Arabidopsis GPAT9 is a highly conserved, single-copy, and essential glycerol-3-phosphate acyltransferase involved in seed oil biosynthesis.
Abstract
The first step in the biosynthesis of nearly all plant membrane phospholipids and storage triacylglycerols is catalyzed by a glycerol-3-phosphate acyltransferase (GPAT). The requirement for an endoplasmic reticulum (ER)-localized GPAT for both of these critical metabolic pathways was recognized more than 60 years ago. However, identification of the gene(s) encoding this GPAT activity has remained elusive. Here, we present the results of a series of in vivo, in vitro, and in silico experiments in Arabidopsis (Arabidopsis thaliana) designed to assign this essential function to AtGPAT9. This gene has been highly conserved throughout evolution and is largely present as a single copy in most plants, features consistent with essential housekeeping functions. A knockout mutant of AtGPAT9 demonstrates both male and female gametophytic lethality phenotypes, consistent with the role in essential membrane lipid synthesis. Significant expression of developing seed AtGPAT9 is required for wild-type levels of triacylglycerol accumulation, and the transcript level is directly correlated to the level of microsomal GPAT enzymatic activity in seeds. Finally, the AtGPAT9 protein interacts with other enzymes involved in ER glycerolipid biosynthesis, suggesting the possibility of ER-localized lipid biosynthetic complexes. Together, these results suggest that GPAT9 is the ER-localized GPAT enzyme responsible for plant membrane lipid and oil biosynthesis.
Glycerol-3-phosphate acyltransferase (GPAT; EC 2.3.1.15) was first characterized biochemically in extracts from animal and plant tissues in a series of reports beginning more than 60 years ago (Kornberg and Pricer, 1953; Weiss et al., 1960; Barron and Stumpf, 1962). GPAT transfers an acyl moiety from either acyl-CoA or acyl-acyl carrier protein (ACP) to the sn-1 position of glycerol-3-phosphate (G3P) to produce 1-acyl-2-lyso-glycerol-3-phosphate (or lysophosphatidic acid). It catalyzes the first step in the synthesis of various types of glycerolipids, including membrane lipids and triacylglycerol (TAG), in all living organisms and, thus, plays an extremely important role in basic cellular metabolism. In addition, the enzymes that esterify fatty acids (FAs) to the glycerol backbone of TAG control the TAG FA composition and, thus, the functional value of plant oils. Acyl-selective isozymes for the last step in TAG assembly have been demonstrated to be crucial for enhancing the control of seed oil content through bioengineering (Liu et al., 2012). Therefore, our ability to produce designer plant oils that will meet the nutritional or industrial needs of the future will require identifying the genes that encode all of the acyltransferase steps in TAG production, including the essential first step catalyzed by GPAT.
Early in vitro biochemical characterization of GPAT activities from various plant tissues demonstrated multiple subcellular locations for GPAT activity, including endoplasmic reticulum (ER; Barron and Stumpf, 1962; Ichihara, 1984; Griffiths et al., 1985), plastid (Joyard and Douce, 1977), and mitochondria (Sparace and Moore, 1979). These and other gene-independent biochemical analyses of plant lipid metabolic activity have in part contributed to the two-pathway hypothesis for plant glycerolipid synthesis that has stood the test of time and has been reviewed extensively (Roughan and Slack, 1982; Ohlrogge and Browse, 1995; Allen et al., 2015). Plants contain two parallel lipid biosynthetic pathways each containing distinct GPAT enzymes to produce glycerolipids. The ER-localized eukaryotic pathway utilizes a membrane-bound acyl-CoA-dependent GPAT for the synthesis of lysophosphatidic acid (LPA). LPA is subsequently acylated to produce phosphatidic acid (PA) by an acyl-CoA-dependent lysophosphatidic acid acyltransferase (LPAAT). PA, or its dephosphorylated diacylglycerol (DAG), provides the glycerolipid backbones for ER-localized phospholipid or TAG synthesis. The three consecutive acyl-CoA-dependent acylations of the glycerol backbone to produce TAG in the ER are commonly called the Kennedy pathway (Weiss et al., 1960; Kennedy, 1961).
In contrast to the eukaryotic glycerolipid assembly pathway, the plastid-localized prokaryotic pathway utilizes a soluble acyl-ACP-dependent GPAT and an acyl-ACP-dependent LPAAT to produce PA for phosphatidylglycerol synthesis in plastids of all plants and the DAG for galactolipid synthesis in some plants. The prokaryotic pathway GPAT has been characterized extensively through both biochemical and molecular genetic approaches, demonstrating species-specific acyl selectivities that control the composition of prokaryotic glycerolipids (Frentzen et al., 1983, 1987; Cronan and Roughan, 1987; Ishizaki et al., 1988; Weber et al., 1991). In addition to prokaryotic galactolipid synthesis in the plastid, plants also utilize DAG derived from the eukaryotic pathway-synthesized membrane lipid phosphatidylcholine (PC) for galactolipid synthesis. The PC-derived DAG is imported into the plastid for galactolipid production and contains a distinctive eukaryotic FA composition (Li-Beisson et al., 2013). Therefore, the ER-localized GPAT of the eukaryotic pathway is vital for lipid assembly within both the ER and the plastid.
The essential nature of the ER GPAT for membrane lipid biosynthesis, and its role in controlling one-third of the FA composition within economically valuable plant oils, have inspired many studies focused on characterizing the ER GPAT enzyme. Microsomal extracts from a wide range of plant tissues have demonstrated a variety of in vitro ER GPAT activities utilizing various common and unusual FAs as substrates (Stobart and Stymne, 1985; Bafor et al., 1990, 1991; Rutter et al., 1997; Manaf and Harwood, 2000; Ruiz-López et al., 2010). However, dissection of the range of potential effects of the ER-localized GPAT on seed oil content and FA composition has still remained unclear, awaiting the identification and molecular characterization of the requisite gene(s) encoding the ER-localized GPAT.
Zheng et al. (2003) were first to classify a family of genes as ER- and mitochondria-localized GPATs in plants. This eight-gene family was identified based on homology to the acyltransferase domains of GPATs from yeast, and most members of this group demonstrated GPAT activity when heterologously expressed. Genetic analysis of Arabidopsis (Arabidopsis thaliana) mutants defective in the first of these genes, designated AtGPAT1, indicated altered tapetal differentiation and frequent abortion of microspores, impacting male fertility and suggesting that AtGPAT1 may be involved in essential membrane lipid production (Zheng et al., 2003). However, more recent biochemical and genetic characterization has revealed that AtGPAT1 to AtGPAT8 are a land plant-specific family of sn-2 GPATs involved in cutin and suberin synthesis (Beisson et al., 2007; Li et al., 2007; Yang et al., 2010, 2012). These GPATs predominantly utilize acyl groups not found in membranes or TAG (e.g. ω-hydroxy and α,ω-dicarboxylic FAs) to produce the sn-2 monoacylglycerol precursors utilized during extracellular polymerization of both the cutin and suberin barriers of land plants (Beisson et al., 2012). Therefore, the GPAT1 to GPAT8 family is likely not involved in the membrane lipid and TAG biosynthesis required in all living organisms.
Another gene, designated AtGPAT9, was identified previously as a candidate for the ER membrane and oil biosynthetic GPAT activity based on substantial sequence identity to mouse and human GPAT3 (Cao et al., 2006; Gidda et al., 2009). The expression profiles and biochemical properties of mGPAT3 and hGPAT3 strongly support a role for these enzymes in TAG biosynthesis within lipid-rich mammalian tissues. However, the definitive determination of the roles of AtGPAT9 in plant lipid metabolism has remained elusive. Complete understanding of plant lipid biosynthesis, and the ability to predictably engineer designer plant oils, await the identification of all the component enzymes (and associated genes) and the determination of how each one functions within the lipid metabolic network to produce the thousands of different possible molecular species of membrane and storage lipids.
The molecular identification of an ER-localized GPAT involved in membrane lipid and/or TAG biosynthesis in plants has not been accomplished until now. Here, we present the results of a series of experiments designed to address the role of AtGPAT9 in lipid biosynthesis. We present strong evidence that (1) AtGPAT9 is a highly conserved, single-copy, and essential gene that likely supplies the LPA necessary for the synthesis of essential membrane lipids in the ER; (2) significant expression of AtGPAT9 is required for normal levels of microsomal GPAT enzymatic activity in seeds and is required for wild-type levels of TAG accumulation in vivo; and (3) the AtGPAT9 protein interacts with other enzymes involved in ER glycerolipid biosynthesis, suggesting the possibility of ER-localized lipid biosynthetic complexes. Together, these results suggest that GPAT9 is the ER-localized GPAT enzyme responsible for plant membrane lipid and TAG biosynthesis.
RESULTS
GPAT9 Protein Sequences Have Been Strongly Conserved across Evolution
The biochemical properties of mouse and human GPAT3 (Cao et al., 2006) matched many of the criteria expected for the Kennedy pathway GPAT in animals. Intriguingly, the sequences of these genes were also homologous to that of AtGPAT9 (At5g60620; Supplemental Fig. S1A). However, simple sequence homology comparisons alone were not deemed sufficient to determine the biochemical/physiological role(s) of GPAT9 in Arabidopsis, given that AtGPAT9 is also homologous to other glycerolipid acyltransferase genes, including AtGPAT7 involved in cutin production (Zheng et al., 2003; Yang et al., 2012) and the lysophospholipid acyltransferases AtLPEAT1 and AtLPEAT2 (Stålberg et al., 2009; Supplemental Fig. S1, B–D). The development of new insights into possible links between the rates of gene evolution and the maintenance of gene copy number (De Smet et al., 2013) provided an additional means by which we could analyze the potential role of AtGPAT9.
Genome evolution often gives rise to large, complex gene families (Shockey and Browse, 2011). Duplicated genes can either be maintained and achieve new function or lost, causing one of the genes in question to revert to singleton status, which often occurs with genes that encode essential functions (Paterson et al., 2006; De Smet et al., 2013). A single GPAT responsible for the majority of extraplastidial membrane lipid and TAG biosynthesis would fit this essential housekeeping role and is supported by digital northern analyses of AtGPAT9 (Toufighi et al., 2005; Gidda et al., 2009), which suggests a ubiquitous expression pattern. From this perspective, we analyzed the evolution of plant GPAT9 genes by comparing the predicted protein sequences of GPAT9s from representative species from each major taxonomic grouping of plants. These included Arabidopsis (Brassicacea), a bryophyte moss (Physcomitrella patens), a lycophyte (Selaginella moellendorffii), a core eudicot (Aquilegia coerulea), a grass (Brachypodium distachyon), and representatives from the dicot families Pentapetalae, Malvidae, and Fabidae (Solanum tuberosum, Salix purpurea, Gossypium raimondii, and Cucumis sativus). The divergence and speciation events separating these plants cover at least 400 million years of evolution. As shown in Supplemental Figure S2, GPAT9 proteins from this diverse group of plants have maintained a remarkable level of sequence identity, with approximately 55% identity and 65% similarity overall, with several individual cross-species pairs at greater than 80% identity and greater than 90% similarity.
To add perspective to these findings, GPAT9 sequences from each of these plants were compared with the corresponding LPEAT1 and LPEAT2 proteins from each species. GPATs and LPEATs utilize the same acyl donor and similar acyl acceptors (in the case of LPEAT1 and LPEAT2, lysophospholipids including LPA, lysophosphatidylcholine, and lysophosphatidylethanolamine), and the sequences of GPAT9, LPEAT1, and LPEAT2 are homologous themselves (Stålberg et al., 2009), making phylogenetic comparisons between them useful. As members of a larger gene family derived at least in part from gene duplication events, LPEAT1 and LPEAT2 display more sequence divergence and are much less likely to encode essential housekeeping functions. LPEAT1 and LPEAT2 sequences from several plant species were aligned and analyzed in a fashion similar to the GPATs (Supplemental Figs. S3 and S4, respectively). The input protein sequences for the two LPEAT groups derive from a much less diverse range of species (including sequences from only three monocots and nine dicots) than those in the GPAT9 group shown in Supplemental Figure S2, yet the sequences within the LPEAT1 and LPEAT2 groups are significantly more diverged, sharing only 34% identity/47% similarity and 31% identity/43% similarity, respectively (Supplemental Figs. S3 and S4). The contrast between the relative rates of evolution among these three families of genes can be seen directly in Figure 1. GPAT9, LPEAT1, and LPEAT2 protein sequences from the three monocot species (Zea mays, Setaria italica, and Oryza sativa) and nine dicot species (Arabidopsis, Citrus clementina, Glycine max, Prunus persica, Populus trichocharpa, Ricinus communis, S. tuberosum, Theobroma cacao, and Vitis vinifera) were aligned and compared phylogenetically. Each of the three gene families forms a distinct clade containing smaller subclades specific to monocots and dicots. However, in all cases, the branch lengths for the GPAT9 family are significantly shorter than those of the members of either LPEAT clade, indicating less evolutionary drift of protein-coding sequences in the GPAT9s relative to either the LPEAT1 or LPEAT2 gene family. Significantly higher constraint on sequence divergence is consistent with the hypothesis that plant GPAT9s encode an essential housekeeping function (Fig. 1).
