Pseudo-response regulators associate with overlapping sets of putative target genes and require G-box-like cis-regulatory elements for transcriptional activity.
Abstract
PSEUDO-RESPONSE REGULATORs (PRRs) play overlapping and distinct roles in maintaining circadian rhythms and regulating diverse biological processes, including the photoperiodic control of flowering, growth, and abiotic stress responses. PRRs act as transcriptional repressors and associate with chromatin via their conserved C-terminal CCT (CONSTANS, CONSTANS-like, and TIMING OF CAB EXPRESSION 1 [TOC1/PRR1]) domains by a still-poorly understood mechanism. Here, we identified genome-wide targets of PRR9 using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and compared them with PRR7, PRR5, and TOC1/PRR1 ChIP-seq data. We found that PRR binding sites are located within genomic regions of low nucleosome occupancy and high DNase I hypersensitivity. Moreover, conserved noncoding regions among Brassicaceae species are enriched around PRR binding sites, indicating that PRRs associate with functionally relevant cis-regulatory regions. The PRRs shared a significant number of binding regions, and our results indicate that they coordinately restrict the expression of target genes to around dawn. A G-box-like motif was overrepresented at PRR binding regions, and we showed that this motif is necessary for mediating transcriptional regulation of CIRCADIAN CLOCK ASSOCIATED 1 and PRR9 by the PRRs. Our results further our understanding of how PRRs target specific promoters and provide an extensive resource for studying circadian regulatory networks in plants.
Eukaryotic circadian clocks consist of complex transcriptional-translational regulatory networks that sustain rhythms with a period length of ∼24 h (Bell-Pedersen et al., 2005). This biochemical oscillator confers a fitness advantage by enabling organisms to anticipate daily and seasonal changes in the environment (Woelfle et al., 2004; Dodd et al., 2005; Yerushalmi and Green, 2009; Yerushalmi et al., 2011). PSEUDO-RESPONSE REGULATORS (PRRs) are key components of green alga and plant circadian networks (Farré and Liu, 2013). These proteins are characterized by sharing a conserved N-terminal pseudo-receiver domain and C-terminal CCT (CONSTANS, CONSTANS-like, and TIMING OF CAB EXPRESSION 1 [TOC1/PRR1]) domain. The protein levels of the five PRRs in Arabidopsis (Arabidopsis thaliana) peak sequentially throughout the day, starting with PRR9 3 to 4 h after dawn, followed by PRR7, PRR5 and PRR3, and TOC1/PRR1 peaking 1 to 3 h after dusk (Fujiwara et al., 2008). PRR9, PRR7, PRR5, and TOC1 have been shown to act as transcriptional repressors and associate with chromatin via their CCT domains by a still-poorly understood mechanism (Nakamichi et al., 2010, 2012; Gendron et al., 2012).
Within the Arabidopsis circadian clock, PRRs are involved in several transcriptional-translational feedback loops. PRR9, PRR7, PRR5, and TOC1 repress clock components CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY), both of which are expressed at dawn (Nakamichi et al., 2010; Gendron et al., 2012). In turn, CCA1 and LHY activate the expression of PRR9 and PRR7 as well as repress the expression of TOC1 and probably PRR5 (Farré et al., 2005; Nagel et al., 2015). REVEILLE 8 (RVE8), which belongs to a subfamily of Myb domain-containing transcription factors that includes CCA1 and LHY, acts as an activator for TOC1 and PRR5 (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013). In addition, a protein complex consisting of LUX ARRHYTHMO, EARLY FLOWERING 4 (ELF4), and ELF3 transcriptionally represses PRR9 and PRR7 during the night period (Dixon et al., 2011; Helfer et al., 2011; Herrero et al., 2012; Mizuno et al., 2014).
Mutant analyses have shown that PRRs play overlapping and distinct functions in maintaining circadian rhythms and regulating diverse biological processes (Farré and Liu, 2013). The identification of PRR7, PRR5, and TOC1 binding regions using chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) has shown that these proteins are involved in the direct regulation of growth, abiotic stress responses, and the photoperiodic control of flowering (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013). To further understand the role of PRRs in regulating gene expression, we identified PRR9 putative target genes using ChIP-seq and performed a comprehensive analysis of PRR bound regions. We observed that PRRs share a large number of binding sites and associate with conserved cis-regulatory regions in open chromatin. We also showed that a G-box-related motif enriched in PRR binding regions is necessary for transcriptional regulation by the PRRs. Since so far there is no evidence that PRRs are able to bind directly to G-box motifs, these results suggest that PRRs associate with other transcription factors to regulate gene expression.
RESULTS
Genome-Wide Identification of PRR9 Binding Regions
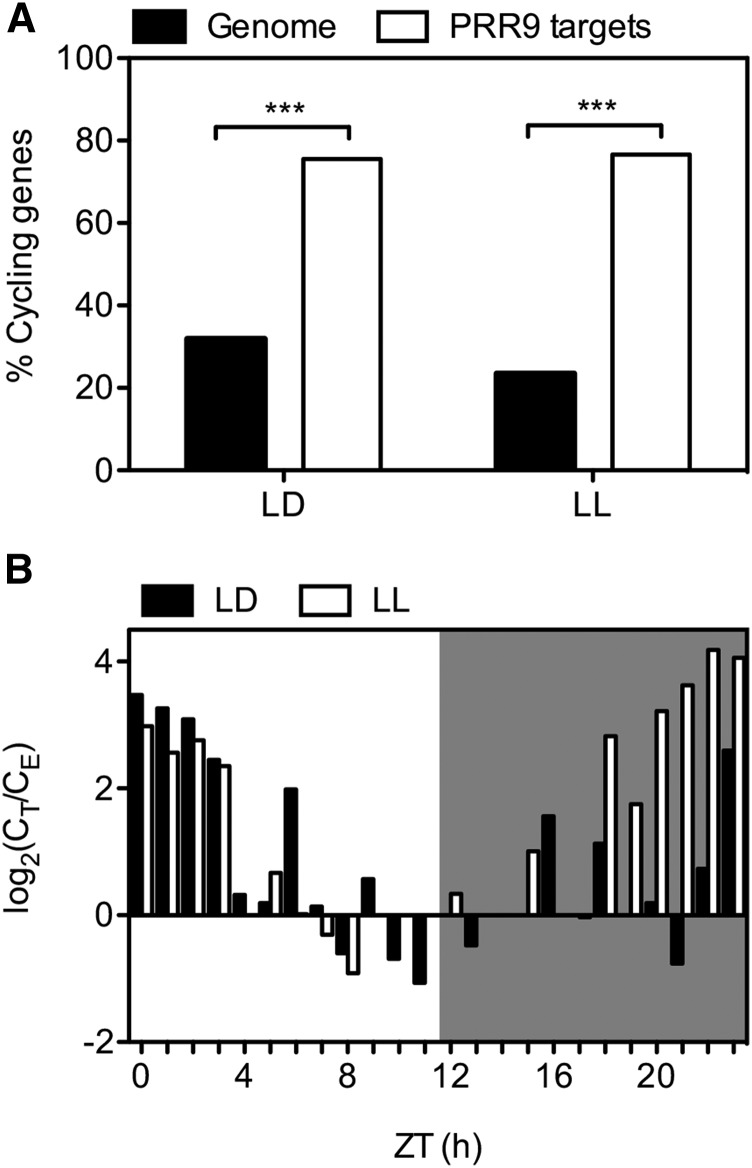
To dissect the contribution of each PRR in regulating transcription, we generated a PRR9 ChIP-seq dataset to compare with available ChIP-seq data for PRR7, PRR5, and TOC1 (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013). The complemented line prr9-1 PRR9::HA-PRR9 CCR2::LUC (Supplemental Fig. S1, A and B) was grown in cycling 12 h light/ 12 h dark for two weeks and harvested 4 h after dawn (Zeitgeber time 4, ZT4), the time at which PRR9 protein levels peak (Fujiwara et al., 2008). Three independent PRR9 chromatin immunoprecipitation (ChIP) experiments were pooled for sequencing, and their quality was confirmed by quantitative PCR, showing an enrichment of PRR9 binding at the CCA1 promoter (Supplemental Fig. S1C; Nakamichi et al., 2010). We identified 150 PRR9 binding regions using the MACS2 algorithm (Zhang et al., 2008), associated each binding region with the nearest transcriptional start site and defined these genes as putative targets. PRR9 putative targets included CCA1 and LHY, known to be regulated by PRR9 (Nakamichi et al., 2010). Approximately 34% of PRR9 putative targets were differentially expressed in the prr5prr7prr9 (prr579) triple mutant compared with wild type (Supplemental Dataset S1; Nakamichi et al., 2009), and all 45 differentially expressed genes, with the exception of one, displayed elevated RNA levels in prr579 (Fisher’s exact test, P value < 0.0001). These findings are consistent with PRR9 functioning as a transcriptional repressor (Supplemental Dataset S1). In addition, approximately 77% of PRR9 putative target genes cycle under constant light conditions (Fig. 1A) and ∼72% of these cycling genes exhibit peak expression around dawn, similar to what has been observed for the putative targets of PRR7, PRR5, and TOC1 (Fig. 1B; Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013).
