tic110185-966 protein may simply adopt a more favorable structure that exposes the relevant domains (i.e. HEAT repeats) for protein interactions. It should be remembered that, in planta, the Tic110 protein is anchored to the envelope by its N-terminal end, whereas in our in vitro system, the His-tag used for attachment to the matrix is located at the C terminus. Regardless of these issues, our results clearly reveal an interaction between Tic110 and Hsp93, and this was confirmed using an in vivo approach (BiFC), which showed that Hsp93 binds directly to Tic110 at the chloroplast envelope. Moreover, the BiFC results show that the N-terminal domain of Hsp93 is sufficient to mediate this interaction, as was previously suggested (Chu and Li, 2012). When considered together, our in vivo and in vitro interaction results also indicate that Hsp93 interacts transiently or weakly with Tic40, which is potentially consistent with the proposed cochaperone role of the latter protein (Chou et al., 2006).
Because of its association with Tic110 and other translocon components, Hsp93 has been assumed to be a component of a motor that drives protein import into chloroplasts (Akita et al., 1997; Nielsen et al., 1997; Chou et al., 2003, 2006), potentially acting downstream of the recently identified 1-MD TIC complex (Kikuchi et al., 2013). However, this notion was challenged by recent findings showing a stable interaction of the Clp protease complex with its Hsp93 partner attached to the envelope (Sjögren et al., 2014). In our work, we attempted to study specifically the function of Hsp93 in the import mechanism by disrupting its association with the Clp proteolytic core using the Hsp93[P-] transgenic lines. Characterization of the transgenic lines indicated that the PBM mutation of Hsp93 effectively disrupts its association with the Clp proteolytic core and, as expected, impairs its degradation activity. We observed slight, partial complementation of the hsp93 phenotype in those transgenic lines showing low expression of Hsp93[P-]. Our initial hypothesis was that this was due to the selective complementation of import-related, ClpP-independent defects of the hsp93 mutants. However, our data were inconsistent with this notion. Instead, we favor an alternative hypothesis in which the complementation effect is linked to partial recovery of Clp proteolytic activity due to the formation of mixed hexameric rings comprising Hsp93[P-] and residual Hsp93-III (we observed no significant increase in the expression of atHSP93-III in our transgenic lines in RT-PCR experiments; data not shown). The fact that high level expression of Hsp93[P-] causes phenotypes that are more severe than that of the hsp93 mutant is consistent with this hypothesis, as this can be interpreted as a dominant-negative effect whereby Hsp93[P-] interferes with Hsp93-III functionality when its expression levels are too high. That the putative positive effect of low-level Hsp93[P-] expression on proteolytic activity was not detected in our degradation assays may simply indicate that these assays lack the sensitivity needed to measure such small changes. Alternatively, the partial complementation mediated by Hsp93[P-] may reveal a ClpP-independent role of the Hsp93 chaperone that contributes in a minor way to chloroplast homeostasis, a function reported for other Hsp100 chaperones (Burton et al., 2001; Weibezahn et al., 2004).
The Hsp93[P-] protein failed to complement the protein import defects caused by the hsp93-V/III-1 and hsp93-V mutations in our transgenic lines (Hsp93[P-] and Hsp93[P-]*, respectively). The inability of Hsp93[P-] to support improved preprotein import is inconsistent with the widely discussed hypothesis that Hsp93 acts as an import motor. Generally, defects in the biogenesis of chloroplasts have been described for other mutants of the Clp core complex. Recently, it was reported that clpr1 mutant chloroplasts are protein import defective (Sjögren et al., 2014). Taking into account recent evidence that some Clp protease complex is localized at the envelope, the protein import defects seen in the Hsp93[P-] transgenic lines (this study) and in the hsp93 and clpr1 mutants (Constan et al., 2004; Sjögren et al., 2004; Kovacheva et al., 2005; Kovacheva et al., 2007) suggest an important role for the Clp protease complex during or immediately following the import of nucleus-encoded proteins. This might take the form of a quality control mechanism serving to remove defective or otherwise unwanted proteins upon arrival. If such a quality control mechanism were closely connected to the protein import mechanism (as is suggested by the direct association of Hsp93 with Tic110), then a defect in quality control might be expected to also disrupt protein import.