Figure 1.
Phylogenetic comparison of GPAT9, LPEAT1, and LPEAT2 from various monocot and dicot plant species. Protein sequences were aligned using ClustalX version 1.8.1 (Thompson et al., 1997). An unrooted phylogenetic tree was created from the alignment, using TreeView version 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/rod.html; Page, 1996). The branch lengths are proportional to the degree of divergence, with the scale of 0.1 representing 10% change.
A gpat9 Knockout Is Homozygous Lethal
The homology of AtGPAT9 to GPATs in lipid-rich mammalian tissues suggests a possible important role in TAG synthesis but does not directly indicate its function within lipid metabolism. Therefore, we initially set out to investigate if T-DNA insertional mutants of GPAT9 display an altered glycerolipid phenotype. The Arabidopsis Information Resource (https://www.arabidopsis.org/) indicates only two available T-DNA insertion lines targeted to exons in the GPAT9 locus, and both were obtained. The gpat9-1 line was a confirmed homozygous Salk line (Salk_052947C) putatively containing a T-DNA insertion in the first exon (Alonso et al., 2003). Initial plantings of gpat9-1 indicated no obvious growth phenotypes, and we confirmed the presence of a homozygous T-DNA insertion near the 5′ end of GPAT9 by PCR of genomic DNA with T-DNA- and gene-specific primers (primer sequences are shown in Supplemental Table S1).
The gpat9-2 (GABI_867A06) line (Kleinboelting et al., 2012) indicated a T-DNA insertion in the fourth exon and contains a sulfadiazine resistance selectable marker. Germination of gpat9-2 seeds on sulfadiazine-containing medium over several generations produced only heterozygous GPAT9-2/gpat9-2 plants, as confirmed by the segregation of resistant and susceptible seedlings and by PCR. Once transferred to soil, there was no obvious difference in the growth of GPAT9-2/gpat9-2 plants from the wild type. The quantitation of seed germination and seedling viability on sulfadiazine (Table I) indicated that, at each generation, approximately 6% to 26% of seeds from GPAT9-2/gpat9-2 plants do not germinate and over 84% of germinated seedlings do not survive on sulfadiazine and, thus, are wild type. All sulfadiazine-resistant seedlings continue to produce sulfadiazine-susceptible offspring, indicating that the parental lines were heterozygous GPAT9-2/gpat9-2. Together, the inability to isolate a homozygous line and the non-Mendelian segregation of sulfadiazine resistance suggest that the gpat9-2 T-DNA insertion line is homozygous lethal.
Table I. Non-Mendelian segregation of GPAT9-2/gpat9-2 under sulfadiazine selection.
n.d., not determined.
| Generation | Seeds Sown | Germinated Seeds | Sulfadiazine Resistant | Percentage Resistant of Germinated | Percentage Not Germinated |
|---|---|---|---|---|---|
| F3 | n.d. | 425 | 66 | 15.5 | n.d. |
| F4 (F3-1) | 376 | 276 | 35 | 12.7 | 26.6 |
| F4 (F3-2) | 126 | 100 | 8 | 8.0 | 20.6 |
| F4 (F3-3) | 285 | 221 | 26 | 11.8 | 22.5 |
| F4 (F3-4) | 327 | 253 | 33 | 13.0 | 22.6 |
| F4 average ± sd | 11.4 ± 2.3 | 23.1 ± 2.5 | |||
| F5 (F3-1-1) | 255 | 228 | 26 | 11.4 | 10.6 |
| F5 (F3-2-1) | 207 | 194 | 17 | 8.8 | 6.3 |
| F5 (F3-3-1) | 242 | 223 | 19 | 8.5 | 7.9 |
| F5 (F3-4-1) | 223 | 203 | 26 | 12.8 | 9.0 |
| F5 average ± sd | 10.4 ± 2.1 | 8.4 ± 1.8 |
The contradictory phenotypes of the gpat9-1 and gpat9-2 insertion lines necessitated further characterization of each mutation. We identified the actual T-DNA insertion locations by sequencing of the T-DNA insertion regions. PCR products were obtained with T-DNA left border primers paired with both upstream and downstream gene-specific GPAT9 primers using genomic DNA from both gpat9-1 and GPAT9-2/gpat9-2 plants; the results indicated the presence of multiple linked inverted T-DNAs at the GPAT9 insertion site in each line. The gpat9-1 T-DNA insertions were located 46 bp upstream of the GPAT9 coding sequence start site, and the gpat9-2 T-DNA insertions were located at the end of exon 4 (Supplemental Fig. S5A). Reverse transcription (RT)-PCR of GPAT9 mRNA from leaves of gpat9-1 indicated that full-length coding sequence was expressed at similar levels to the wild type (Supplemental Fig. S5B). Quantitative RT-PCR of GPAT9 mRNA from leaves of GPAT9-2/gpat9-2 demonstrated that GPAT9 expression was reduced to approximately 50% of that in the wild type (Supplemental Fig. S5C). Given the minor effect on transcription of GPAT9 in gpat9-1, we concentrated on the sole exon-targeted mutant, gpat9-2 (GABI_867A06), for the remainder of our studies.
The gpat9-2 T-DNA Mutation Produces Pollen Lethality and Partial Female Gametophyte Lethality Phenotypes
The expected Mendelian segregation ratios from a self-fertilized heterozygous parent that produces embryonic lethal mutants is approximately 67% heterozygous and 33% wild type. Single gametophyte lethality mutants are expected to produce 50% heterozygous and 50% wild-type offspring. That GPAT9-2/gpat9-2 plants produced less than 30% heterozygous seeds and more than 70% wild-type seeds (Table I) suggests that the gpat9-2 mutation may have reduced viability of both gametophytes. Therefore, we analyzed the transmittance of the gpat9-2 mutation (by sulfadiazine resistance) through reciprocal crosses with the wild type (Table II). When GPAT9-2/gpat9-2 flowers were used as the pollen donor with Columbia-0 (Col-0) pistils as the pollen acceptor, no sulfadiazine-resistant seedlings were obtained. However, when Col-0 was the pollen donor with GPAT9-2/gpat9-2 pistils, approximately 14% to 22% of the germinated seedlings were resistant to sulfadiazine, similar to the transmittance of sulfadiazine resistance from selfed GPAT9-2/gpat9-2 in Table I. These results suggest that gpat9-2 pollen is not viable and that most gpat9-2 female gametophytes do not survive until fertilization with pollen containing wild-type GPAT9.
Table II. Transmittance of sulfadiazine resistance from reciprocal crosses of GPAT9-2/gpat9-2 (G9 ht) and Col-0 (wild type).
| Cross | Male | Female | Germinated Seedlings | Sulfadiazine Resistant | Percentage Resistant | χ2 to the Expected 1:1 Ratio if gpat9-2 Is Nonlethal |
|---|---|---|---|---|---|---|
| 1-1 | Wild type | G9 ht | 90 | 20 | 22.2 | 44.5, P ≤ 0.0001 |
| 1-2 | G9 ht | Wild type | 113 | 0 | 0 | 113, P ≤ 0.0001 |
| 2-2 | Wild type | G9 ht | 139 | 20 | 14.4 | 89.1, P ≤ 0.0001 |
| 2-2 | G9 ht | Wild type | 242 | 0 | 0 | 242, P ≤ 0.0001 |
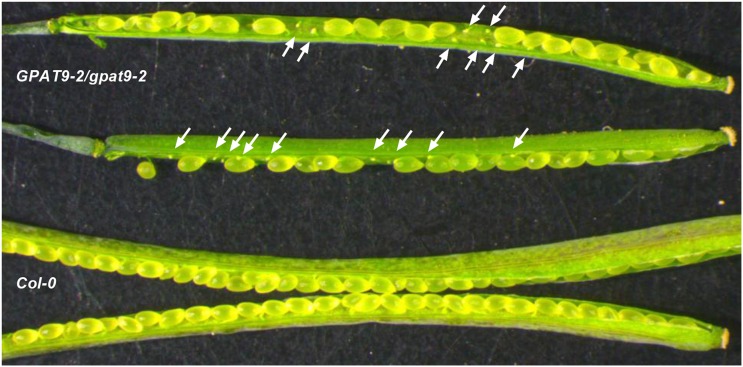
In support of a partial female gametophyte lethality phenotype of gpat9-2, aborted ovules were observed in siliques of GPAT9-2/gpat9-2 but not in Col-0 (Fig. 2). Quantitation of the developing seeds and aborted ovules indicated that GPAT9-2/gpat9-2 siliques contained approximately 28% to 43% aborted ovules, with an average of 35% (Table III). Siliques from heterozygous plants should produce wild-type and mutant ovules at a ratio of 50:50. If 35 of the 50 gpat9-2 ovules abort, the segregation of gpat9-2:wild-type ovules is 15:50, or approximately 23% gpat9-2. Considering that no pollen from a selfed GPAT9-2/gpat9-2 plant can transmit the gpat9-2 mutation (Table II), all viable ovules will be fertilized with wild-type pollen, producing approximately 23% heterozygotes and 77% wild-type seeds. Therefore, the proportion of aborted ovules in GPAT9-2/gpat9-2 siliques also supports the segregation results obtained from selfing GPAT9-2/gpat9-2 (Table I) and the reciprocal crosses (Table II).
Figure 2.
Aborted ovules in GPAT9-2/gpat9-2 siliques. The white arrows indicate aborted ovules in GPAT9-2/gpat9-2 siliques. The Col-0 silique did not contain aborted ovules.
Table III. Ovule viability within GPAT9-2/gpat9-2 half siliques.
| Plant-Silique | Developing Seeds | Aborted Ovules | Percentage Aborted |
|---|---|---|---|
| 1-1 | 21 | 9 | 30.0 |
| 1-2 | 21 | 8 | 27.6 |
| 1-3 | 18 | 11 | 37.9 |
| 1-4 | 20 | 11 | 35.5 |
| 1-5 | 20 | 14 | 41.2 |
| 2-1 | 22 | 14 | 38.9 |
| 2-2 | 20 | 15 | 42.9 |
| 2-3 | 23 | 10 | 30.3 |
| 2-4 | 20 | 9 | 31.0 |
| 2-5 | 21 | 13 | 38.2 |
| Average ± sd | 20.6 ± 1.3 | 11.4 ± 2.5 | 35.4 ± 5.3 |
The gpat9-2 Pollen Is Smaller and Does Not Produce Pollen Tubes
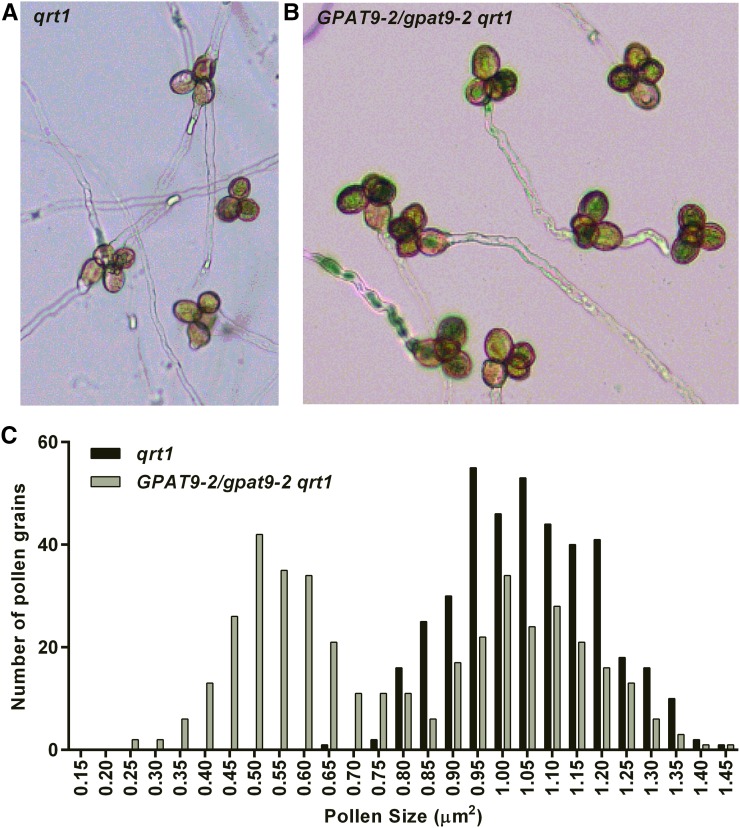
The reciprocal crosses (Table II) suggested a male gametophyte defect. To investigate pollen lethality, we utilized histochemical staining for pollen abortion and pollen viability with Alexander (Peterson et al., 2010) and nitroblue tetrazolium (Regan and Moffatt, 1990) staining, respectively (Supplemental Fig. S6). To our surprise, we did not detect any quantitative differences in histochemical staining from pollen of GPAT9-2/gpat9-2 plants compared with that of wild-type plants. TAG production in developing pollen is required for fertilization (Zhang et al., 2009). Therefore, we hypothesized that the gpat9-2 mutation may produce pollen with viable cells; however, if TAG production is reduced due to the gpat9-2 mutation, it may limit pollen tube growth and the transmittance of the gpat9-2 genotype. To investigate this hypothesis, we crossed GPAT9-2/gpat9-2 into the qrt1-4 background, where microspores from a single meiosis event remain attached throughout development (Preuss et al., 1994; Rhee and Somerville, 1998). The qrt1 mutant background facilitates the analysis of pollen defects because a heterozygous parent will produce tetrad pollen containing two affected and two unaffected pollen grains. Tetrads from qrt1 GPAT9/gpat9-2 displayed two larger and two smaller pollen grains (Fig. 3, A and B), consistent with the segregation of the mutant gpat9-2 allele producing aberrant pollen. While all the pollen from the qrt1 parent was close to the same size and followed a Gaussian distribution, the pollen from qrt1 GPAT9/gpat9-2 plants showed a bimodal distribution, with the small pollen roughly half the size of the larger pollen (Fig. 3C).