Figure 1.
Cycling gene expression patterns of PRR9 putative target genes determined by ChIP-seq. A, Percentage of PRR9 putative target genes that cycle in light/dark (LD; Bläsing et al., 2005) and constant light conditions (LL; Edwards et al., 2006) compared with genome-wide cyclic gene expression patterns. Cycling gene expression is defined as having an mbpma > 0.8. Fisher's exact test: ***, P value < 0.0001. B, Phase of PRR9 putative target gene expression under LD (black bars) and LL (white bars). Shaded area indicates dark or subjective dark period in LD or LL, respectively. PHASER analysis: mbpma > 0.8 (Michael et al., 2008). CE, count of expected; CT, count of targets; H, hours; ZT, Zeitgeber.
PRRs Bind to the Same DNA Regions to Regulate Common Target Genes
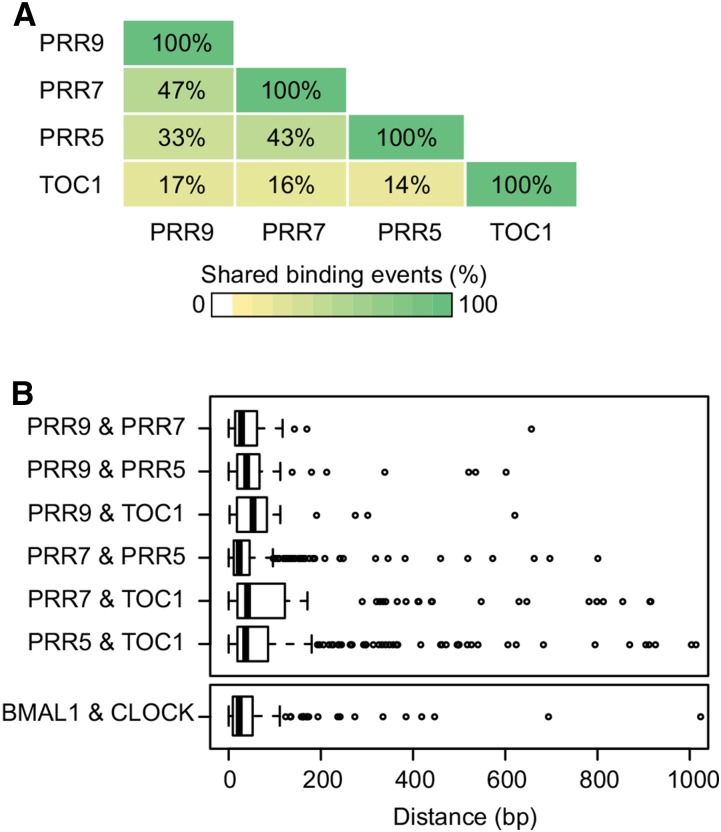
To compare the binding regions of different PRRs, raw data from PRR7, PRR5, and TOC1 ChIP-seq experiments (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013) were uniformly processed in parallel to PRR9. Between 37% and 52% of PRR binding summits were located within 500 bp of transcriptional start sites (Supplemental Fig. S2, A–D). Most PRR9 (86%), PRR7 (80.7%), and to a lesser degree PRR5 (61%) binding summits were located upstream, whereas a large number of TOC1 (30.4%) binding summits were located in the first exon (Supplemental Fig. S2, E–H). We then asked whether the PRRs bind to the same regions of DNA. To determine shared binding events, we performed pairwise analyses of the proximity between PRR binding regions using IntervalStats (Chikina and Troyanskaya, 2012). Focusing on the top-ranked 150 binding regions for each of the PRRs, we observed that PRR9, PRR7, and PRR5 have the most binding events in common (P < 0.05; Fig. 2A). TOC1 was more distinct and shared fewer binding events with each of the other PRRs (Fig. 2A). This difference might be due to the preferential association of TOC1 to exonic regions (Supplemental Fig. S2H). We also calculated the distance between binding summits of PRRs with shared target genes. The median distance between binding summits in each of the PRR pairwise comparisons ranged from 23 to 53 bp (Fig. 2B). To compare these values with those for proteins that bind to the same cis-regulatory regions, we analyzed the positions of mouse BMAL1 (Brain and Muscle ARNT-like 1) and CLOCK (Circadian Locomotor Output Cycles Kaput) binding summits determined by ChIP-seq. These clock components are bHLH transcription factors that form heterodimers to regulate gene expression by associating with the same cis-regulatory element (CRE; Ko and Takahashi, 2006; Hatanaka et al., 2010; Yoshitane et al., 2014; Fig. 2B). The median distance between BMAL1 and CLOCK binding summits is 23 bp, which is comparable to the values calculated for the PRR pairwise comparisons, particularly PRR9-PRR7 (28 bp) and PRR7-PRR5 (23 bp). Taken together, our results indicate that PRRs associate with the same regions of DNA to regulate shared target genes.
Figure 2.
PRRs associate to the same chromatin regions. A, Mean percentage of shared binding events from reciprocal pairwise comparisons of the top 150 binding regions in each PRR dataset determined by IntervalStats (Chikina and Troyanskaya, 2012; P value < 0.05). B, Tukey boxplots of the distance between PRR binding summits with shared putative target genes. For graphical clarity, data points > 1000 bp were omitted, which included one PRR5 & TOC1 data point and 102 Mus musculus BMAL1 & CLOCK data points (Hatanaka et al., 2010; Yoshitane et al., 2014).
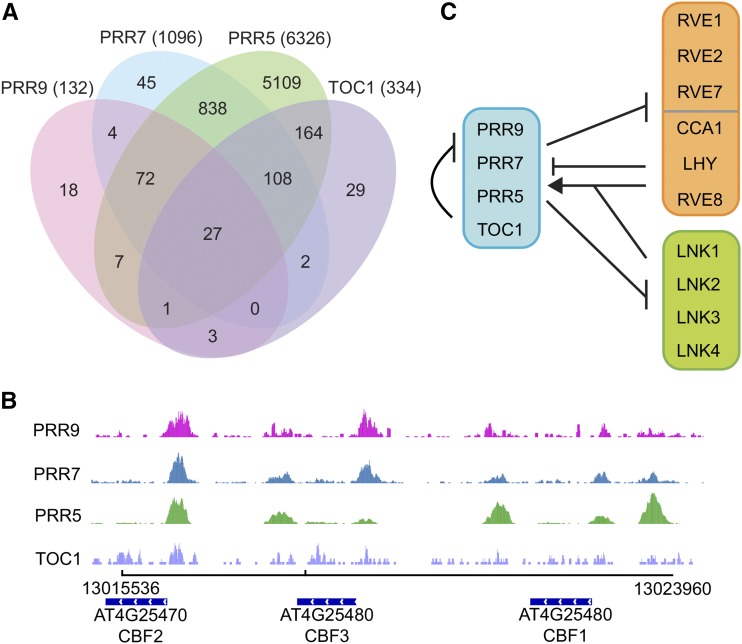
Although only 27 putative target genes were shared among all four PRRs, a larger percentage of genes were shared among three or two PRRs (Fig. 3A). For example, 75% of PRR9 putative target genes were in common with PRR7 and PRR5, and of these genes, 38.4% were upregulated in prr579 (Supplemental Dataset S2). Moreover, pairwise comparisons showed that 95.4% of PRR7 and 89.8% of TOC1 putative target genes were in common with PRR5 (Supplemental Dataset S2). Among all four PRRs, common enriched biological process Gene Ontology (GO) terms were related to flowering and responses to various abiotic stresses (Supplemental Fig. S3A; Supplemental Dataset S3). The PRRs were also significantly enriched for molecular function GO terms associated with DNA binding and transcription factor activity (Supplemental Fig. S3B; Supplemental Dataset S3), reflecting the role of PRRs in orchestrating rhythmic transcription.