Recent reports have revealed the central involvement of Hsp70 in protein translocation into chloroplasts, in organisms from moss to higher plants (Shi and Theg, 2010; Su and Li, 2010), and have shown that the energetics of protein import are dominated by the stromal Hsp70 (Liu et al., 2014). A stromal Hsp90 chaperone is also involved in protein import (Inoue et al., 2013). Thus, various stromal chaperones contribute to the protein import mechanism, and they may act at different stages in the process. As Hsp93 appears not to confer the ATP-dependent force that drives preprotein import (a function apparently fulfilled by Hsp70), a role during a downstream step in the process (such as quality control) seems likely. This view is in accordance with the proposal that Hsp93 and Hsp70 act independently in the import process (Su and Li, 2010).
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants were Columbia-0 ecotype. The hsp93-V-1, hsp93-III-2, hsp93-III-1, and double hsp93-V/hsp93-III mutants have been described previously (Kovacheva et al., 2005, 2007). In vitro plant growth employed Murashige and Skoog medium with a long-day photoperiod (16 h light/8 h dark) according to published procedures (Kubis et al., 2008). Plants grown on soil were kept under standard greenhouse conditions with long-day photoperiods. When necessary, antibiotics were included in the medium: 15 μg/mL hygromycin B (Duchefa) for the selection of transformants, 10 μg/mL dl-phosphinothricin (Duchefa) to select for hsp93-V-1 and hsp93-III-1, and 11.25 g/mL sulfadiazine (Sigma-Aldrich) to select for hsp93-III-2.
Expression and Purification of Recombinant Proteins and Protein Pull-Down Assays
To generate the following soluble domain constructs, the selected sequences were amplified by PCR using cDNA as template: pET23d::atTic11093-602 and pET23d::atTic110603-966 (residues 93–602 and 603–966 of the mature protein respectively) (atTIC110 accession number At1g06950), pET23d15b::atTic40ΔTM (residues 131–447 of the mature protein), and pET23d::atTic55ΔTM (residues 49–481 of the mature protein) (atTIC40 and atTIC55 accession numbers At5g16620 and At2g24820, respectively). The PCR products were digested, cloned into the 5′-NcoI and 3′-XhoI or 3′-SalI sites of pET23d, or the 5′-NdeI and 3′-BamHI sites of pET15b, as indicated. Finally, constructs were sequenced. The primers used were as follows: Tic110 +93 BspHI forward 5′-GGTCATGATGGTACCGGAGGTAGCTG-3′; Tic110 +602 XhoI reverse 5′-CCCTCGAGATCAGCCACCATTTCAGT-3′; Tic110 +603 BspHI forward 5′-GGTCATGATGATCAAGGGAGAATCTT-3′; Tic110 +966 XhoI reverse 5′-CCCTCGAGAAAGACGAAATTGCCCTC-3′; Tic40 NdeI forward 5′-ACATATGAAATATGCAATGCAAACAGC-3′ and Tic40 BamHI reverse 5′-AGGATCCTTTTCAACCCGTCATTCCTG-3′; Tic55 +49 NcoI forward 5′-TACCATGGCTCGCTCCGCCGTCGCTG-3′; and Tic55 +481 SalI reverse 5′-TTGTCGACTTTCCAGAGCTCAAAAGA-3′.
The Tic110 domains Tic11093-966 and Tic110185-966 were expressed and purified as previously described (Inaba et al., 2003). The protein domains Tic11093-602, Tic110603-966, Tic40ΔTM, and Tic55ΔTM were expressed in Escherichia coli BL21 (Tuner) or BL21 (DE3) for Tic40ΔTM. Bacterial cultures were grown in 200 mL Luria-Bertani medium supplemented with ampicillin (100 μg/mL) and Glc 1% (w/v), at 37°C, 250 rpm, until they reached an OD600 of approximately 0.6 to 0.7. Expression was induced with isopropylthio-β-galactoside (0.5 mm for Tic110 domains, 1 mm for Tic40ΔTM, or 0.2 mm for Tic55ΔTM) for 4 to 5 h (37°C) (for Tic110 domains) or overnight (20°C) (for Tic40ΔTM and Tic55ΔTM) at 250 rpm. Pelleted cells were resuspended in 20 mL purification buffer: 40 mm Tris-HCl, pH 8.0, 1 m NaCl, 5 mm MgCl2, and 10 mm imidazole, supplemented with 1 mg/mL lysozyme and protease inhibitor cocktail (Complete EDTA-free tablets; Roche) and lysed by sonication. Cleared cell lysates were recovered by centrifugation at 20,000g for 20 min, 4°C, and incubated in batches with 200 μL Ni-NTA agarose (Qiagen) for 2 to 3 h at 4°C. After washing with 20 mL of purification buffer (20 mm imidazole), proteins were eluted with elution buffer (purification buffer supplemented with 200 mm imidazole and 10% [v/v] glycerol), in 200-μL fractions. These fractions were desalted in Sephadex G-50 columns, as described (Rosano et al., 2011). Protein concentration was quantified by Bradford assay.