Figure 3.
Reduced pollen size and pollen tube growth of GPAT9-2/gpat9-2 pollen in the qrt1 background. A, Tetrad pollen in the qrt1 mutant can germinate to produce up to four pollen tubes, one from each attached pollen grain. B, Tetrad pollen in the qrt1 GPAT9-2/gpat9-2 line does not produce more than two pollen tubes per tetrad. C, Size of individual tetrad pollen grains from qrt1 and qrt1 GPAT9-2/gpat9-2 plants. Pollen grain length and width were measured to calculate the apparent surface area of each pollen grain.
To determine if gpat9-2 pollen can produce pollen tubes, we analyzed tetrad pollen germination (Table IV). In the qrt1 background, 5.5% of approximately 4,800 tetrads produced three or four pollen tubes (Fig. 3A; Table IV). However, when over 6,800 tetrads from the qrt1 GPAT9/gpat9-2 background were analyzed, no tetrads with three or four pollen tubes were observed (Table IV; Fig. 3B). These results indicate that the gpat9-2 pollen grains do not produce pollen tubes and support the lack of transmittance of gpat9-2 through pollen in the reciprocal crosses (Table II).
Table IV. Pollen germination and tube growth of GPAT9-2/GPAT9-2-2 within the qrt1 background.
| Experiment |
qrt1 |
qrt1 GPAT9-2/gpat9-2 |
|||||
|---|---|---|---|---|---|---|---|
| Pollen | Three to Four Tubes per Tetrad | Pollen | Three to Four Tubes per Tetrad | ||||
| tetrads | n | % | tetrads | predicteda | n | % | |
| 1b | 1,285 | 52 | 4 | 1,256 | 50 | 0 | 0 |
| 2b | 694 | 15 | 2.2 | 1,380 | 30 | 0 | 0 |
| 3c | 991 | 52 | 5.2 | 1,830 | 95 | 0 | 0 |
| 4c | 608 | 108 | 17.8 | 836 | 149 | 0 | 0 |
| 5c | 1,282 | 41 | 3.2 | 1,543 | 49 | 0 | 0 |
Predicted tube growth based on percentage growth in qrt1 during that experiment. bGermination medium from Zhu et al. (2013). cGermination medium from Boavida and McCormick (2007).
Functional Plant GPAT9 Genes Complement the Gametophytic Lethality Phenotypes of the Heterozygous GPAT9/gpat9-2 Mutant
Wild-type copies of GPAT9 open reading frames (ORF) from Arabidopsis and tung tree (Vernicia fordii) driven by the AtGPAT9 promoter were used to test for complementation of the GPAT9/gpat9-2 phenotypes. We used the AtGPAT9 promoter in an attempt to match the tissue- and cell type-specific transgenic expression of GPAT9s as closely as possible to that of the native gene, instead of using other, better known constitutive promoters (e.g. cauliflower mosaic virus 35S) that do not express well in pollen (Kay et al., 1987; Wilkinson et al., 1997) and, therefore, would be unlikely to complement the gpat9-2 phenotypes. A population of sulfadiazine-resistant GPAT9/gpat9-2 seedlings were transformed with Agrobacterium tumefaciens bearing either empty binary plasmid or the complementation plasmid for AtGPAT9 or VfGPAT9. Transgenic plants containing the complementation constructs were selected using cassava vein mosaic virus promoter-driven expression of the DsRed fluorescent protein as a selectable marker (Verdaguer et al., 1996; Stuitje et al., 2003).Very few red seeds from the empty vector line were sulfadiazine resistant (approximately 4%; Supplemental Fig. S7, A and D), similar to the segregation analysis of GPAT9/gpat9-2 in Table I. However, the homozygous red fluorescent seeds from the AtGPAT9 and VfGPAT9 complementation lines effectively germinated and established photosynthetic competency on sulfadiazine agar medium (Supplemental Fig. S7, B/E and C/F, respectively). This result confirms that the AtGPAT9 gene is essential and responsible for the observed segregation and gametophytic lethality phenotypes in mutant plants. Additionally, GPAT9 genes from distantly related plant species can complement the Arabidopsis mutant lethality phenotype, supporting the conserved nature of GPAT9 enzymatic activity within plant metabolism.
Seed-Specific Knockdown of GPAT9 Reduces Oil Content and Alters FA Composition
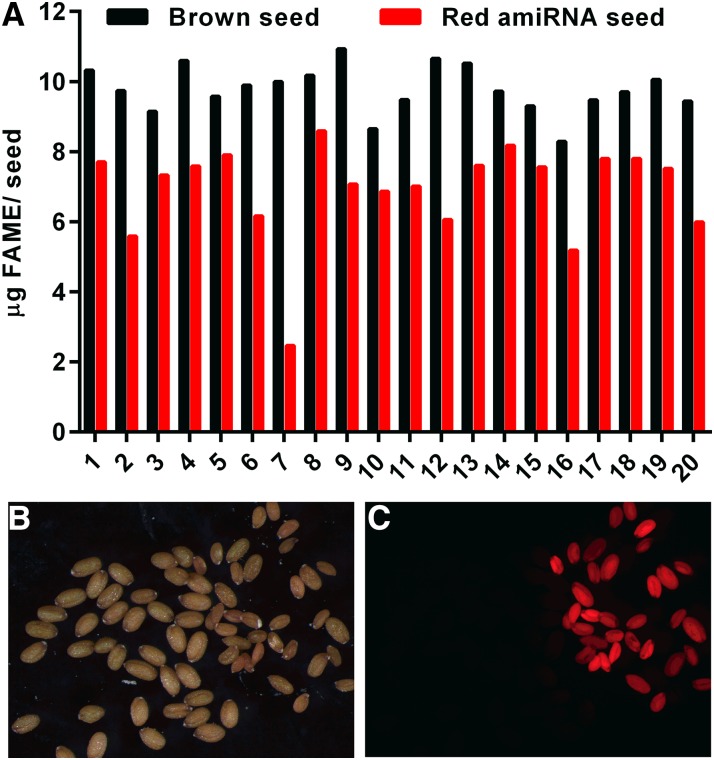
Our analysis of GPAT9/gpat9-2 suggests that AtGPAT9 is an essential gene and that a homozygous gpat9 knockout cannot be obtained. Analysis of lipid content in heterozygous GPAT9/gpat9-2 plants indicated no change from the wild type in leaves (Supplemental Fig. S8A) or seeds (Supplemental Fig. S8, B and C). The latter was as expected, since most seeds produced from GPAT9/gpat9-2 are wild type (Table I). Therefore, a single copy of GPAT9 is sufficient for vegetative growth, and a different approach must be taken to investigate the role of GPAT9 in lipid metabolism. A strong, constitutive knockdown of GPAT9 expression would be expected to severely reduce plant growth, complicating the characterization of a lipid phenotype associated with aberrant GPAT9 expression. Therefore, we chose to investigate the role of GPAT9 in TAG biosynthesis in seeds (which has high flux through ER GPAT activity compared with other tissues) by creating seed-specific GPAT9 knockdown Arabidopsis lines. An artificial microRNA (amiRNA) construct for GPAT9 was expressed under the control of the seed-specific Phaseolus vulgaris phaseolin promoter (Sengupta-Gopalan et al., 1985), with the fluorescent protein DsRed as a selectable marker (Stuitje et al., 2003). Heterozygous transformed (red) T1 seeds were planted, and the corresponding segregating T2 seeds were harvested from 20 individual T1 transformants. Red T2 seeds were separated from the segregating untransformed brown seeds, and each set was analyzed for oil content (Fig. 4). Oil content varies considerably between individual Arabidopsis plants (Li et al., 2006), and the cosegregating nontransformed seed acts as a plant-specific wild-type control for seed oil levels. Red seeds from each individual transformed line had a 15% to 75% reduction in total lipid relative to the plant-specific brown seeds (Fig. 4A). In the lines with the largest reduction in oil content, the red seeds were smaller and displayed a wrinkled phenotype compared with the corresponding brown seeds (Fig. 4, B and C). A wrinkled seed phenotype is typical for Arabidopsis seeds with very low oil content (Focks and Benning, 1998).
Figure 4.
Segregating red and brown T2 GPAT9 amiRNA seed oil content and size. A, Oil content of segregating brown (not transformed; black bars) and red (transformed; red bars) T2 seeds from 20 individual T1 plant lines. Twenty red or brown seeds were used per analysis. FAME, FA methyl ester. B, Line 7 seeds under white light. C, The same seeds as in B under green light with a red filter. In B and C, the red GPAT9 amiRNA seeds are smaller than the brown untransformed seeds.
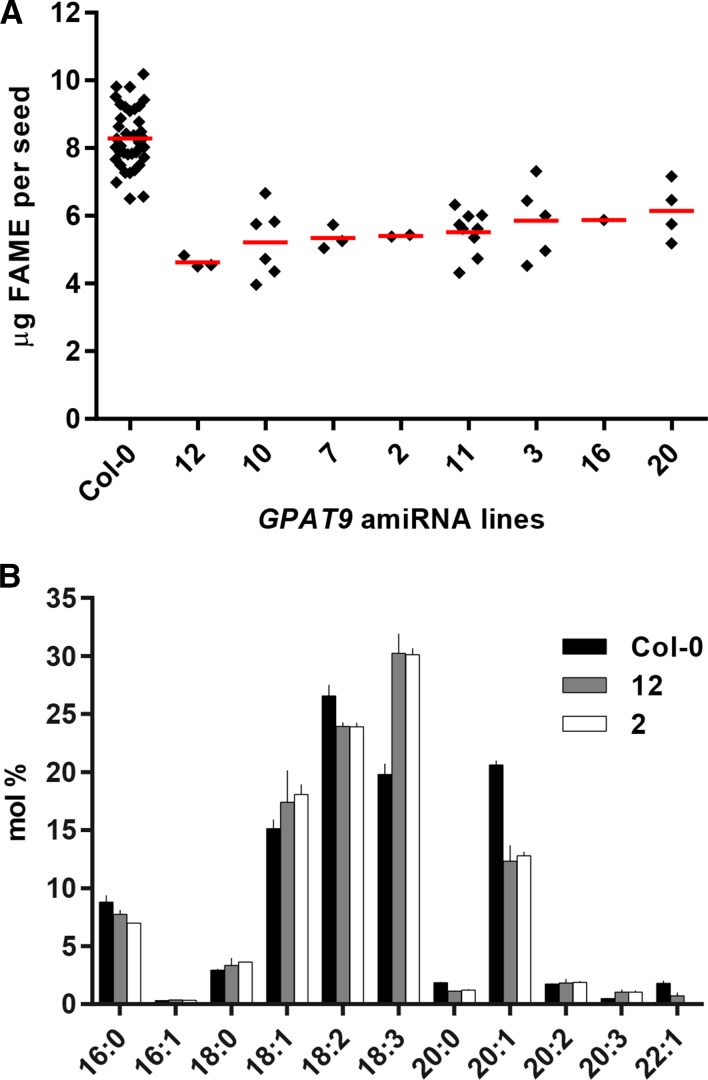
Eight GPAT9 amiRNA lines with low oil content were identified to have single T-DNA inserts based on 3:1 segregation ratios of red and brown seeds, and they were propagated further. Homozygous red T3 seeds were analyzed for seed oil content and composition (Fig. 5). The oil content was reduced by 26% to 44% (Fig. 5A), and seed lipids from all eight amiRNA lines also contained altered FA compositions (Supplemental Fig. S9). The FA compositions of two GPAT9 amiRNA knockdown lines chosen for further experiments (lines 2 and 12) are demonstrated in Figure 5B. Both GPAT9 knockdown lines have significant increases in total polyunsaturated FAs, from approximately 48.5% in the wild type to approximately 57% in the knockdown lines, including a dramatic shift in the 18:3/18:2 ratio, which increases from 0.75 to 1.26. Analysis of GPAT9 gene expression by quantitative PCR in developing T4 seeds from lines 2 and 12 indicated that GPAT9 transcript is reduced more than 88% from the wild type in each line (Supplemental Fig. S10). Together, these results indicate that reduced GPAT9 expression impacts the amount and composition of TAG in seeds.