Figure 3.
Shared putative target genes among PRR9, PRR7, PRR5, and TOC1. A, Venn diagram of overlapping and distinct PRR target genes determined by ChIP-seq. The total number of target genes for each PRR is shown in parentheses. Putative target genes were defined by associating each binding region to the closest transcriptional start site. PRR9 is in pink, PRR7 in blue, PRR5 in green, and TOC1 in purple. B, Binding profile of the PRRs at the CBF regulon. ChIP-seq reads (IP) were visualized using the Integrative Genomics Viewer (Robinson et al., 2011). C, Model of transcriptional regulation between the PRRs (blue), CCA1/LHY/RVEs (orange), and LNKs (green). Arrows and tacks represent transcriptional activation and repression, respectively.
The prr579 triple mutant is arrhythmic, late flowering, resistant to cold and drought, and has a long hypocotyl (Nakamichi et al., 2007, 2009; Niwa et al., 2009). Accordingly, we found that PRR9, PRR7, and PRR5 were bound to regions upstream of CYCLING DOF FACTOR 2, which encodes a repressor of CONSTANS (Fornara et al., 2009), a key component in the photoperiodic control of flowering. We also found these PRRs associated with multiple sites in the CBF (C-REPEAT/DRE BINDING) regulon (Fig. 3B) and to the promoter of a gene involved in abscisic acid biosynthesis ABA DEFICIENT 1. Moreover, master regulators of growth and development, such as ELONGATED HYPOCOTYL 5 (HY5), HY5 HOMOLOG, PHYTOCHROME INTERACTING FACTOR 4 (PIF4), and PIF5 were also targets of multiple PRRs (Supplemental Dataset S1).
Our results demonstrate a tight interconnected network between members of several families of circadian clock-related genes, such as the PRRs, CCA1/LHY/RVEs, and LNKs (NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED). All four PRRs associated with the promoters of CCA1 and LHY, along with the promoters of at least two other PRRs (Supplemental Dataset S1). Due to the presence of a small open reading frame in the PRR9 promoter (AT2G46787), PRR9 was not initially identified as a PRR target in the ChIP-seq datasets, but its RNA level is reduced in PRR overexpressing lines (Makino et al., 2002; Sato et al., 2002; Liu et al., 2013). We found that PRR9, PRR7, and PRR5 were also bound within 500 bp of the transcriptional start sites of RVE8, RVE1, RVE2/EARLY-PHYTOCHROME-RESPONSIVE 1, and RVE7. The expression of RVE8, RVE1, and RVE7 is upregulated in prr579, and the expression of RVE2 is repressed in a PRR7 overexpressing line (Rawat et al., 2011; Liu et al., 2013), indicating that the association is functional. In turn, it has been shown that CCA1/LHY/RVE transcription factors regulate the expression of several PRRs (Fig. 3C; Alabadí et al., 2001; Farré et al., 2005; Rawat et al., 2011; Hsu et al., 2013). Finally, we observed PRR9 and other PRRs at the promoters of various LNK genes. LNK proteins associate with CCA1/LHY/RVE to regulate gene expression (Xie et al., 2014). For example, LNK1 and LNK2 are necessary for RVE8 activation of PRR5 (Xie et al., 2014). The expression of all four LNK genes is upregulated in the prr579 mutant (Supplemental Dataset S1; Nakamichi et al., 2010), and the RNA levels of LNK1 and LNK2 are elevated in toc1 mutants (Rugnone et al., 2013). These results indicate strong reciprocal regulation among families of transcription regulators involved in circadian control.
PRRs Associate with Conserved cis-Regulatory Regions
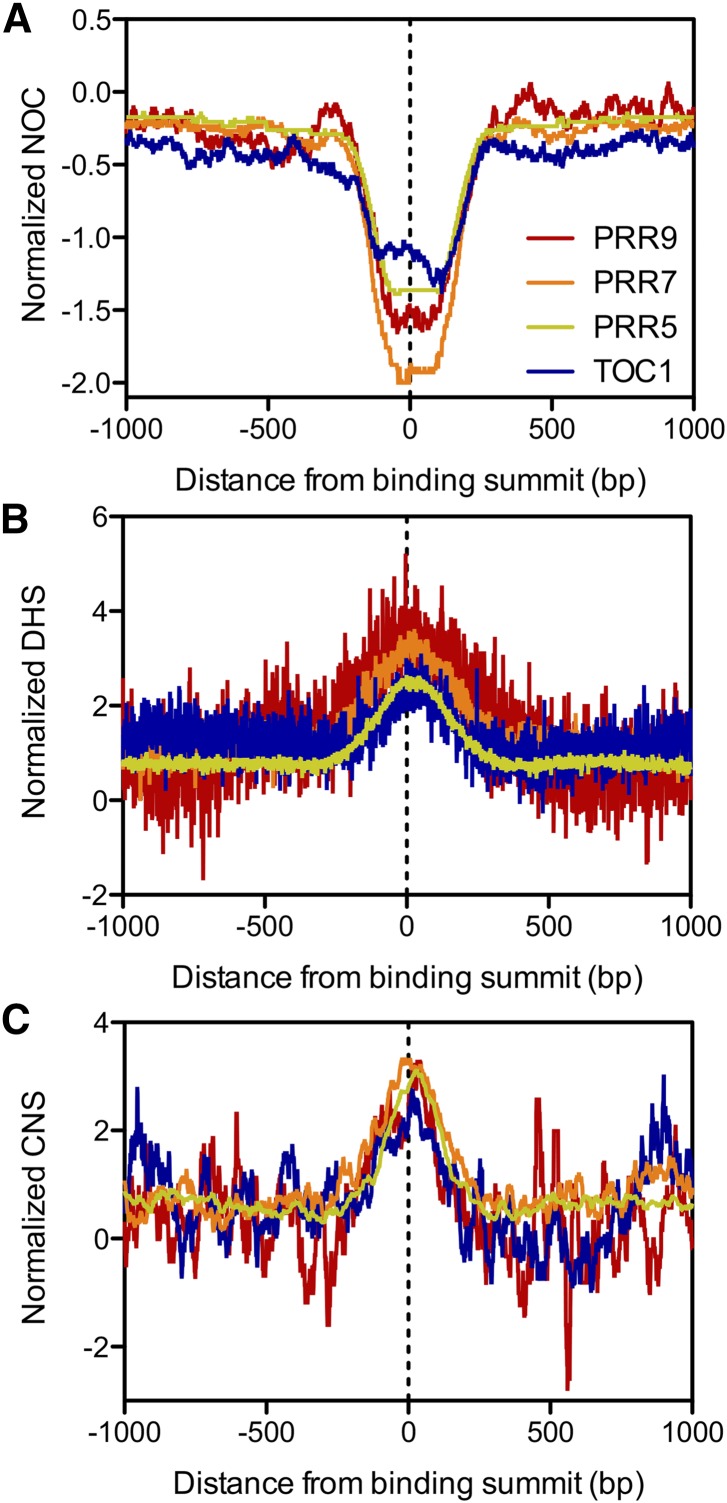
It is unknown whether PRRs bind to cis-regulatory regions to influence gene expression. The positions of active cis-regulatory sites correlate with sections of open chromatin and low nucleosome occupancy (NOC) in plants and animals (Bell et al., 2011). To investigate whether the PRRs associate with regulatory sites, we examined the chromatin landscape of PRR binding regions. We first quantified NOC around PRR binding summits using micrococcal nuclease digestion combined with sequencing data on plants harvested at ZT4 (Liu et al., 2015). Our analyses revealed that PRR binding summits coincide with regions of low NOC compared with background (Fig. 4A). In addition, open chromatin is susceptible to DNase I cleavage, and DNase I hypersensitive sites (DHSs) also correlate with transcription factor-bound DNA in eukaryotes, including plants (Bell et al., 2011; Jiang, 2015). Analysis of recently published DHS data on seedlings grown in light/dark cycles (Sullivan et al., 2014) showed that PRR binding summits are located within DHSs, further establishing that PRRs bind to regulatory regions of DNA (Fig. 4B).