Pull-down assays using recombinant proteins were performed in binding buffer (20 mm HEPES, pH 7.6, 150 mm KCl, 5 mm MgCl2, 10 mm imidazole, 0.005% [v/v] Tween 20, 10% [v/v] glycerol, and 1 mm DTT). First, pelleted isolated chloroplasts (approximately 100 million) were lysed in 1 mL hypotonic lysis buffer for 1 h at 4°C (see below). Then, the lysate was centrifuged at 20,000g for 30 min at 4°C and the supernatant recovered. Finally, protein concentration was quantified. In parallel, 175 pmol of each His-tagged protein was immobilized in 15 μL of Ni-NTA agarose for 1 h with rotation, at 4°C in binding buffer. To pull down the Hsp93/ClpC protein, approximately 200 μg of soluble chloroplast protein was incubated with the immobilized recombinant protein, overnight, with rotation at 4°C, in a final volume of 1 mL of binding buffer. Then, the agarose was carefully loaded on a column and washed with 3 mL of binding buffer. The bound proteins were eluted with 60 μL binding buffer supplemented with 300 mm imidazole after a short 5-min incubation. A similar assay was conducted in the absence of the His-tagged protein in order to assess for any nonspecific interaction between chloroplast protein and the Ni-NTA agarose. Eluted proteins were analyzed by immunoblotting and by Coomassie Brilliant Blue staining.
BiFC Analysis
To generate full-length YFP and half-YFP (NY and CY) fusions, the selected sequences were amplified by PCR using cDNA as template, cloned into the pGEM-T Easy (Promega) vector, sequenced, and then subcloned into the 5′-XhoI and 3′-KpnI sites of the pWEN18 (Kost et al., 1998), pWEN-NY, and pWEN-CY vectors (Maple et al., 2005). The following primers were used: Hsp93-V SalI forward 5′-AGTCGACAGTCATGGCTATGGCCAC-3′; Hsp93-V KpnI reverse 5′-AGGTACCAGCAACAGGGAGAGAATC-3′; Hsp93-VΔC KpnI reverse 5′-AAGGTACCCTCACCCACCATGCGTATCA-3′; Tic55 XhoI forward 5′-CCCTCGAGCATGGCTGTTCCATTTCTAA-3′; and Tic55 KpnI reverse 5′-GGGTACCTAGTCTTCTGTGTGTTCTAAT-3′ (atHSP93-V accession number At5g50920). The constructs for atTic110 and atTic40 were previously described (Bédard et al., 2007).
Protoplasts from 4- to 5-week-old wild-type plants grown on soil were isolated using the tape-Arabidopsis-sandwich method (Wu et al., 2009). Approximately 1 × 105 protoplasts and 5 μg of plasmid DNA were used per (co)transfection in 40% (w/v) polyethylene glycol 4000 (Wu et al., 2009). Samples were analyzed after 16 to 18 h of (co)transfection using a Zeiss LSM 510 META laser-scanning confocal microscope using a C-Apochromat 40×/1.2 W Korr objective. To detect YFP, 514-nm excitation from a 5-mW argon ion laser with an HFT 458/514 primary dichroic mirror (Carl Zeiss) and a 535- to 590-nm emission filter was used. To simultaneously detect chlorophyll autofluorescence, a NFT 635 Vis long-pass filter was used. The frequency of protoplasts that successfully express YFP fluorescence was estimated by counting the number of positives and the total number of protoplasts observed per microscope field, in 10 to 15 microscope fields per (co)transfection. The frequency of YFP-expressing protoplasts in the cotransfections was normalized to the frequency of its respective full-length YFP fusion, which was used as a control. Images were processed with the Zeiss LSM Image Browser software. Each (co)transfection was conducted three times, with the same result, and typical images are shown.