Figure 5.
Oil quantity and composition of homozygous T3 GPAT9 amiRNA lines. A, Distribution of whole-seed FA methyl ester (FAME) content among the wild type (Col-0) and different knockdown lines. Each diamond represents a 50-seed sample from an individual plant. Red bars indicate averages. B, FA composition of seed lipid analysis from A. Wild-type Col-0 (n = 36) and amiRNA knockdown lines 12 (n = 3) and 2 (n = 2) were chosen for further analysis. Values are averages ± sd.
Seeds from GPAT9 Knockdown Lines Have Reduced GPAT Activity
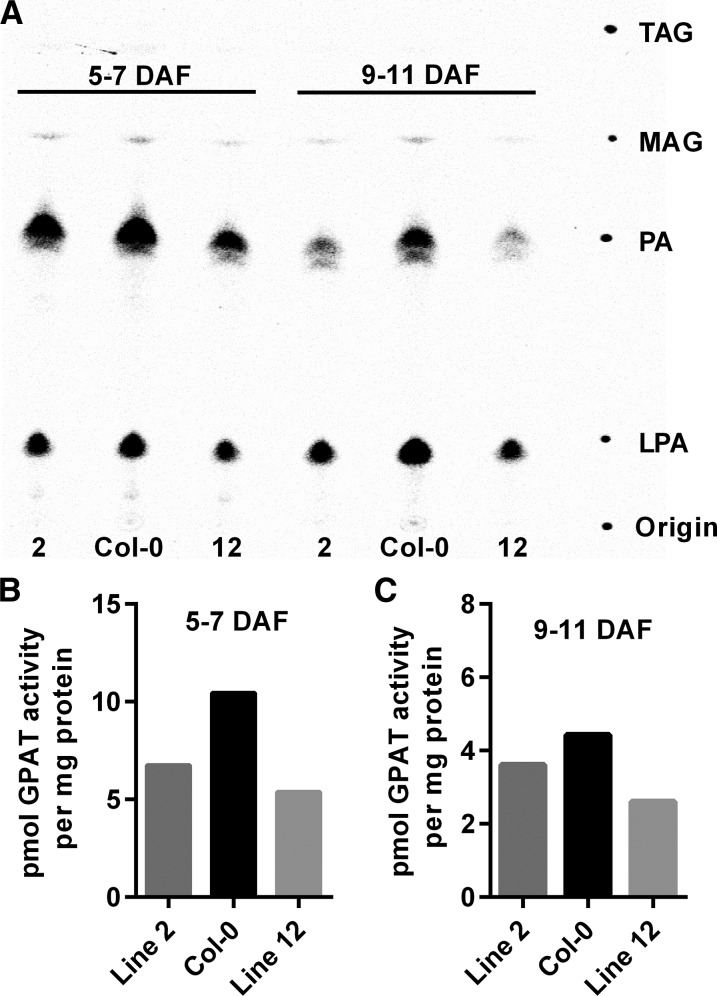
The sequence of AtGPAT9 closely resembles mammalian GPATs that contribute to TAG synthesis (Cao et al., 2006). However, AtGPAT9 displays high sequence homology to multiple classes of plant lysophospholipid acyltransferases (including LPEAT1 and LPEAT2, which also use LPA as a substrate; Fig. 1). Additionally, plant GPAT9s in general more closely match plant LPAATs in protein length (approximately 375–390 amino acid residues for both enzyme classes; Maisonneuve et al., 2010) than they do the land plant-specific sn-2 GPAT family (500–585 residues). Reduction of either GPAT or LPAAT activity in the amiRNA knockdown lines has the possibility to affect seed oil content. Therefore, to determine which enzymatic activity is affected in the knockdown lines, we performed in vitro GPAT and LPAAT assays from microsomes isolated from developing siliques of wild-type and homozygous T4 GPAT9 amiRNA lines (Figs. 6 and 7). Figure 6A demonstrates that silique microsome GPAT assays utilizing [14C]G3P and palmitoyl-CoA as substrates produce LPA and PA as the major products. For both wild-type and amiRNA lines, [14C]G3P distribution between LPA and PA was approximately 3:7 LPA:PA at 5 to 7 DAF and 7:3 LPA:PA at 9 to 11 DAF (Fig. 6A). However, the total amount of [14C]G3P incorporated into lipids differed between the wild-type and knockdown lines. [14C]G3P is incorporated into PA only after LPA has been formed; therefore, the quantification of both LPA and PA represents total GPAT activity. Total GPAT activity was reduced approximately 19% to 49% at the two different stages of silique development (Fig. 6, B and C). To confirm that the reduced GPAT activity measured from whole-silique microsomes is due to the seed-specific knockdown of ER GPAT activity in GPAT9 amiRNA lines, we also performed GPAT assays on microsomes isolated from developing seeds dissected out of 9- to 11-DAF siliques (Supplemental Fig. S11). Similar to the whole-silique results, the amiRNA lines had approximately 25% to 55% reduction in seed microsome GPAT activity from the wild type.
Figure 6.
Developing silique microsome GPAT activity. GPAT assays were done with microsomes isolated from whole developing siliques 5 to 7 and 9 to 11 d after flowering (DAF) from Col-0 and GPAT9 amiRNA knockdown lines 2 and 12. The assay utilized 3.55 nmol of [14C]G3P and 25 nmol of palmitoyl-CoA for 15 min at 24°C. A, Phosphor image of a thin-layer chromatography (TLC) plate indicating that [14C]LPA and [14C]PA are the major products of whole-silique microsome GPAT assays utilizing palmitoyl-CoA as an acyl donor. B and C, Quantification of total products from GPAT activity in whole-silique microsomes. Two representative GPAT assays from a total of six GPAT assays were performed with different batches of plants grown at different times. Total GPAT activity was dependent on growth conditions and stage of development. However, with each batch of plants or stage of development, the GPAT activity of amiRNA lines was always less than that of the wild-type control. An additional two representative GPAT assays are shown in Supplemental Figure S11. MAG, Monoacylglycerol.
Figure 7.
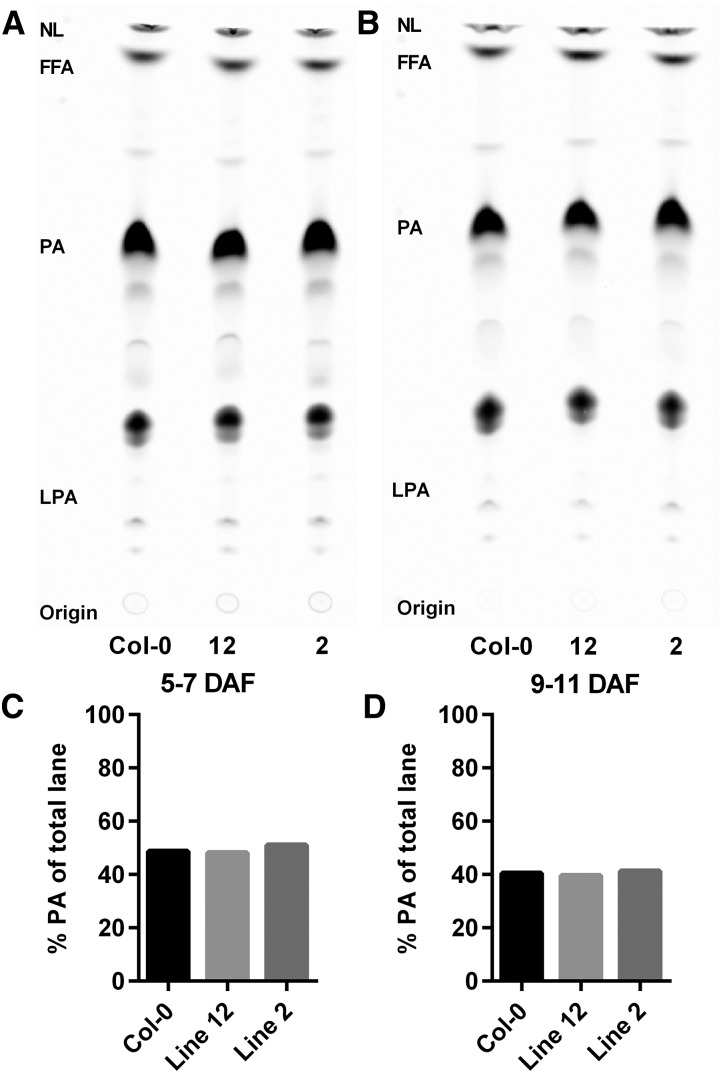
Silique microsome LPAAT assays. LPAAT assays were done with microsomes isolated from whole developing siliques of Col-0 and GPAT9 amiRNA knockdown lines 2 and 12. Two assays were done with siliques aged 5 to 7 (left) and 9 to 11 (right) DAF. The assays utilized the same conditions as the GPAT assays in the text except that 3.33 nmol of [14C]oleoyl-CoA was the acyl donor and 25 nmol of 18:1-LPA was used as the acyl acceptor for 15 min at 24°C. The [14C]oleoyl-CoA can also be used by other acyltransferases and endogenous lipid acceptors found within the microsomes. However, PA was the major product of the assay in all samples. A and B, Phosphor images of TLC separation of products. C and D, Quantitation of PA within each lane of the TLC image. NL, Neutral lipids; FFA, free fatty acids.
LPAAT assays were performed with the same set of whole-silique microsomes as for GPAT assays (Fig. 6) utilizing LPA and [14C]oleoyl-CoA as substrates. The [14C]oleoyl-CoA can also be used by other acyltransferases and endogenous lipid acceptors found within the microsomes. However, since only LPA was added as an exogenous acyl acceptor, PA was the major product of the assay in all samples (Fig. 7, A and B). No differences in LPAAT activity between the wild type and the GPAT9 amiRNA lines were detected (Fig. 7, C and D). The reduction of GPAT enzymatic activity in seeds of GPAT9 amiRNA knockdown lines (Fig. 6; Supplemental Fig. S11), and the corresponding lack of a change in LPAAT activity (Fig. 7), together support the conclusion that the reduced oil accumulation in these transgenic lines (Figs. 4 and 5) is due to a reduction in seed GPAT activity, specifically caused by the GPAT9 mRNA knockdown.