Figure 4.
Chromatin profile of PRR binding regions. A to C, (A) NOC, (B) DHSs, and (C) presence of CNS around PRR binding summits. Binding summits were centered at zero on the x axis. Normalized values represent the log2 ratio between the median (NOC) and average (DHS, CNS) score per base from −1 kb upstream to +1 kb downstream of the binding summit, to those with the same coordinates at randomly selected genes. All binding summits were used for NOC and DHS analyses, whereas binding summits located in noncoding regions were used for the CNS analysis. NOC data are from (Liu et al., 2015), DHS data from (Sullivan et al., 2014), and CNS data from (Haudry et al., 2013).
PRRs are conserved among plants (Farré and Liu, 2013), and many orthologous genes in angiosperms exhibit similar diurnal and circadian gene expression patterns (Filichkin et al., 2011). Therefore, we hypothesized that functionally relevant PRR binding regions would display a high degree of conservation. We examined publicly available data on conserved noncoding sequences (CNSs) among nine Brassicaceae species and compared regions surrounding the PRR binding summits to the same coordinates of randomly selected genes (Haudry et al., 2013). CNSs are enriched around PRR binding summits (Fig. 4C), further supporting that PRRs associate with regulatory regions of DNA.
G-boxes Are Necessary for Transcriptional Regulation by PRRs
Uncovering the CREs necessary for PRR association to DNA will provide insight on how PRRs directly target genes to regulate their expression. We identified CREs de novo using MEME (Machanick and Bailey, 2011) by analyzing binding regions located upstream of transcriptional start sites. As previously observed for PRR7, PRR5, and TOC1 (Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013), a G-box (CACGTG)-related motif was also the most enriched element found at PRR9 binding regions (Fig. 5A). To determine the similarity between each of the G-box motifs, a distance measure of 1− Pearson Correlation Coefficient (PCC) was calculated for each pair of PRR-associated motifs. We found that the motifs identified for each of the PRRs were significantly more similar to each other than randomly expected (Fig. 3C).
Figure 5.
PRR binding regions are enriched with G-box-containing motifs. A, Enriched motifs in PRR binding regions located upstream of transcriptional start sites. Motifs identified de novo using MEME (Machanick and Bailey, 2011) with corresponding e-values, the percentage of upstream binding regions containing at least one motif, and distance matrix between PRR motifs (values represent 1– PCC). B and C, Chromatin profiles of all four PRRs binding at the (B) CCA1 and (C) PRR9 promoters. ChIP-seq reads (IP) were visualized for each of the PRRs (blue background), and magnification of the DHS footprints within the PRR binding regions (yellow background) show G-box elements (orange). Chr2, Chromosome 2.
We observed a statistically significant enrichment of G-box-like elements at PRR binding regions located upstream of transcriptional start sites. However, a large percentage of TOC1 binding regions are located in exons (Supplemental Fig. S2). When we performed motif enrichment analyses using TOC1 binding regions located within genes, we did not identify any overrepresented motifs. Therefore, we hypothesize that TOC1 binding within genes may represent a different mechanism of regulation as opposed to the mechanism mediated by G-box-like elements located in the upstream regions.
To investigate the role of G-boxes in mediating transcriptional regulation by PRRs, we focused on the promoters of PRR9 and CCA1, which were bound by all four PRRs (Fig. 5, B and C). In Arabidopsis, the PRR9 promoter contains two G-box motifs (−286, −214), whereas the CCA1 promoter contains one G-box motif (−296). These sequences are located within DHSs, and the CCA1 G-box is located within a CNS (Fig. 5, B and C). The G-boxes in the PRR9 and CCA1 promoters, along with other circadian-associated motifs, are conserved across different Brassicaceae species (Supplemental Fig. S4).
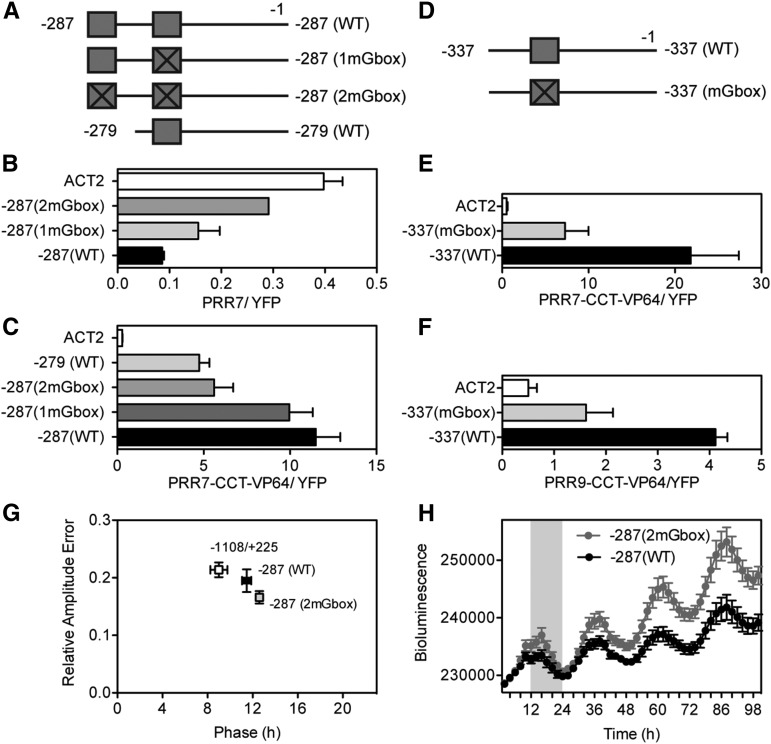
We used CCA1 and PRR9 promoter fragments driving a luciferase reporter gene in Arabidopsis protoplasts to investigate the role of G-boxes in vivo. A short fragment of the PRR9 promoter (−287/−1 bp) was sufficient to mediate repression by full-length PRR7 (Fig. 6, A and B). This activity was reduced when the G-boxes were mutated. To provide further evidence that PRR7 directly regulates gene expression, we used the PRR7-CCT domain fused to the herpes simplex viral protein 16 (VP16) transactivation domain in our transfection assays. The CCT domain by itself cannot mediate transcriptional repression but can be converted to an activator when fused to the VP16 domain (Nakamichi et al., 2012). Therefore, if the transcriptional regulation by the PRRs is direct, we expect to see opposite activities between the full-length protein (repression) and the CCT-VP16 fusion protein (activation). Accordingly, the PRR7-CCT domain fused to the VP16 transactivation domain led to transcriptional activation in a G-box-dependent manner (Fig. 6C), indicating that this effect may be direct. The PRR9 G-box located at the −286 position appeared to mediate most of the activity, since the absence of this motif in the −279/−1 fragment caused the same loss of activity as mutations in both G-boxes (Fig. 6C). Notably, this G-box is conserved in the promoter of a putative PRR9 ortholog that displays a morning phase of expression in the distantly related Carica papaya (Supplemental Fig. S4A; Zdepski et al., 2008). Moreover, a −337/−1 fragment of the CCA1 promoter (Fig. 6D) was also sufficient to mediate activation by PRR7-CCT-VP16 and PRR9-CCT-VP16 in a G-box dependent manner (Fig. 6, E and F).
Figure 6.