Construction of Hsp93 PBM Expression Constructs
The HSP93-V promoter (1405 bp of sequence upstream of the translation start site including the 5′ untranslated region exon and intron sequences) was amplified from Arabidopsis genomic DNA using the primers Hsp93-V Pro SacI F 5′-TGAGCTCGACACATCATTGCTCTCTAGG-3′ and Hsp93-V Pro AvrII R 5′-TCCTAGGGACTTCCTAAATAAATTCAACAAG-3′, introducing SacI and AvrII restriction sites upstream and downstream of the promoter sequence, respectively. At the same time, the Gateway cassette followed by a C-terminal FLAG tag and the Octopine Synthase terminator (OCS 3′) from the pEarlyGate 302 binary vector (Earley et al., 2006) was amplified using the attR1 NheI F 5′-AGCTAGCATACAAGTTTGTACAAAAAAGC-3′ and the OCS 3′ HindIII R 5′-TTTCGAAGTCCTGCTGAGCCTCGAC-3′ primers, introducing a novel NheI restriction site upstream of the Gateway cassette.
The pH2GW7-Hsp93Pro-Flag Gateway destination vector was then generated by first excising the entire 35S Cauliflower mosaic virus promoter sequence, Gateway cassette, and 35S Cauliflower mosaic virus terminator sequence from the pH2GW7 vector (Karimi et al., 2002) by SacI and HindIII digestion. These elements were then replaced by the SacI-AvrII restriction fragment containing the HSP93-V promoter and the NheI-HindIII restriction fragment containing the pEarlyGate 302 cassette by directional triple ligation. The resulting pH2GW7 Hsp93-V Pro Flag binary vector was used to generate constructs that drive endogenous levels of expression of the Hsp93-V proteins with or without a C-terminal Flag tag.
The Hsp93[P+] coding sequences with and without a stop codon were amplified using primers Hsp93-V attB1 5′-AAAAAGCAGGCTATGGCTATGGCCACAACG-3′ and Hsp93-V stop attB2 5′-AGAAAGCTGGGTTTAAGCAACAGGGAGAGA-3′, or Hsp93-V attB2 5′-AGAAAGCTGGGTTAGCAACAGGGAGAGAATC-3′, respectively, from Arabidopsis cDNA (AY102125). These PCR fragments were extended by further amplification with the attB1 and attB2 primers (Gateway Technology; Invitrogen, Life Technologies) to add the full-length attB recombination sequences and were then transferred to the pH2GW7-Hsp93Pro-Flag vector by Gateway cloning (Life Technologies) to produce the Hsp93[P+]* (nontagged) and Hsp93[P+] (FLAG-tagged) constructs, respectively.
The Hsp93[P-] coding sequences, again with and without a stop codon, were generated by using a splicing by overhang extension (SOEing) approach (Ho et al., 1989; Horton et al., 1989) to introduce the I772E mutation in Hsp93-V. Initial PCR fragments were generated using primers Hsp93-V attB1 with Hsp93-V I772E R 5′-GTCGAATCCTTCACGTCTTCCTCCTTTCTC-3′ (N-terminal) and Hsp73-V I772E F 5′-GGAAGACGTGAAGGATTCGACTTAGACTAC-3′ with Hsp93-V stop attB2 (C-terminal stop) or Hsp93-V attB2 (C-terminal) from the Arabidopsis HSP93-V cDNA (AY102125). The N-terminal and C-terminal stop as well as the N-terminal and C-terminal fragments were fused together by amplification with the attB1 and attB2 primers. The resulting PCR products were, as before, transferred to the pH2GW7-Hsp93Pro-Flag vector by Gateway cloning to produce the Hsp93[P-]* (nontagged) and Hsp93[P-] (FLAG-tagged) constructs, respectively.
Generation of Transgenic Plants
Constructs were introduced into Agrobacterium tumefaciens (GV103::pMP90) and subsequently transferred to Arabidopsis hsp93-V-1 plants using the floral-dip method (Clough and Bent, 1998). Transformants were identified on selective medium and then transferred to soil and allowed to set seed. Single-insertion lines were identified by T2 segregation analysis, and homozygous T3 plants were propagated for further analysis.