AtGPAT9 Interacts Physically with Other Enzymes in the Kennedy Pathway and the Acyl-Editing Cycle
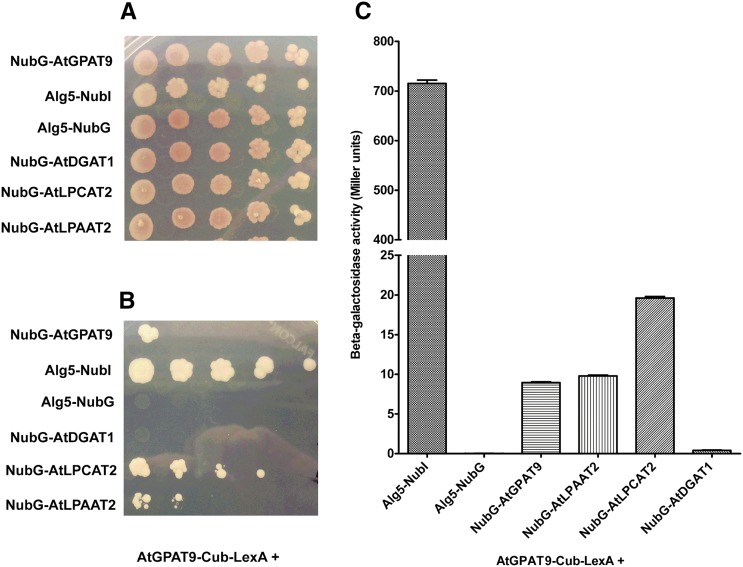
Enzymes that make up various biochemical pathways, including some lipid biosynthetic pathways, often form multicomponent complexes (Roughan and Ohlrogge, 1996; Mo and Bard, 2005). Such complexes allow for rapid and efficient transfer of metabolites to downstream enzymes in the pathway. Given the possibility of metabolic networking between the enzymes of the Kennedy pathway to assemble glycerolipids and the enzymes of acyl editing, which provides polyunsaturated FAs for lipid assembly (Bates et al., 2013b; Li-Beisson et al., 2013; Allen et al., 2015), we sought to explore potential protein-protein interactions between AtGPAT9 and other potential partners using the split-ubiquitin yeast two-hybrid system (Johnsson and Varshavsky, 1994). The split-ubiquitin system eliminates the need for the importation of target enzymes and proteins into the nucleus of the yeast cell, as in traditional yeast two-hybrid systems (Chien et al., 1991), which is especially problematic for integral membrane proteins such as GPAT9 and other ER-localized acyltransferases. This system has been used previously to search for interacting partners of glycerolipid synthesis enzymes from tung tree (Gidda et al., 2011). As shown in Figure 8, AtGPAT9 interacted with itself, AtLPAAT2, and AtLPCAT2 (an acyl-CoA:lysophosphatidylcholine acyltransferase, LPCAT), but not with AtDGAT1 (an acyl-CoA:diacylglycerol acyltransferase, DGAT), the dominant TAG biosynthetic DGAT isozyme in Arabidopsis (Zou et al., 1999). AtDGAT1 did weakly interact with both AtLPAAT2 and AtLPCAT2, however, as shown in Supplemental Figure S12. The homomeric and heteromeric interactions between AtGPAT9 and AtLPAAT2 or AtLPCAT2 were approximately 8- to 20-fold stronger than the corresponding interactions with AtDGAT1. Together, these results suggest that AtGPAT9 likely interacts in vivo with AtLPAAT2 (the next step in glycerolipid assembly after GPAT) and with AtLPCAT2, the main enzyme involved in acyl editing, which provides polyunsaturated FAs to the acyl-CoA pool for incorporation into de novo glycerolipid synthesis (Stymne and Stobart, 1984; Bates et al., 2012, 2013b; Wang et al., 2012b; Lager et al., 2013; Pan et al., 2015).
Figure 8.
Testing of protein-protein interactions between AtGPAT9 and other lipid acyltransferases. Coding sequences for each of two different membrane proteins of interest were ligated in frame to either the C-terminal half of ubiquitin (Cub)-LexA transcription factor protein fusion or the N-terminal half of ubiquitin (Nub). Nub may be represented as either native polypeptide sequence (NubI) or a mutant form of Nub containing an Ile/Gly conversion point mutation (NubG). NubI strongly interacts with Cub. NubG has very weak affinity for Cub and must be brought into close proximity to Cub to allow for interaction of the two halves of ubiquitin, release of the LexA transcription factor, and, finally, activation of the reporter genes (His and adenine prototrophic markers and β-galactosidase, for quantitative analysis). A and B, Prototrophic growth assay of yeast strains containing various combinations of AtGPAT9 bait plasmid coexpressed with potential prey NubG-acyltransferase plasmids. Serial dilutions of cells expressing different bait-prey combinations were plated on nonselective (A) or selective (B) medium conditions. C, Quantitative measurement of β-galactosidase activity from cell lysates of the strains used in the serial dilution assays. LPCAT, Acyl-CoA:Lysophosphatidylcholine acyltransferase; DGAT, acyl-CoA:Diacylglycerol acyltransferase.
DISCUSSION
The genetic dissection of the Kennedy pathway with the ultimate goal of complete biotechnological control of plant oil synthesis began in earnest more than 15 years ago, with the identification of the genes that encode what are now known as AtDGAT1 (Routaboul et al., 1999; Zou et al., 1999) and AtDGAT2 (Lardizabal et al., 2001). Meaningful progress has been made in the years since toward the identification and functional characterization of these and many other enzymes that contribute to plant membrane phospholipid and storage TAG biosynthesis. However, despite such progress on many fronts, definitive isolation of the gene or genes that encode for the initial G3P acylation reaction that feeds both the ER-localized membrane lipid and TAG biosynthesis pathways has eluded the plant lipid biotechnology community. Evidence is presented here that firmly supports the assignment of AtGPAT9 to that role.
AtGPAT9 Is the GPAT Involved in TAG Biosynthesis
The results presented here establish that AtGPAT9 is a single-copy and essential gene, demonstrated by both male and female gametophyte lethality phenotypes of the gpat9-2 mutant and the inability to obtain a homozygous mutant (Figs. 2 and 3; Tables I–IV). The reduction in oil content of seed-specific GPAT9 knockdowns (Figs. 4 and 5), and the corresponding reduction in GPAT activity (Fig. 6) but not LPAAT activity (Fig. 7), demonstrate that AtGPAT9 is a GPAT involved in seed oil accumulation. This result could not be demonstrated with any of the genes in the AtGPAT1 to AtGPAT8 family, which further supports their vastly different roles within plant metabolism. Since GPAT9 is essential for gametophyte function, the GPAT activity involved in seed TAG biosynthesis is likely also required for ER membrane lipid synthesis in other tissues.
AtGPAT9 Knockdown Seed Oil Phenotype, and Protein Interactors Help to Further Define the Plant Oil Synthesis Metabolic Network
Protein-protein interaction analysis (Fig. 8) provided insights that strongly suggest the proper placement of AtGPAT9 at an intersection between the Kennedy de novo glycerolipid biosynthetic pathway and the acyl-editing cycle, supporting current models of the Arabidopsis lipid biosynthetic network (Fig. 9) based on both molecular genetic and in vivo metabolic labeling approaches (Bates et al., 2013b; Li-Beisson et al., 2013; Allen et al., 2015). In this study, AtGPAT9 was found to interact with itself, AtLPAAT2, and AtLPCAT2 but not with AtDGAT1. How each interaction fits into current models of TAG biosynthesis is discussed below.
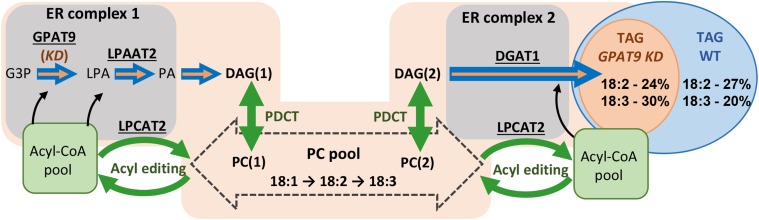
Figure 9.
Models of GPAT9-dependent glycerol flux and the spatial organization of TAG biosynthesis. The model is based on our results and previous in vivo labeling experiments (Allen et al., 2015). Wild-type (WT) TAG biosynthesis is dependent on the flux of G3P through GPAT9 for the initial incorporation of the glycerol backbone into glycerolipids (wide blue arrows). GPAT9, LPAAT2, and LPCAT2 are localized together for the efficient flux of acyl groups out of PC into de novo glycerolipid synthesis (ER complex 1). De novo DAG [DAG(1)] is utilized to produce PC. After traversing through the ER membrane, where desaturation can take place (large dashed arrow), PC is converted to PC-derived DAG [DAG(2)] and is incorporated into TAG through DGAT1. DGAT1 is spatially separated from GPAT9 but also associates with LPCAT2 in ER complex 2. Here, de novo DAG and PC-derived DAG are spatially separated and localized within ER complexes 1 and 2, respectively. The amount of 18:2/18:3 in TAG is dependent on the residence time of acyl groups in PC for desaturation versus the flux out of PC for incorporation into TAG. In GPAT9 amiRNA knockdown (KD) lines, the flux of G3P into glycerolipid synthesis is reduced (thin orange arrows embedded in large blue arrows), lowering total TAG accumulation. However, the rate of acyl exchange through acyl editing, and DAG exchange into/out of PC by phosphatidylcholine:diacyglycerol cholinephosphotransferase (PDCT), is not changed (green arrows). Therefore, the residence time of acyl groups in PC for desaturation increases as the overall flux of DAG to TAG slows down, leading to both higher overall levels of polyunsaturated FAs and a significantly higher ratio of 18:3 to 18:2 in the knockdown lines.
The self-interaction of AtGPAT9 is consistent with the discovery of the oligomerization of yeast-expressed Erysimum asperum plastidial GPAT (Chen et al., 2014). The self-associating properties of EaGPAT may suggest that it undergoes self-allosteric regulation, since many allosteric enzymes exhibit quaternary structure. Future studies will address possible correlations between the homomeric assembly of AtGPAT9 subunits and the allosteric regulation of GPAT activity. The interaction between AtGPAT9 and AtLPAAT2 (the activity expected to occur immediately downstream of GPAT in the lipid biosynthetic pathway) suggests metabolic channeling between Kennedy pathway enzymes within lipid biosynthesis (Fig. 9) and supports previous studies that found lipid biosynthetic enzymes colocalized to specific microdomains of the ER (Shockey et al., 2006; Gidda et al., 2011). Together, these results suggest the possibility of lipid metabolic complexes that localize to specific regions within the ER membrane that may act to control lipid biosynthesis from a select pool of metabolites. Going forward, additional in planta protein-protein characterization, such as using coimmunoprecipitation analysis of extracts from tobacco (Nicotiana tabacum) leaves that transiently coexpress relevant pairs of lipid metabolic genes, as described by Fan et al. (2015), will further enhance our understanding of the spatial organization of lipid metabolism within plant tissues.
LPCAT works in both the forward and reverse directions (Stymne and Stobart, 1984; Lager et al., 2013). The interaction of AtGPAT9 with AtLPCAT2 fits with current metabolic models from in vivo lipid flux analysis, which indicate that the acyl groups removed from PC by acyl editing are the major source of acyl-CoA for de novo glycerolipid synthesis by Kennedy pathway enzymes (Bates et al., 2007, 2009; Bates and Browse, 2012; Allen et al., 2015). The combined loss of AtLPCAT1 and AtLPCAT2 stops acyl editing in Arabidopsis seeds and causes a dramatic shift in acyl flux such that newly synthesized FAs are directly incorporated into de novo glycerolipid synthesis through GPAT and LPAAT rather than the direct incorporation into PC through acyl editing as in the wild type (Bates et al., 2012; Wang et al., 2012b). The interaction of AtGPAT9 and AtLPCAT2 demonstrated here supports that nascent acyl groups destined for AtLPCAT2-mediated acyl editing in the wild type can be readily shifted to AtGPAT9 within the lpcat1/2 double mutant for enhanced de novo glycerolipid synthesis.
Importantly, the lack of interaction between AtGPAT9 and AtDGAT1 fits with current models of TAG biosynthesis from in vivo labeling studies that indicate a kinetic separation of de novo glycerolipid synthesis and TAG synthesis. [14C]Glycerol kinetic labeling in Arabidopsis and soybean (Glycine max) indicates that DAG synthesized de novo from the GPAT, LPAAT, and PA phosphatase activities of the Kennedy pathway is not directly utilized for TAG synthesis. Instead, de novo DAG [Fig. 9, DAG(1)] is utilized to synthesize PC, and DAG for TAG synthesis is later derived from PC [Fig. 9, DAG(2); Bates et al., 2009; Bates and Browse, 2011]. This flux of DAG through PC, combined with the acyl-editing cycle, enhances the residence time of acyl groups in PC for the production of polyunsaturated FAs (Bates and Browse, 2012; Allen et al., 2015). In Arabidopsis, the fluxes of de novo DAG into PC and PC-derived DAG out of PC are predominantly controlled by PDCT (Lu et al., 2009). Since PDCT activity does not involve the net synthesis or turnover of PC, only a head-group exchange reaction, a reduction in glycerol flux into the TAG biosynthetic pathway by reduced GPAT activity should not affect the rate of PDCT action (Fig. 9).
AtLPCAT2 interacted with both AtGPAT9 and AtDGAT1 (Fig. 8; Supplemental Fig. S12). LPCAT activity has been demonstrated within multiple cellular localizations (Larsson et al., 2007; Tjellström et al., 2012). Together, these results suggest that there may be multiple sites for acyl editing within the cell (Fig. 9) and strongly suggest that both the first and last glycerol acylation reactions may be fed at least in part with acyl-CoA derived directly from PC by AtLPCAT2. These results are also consistent with recent coexpression and biochemical analyses using flax (Linum usitatissimum) DGAT1 and LPCAT (Pan et al., 2015). Those authors showed that LPCAT likely mediates the direct channeling of PC-derived acyl-CoA to DGAT1, thus accounting for the elevated polyunsaturated FA content in flax oil, while also helping to overcome the thermodynamically unfavorable reverse reaction of LPCAT (Pan et al., 2015). The results demonstrated here (Fig. 8; Supplemental Fig. S12) strongly suggest that a high degree of channeling also occurs in Arabidopsis (and likely many other plants as well), with at least two entry points for PC-derived acyl-CoA into glycerolipid assembly (Fig. 9).