G-box motifs are necessary for transcriptional regulation by PRRs. A, Graphic representation of PRR9 promoter fragments. Boxes indicate G-box motifs; crossed boxes indicate mutated G-box motifs; numbers indicate the position with respect to the transcriptional start site. WT, wild type; 1mGbox, one mutated G-box; 2mGbox, two mutated G-boxes. B and C, Expression of PRR9::LUC constructs using (B) 35S::PRR7 as the effector in prr579 protoplasts or (C) 35S::PRR7-CCT-VP16 as the effector in wild-type protoplasts. D, Graphic representation of CCA1 promoter fragments. Labeling as in A; mGbox, mutated G-box. E and F, Relative expression of different CCA1::LUC constructs after the addition of (E) 35S::PRR7-CCT-VP16 or (F) 35S::PRR9-CCT-VP16 in wild-type protoplasts. In (B), (C), (E), and (F), the luciferase expression was normalized to the 35S::REN transformation control and to the 35S::YFP vector control. Values are the average ± sem of 2 to 4 independent experiments. G, Rhythms of seedlings expressing PRR9::LUC fragments. Relative Amplitude Error is a measure of rhythmic strength that varies between 0 (perfect fitted rhythm) and 1 (no significant rhythm) and was determined under constant light. Phase was determined during the light/dark cycle. H, Bioluminescence (counts/seedling/20 min) of PRR9::LUC-expressing seedlings. The shaded area indicates dark period. For (G) and (H), values are the average ± sem of five −287 (WT) and ten −287 (2mGbox) independently transformed lines; 2 to 4 T2 seedlings were analyzed per line. Eight homozygous seedlings were analyzed for the −1108/+225 line. Similar results were observed in three additional experiments.
We also analyzed plants expressing PRR9 promoter reporter constructs harboring wild-type or mutated G-boxes. The −287/−1 constructs retained strong rhythmicity under constant light with a phase slightly later than a longer PRR9 promoter construct (−1108/+225) that includes the 5′ untranslated region (Para et al., 2007; Fig. 6G). The absence of G-boxes in the context of the −287/−1 fragment led to a slight delay in phase, to a higher expression level during the end of the day under light/dark cycles, and overall higher expression under constant light conditions (Fig. 6, G and F). These results indicate that these G-boxes mediate the association of a transcriptional repressor during the end of the day.
DISCUSSION
We performed a comprehensive analysis of PRR binding regions showing that the PRRs share a large number of target genes. Our findings provide an explanation of their partially redundant functions in regulating the circadian clock, growth, development, and responses to abiotic stimuli. Most PRR binding regions are shared by at least two PRRs, which could explain the similarity between the overall phase distributions among the target genes of different PRRs (Fig. 3A; Huang et al., 2012; Nakamichi et al., 2012; Liu et al., 2013). As recently reported for CCA1 (Nagel et al., 2015), we also found noncycling genes among PRR targets. This could be due to differences in the phase of expression in different tissues or masking of transcriptional regulation caused by changes in RNA stability (Gutierrez et al., 2002; Endo et al., 2014). However, it may also reflect a broader role of clock components as modulators of signaling processes. For example, one of the genes bound by PRR9, PRR7, and PRR5 is ELIP1 (EARLY LIGHT INDUCED PROTEIN 1), which is upregulated in the prr579 mutant, but does not cycle in constant light (Edwards et al., 2006; Nakamichi et al., 2010). ELIP1 is also directly regulated by CCA1 (Nagel et al., 2015) and the Evening Complex, which is composed of LUX, ELF4, and ELF3 (Takeuchi et al., 2014). In addition, ELIP1 RNA levels cycle under light/dark conditions and are induced by visible (Harari-Steinberg et al., 2001) and UV-B light (Takeuchi et al., 2014). Thus, in some cases, regulation by clock components may be restricted to provide a gated response to light signals.
Our results show that there is a tight regulatory network among PRR, CCA1/RVE, and LNK protein families. The PRRs are conserved among the green lineage, and a single feedback loop between a PRR and a CCA1-like gene forms the basis of the clock in the green algae, Ostreococcus tauri (Corellou et al., 2009). The PRR and CCA1 families of transcription regulators have expanded in Angiosperms (Takata et al., 2009, 2010), and some of these genes have retained their function as circadian clock components, such as the PRRs, CCA1, LHY, RVE8, RVE4, and RVE6 (Hsu and Harmer, 2014). However, others have lost their role in the regulation of circadian rhythms; for example, RVE1 has been shown to regulate auxin biosynthesis (Rawat et al., 2011). The recently characterized LNK proteins act as transcriptional coregulators with RVE8 and possibly CCA1 and LHY (Rugnone et al., 2013; Xie et al., 2014). We observed several PRRs bound to the promoters of CCA1/LHY/RVE and LNK genes (Supplemental Dataset S1). In turn, CCA1 has been found to associate with promoter regions of several PRRs, LNK3, RVE1, RVE2, RVE7, LHY, and its own promoter (Nagel et al., 2015). These results show that transcriptional regulation has been maintained during the expansion of these gene families in spite of some functional divergence. Moreover, they confirm our observation that genes tend to maintain cyclic regulation after duplication (Panchy et al., 2014) and emphasize the importance of cyclic expression for gene function.
We observed that the PRRs were bound to conserved regions in open chromatin, indicating that they associate with specific regulatory sites. Our results also suggest that PRRs are able to bind to the same cis-regulatory regions and therefore might compete for certain binding sites. Differences in time and tissue-specific expression could reduce the direct competition for binding at the cellular level between the PRRs. Some differences in tissue-specific expression of the PRRs have been reported (Para et al., 2007; Endo et al., 2014), but the implications for the regulation of target gene expression are not well understood. PRR binding to target promoters appear to correlate with protein abundance (Nakamichi et al., 2010; Huang et al., 2012). However, the lack of correlation between PRR5 and PRR7 protein abundance and their binding to the CCA1 and LHY promoters at certain times of day (Nakamichi et al., 2010) indicate that other factors might influence PRR association to DNA.
The mechanism by which PRRs associate with target promoters is poorly understood. It has been shown that the C-terminal CCT domain is necessary and sufficient for PRR association to DNA (Gendron et al., 2012; Nakamichi et al., 2012). CCT domains share homology with the DNA binding domain of HEME ACTIVATOR PROTEIN 2 (HAP2; Wenkel et al., 2006). HAP2 is able to bind DNA when forming a complex with other HAP proteins in yeast, (Olesen et al., 1987; Forsburg and Guarente, 1988) and the CCT domain containing protein CONSTANS binds HAP proteins in Arabidopsis (Wenkel et al., 2006). The interaction of PRRs with HAP proteins remains to be studied. It has been shown that the CCT domains of TOC1, PRR5, PRR7, and PRR9 are able to bind a region within the CCA1 gene in vitro (Gendron et al., 2012). However, we showed that a different region located in the CCA1 promoter is sufficient to maintain cyclic expression and regulation by PRRs in vivo. It is possible that tethering or cobinding of the PRRs with other transcription factors might be sufficient for transcriptional activity at specific promoters.
G-box-containing elements are enriched among PRR target genes, and we showed that these elements are necessary to mediate PRR transcriptional regulation of PRR9 and CCA1 promoters. PRRs share a significant number of target genes with G-box binding factors, including HY5 and PIF1, and both PRR7 and PRR5 share binding regions with PIF4 and PIF5 (Liu et al., 2013; Heyndrickx et al., 2014). In addition, PRRs have been shown to interact with some G-box binding proteins, such as TOC1 with PIF7 (Kidokoro et al., 2009). However, not all PRR binding regions contain G-box-like motifs (Fig. 5A). In a similar manner, a recent study of Arabidopsis transcription factors showed that single motifs are rarely present in more than one-half of transcription factor binding regions determined by ChIP-seq (Heyndrickx et al., 2014). This study also showed that some PRR5 and PRR7 binding regions contain FHY3 (FAR RED ELONGATED HYPOCOTYLS 3) and FAR1 (FAR RED-IMPAIRED RESPONSE) binding sites, in agreement with significant overlaps in PRR5 and PRR7 with FHY3 target genes (Heyndrickx et al., 2014). Future analyses of PRR interactions with other transcription factors will aid in understanding the role of different cis-regulatory sites for determining promoter specificity.