Selected single-insertion plants (T2) were crossed to hsp93-III-1 single mutants to assess for complementation. Putative F1 individuals carrying both hsp93-V-1 and hsp93-III-1 mutations, and the relevant Hsp93[P] construct, were identified on selective medium, transferred to soil, and allowed to set seed. Segregating F2 plants were also selected on medium and hsp93 genotypes were further confirmed by PCR analysis; briefly, genomic DNA isolated according to Edwards et al. (1991) was analyzed using gene-specific primers described by Kovacheva et al. (2007). Positive plants were propagated (F3) for further analysis, and the homozygous condition of the relevant Hsp93[P] construct was confirmed on selective medium.
Characterization of Transgenic Lines
Analyses of transcripts and protein levels, and chloroplast isolations and protein import assays were all performed using previously described procedures (Kovacheva et al., 2005), with the following minor changes. Briefly, total RNA was isolated from seedlings using the RNeasy Plant Mini Kit (Qiagen). DNase-treated RNA samples (4 μg) were used for first-strand cDNA synthesis using the SuperScript II Reverse Transcriptase Kit (Invitrogen). We used atHSP93-V and eIF4E1 gene-specific primers (Kovacheva et al., 2005) for the production of a nonsaturating amount of RT-PCR product (15 cycles and 20 cycles for atHSP93-V and eIF4E1, respectively). One-half of the total PCR product was separated on 1% agarose gel and visualized using an LAS-4000 image analyzer (Fujifilm; GE Healthcare). Total plant protein was extracted and quantified according to standard procedures (Kovacheva et al., 2005), and 20-μg samples were separated by SDS-PAGE prior to Coomassie Brilliant Blue R250 staining or immunoblotting. Protein import experiments were analyzed by phosphor imaging and quantified using the AIDA v. 4.27 software (Raytest); mean values shown in the relevant figures derive from the indicated number of replicates. To determine chlorophyll concentrations in adult plants, measurements were performed on plants grown on soil using a Konica-Minolta SPAD-502 m (Ling et al., 2011).
Purification of Chloroplasts Fractions and Blue Native PAGE
Pelleted fresh isolated chloroplasts (approximately 150 million), from 14- or 18-d-old plants grown in vitro, were resuspended in 1 mL hypotonic lysis buffer (25 mm HEPES, pH 8.0, supplemented with 1% [v/v] Sigma-Aldrich protease inhibitor cocktail for plant cells) and incubated for 1 h at 4°C with rotation before loading the lysate onto a three-layer Suc gradient (1.2, 1.0, and 0.46 m Suc, 25 mm HEPES, pH 8.0; Bruce et al., 1994). Stromal and intermembrane space, and thylakoid and envelope fractions were separated by ultracentrifugation at 200,000g for 1 h, at 4°C. The envelope fraction (bottom band) was recovered and washed with lysis buffer. The soluble fraction (stroma and intermembrane space) was recovered and precipitated with 20% TCA (w/v), washed twice with ice-cold 100% acetone, and resuspended in lysis buffer. Purified stroma and envelope fractions were analyzed by immunoblotting in equivalent amounts (5% of total isolated). Additional envelope membrane fractions were separated by blue native PAGE as previously described (Sjögren et al., 2014).
Anti-FLAG Immunoprecipitation
For anti-FLAG immunoprecipitations, freshly isolated chloroplasts (approximately 60 million) were crossed-linked in a 15-min incubation step in the presence DSP (0.25 mM), on ice. Reactions were subsequently quenched with 50 mm Gly prepared in HMS buffer (Kubis et al., 2008), for 15 min before the chloroplasts were pelleted at 400g at 4°C, and washed once with HMS buffer. Pelleted chloroplasts were solubilized by rotation for 20 min in a turning wheel, at 4°C, in 1 mL solubilization buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10% [v/v] glycerol, 1% [w/v] DDM, and 0.5% [v/v] Sigma-Aldrich protease inhibitor cocktail for plant cells). The soluble fraction was separated by a centrifugation step, at 19,000g for 30 min, at 4°C, and used immediately in the following step. A volume of 20 μL (slurry, per sample) of Anti-FLAG M2 Affinity Gel (Sigma-Aldrich; preequilibrated in solubilization buffer) was added to the chloroplast soluble fraction and incubated for 2 h, in a rotating wheel, at 4°C. Next, the Anti-FLAG M2 Affinity Gel was washed five times with a total of 3 mL (150 gel volumes) of washing buffer (0.3% [w/v] DDM in solubilization buffer). Anti-FLAG immunoprecipitated proteins were eluted by adding 50 μL of 2× SDS-PAGE sample loading buffer (supplemented with 100 mM dithiothreitol) and incubating for 5 min at 90°C. Samples were analyzed by immunoblotting.