Finally, a consideration of TAG biosynthesis as a metabolic network involving multiple highly active exchange reactions in/out of PC that are independent of total acyl flux through the network (Fig. 9, green arrows) can be used to explain how reduced GPAT activity leads to increased levels of polyunsaturated FAs in the seed oil of GPAT9 amiRNA knockdown lines (Fig. 5B; Supplemental Fig. S9). Acyl editing involves a cycle of PC deacylation and LPC reacylation by LPCAT enzymes in which the rate of acyl exchange on/off PC can be up to 15 times the rate of de novo glycerolipid synthesis (Bates et al., 2007, 2009, 2013b). Likewise, PDCT activity can be independent of the total rate of DAG and PC synthesis and turnover, leading to a rapid exchange of DAG in/out of PC. The reduction of GPAT activity in GPAT9 amiRNA lines slows the total rate of glycerol flux (and thus DAG flux) through the lipid metabolic network into oil (Fig. 9, thin orange arrows [GPAT9 knockdown flux] embedded within the wide blue arrows [wild-type flux]). However, the flux through the acyl and head-group exchange reactions with PC is independent of the total rate of glycerolipid biosynthesis, allowing FAs and DAGs to continually cycle in/out of PC, enhancing the total residence time in PC prior to incorporation into TAG. The longer time an acyl group spends within the PC pool allows for greater access to the desaturases and a higher probability that oleate will be fully desaturated to linolenate prior to incorporation into TAG (Bates and Browse, 2012; Allen et al., 2015). Therefore, a slower rate of TAG assembly, combined with no change in the exchange reactions, explains the significant increase in total polyunsaturates and the 18:3-to-18:2 ratio of TAG in the GPAT9 amiRNA knockdown lines.
Strict Maintenance of Single-Copy GPAT9 Genes in Plants Reinforces Their Essentiality
Plant genomes have been subjected to enormous evolutionary pressures over time, including whole- or partial-genome duplications (Simillion et al., 2002) and small-scale duplication events (Shockey and Browse, 2011), which often result in sets of duplicated genes that help an organism to meet new metabolic requirements. Some duplicated genes are retained and ultimately achieve specialized functions via changes in temporal or tissue/organ-specific gene expression profiles, subcellular targeting, enzyme substrate specificity, etc. (De Smet et al., 2013). However, duplicated genes from genome duplication events can revert back to singleton status due to pressures from factors such as functional redundancy, epigenetic silencing, and chromosomal instability (Paterson et al., 2006). Many such singleton genes have been found to encode for essential housekeeping functions (Paterson et al., 2006; De Smet et al., 2013), and the same appears to be true for AtGPAT9.
The various functional analyses shown here reveal that AtGPAT9 (and likely, by extension, GPAT9 genes from other plants in general) is not genetically redundant (Figs. 1–3; Tables I–IV; Supplemental Fig. S7), unlike DGATs, LPAATs, and several other classes of plant lipid metabolic enzymes (Li-Beisson et al., 2013). Most plants contain a single copy of GPAT9, or at least a single copy per diploid genome, and appear to have been subjected to selection pressures that maintain the respective GPAT9 genes as singletons (Paterson et al., 2006; De Smet et al., 2013). Searches across a wide array of sequenced plant genomes, covering many of the major branch points in plant evolution, found a very high number of single-copy GPAT9 genes (Supplemental Fig. S2). G. raimondii (Pima cotton) is the only one of nine diverse species shown in Supplemental Figure S2 that contained two copies of GPAT9, as per the data available in Phytozome 10.3. The ancestor to G. raimondii and other Gossypium spp. underwent a cotton-specific whole-genome duplication event approximately 16 to 17 million years ago (Wang et al., 2012a); the existing tetraploid status of cotton, and the relatively recent occurrence of this duplication event, may not yet have allowed for enough selection pressure or provided enough evolutionary time for resolution of the ultimate fate of the second copy of GraGPAT9.
GPAT9s are also highly conserved across broader sections of the tree of life, suggesting a role in essential cellular metabolism. The homology between AtGPAT9 and mouse GPAT3 (which contains minimal sequence identity to any of the other eight known Arabidopsis extraplastidial GPATs) was one of the initial indicators that AtGPAT9 might be an actual sn-1 GPAT (Cao et al., 2006; Gidda et al., 2009). Also unlike the larger family of sn-2 GPATs, AtGPAT9 is highly conserved across Animalia in general. AtGPAT9 is closely related to representative proteins from hundreds of animal species, the closest of which is shown in Supplemental Figure S13. A GPAT from killer whale (Orcinus orca) shares 39% identity and 58% similarity with the amino acid sequence of AtGPAT9 over a 360-amino acid region of the protein (nearly the full length of AtGPAT9). In contrast, the top Animalia BLASTP hit for AtGPAT1 is an uncharacterized protein from moonfish (Xiphophorus maculates), which is only 27% identical and 42% similar, over a 256-residue portion of the protein (less than half of the AtGPAT1 protein). Conversely, O. orca does not possess a GPAT1, while X. maculates GPAT9 still retains 36% identity and 54% amino acid similarity to AtGPAT9 (Supplemental Fig. S13).
Many of the genes in the duplication-resistant and mostly single-copy categories described by De Smet et al. (2013) encode essential housekeeping functions. That AtGPAT9 is essential was proven by our inability to recover a homozygous gpat9-2 mutant plant, even after germination of heterozygous mutant seeds on solid medium containing vitamins and sugars. The abnormally low recovery of sulfadiazine-resistant progeny from heterozygous gpat9-2 parents also indicates that AtGPAT9 mutants are gametophytic lethal. The incomplete gametophytic lethality of the gpat9-2 female gamete may arise from minimally sufficient transfer of residual GPAT9 protein and GPAT9 mRNA between cells during cell division to maintain the viability of a few eggs until fertilization with wild-type pollen, as has been demonstrated for other genes (Muralla et al., 2011). The complementation of the low penetrance of the GPAT9/gpat9-2 heterozygous phenotype with both transgenic Arabidopsis and tung tree GPAT9 constructs (Supplemental Fig. S7) proved that the gpat9-2 mutation was indeed responsible for the observed mutant phenotypes and that somewhat distantly related plant GPAT9s can functionally complement one another. This latter result may be key to finally controlling the FA composition at each spot within the TAG backbone by isolating acyl-selective GPAT enzymes from different species.
CONCLUSION
Taken together, the results presented here confirm that AtGPAT9 is an ancient gene that is essential in Arabidopsis and likely other plants as well. Experimental evidence suggests that GPAT9 fulfills an indispensable role in the catalysis of the first step in the synthesis of storage and membrane lipids required for life. This role may help to explain why GPAT9 genes have been maintained throughout most of the course of evolution of life on Earth and why the sequences of GPAT9s have evolved so conservatively. Finally, the characterization of GPAT9 at the intersection of de novo glycerolipid synthesis and acyl editing, and our demonstration that a distantly related plant GPAT9 can replace the essential AtGPAT9, suggest that bioengineering strategies around GPAT9 may be valuable for the production of the designer vegetable oils of the future.
MATERIALS AND METHODS
Phylogenetic Analyses
Arabidopsis (Arabidopsis thaliana) GPAT9 and LPEAT protein sequences were identified from species-specific searches of the Phytozome version 10.3 server (http://phytozome.jgi.doe.gov/pz/portal.html#). Phylogenetic comparisons were carried out using alignment (.aln) files generated using the default settings provided in ClustalX version 1.8 (Thompson et al., 1997). Neighbor-joining dendrograms were created in the TreeView program (Saitou and Nei, 1987; Page, 1996) using the Boxshade tool at the ExPASy Bioinformatics Resource Portal (http://www.ch.embnet.org/software/BOX_form.html).
Genotyping of T-DNA Mutants
Mutant genotyping utilized gene- and T-DNA-specific primers (Supplemental Table S1) and GoTaq Green (Promega; www.promega.com) with the manufacturer’s recommended protocol. For insertion site sequencing, the left border-gene junction was amplified by PCR with GoTaq Flexi (Promega), gel purified with the QIAquick Gel Extraction Kit (Qiagen; www.qiagen.com), and sequenced from both the left border primer and a gene-specific primer in the flanking sequence by Eurofins (http://www.eurofinsgenomics.com/).
Gene Expression Analysis
Leaf RNA was extracted with the RNeasy Plant Mini Kit (Qiagen) and treated with RNase-free DNase (Qiagen) using on-column DNase digestion. RNA was quantified using a Nanophotometer (Implen). Complementary DNA was synthesized using the SuperScript III First-Strand Synthesis System for RT-PCR (Life Technologies). RT-PCR utilized standard conditions as per the GoTaq Flexi (Promega) protocol with 30 cycles of amplification. Transcript levels were analyzed by quantitative PCR using Platinum SYBR Green qPCR SuperMix-UDG (Life Technologies) and the Stratagene Mx3005P Quantitative PCR system (Agilent Technologies). For developing seed transcript analysis, frozen developing seeds (9–11 DAF) were collected from liquid N2 frozen siliques (Bates et al., 2013a). Approximately 50 to 100 mg of frozen seeds was mechanically pulverized to a fine powder with steel beads (TissueLyser LT; Qiagen), RNA was extracted (Suzuki et al., 2004), and DNA was removed (DNA-Free RNA Kit; Zymo Research; www.zymoresearch.com/). Complementary DNA synthesis and quantification were performed as described above. Normalized relative quantity was calculated (Rieu and Powers, 2009) against TIP41-like (Czechowski et al., 2005).
Materials, Plants, and Growth Conditions
Unless specified otherwise, all chemicals were from Fisher Scientific (https://www.fishersci.com), and solvents were HPLC grade or higher. Arabidopsis lines gpat9-1 (Salk_052947C; Alonso et al., 2003) and qrt1-4 (CS25041; Francis et al., 2006) were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/), and gpat9-2 (GABI_867A06; Kleinboelting et al., 2012) was obtained from Gabi-kat (https://www.gabi-kat.de/). Control plants were in the Arabidopsis Col-0 ecotype. Seeds were surface sterilized (30% [v/v] ethanol, 10% [v/v] bleach, and 0.1% [w/v] SDS) for 5 min, rinsed five times with sterile water, and plated onto germination medium (2.5 mm MES, pH 5.7, 1% [w/v] Suc, 1× Murashige and Skoog Plant Salt Mix [MP Biomedicals; http://www.mpbio.com/], and 0.8% [w/v] agar). Seeds from GPAT9-2/gpat9-2 plants were always germinated on the plate medium containing an additional 5.25 mg L−1 sulfadiazine (Sigma; www.sigmaaldrich.com/) to select for the heterozygotes. Plated seeds were stratified for 4 d at 4°C, prior to moving to a growth chamber. After 10 d on the plate, seedlings were transferred to soil. Plants were grown in growth chambers under continuous white light at approximately 130 to 170 µmol photons m−2 s−1 and at 22°C to 24°C. Silique ages were determined by trimming each plant to one main shoot and counting the new open flowers/siliques produced each day.
DNA Manipulation and Plasmid Construction
Binary complementation plasmids were generated as follows. The phaseolin promoter from cloning vector pK8 (Shockey et al., 2015) was removed by KpnI and NotI digestion and replaced with a 1,594-bp portion of the 5′ upstream region of the AtGPAT9 gene, resulting in the synthesis of cloning vector pK51. The open reading frames of AtGPAT9 and tung tree (Vernicia fordii) GPAT9 (VfGPAT9) were cloned into the NotI and SacII sites of pK51, resulting in plant shuttle plasmids pB447 and pB448, respectively. The AscI fragments bearing the respective AtGPAT9 promoter:GPAT9 open reading frame:35S terminator cassettes from pB447 and pB448 were purified by gel electrophoresis and ligated into the AscI site of the DsRed-selectable binary vector pB110 (Shockey et al., 2015), resulting in the final plant transformation binary plasmids pE434 and pE437. Binary plasmids were transformed into Agrobacterium tumefaciens strain GV3101 via electroporation and selection on solid medium containing 50 μg mL−1 each kanamycin and gentomycin. Individual A. tumefaciens colonies were inoculated into liquid medium containing the same antibiotics and cultured overnight at 28°C prior to plant transformation by floral dip, as described previously (Clough and Bent, 1998).