MATERIALS AND METHODS
Generation of prr9-1 PRR9::HA-PRR9 Lines
The construct pENTR HA-PRR9 containing the PRR9 coding sequence was used to introduce HA-PRR9 into pMDC32 (Curtis and Grossniklaus, 2003) via Gateway technology. The primers 5′-caccatgtacccatacgatgttccagattacgctatgggggagattgtggttt-3′ and 5′-tgattttgtagacgcgtctga-3′ were used to amplify the PRR9 coding region and introduce an HA-tag at the amino terminus of PRR9. The PRR9 promoter/5′ untranslated region (-1332/+225) was amplified using the primers 5′-cactcctgcaggtcaaccaagaatccgttca-3′ and 5′-catcggtaccagactcagacctcaaaaca-3′ and cloned into the pCRBlunt II and pMDC32 HA-PRR9 vector using Sse8387I and BamHI to exchange the 35S promoter. The final PRR9::HA-PRR9 construct was used to transform prr9-1 CCR2::LUC (prr9; Farré et al., 2005). Circadian rhythms were monitored as described previously (Liu et al., 2013) and analyzed using FFT-NLLS implemented in BRASS (http://millar.bio.ed.ac.uk/Downloads.html; Plautz et al., 1997). Three independent lines that displayed similar RNA levels to wild type were selected (Supplemental Fig. S1A). Gene expression was determined as described previously (Liu et al., 2013). The line PRR9::HA-PRR9 #109 (HA9) complemented the long period phenotype of prr9 under constant light (Supplemental Fig. S1B) and was chosen for further studies.
ChIP, Library Preparation, and Sequencing
Plants were grown on Murashige and Skoog (MS) medium with 0.8% agar and 2% Suc under cycling 12-h light (70 µmol m−2 s−1 white light)/12-h dark at 22°C for 2 weeks before harvesting at 4 h after the onset of light. ChIPs were performed on the complemented HA9 line and prr9 parental control as described earlier (Liu et al., 2013). We pooled three independent ChIPs and confirmed their quality via qPCR of DNA from the immunoprecipitated (IP) fraction normalized to the input control using previously described primers to check the CCA1 promoter (Supplemental Fig. S1C; Liu et al., 2013). The Research Technology Sequencing Facility at MI State University prepared the libraries using the ThruPLEX-FD Prep Kit (Rubicon Genomics) following the manufacturer’s protocol for multiplexing. The DNA size and quality were checked using the BioAnalyser DNA high sensitivity kit (Agilent, Santa Clara, CA) and Fluorometer Qubit (Invitrogen, Carlsbad, CA). DNA sequencing of 50-bp, single-end reads with the Illumina Hi-SEquation 2500 yielded a depth of coverage of 4.8 for the HA9 input and 9.5 for the HA9 IP.
ChIP-seq Data Analysis
The NCBI SRA Toolkit was used to convert PRR5 and TOC1 ChIP-seq data acquired from the NCBI Gene Expression Omnibus to fastq format. ChIP-seq data for PRR9 and PRR7 were generated in our laboratory and were already in fastq format. All sequences were preprocessed using the fastx_quality_trimmer from the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) with a Phred quality score threshold of 20 (−t 20) and a minimum length of 30 nucleotides (−l 30). Quality filtered reads were aligned to the Arabidopsis (Arabidopsis thaliana) TAIR10 genome using Bowtie with the option of suppressing all alignments for reads with more than 1 reportable alignment (−m 1; Langmead et al., 2009). Alignments were visualized using Samtools (Li et al., 2009) and the Integrative Genomics Viewer (Robinson et al., 2011). MACS2 (Zhang et al., 2008) was used to identify binding regions with the broad peak calling parameter (−broad). In each experiment, the IP sample was compared with their respective input as the negative control, except for the PRR7 ChIP-seq experiment, in which the prr7-3 PRR7::HA-PRR7 IP was compared with the prr7-3 IP as the negative control. ChIPpeakAnno (Zhu et al., 2010) was used to associate binding regions to the nearest transcriptional start site as well as for the GO enrichment tests (Benjamini Hochberg adjusted P value < 0.05). Binding regions located between 5 kb upstream to 500 bp downstream of genes on chromosomes 1–5 captured more than 90% of PRR binding regions and were used for further analysis. Bedtools (Quinlan and Hall, 2010) was used to acquire DNA sequences based on bed files. Sequence motifs were identified de novo using MEME-ChIP (Machanick and Bailey, 2011) on PRR binding regions located upstream. The JASPAR CORE Plantae 2014 database (Mathelier et al., 2014) was used for TOMTOM (Gupta et al., 2007) to find known motifs and CentriMo (Bailey and Machanick, 2012; -db JASPAR_CORE_2014_plants.meme) to examine the distribution of the best-matched known motif. Each PRR had one significantly enriched motif, and FIMO (Grant et al., 2011) was used to determine the percentage of upstream PRR binding regions that contained at least one instance of the corresponding PRR motif. The MEME position weight matrices were converted to TAMO format using meme2tamo.py from the Fraenkel Lab (Gordon et al., 2005). PCC distance (1-PCC; Zou et al., 2011) was used to determine the extent to which the position weight matrices of PRR motifs differ (Zou et al., 2011). The threshold (0.38) was determined using the fifth percentile of the distances between binding motifs from different families of transcription factors (Franco-Zorrilla et al., 2014).
Expression Analysis
Data on cyclic expression are from (Mockler et al., 2007), and genes with an model-based, pattern-matching algorithm (mbpma) > 0.8 were defined as cycling. LD data are from (Bläsing et al., 2005) and LL data from (Edwards et al., 2006). PHASER (Michael et al., 2008) was used to analyze peak gene expression of cycling genes with an mbpma > 0.8. The prr579 expression data compared with wild type is from (Nakamichi et al., 2009), the PRR5-VP16 expression data are from (Nakamichi et al., 2012), and the alcohol-inducible ALC::TOC1 expression data are from (Gendron et al., 2012).
Analysis of NOC, DHSs, and Conserved Noncoding Regions
NOC data are from (Liu et al., 2015), DHS data from (Sullivan et al., 2014), and CNS data from (Haudry et al., 2013). Normalized values represent the log2 ratio of the average (NOC) or median (DHS and CNS) score per base from −1 kb upstream to +1 kb downstream of the binding summit, to the average (NOC) or median (DHS and CNS) score at the same coordinates of randomly selected genes. For the CNS reference file, the presence of a CNS was given a value of one, whereas the absence of a CNS was given a value of zero. To focus on binding regions located in noncoding regions, binding summits located in exons were omitted for the CNS analyses.
Generation of Constructs for Protoplast Transformation
The PRR full-length or CCT domain coding sequence was amplified and cloned into pENTR/D-TOPO (Invitrogen) and transferred into pRTL2-35S-GW or pRTL2-GW-VP64, respectively. The PRR7 CCT domain was amplified using the primers 5′-caccatgaataagatctctcaaagggaa-3′ and 5′-gctatcctcaatgttttttatgt-3′ and the PRR9 CCT domain using 5′-caccatgtggagtagaagccagagag-3′ and 5′-tgattttgtagacgcgtctgaatt-3′. To generate the pRTL2-35S-GW vector, the gateway cassette was amplified from pESpyce (Berendzen et al., 2012) using the primers XhoI-GW-F 5′-cacactcgagacaagtttgtacaaaaaagc-3′ and GWs-XbaI-R 5′-tgtgtctagattaaaccactttgtac-3′ and inserted into vector pRTL2-35S-NYFP (Strayer et al., 2000) using the XhoI and XbaI sites. The pRTL2-35S-GW-VP64 vector was used to express the PRR-CCT domains fused to four copies of the VP16 transactivation domain. The GW-VP64 insert was amplified from pB7WG2-VP64 (Helfer et al., 2011) using the primers XhoI-GW-F and VP64s-XbaI-R 5′-tgtgtctagattagttaattaacatatcg-3′ and cloned into pRTL2-NYFP using XhoI and XbaI. In both cases the NYFP was removed from pRTL2.