Protein Degradation Assay
Intact chloroplasts from wild-type Arabidopsis and the various transgenic lines were isolated according to Sjögren et al. (2006). The in organello protein degradation assay was performed as previously described (Sjögren et al., 2014). At selected time points, stromal proteins were fractionated from the chloroplasts and separated by SDS-PAGE on 3 to 8% Tris-acetate gels. Proteins were visualized by staining with Coomassie Brilliant Blue G 250. The amount of EF-Ts protein was quantified using the ChemiGenius2 imaging system with associated software (Syngene).
Immunoblotting
Protein samples were diluted in 2× SDS-PAGE sample buffer and separated by SDS-PAGE (Laemmli, 1970). Separated proteins were blotted onto Hybond ECL membrane (GE Healthcare). Primary antibodies were polyclonal sera raised in rabbits against Hsp93/ClpC (antigen from Synechococcus sp.) (Porankiewicz and Clarke, 1997); Tic110 (Aronsson et al., 2010), ClpP6, ClpR4 (antigens from Arabidopsis) (Zheng et al., 2002); chloroplast GAPDH (glyceraldehyde-3-phosphate dehydrogenase, subunits GapA and GapB) (antigen from spinach) (Howard et al., 2008); LHCP (antigen from pea; Huang et al., 2011); FLAG-tag (raised in mouse) (Sigma-Aldrich, A9469); H3 (histone H3) (antigen from human) (Abcam, ab1791); chloroplast Hsp70 (antigen from Arabidopsis) (Agrisera, AS08 348); and plastid Cpn60β (antigen from Arabidopsis) (Suzuki et al., 2009). Secondary antibodies were antirabbit IgG conjugated with horseradish peroxidase (Santa Cruz Biotechnology) or with alkaline phosphatase (Sigma-Aldrich, A3687) and antimouse IgG conjugated with horseradish peroxidase (GE Healthcare) (for the anti-FLAG-tag primary antibody). The chemiluminescent signal was detected using the EZ-ECL chemiluminescence detection kit for HRP (Biological Industries) and an LAS-4000 (Fujifilm; GE Healthcare) image analyzer. Where indicated, quantification was done with the AIDA v. 4.27 software (Raytest). When using alkaline phosphatase secondary antibody, the detection reagent was 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium alkaline phosphatase substrate (Sigma-Aldrich).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers atTIC110, At1g06950; atTIC40, At5g16620; atTIC55, At2g24820; and atHSP93-V, At5g50920.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pull-down and BiFC controls, and quantitative analyses associated with the BiFC studies.
Supplemental Figure S2. Hsp93 PBM mutant complementation analysis using non-tagged constructs.
Supplemental Figure S3. High-level expression of the Hsp93[P-] protein in transgenic plants negatively affects plant vigor.
Supplementary Material
Acknowledgments
We thank Sean Maguire, Ramesh Patel, and Lorenza Francescut for technical support. We are grateful to Danny Schnell for providing Tic110 constructs and to Shin-ya Miyagishima for providing the ptCpn60β antibody.
Glossary
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- BiFC
bimolecular fluorescence complementation
- EF-T
elongation factor-T
- DDM
dodecyl maltoside
- DSP
dithiobis succinimidylpropionate
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grants BB/J017256/1, BB/J009369/1, and BB/F020325/1 to P.J.) and by grants from the Swedish Research Council (to A.C.).
Articles can be viewed without a subscription.