Mutant Complementation
Approximately 100,000 T1 seeds from each of the three transformations were sown on flats of soil for approximately 10 d, then heavily misted with a solution of 500 mg L−1 sulfadiazine containing 0.03% (w/v) of the surfactant Silwet L-77, as described by Thomson et al. (2011). Plants that survived the sulfadiazine application and displayed red fluorescence were transferred to pots of untreated soil and grown to maturity. At the end of this experiment, nine, 25, and 30 individual T1 transgenic plants were identified for B110 control, E434, and E437, respectively. Representative lines from each transgenic genotype were propagated to the T3/T4 generations, with sulfadiazine selection, until plants producing homozygous red fluorescent seeds were identified. Batches of seeds from these plants, representing empty vector control B110 and GPAT9 overexpressor E434 and E437 lines, were surface sterilized and sown on sulfadiazine agar plates for complementation testing.
GPAT9-2/gpat9-2 Gametophyte Viability Analysis
To determine the rate of gpat9-2 ovule abortion, aborted ovules were counted in opened half siliques from Col-0 and GPAT9-2/gpat9-2 and imaged with a Leica DM2000 microscope equipped with a DFC295 camera. Alexander staining of aborted pollen, and nitroblue tetrazolium staining for pollen viability, were done by previously optimized methods (Regan and Moffatt, 1990; Peterson et al., 2010).
Analysis of qrt1-4 GPAT9-2/gpat9-2 Pollen
GPAT9-2/gpat9-2 was crossed with the qrt1-4 homozygous mutant as the pollen donor. F1 seeds were germinated on sulfadiazine to identify gpat9-2 heterozygotes. F1 plants were selfed, and F2 seeds again were selected on sulfadiazine. Pollen from F2 plants was examined visually for the tetrad pollen phenotype to identify those homozygous for the qrt1-4 mutation, and genotype was confirmed with PCR. Pollen tube germination was measured by dusting pollen from qrt1 GPAT9-2/gpat9-2 and qrt1 plants onto one of two different optimized pollen germination media (Boavida and McCormick, 2007; Zhu et al., 2013), except that the pH of Boavida and McCormick medium was adjusted with 1 m KOH instead of NaOH. Pollen was then prehydrated and allowed to germinate for 8 h at 22°C as described previously (Boavida and McCormick, 2007). Images of germinated quartets were captured using Leica Application Suite (version 4.2.0) software using a Leica DM2000 microscope equipped with a DFC295 camera. Pollen size was measured using ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij). To analyze the small pollen phenotype, tetrads were analyzed only if all four pollen grains were in the plane of focus. The longest apparent diameter (a) and the diameter perpendicular to the longest diameter (b) of each pollen grain of 100 tetrads were measured, and the apparent two-dimensional area of the pollen grain was calculated using the formula for an ellipse: 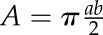 .
.
Seed and Leaf Lipid Composition and Quantification
Whole seed (or leaf) lipids were converted to FA methyl esters in 5% (v/v) sulfuric acid in methanol, to which was added 0.2 mL of toluene containing 20 µg of tri-17:0 TAG (Nucheck Prep; www.nu-chekprep.com/) as an internal standard, for 1.5 h at 85°C. FA methyl esters were quantified on an Agilent gas chromatograph with flame ionization detection on a wax column (30 m × 0.53 mm i.d. × 1.20 µm; EC Wax; Alltech).
Split Ubiquitin-Based Membrane Yeast Two-Hybrid Assay
Protein-protein interactions between AtGPAT9 and other lipid metabolic enzymes, including those representing other steps in the Kennedy pathway, were characterized using the DUALmembrane split-ubiquitin system (Dualsystems Biotech), essentially as described by Gidda et al. (2011). Plasmids were constructed for AtGPAT9, AtLPAAT2 (Kim et al., 2005), AtDGAT1 (Zou et al., 1999), and AtLPCAT2 (Ståhl et al., 2008). Primarily, only bait fusion proteins containing Cub-LexA fused to the C terminus of proteins, and prey fusion proteins containing NubG fused to the N terminus of proteins, were used in this study to avoid false-negative results, as explained by Gidda et al. (2011). Basic analysis for interactions was conducted via dilution assays on solid agar medium lacking Leu, His, Trp, and adenine, using multiple serial 1:5 dilutions of cell cultures starting from an optical density at 600 nm of 0.5. The strength of specific bait-prey combinations was measured by quantitative β-galactosidase activity from cell lysates using the β-Gal Assay Kit from Pierce Protein Research Products (Thermo Scientific).
amiRNA Knockdown of AtGPAT9 Expression
The amiRNAs were designed using Web microRNA Designer (http://wmd3.weigelworld.org). AT5G60620.1 (GPAT9) was input as the target gene into the form on the Designer section of WMD3 and compared against The Arabidopsis Information Resource 8 to get recommendations of amiRNAs. The minimum number of targets was one, and accepted off-site targets was zero. We selected nucleotide region +191 to 211 (5′-TAGATGTCTAGCAAATCGCGC-3′) for cloning. Using the GPAT9-specific amiRNA sequences, oligonucleotide sequences were generated by WMD3 for site-directed mutagenesis to introduce the sequences of interest in the microRNA precursor gene MIR319a. PCR was conducted using these oligonucleotides and pRS300 (MIR319a) as template and following the instructions described on WMD3. Using Gateway cloning, the final PCR products were cloned into the pENTR vector and eventually introduced into the pDS-Red-PHAS binary vector under the control of the Phaseolus vulgaris phaseolin promoter (Sengupta-Gopalan et al., 1985). Both constructs were transformed into A. tumefaciens strain GV3101, which was used to transform Col-0 wild-type plants.
GPAT Enzyme Activity Assays
GPAT assays were performed with microsomes collected from 9- to 11-DAF developing seeds, or whole siliques, from the wild type and amiRNA knockdown lines. Microsomes were prepared as described previously (Guan et al., 2014). In brief, approximately 100 whole siliques, or 0.1 mL volume of developing seeds dissected from siliques, were homogenized in 0.1 m potassium phosphate buffer, pH 7.2, 1% (w/v) bovine serum albumin, 1,000 units mL−1 catalase, 0.33 m Suc, and 1× Thermo Scientific Halt Protease Inhibitor Cocktail, EDTA-free (100×), with a Kinematica Polytron PT-MR 2100 on ice. The homogenate was filtered through two layers of buffer-soaked Miracloth (EMD Millipore) and centrifuged at 15,000g for 10 min at 4°C. The supernatant was centrifuged at 105,000g for 90 min at 4°C. The pellet was rinsed with phosphate buffer, pH 7.2, and then resuspended in 0.1 mL of phosphate buffer, pH 7.2, containing 1,000 units of catalase. Microsomes were quantified using the Thermo Scientific Pierce BCA Protein Assay Kit. Approximately 0.3 to 0.6 mg of total protein of isolated microsomes was utilized for GPAT assays, which were performed in 0.1 mL of 0.1 m Tris, pH 7.2, 4 mm MgCl2, 1% (w/v) bovine serum albumin, and 1 mm dithiothreitol (Yang et al., 2012), with 3.55 nmol [14C]G3P (161 mCi mmol−1; www.perkinelmer.com) as acyl acceptor and 25 nmol palmitoyl-CoA (Sigma) as acyl donor, at 24°C with constant mixing (1,250 rpm) for 15 min. The reaction was terminated by adding 0.12 mL of 0.15 m acetic acid with 400 nmol of unlabeled G3P and 1.2 mL of CHCl3:methanol (1:2). Approximately 25 µg of carrier LPA and PA in 0.4 mL of CHCl3 was added as a carrier, and lipids were extracted (Bligh and Dyer, 1959). In brief, the organic layer was collected after phase separation, followed by two back extractions of the aqueous phase with CHCl3. The combined chloroform extracts were rinsed with water:methanol (1:1) to remove any remaining labeled G3P and evaporated under N2. The lipids were resuspended in 0.1 mL of CHCl3:methanol (9:1), total radioactivity in 10 µL was quantified by liquid scintillation counting on a Beckman Coulter LS 6500 Multi-Purpose Scintillation Counter, and the remaining extract was loaded onto Merck 20- × 20-cm silica gel 60 plates developed with the solvent system CHCl3:methanol:acetic acid:water, 75:15:10:3.5 (v/v/v/v). Radioactivity on TLC plates was quantified with a GE Typhoon FLA 7000 and ImageQuant TL Image Analysis Software version 8.1.
Software and Statistics
Graphs and statistical analyses indicated in each figure were produced with GraphPad Prism (http://www.graphpad.com/).
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: AtGPAT9 (At5g60620), AtLPEAT1 (At1g80950), and AtLPEAT2 (At2g45670). The GenBank accession numbers for RcGPAT9 and VfGPAT9 are ACB30546 and FJ479751, respectively. All other gene identifiers are as described in the species-specific data subsets in the Phytozome database (version 10.3; http://phytozome.jgi.doe.gov/pz/portal.html#). All identifiers are listed individually in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Sequence comparisons between AtGPAT9 and other plant and mammalian proteins.
Supplemental Figure S2. Sequence comparison of selected GPAT9 enzymes from plants.
Supplemental Figure S3. Sequence alignment and identity/similarity shading of various plant LPEAT1 proteins.
Supplemental Figure S4. Sequence alignment and identity/similarity shading of various plant LPEAT2 proteins.
Supplemental Figure S5. Characterization of GPAT9 T-DNA insertions.
Supplemental Figure S6. Analysis of viability in wild-type and GPAT9-2/gpat9-2 pollen.
Supplemental Figure S7. Genetic complementation of gpat9-2 mutants with transgenic plant GPAT9.
Supplemental Figure S8. Lipid characterization of wild-type and GPAT9-2/gpat9-2 plants.
Supplemental Figure S9. Seed fatty acid Composition of GPAT9 amiRNA knockdown lines.
Supplemental Figure S10. Expression of GPAT9 in developing seeds of wild-type and GPAT9 amiRNA knockdown lines.
Supplemental Figure S11. Developing seed microsome GPAT assays of wild-type and amiRNA knockdown lines.
Supplemental Figure S12. Testing of protein:protein interactions between AtLPAAT2, AtDGAT1, and AtLPCAT2.
Supplemental Figure S13. Sequence identity comparisons between AtGPAT1 and AtGPAT9 to the animalia subset of the NCBI protein database.
Supplemental Table S1. Primers utilized for PCR, RT-PCR, and qPCR.
Supplemental Table S2. Gene identifiers for GPAT9, LPEAT1, and LPEAT2 sequences.
Supplementary Material
Acknowledgments
We thank Catherine Mason (USDA-ARS, SRRC, New Orleans) for expert technical assistance and for proofreading the article and Dr. Charles Miller III (Tulane University Medical Center) for the generous gift of the complete set of pBEVY yeast expression plasmids.
Glossary
- TAG
triacylglycerol
- FA
fatty acid
- ER
endoplasmic reticulum
- LPA
lysophosphatidic acid
- PA
phosphatidic acid
- DAG
diacylglycerol
- PC
phosphatidylcholine
- T-DNA
transfer DNA
- RT
reverse transcription
- Col-0
Columbia-0
- amiRNA
artificial microRNA
- G3P
glycerol-3-phosphate
- DAF
days after flowering
- TLC
thin-layer chromatography
Footnotes
This work was supported by the National Science Foundation (grant nos. EPS 0903787 and EPS 1006883 and Plant Genome Research Program grant no. IOS–1339385), Mississippi INBRE, funded by an Institutional Development Award of the National Institute of General Medical Sciences, National Institutes of Health (grant no. P20GM103476), a National Institutes of Health/National Institute of General Medical Sciences-funded predoctoral fellowship (T32 GM008336) to K.C., and the University of Southern Mississippi Department of Chemistry and Biochemistry New Faculty Start-Up program.
Articles can be viewed without a subscription.