To generate pRTL2-GW-Luc, which was used to clone the different promoter fragments, the GW-Luc-Nos insert was amplified from pMDC140-Luc+-HA (Farré and Kay, 2007) using primers pRTL2-GW-F 5′-ctatgaccatgattacgccaatcaacaagtttgtacaaaaaagc-3′ and Nos-pRTL2-R 5′-cgacggccagtgccaagctagtaacatagatgacacc-3′. Gibson Assembly (NEB) was used to clone the insert into the pRTL2-NYFP vector after digestion with HindIII. Promoter fragments of −287/−1 bp and −337/−1 bp for PRR9 and CCA1, respectively, were amplified from ecotype Columbia of Arabidopsis (Col-0) genomic DNA using the primers shown in Supplemental Table S1. Promoter fragments were cloned into pENTR/D-TOPO (Invitrogen) and transferred to pRTL2-GW-Luc-Nos. Fragments with the G-boxes mutated for both CCA1 and PRR9 promoters were made by changing the G-box sequence from CACGTG to ACATGT or TGTACA using primers shown in Supplemental Table S1. The fragments were cloned using the mega-primer strategy. The ACT2 promoter (−473/−1) was used as the promoter negative control, the pRTL2-35S-NYFP construct was used as the effector negative control, and the Renilla reniformis luciferase (Helfer et al., 2011) expressing vector pRTL2-35S-Renilla as the transformation control.
Protoplast Transient Transformation Assays
Arabidopsis (Arabidopsis thaliana) Col-0 or prr579 (Liu et al., 2013) seeds were sown on soil (Sure-Mix) and placed at 4°C for 3 to 4 d. They were grown for 3 to 4 weeks at about 20°C under 16-h-light (110 μmol m−2s−1 white light)/8-h-dark cycles. Leaves 5 to 7 were removed from 25 to 45 seedlings and cut into thin strips. The leaf strips were placed in 10 mL digestion buffer (400 mm mannitol, 20 mm potassium chloride, 20 mm MES pH 5.7, 15 mg mL−1 cellulase, 4 mg mL−1 macerozyme, 10 mm calcium chloride, 0.1% nuclease-free bovine serum albumin) and incubated in the dark for 3 h. After lightly shaking the leaf strips to release the protoplasts, 10 mL of W5 buffer (2 mm MES pH 5.7, 125 mm calcium chloride, 154 mm sodium chloride, 5 mm potassium chloride) was added and protoplasts were filtered through a 75-μm nylon mesh. The protoplasts were centrifuged at 100g and 4°C for 2 min. The protoplast pellet was resuspended in 5 mL chilled W5 buffer and centrifuged again at 100g and 4°C for 2 min. The pellet was resuspended in 5 mL chilled W5 buffer and allowed to settle for 30 min on ice. The protoplasts were resuspended in MMG buffer (400 mm mannitol, 15 mm magnesium chloride, 4 mm MES pH 5.7) to 2.5 million protoplasts mL−1. Ten micrograms of DNA was added to 250,000 protoplasts. The DNA mixture consisted of 5 μg effector, 4 μg target pRTL2-promoter-Luc, and 1 μg pRTL2-35S-Renilla. To transfect the protoplasts, 110 μL polyethylene glycol solution (200 mm mannitol, 100 mm calcium chloride, and 40% polyethylene glycol 4000) was added, and samples were incubated at room temperature for 30 min. To stop the transfection, 440 µL of W5 buffer was added. Samples were centrifuged at 100g and 22°C for 2 min. Protoplasts were washed twice with 1 mL W5 buffer and centrifuged again and spread in a thin layer on a plate treated with 5% BSA in MMG buffer. Protoplasts were left under 15 μmol m−2 s−1 light for ∼16 h. Transfected protoplasts were centrifuged at 200g and 22°C for 20 min and assayed using the Promega Dual Luciferase kit (Promega). The protoplast pellet was resuspended in 50 μL extraction buffer (1× passive lysis buffer, 1× protease inhibitor, 5 mm benzamidine, 1 mm phenylmethylsulfonyl fluoride) and incubated on ice for 15 min. Samples were centrifuged for 10 min at 18,000g and 4°C. The supernatant was centrifuged for 5 min at 18,000g and 4°C. To measure the firefly luciferase activity, 25 μL of Lar II was added to 5 μL of the protein extract, and the luminescence was measured for 0.05 s. To measure renilla luciferase activity, 25 μL of Stop and Glow was added, and the luminescence was again measured for 0.05 s using a Berthold LB960XS3 luminometer.
In Planta Luciferase Expression of PRR9 Promoter Fragments
The promoter fragments in pENTR/D-TOPO (Invitrogen) described above were transferred to the gateway compatible pFlash vector (Gendron et al., 2012) and used to transform Arabidopsis Col-0 (Clough and Bent, 1998). Transgenic seedlings were selected on MS medium containing 0.8% agar and gentamycin (100 µg mL−1). For imaging, 12-d-old T2 plants grown on gentamycin under 12 h white light/12 h dark were transferred to MS medium containing 0.8% agar and 2% Suc and treated with 5 mm luciferin (Gold Biotechnology) in 0.01% Silwett. The next day, plants were transferred to 150 µmol m−2 s−1 light (25% blue light LED, 75% red light LED) and imaged using an Andor iKon-M DU-934N-BV camera every 2 h for 20 min. Two to four seedlings per line were analyzed. Phase and relative amplitude error were quantified using BRASS. The PRR9::LUC construct −1108/+225 has been described previously (Para et al., 2007).
Accession Numbers
Sequence data from this article can be found in the NCBI GEO data libraries under accession numbers GSE35952 for the TOC1 ChIP-seq, GSE36361 for the PRR5 ChIP-seq, GSE49282 for the PRR7 ChIP-seq, and GSE71397 for both the PRR9 ChIP-seq and the processed data from this study.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of prr9-1 PRR9::HA-PRR9 CCR2::LUC and quality testing of HA-PRR9 ChIPs.
Supplemental Figure S2. Relative positions of PRR binding summits determined by ChIP-seq.
Supplemental Figure S3. Common overrepresented GO terms among PRR putative target genes identified by ChIP-seq.
Supplemental Figure S4. Conservation of the PRR9 and CCA1 promoters.
Supplemental Table S1. Primers used to generate promoter fragments.
Supplemental Dataset S1. PRR binding sites and associated genes.
Supplemental Dataset S2. Expression of putative PRR target genes identified by ChIP-seq and overlap of PRR target genes.
Supplemental Dataset S3. Enriched GO terms in putative PRR target genes.
Supplementary Material
Acknowledgments
We thank the Shiu laboratory members for helpful discussions, especially Sahra Uygun. We are grateful to Jeff Landgraf and the rest of the Research Technology Sequencing Facility at MSU for handling the library preparation and sequencing.
Glossary
- ChIP-seq
chromatin immunoprecipitation followed by high-throughput sequencing
- ChIP
chromatin immunoprecipitation
- CNS
conserved noncoding sequence
- CRE
cis-regulatory elements
- DHS
DNase I hypersensitive site
- GO
Gene Ontology
- IP
immunoprecipitated
- mbpma
model-based, pattern-matching algorithm
- NOC
nucleosome occupancy
- PCC
Pearson Correlation Coefficient
Footnotes
This work was funded by the National Science Foundation (IOS-1054243 to E.F. and MCB-1119778 and IOS-1126998 to S.-H.S.); T.L. was also supported by graduate fellowships from Michigan State University.
Articles can be viewed without a subscription.