References
- Akita M, Nielsen E, Keegstra K (1997) Identification of protein transport complexes in the chloroplastic envelope membranes via chemical cross-linking. J Cell Biol 136: 983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134: 117–131 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Combe J, Patel R, Agne B, Martin M, Kessler F, Jarvis P (2010) Nucleotide binding and dimerization at the chloroplast pre-protein import receptor, atToc33, are not essential in vivo but do increase import efficiency. Plant J 63: 297–311 [DOI] [PubMed] [Google Scholar]
- Bédard J, Kubis S, Bimanadham S, Jarvis P (2007) Functional similarity between the chloroplast translocon component, Tic40, and the human co-chaperone, Hsp70-interacting protein (Hip). J Biol Chem 282: 21404–21414 [DOI] [PubMed] [Google Scholar]
- Biegert A, Söding J (2008) De novo identification of highly diverged protein repeats by probabilistic consistency. Bioinformatics 24: 807–814 [DOI] [PubMed] [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K (1994) In vitro import of proteins into chloroplasts. In Gelvin SB, Schilperoot RA, eds, Plant Molecular Biology Manual. Kluwer Academic Publishers, London, pp 1-15 [Google Scholar]
- Burton BM, Williams TL, Baker TA (2001) ClpX-mediated remodeling of mu transpososomes: selective unfolding of subunits destabilizes the entire complex. Mol Cell 8: 449–454 [DOI] [PubMed] [Google Scholar]
- Chou ML, Chu CC, Chen LJ, Akita M, Li HM (2006) Stimulation of transit-peptide release and ATP hydrolysis by a cochaperone during protein import into chloroplasts. J Cell Biol 175: 893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou ML, Fitzpatrick LM, Tu SL, Budziszewski G, Potter-Lewis S, Akita M, Levin JZ, Keegstra K, Li HM (2003) Tic40, a membrane-anchored co-chaperone homolog in the chloroplast protein translocon. EMBO J 22: 2970–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Li HM (2012) The amino-terminal domain of chloroplast Hsp93 is important for its membrane association and functions in vivo. Plant Physiol 158: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Constan D, Froehlich JE, Rangarajan S, Keegstra K (2004) A stromal Hsp100 protein is required for normal chloroplast development and function in Arabidopsis. Plant Physiol 136: 3605–3615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P (2013) Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta 1833: 332–340 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW (2005) AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59 [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77: 61–68 [DOI] [PubMed] [Google Scholar]
- Howard TP, Metodiev M, Lloyd JC, Raines CA (2008) Thioredoxin-mediated reversible dissociation of a stromal multiprotein complex in response to changes in light availability. Proc Natl Acad Sci USA 105: 4056–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Huang W, Ling Q, Bédard J, Lilley K, Jarvis P (2011) In vivo analyses of the roles of essential Omp85-related proteins in the chloroplast outer envelope membrane. Plant Physiol 157: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Alvarez-Huerta M, Li M, Bauer J, Ewers C, Kessler F, Schnell DJ (2005) Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17: 1482–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Li M, Alvarez-Huerta M, Kessler F, Schnell DJ (2003) atTic110 functions as a scaffold for coordinating the stromal events of protein import into chloroplasts. J Biol Chem 278: 38617–38627 [DOI] [PubMed] [Google Scholar]
- Inoue H, Li M, Schnell DJ (2013) An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc Natl Acad Sci USA 110: 3173–3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DT, Froehlich JE, Keegstra K (1998) The hydrophilic domain of Tic110, an inner envelope membrane component of the chloroplastic protein translocation apparatus, faces the stromal compartment. J Biol Chem 273: 16583–16588 [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E (2013) Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol 14: 787–802 [DOI] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT (2004) Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol 11: 404–411 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kim DY, Kim KK (2003) Crystal structure of ClpX molecular chaperone from Helicobacter pylori. J Biol Chem 278: 50664–50670 [DOI] [PubMed] [Google Scholar]
- Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA (2001) Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nat Struct Biol 8: 230–233 [DOI] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Patel R, Dudley P, Twell D, Ríos G, Koncz C, Jarvis P (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428 [DOI] [PubMed] [Google Scholar]
- Kovacheva S, Bédard J, Wardle A, Patel R, Jarvis P (2007) Further in vivo studies on the role of the molecular chaperone, Hsp93, in plastid protein import. Plant J 50: 364–379 [DOI] [PubMed] [Google Scholar]
- Kovács-Bogdán E, Soll J, Bölter B (2010) Protein import into chloroplasts: the Tic complex and its regulation. Biochim Biophys Acta 1803: 740–747 [DOI] [PubMed] [Google Scholar]
- Kubis SE, Lilley KS, Jarvis P (2008) Isolation and preparation of chloroplasts from Arabidopsis thaliana plants. Methods Mol Biol 425: 171–186 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Leister D. (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56 [DOI] [PubMed] [Google Scholar]
- Ling Q, Huang W, Jarvis P (2011) Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth Res 107: 209–214 [DOI] [PubMed] [Google Scholar]
- Liu L, McNeilage RT, Shi LX, Theg SM (2014) ATP requirement for chloroplast protein import is set by the Km for ATP hydrolysis of stromal Hsp70 in Physcomitrella patens. Plant Cell 26: 1246–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Møller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Nakai M. (2015) The TIC complex uncovered: The alternative view on the molecular mechanism of protein translocation across the inner envelope membrane of chloroplasts. Biochim Biophys Acta 1847: 957–967 [DOI] [PubMed] [Google Scholar]
- Nielsen E, Akita M, Davila-Aponte J, Keegstra K (1997) Stable association of chloroplastic precursors with protein translocation complexes that contain proteins from both envelope membranes and a stromal Hsp100 molecular chaperone. EMBO J 16: 935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, van Wijk KJ (2015) Organization, function and substrates of the essential Clp protease system in plastids. Biochim Biophys Acta 1847: 915–930 [DOI] [PubMed] [Google Scholar]
- Ohad N, Shichrur K, Yalovsky S (2007) The analysis of protein-protein interactions in plants by bimolecular fluorescence complementation. Plant Physiol 145: 1090–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paila YD, Richardson LG, Schnell DJ (2015) New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J Mol Biol 427: 1038–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porankiewicz J, Clarke AK (1997) Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol 179: 5111–5117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano GL, Bruch EM, Ceccarelli EA (2011) Insights into the Clp/HSP100 chaperone system from chloroplasts of Arabidopsis thaliana. J Biol Chem 286: 29671–29680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer E, Delbac F, Tora L, Moras D, Romier C (2012) TFIID TAF6-TAF9 complex formation involves the HEAT repeat-containing C-terminal domain of TAF6 and is modulated by TAF5 protein. J Biol Chem 287: 27580–27592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanklin J, DeWitt ND, Flanagan JM (1995) The stroma of higher plant plastids contain ClpP and ClpC, functional homologs of Escherichia coli ClpP and ClpA: an archetypal two-component ATP-dependent protease. Plant Cell 7: 1713–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2010) A stromal heat shock protein 70 system functions in protein import into chloroplasts in the moss Physcomitrella patens. Plant Cell 22: 205–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013a) The chloroplast protein import system: from algae to trees. Biochim Biophys Acta 1833: 314–331 [DOI] [PubMed] [Google Scholar]
- Shi LX, Theg SM (2013b) Energetic cost of protein import across the envelope membranes of chloroplasts. Proc Natl Acad Sci USA 110: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR (2001) Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem 276: 29420–29429 [DOI] [PubMed] [Google Scholar]
- Sjögren LL, Clarke AK (2011) Assembly of the chloroplast ATP-dependent Clp protease in Arabidopsis is regulated by the ClpT accessory proteins. Plant Cell 23: 322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, MacDonald TM, Sutinen S, Clarke AK (2004) Inactivation of the clpC1 gene encoding a chloroplast Hsp100 molecular chaperone causes growth retardation, leaf chlorosis, lower photosynthetic activity, and a specific reduction in photosystem content. Plant Physiol 136: 4114–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Stanne TM, Zheng B, Sutinen S, Clarke AK (2006) Structural and functional insights into the chloroplast ATP-dependent Clp protease in Arabidopsis. Plant Cell 18: 2635–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren LL, Tanabe N, Lymperopoulos P, Khan NZ, Rodermel SR, Aronsson H, Clarke AK (2014) Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J Biol Chem 289: 11318–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LL, Koussevitzky S, Clarke AK (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Nakanishi H, Bower J, Yoder DW, Osteryoung KW, Miyagishima SY (2009) Plastid chaperonin proteins Cpn60 alpha and Cpn60 beta are required for plastid division in Arabidopsis thaliana. BMC Plant Biol 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K (1989) Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J Biol Chem 264: 6730–6736 [PubMed] [Google Scholar]
- Tryggvesson A, Ståhlberg FM, Mogk A, Zeth K, Clarke AK (2012) Interaction specificity between the chaperone and proteolytic components of the cyanobacterial Clp protease. Biochem J 446: 311–320 [DOI] [PubMed] [Google Scholar]
- Tsai JY, Chu CC, Yeh YH, Chen LJ, Li HM, Hsiao CD (2013) Structural characterizations of the chloroplast translocon protein Tic110. Plant J 75: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B (2004) Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell 119: 653–665 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS (2009) Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Halperin T, Hruskova-Heidingsfeldova O, Adam Z, Clarke AK (2002) Characterization of Chloroplast Clp proteins in Arabidopsis: Localization, tissue specificity and stress responses. Physiol Plant 114: 92–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.