References
- Alabadí D, Oyama T, Yanovsky MJ, Harmon FG, Más P, Kay SA (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40: e128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Tiwari VK, Thomä NH, Schübeler D (2011) Determinants and dynamics of genome accessibility. Nat Rev Genet 12: 554–564 [DOI] [PubMed] [Google Scholar]
- Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, Zoran MJ (2005) Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet 6: 544–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen KW, Böhmer M, Wallmeroth N, Peter S, Vesić M, Zhou Y, Tiesler FK, Schleifenbaum F, Harter K (2012) Screening for in planta protein-protein interactions combining bimolecular fluorescence complementation with flow cytometry. Plant Methods 8: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina MD, Troyanskaya OG (2012) An effective statistical evaluation of ChIPseq dataset similarity. Bioinformatics 28: 607–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Corellou F, Schwartz C, Motta JP, Djouani-Tahri B, Sanchez F, Bouget FY (2009) Clocks in the green lineage: comparative functional analysis of the circadian architecture of the picoeukaryote ostreococcus. Plant Cell 21: 3436–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, Knox K, Kozma-Bognar L, Southern MM, Pokhilko A, Millar AJ (2011) Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr Biol 21: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR (2005) Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633 [DOI] [PubMed] [Google Scholar]
- Edwards KD, Anderson PE, Hall A, Salathia NS, Locke JC, Lynn JR, Straume M, Smith JQ, Millar AJ (2006) FLOWERING LOCUS C mediates natural variation in the high-temperature response of the Arabidopsis circadian clock. Plant Cell 18: 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Shimizu H, Nohales MA, Araki T, Kay SA (2014) Tissue-specific clocks in Arabidopsis show asymmetric coupling. Nature 515: 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P (2011) Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J 66: 318–329 [DOI] [PubMed] [Google Scholar]
- Farré EM, Harmer SL, Harmon FG, Yanovsky MJ, Kay SA (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr Biol 15: 47–54 [DOI] [PubMed] [Google Scholar]
- Farré EM, Kay SA (2007) PRR7 protein levels are regulated by light and the circadian clock in Arabidopsis. Plant J 52: 548–560 [DOI] [PubMed] [Google Scholar]
- Farré EM, Liu T (2013) The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol 16: 621–629 [DOI] [PubMed] [Google Scholar]
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Michael TP, Chory J, Kay SA, Mockler TC (2011) Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS One 6: e16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Rühl M, Jarillo JA, Coupland G (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell 17: 75–86 [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Guarente L (1988) Mutational analysis of upstream activation sequence 2 of the CYC1 gene of Saccharomyces cerevisiae: a HAP2-HAP3-responsive site. Mol Cell Biol 8: 647–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R (2014) DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci USA 111: 2367–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DB, Nekludova L, McCallum S, Fraenkel E (2005) TAMO: a flexible, object-oriented framework for analyzing transcriptional regulation using DNA-sequence motifs. Bioinformatics 21: 3164–3165 [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari-Steinberg O, Ohad I, Chamovitz DA (2001) Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol 127: 986–997 [PMC free article] [PubMed] [Google Scholar]
- Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, Kanai A, Suzuki Y, Sassone-Corsi P, Aburatani H, Sugano S, Takumi T (2010) Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol 30: 5636–5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, Dewar K, Stinchcombe JR, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Helfer A, Nusinow DA, Chow BY, Gehrke AR, Bulyk ML, Kay SA (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr Biol 21: 126–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, Webb A, Gonçalves J, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyndrickx KS, Van de Velde J, Wang C, Weigel D, Vandepoele K (2014) A functional and evolutionary perspective on transcription factor binding in Arabidopsis thaliana. Plant Cell 26: 3894–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: the plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Mas P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79 [DOI] [PubMed] [Google Scholar]
- Jiang J. (2015) The ‘dark matter’ in the plant genomes: non-coding and unannotated DNA sequences associated with open chromatin. Curr Opin Plant Biol 24: 17–23 [DOI] [PubMed] [Google Scholar]
- Kidokoro S, Maruyama K, Nakashima K, Imura Y, Narusaka Y, Shinwari ZK, Osakabe Y, Fujita Y, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2009) The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol 151: 2046–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS (2006) Molecular components of the mammalian circadian clock. Hum Mol Genet 15: R271–R277 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MJ, Seddon AE, Tsai ZT, Major IT, Floer M, Howe GA, Shiu SH (2015) Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res 25: 1182–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Carlsson J, Takeuchi T, Newton L, Farré EM (2013) Direct regulation of abiotic responses by the Arabidopsis circadian clock component PRR7. Plant J 76: 101–114 [DOI] [PubMed] [Google Scholar]
- Machanick P, Bailey TL (2011) MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 27: 1696–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, Lim J, Shyr C, et al. (2014) JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42: D142–D147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM, Givan SA, Yanovsky M, et al. (2008) Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet 4: e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72: 353–363 [DOI] [PubMed] [Google Scholar]
- Nagel DH, Doherty CJ, Pruneda-Paz JL, Schmitz RJ, Ecker JR, Kay SA (2015) Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc Natl Acad Sci USA 112: E4802–E4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kita M, Niinuma K, Ito S, Yamashino T, Mizoguchi T, Mizuno T (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol 48: 822–832 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kusano M, Fukushima A, Kita M, Ito S, Yamashino T, Saito K, Sakakibara H, Mizuno T (2009) Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Kamioka M, Suzuki T, Yamashino T, Higashiyama T, Sakakibara H, Mizuno T (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc Natl Acad Sci USA 109: 17123–17128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Olesen J, Hahn S, Guarente L (1987) Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell 51: 953–961 [DOI] [PubMed] [Google Scholar]
- Panchy N, Wu GX, Newton L, Tsai CH, Chen J, Benning C, Farre EM, Shiu SH (2014) Prevalence, evolution, and cis-regulation of diel transcription in Chlamydomonas reinhardtii. G3 4: 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, Phinney BS, Harmer SL (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone ML, Faigón Soverna A, Sanchez SE, Schlaen RG, Hernando CE, Seymour DK, Mancini E, Chernomoretz A, Weigel D, Más P, Yanovsky MJ (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc Natl Acad Sci USA 110: 12120–12125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato E, Nakamichi N, Yamashino T, Mizuno T (2002) Aberrant expression of the Arabidopsis circadian-regulated APRR5 gene belonging to the APRR1/TOC1 quintet results in early flowering and hypersensitiveness to light in early photomorphogenesis. Plant Cell Physiol 43: 1374–1385 [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Sullivan AM, Arsovski AA, Lempe J, Bubb KL, Weirauch MT, Sabo PJ, Sandstrom R, Thurman RE, Neph S, Reynolds AP, Stergachis AB, Vernot B, et al. (2014) Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Reports 8: 2015–2030 [DOI] [PubMed] [Google Scholar]
- Takata N, Saito S, Saito CT, Nanjo T, Shinohara K, Uemura M (2009) Molecular phylogeny and expression of poplar circadian clock genes, LHY1 and LHY2. New Phytol 181: 808–819 [DOI] [PubMed] [Google Scholar]
- Takata N, Saito S, Saito CT, Uemura M (2010) Phylogenetic footprint of the plant clock system in angiosperms: evolutionary processes of pseudo-response regulators. BMC Evol Biol 10: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Newton L, Burkhardt A, Mason S, Farré EM (2014) Light and the circadian clock mediate time-specific changes in sensitivity to UV-B stress under light/dark cycles. J Exp Bot 65: 6003–6012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18: 2971–2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH (2004) The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol 14: 1481–1486 [DOI] [PubMed] [Google Scholar]
- Xie Q, Wang P, Liu X, Yuan L, Wang L, Zhang C, Li Y, Xing H, Zhi L, Yue Z, Zhao C, McClung CR, et al. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26: 2843–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi S, Green RM (2009) Evidence for the adaptive significance of circadian rhythms. Ecol Lett 12: 970–981 [DOI] [PubMed] [Google Scholar]
- Yerushalmi S, Yakir E, Green RM (2011) Circadian clocks and adaptation in Arabidopsis. Mol Ecol 20: 1155–1165 [DOI] [PubMed] [Google Scholar]
- Yoshitane H, Ozaki H, Terajima H, Du NH, Suzuki Y, Fujimori T, Kosaka N, Shimba S, Sugano S, Takagi T, Iwasaki W, Fukada Y (2014) CLOCK-controlled polyphonic regulation of circadian rhythms through canonical and noncanonical E-boxes. Mol Cell Biol 34: 1776–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdepski A, Wang W, Priest HD, Ali F, Alam M, Mockler TC, Michael TP (2008) Conserved daily transcriptional programs in Carica papaya. Trop Plant Biol 1: 236–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR (2010) ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Sun K, Mackaluso JD, Seddon AE, Jin R, Thomashow MF, Shiu SH (2011) Cis-regulatory code of stress-responsive transcription in Arabidopsis thaliana. Proc Natl Acad Sci USA 108: 14992–14997 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.