References
- Allen DK, Bates PD, Tjellström H (2015) Tracking the metabolic pulse of plant lipid production with isotopic labeling and flux analyses: past, present and future. Prog Lipid Res 58: 97–120 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bafor M, Jonsson L, Stobart AK, Stymne S (1990) Regulation of triacylglycerol biosynthesis in embryos and microsomal preparations from the developing seeds of Cuphea lanceolata. Biochem J 272: 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafor M, Smith MA, Jonsson L, Stobart K, Stymne S (1991) Ricinoleic acid biosynthesis and triacylglycerol assembly in microsomal preparations from developing castor-bean (Ricinus communis) endosperm. Biochem J 280: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron EJ, Stumpf PK (1962) Fat metabolism in higher plants. XIX. The biosynthesis of triglycerides by avocado-mesocarp enzymes. Biochim Biophys Acta 60: 329–337 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2011) The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J 68: 387–399 [DOI] [PubMed] [Google Scholar]
- Bates PD, Browse J (2012) The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front Plant Sci 3: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Durrett TP, Ohlrogge JB, Pollard M (2009) Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol 150: 55–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Fatihi A, Snapp AR, Carlsson AS, Browse J, Lu C (2012) Acyl editing and headgroup exchange are the major mechanisms that direct polyunsaturated fatty acid flux into triacylglycerols. Plant Physiol 160: 1530–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Jewell JB, Browse J (2013a) Rapid separation of developing Arabidopsis seeds from siliques for RNA or metabolite analysis. Plant Methods 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PD, Ohlrogge JB, Pollard M (2007) Incorporation of newly synthesized fatty acids into cytosolic glycerolipids in pea leaves occurs via acyl editing. J Biol Chem 282: 31206–31216 [DOI] [PubMed] [Google Scholar]
- Bates PD, Stymne S, Ohlrogge J (2013b) Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol 16: 358–364 [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB (2007) The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. Plant Cell 19: 351–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M (2012) Solving the puzzles of cutin and suberin polymer biosynthesis. Curr Opin Plant Biol 15: 329–337 [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Boavida LC, McCormick S (2007) Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J 52: 570–582 [DOI] [PubMed] [Google Scholar]
- Cao J, Li JL, Li D, Tobin JF, Gimeno RE (2006) Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc Natl Acad Sci USA 103: 19695–19700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Miles R, Snyder C, Truksa M, Zhang J, Shah S, Weselake RJ (2014) Possible allostery and oligomerization of recombinant plastidial sn-glycerol-3-phosphate acyltransferase. Arch Biochem Biophys 554: 55–64 [DOI] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S (1991) The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA 88: 9578–9582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cronan JE, Roughan PG (1987) Fatty acid specificity and selectivity of the chloroplast sn-glycerol 3-phosphate acyltransferase of the chilling sensitive plant, Amaranthus lividus. Plant Physiol 83: 676–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Adams KL, Vandepoele K, Van Montagu MC, Maere S, Van de Peer Y (2013) Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc Natl Acad Sci USA 110: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Zhai Z, Yan C, Xu C (2015) Arabidopsis TRIGALACTOSYLDIACYLGLYCEROL5 interacts with TGD1, TGD2, and TGD4 to facilitate lipid transfer from the endoplasmic reticulum to plastids. Plant Cell 27: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142: 1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzen M, Heinz E, McKeon TA, Stumpf PK (1983) Specificities and selectivities of glycerol-3-phosphate acyltransferase and monoacylglycerol-3-phosphate acyltransferase from pea and spinach chloroplasts. Eur J Biochem 129: 629–636 [DOI] [PubMed] [Google Scholar]
- Frentzen M, Nishida I, Murata N (1987) Properties of the plastidial acyl-(acyl-carrier-protein):glycerol-3-phosphate acyltransferase from the chilling-sensitive plant squash (Cucurbita moschata). Plant Cell Physiol 28: 1195–1201 [Google Scholar]
- Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT (2011) Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12: 452–472 [DOI] [PubMed] [Google Scholar]
- Gidda SK, Shockey JM, Rothstein SJ, Dyer JM, Mullen RT (2009) Arabidopsis thaliana GPAT8 and GPAT9 are localized to the ER and possess distinct ER retrieval signals: functional divergence of the dilysine ER retrieval motif in plant cells. Plant Physiol Biochem 47: 867–879 [DOI] [PubMed] [Google Scholar]
- Griffiths G, Stobart AK, Stymne S (1985) The acylation of sn-glycerol 3-phosphate and the metabolism of phosphatidate in microsomal preparations from the developing cotyledons of safflower (Carthamus tinctorius L.) seed. Biochem J 230: 379–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Lager I, Li X, Stymne S, Zhu LH (2014) Bottlenecks in erucic acid accumulation in genetically engineered ultrahigh erucic acid Crambe abyssinica. Plant Biotechnol J 12: 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara K. (1984) sn-Glycerol-3-phosphate acyltransferase in a particulate fraction from maturing safflower seeds. Arch Biochem Biophys 232: 685–698 [DOI] [PubMed] [Google Scholar]
- Ishizaki O, Nishida I, Agata K, Eguchi G, Murata N (1988) Cloning and nucleotide sequence of cDNA for the plastid glycerol-3-phosphate acyltransferase from squash. FEBS Lett 238: 424–430 [DOI] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA 91: 10340–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Douce R (1977) Site of synthesis of phosphatidic acid and diacyglycerol in spinach chloroplasts. Biochim Biophys Acta 486: 273–285 [DOI] [PubMed] [Google Scholar]
- Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236: 1299–1302 [DOI] [PubMed] [Google Scholar]
- Kennedy EP. (1961) Biosynthesis of complex lipids. Fed Proc 20: 934–940 [PubMed] [Google Scholar]
- Kim HU, Li YB, Huang AHC (2005) Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell 17: 1073–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B (2012) GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res 40: D1211–D1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg A, Pricer WE Jr (1953) Enzymatic esterification of alpha-glycerophosphate by long chain fatty acids. J Biol Chem 204: 345–357 [PubMed] [Google Scholar]
- Lager I, Yilmaz JL, Zhou XR, Jasieniecka K, Kazachkov M, Wang P, Zou J, Weselake R, Smith MA, Bayon S, et al. (2013) Plant acyl-CoA:lysophosphatidylcholine acyltransferases (LPCATs) have different specificities in their forward and reverse reactions. J Biol Chem 288: 36902–36914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, Hawkins DJ (2001) DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. J Biol Chem 276: 38862–38869 [DOI] [PubMed] [Google Scholar]
- Larsson KE, Kjellberg JM, Tjellström H, Sandelius AS (2007) LysoPC acyltransferase/PC transacylase activities in plant plasma membrane and plasma membrane-associated endoplasmic reticulum. BMC Plant Biol 7: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc Natl Acad Sci USA 104: 18339–18344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161 doi: 10.1199/tab.0133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Siloto RMP, Lehner R, Stone SJ, Weselake RJ (2012) Acyl-CoA:diacylglycerol acyltransferase: molecular biology, biochemistry and biotechnology. Prog Lipid Res 51: 350–377 [DOI] [PubMed] [Google Scholar]
- Lu C, Xin Z, Ren Z, Miquel M, Browse J (2009) An enzyme regulating triacylglycerol composition is encoded by the ROD1 gene of Arabidopsis. Proc Natl Acad Sci USA 106: 18837–18842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ (2010) Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol 152: 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaf AM, Harwood JL (2000) Purification and characterisation of acyl-CoA:glycerol 3-phosphate acyltransferase from oil palm (Elaeis guineensis) tissues. Planta 210: 318–328 [DOI] [PubMed] [Google Scholar]
- Mo C, Bard M (2005) A systematic study of yeast sterol biosynthetic protein-protein interactions using the split-ubiquitin system. Biochim Biophys Acta 1737: 152–160 [DOI] [PubMed] [Google Scholar]
- Muralla R, Lloyd J, Meinke D (2011) Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6: e28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Pan X, Chen G, Kazachkov M, Greer MS, Caldo KM, Zou J, Weselake RJ (2015) In vivo and in vitro evidence for biochemical coupling of reactions catalyzed by lysophosphatidylcholine acyltransferase and diacylglycerol acyltransferase. J Biol Chem 290: 18068–18078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Chapman BA, Kissinger JC, Bowers JE, Feltus FA, Estill JC (2006) Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet 22: 597–602 [DOI] [PubMed] [Google Scholar]
- Peterson R, Slovin JP, Chen C (2010) A simplified method for differential staining of aborted and non-aborted pollen grains. International Journal of Plant Biology 1: e13 [Google Scholar]
- Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264: 1458–1460 [DOI] [PubMed] [Google Scholar]
- Regan SM, Moffatt BA (1990) Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell 2: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Somerville CR (1998) Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J 15: 79–88 [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Ohlrogge JB (1996) Evidence that isolated chloroplasts contain an integrated lipid-synthesizing assembly that channels acetate into long-chain fatty acids. Plant Physiol 110: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roughan PG, Slack CR (1982) Cellular-organization of glycerolipid metabolism. Annu Rev Plant Physiol Plant Mol Biol 33: 97–132 [Google Scholar]
- Routaboul JM, Benning C, Bechtold N, Caboche M, Lepiniec L (1999) The TAG1 locus of Arabidopsis encodes for a diacylglycerol acyltransferase. Plant Physiol Biochem 37: 831–840 [DOI] [PubMed] [Google Scholar]
- Ruiz-López N, Garcés R, Harwood JL, Martínez-Force E (2010) Characterization and partial purification of acyl-CoA:glycerol 3-phosphate acyltransferase from sunflower (Helianthus annuus L.) developing seeds. Plant Physiol Biochem 48: 73–80 [DOI] [PubMed] [Google Scholar]
- Rutter AJ, Sanchez J, Harwood JL (1997) Glycerolipid synthesis by microsomal fractions from Olea europaea fruits and tissue cultures. Phytochemistry 46: 265–272 [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sengupta-Gopalan C, Reichert NA, Barker RF, Hall TC, Kemp JD (1985) Developmentally regulated expression of the bean β-phaseolin gene in tobacco seed. Proc Natl Acad Sci USA 82: 3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey J, Browse J (2011) Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J 66: 143–160 [DOI] [PubMed] [Google Scholar]
- Shockey J, Mason C, Gilbert M, Cao H, Li X, Cahoon E, Dyer J (2015) Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues. Plant Mol Biol 89: 113–126 [DOI] [PubMed] [Google Scholar]
- Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM (2006) Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18: 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparace SA, Moore TS (1979) Phospholipid metabolism in plant mitochondria: submitochondrial sites of synthesis. Plant Physiol 63: 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl U, Stalberg K, Stymne S, Ronne H (2008) A family of eukaryotic lysophospholipid acyltransferases with broad specificity. Febs Letters 582: 305–309 [DOI] [PubMed] [Google Scholar]
- Stålberg K, Ståhl U, Stymne S, Ohlrogge J (2009) Characterization of two Arabidopsis thaliana acyltransferases with preference for lysophosphatidylethanolamine. BMC Plant Biol 9: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobart AK, Stymne S (1985) The regulation of the fatty-acid composition of the triacylglycerols in microsomal preparations from avocado mesocarp and the developing cotyledons of safflower. Planta 163: 119–125 [DOI] [PubMed] [Google Scholar]
- Stuitje AR, Verbree EC, van der Linden KH, Mietkiewska EM, Nap JP, Kneppers TJA (2003) Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol J 1: 301–309 [DOI] [PubMed] [Google Scholar]
- Stymne S, Stobart AK (1984) Evidence for the reversibility of the acyl-CoA:lysophosphatidylcholine acyltransferase in microsomal preparations from developing safflower (Carthamus tinctorius L.) cotyledons and rat liver. Biochem J 223: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Kawazu T, Koyama H (2004) RNA isolation from siliques, dry seeds, and other tissues of Arabidopsis thaliana. Biotechniques 37: 542–544 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JG, Cook M, Guttman M, Smith J, Thilmony R (2011) Novel sulI binary vectors enable an inexpensive foliar selection method in Arabidopsis. BMC Research Notes 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H, Yang Z, Allen DK, Ohlrogge JB (2012) Rapid kinetic labeling of Arabidopsis cell suspension cultures: implications for models of lipid export from plastids. Plant Physiol 158: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ (2005) The Botany Array Resource: e-northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Verdaguer B, de Kochko A, Beachy RN, Fauquet C (1996) Isolation and expression in transgenic tobacco and rice plants, of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Wang K, Wang Z, Li F, Ye W, Wang J, Song G, Yue Z, Cong L, Shang H, Zhu S, et al. (2012a) The draft genome of a diploid cotton Gossypium raimondii. Nat Genet 44: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Wang L, Shen W, Kazachkov M, Chen G, Chen Q, Carlsson AS, Stymne S, Weselake RJ, Zou J (2012b) Metabolic interactions between the Lands cycle and the Kennedy pathway of glycerolipid synthesis in Arabidopsis developing seeds. Plant Cell 24: 4652–4669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S, Wolter FP, Buck F, Frentzen M, Heinz E (1991) Purification and cDNA sequencing of an oleate-selective acyl-ACP:sn-glycerol-3-phosphate acyltransferase from pea chloroplasts. Plant Mol Biol 17: 1067–1076 [DOI] [PubMed] [Google Scholar]
- Weiss SB, Kennedy EP, Kiyasu JY (1960) The enzymatic synthesis of triglycerides. J Biol Chem 235: 40–44 [PubMed] [Google Scholar]
- Wilkinson JE, Twell D, Lindsey K (1997) Activities of CaMV 35S and nos promoters in pollen: implications for field release of transgenic plants. J Exp Bot 48: 265–275 [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol 160: 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21: 3885–3901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Kang E, Xu Q, Wang M, Rui Y, Liu B, Yuan M, Fu Y (2013) MAP18 regulates the direction of pollen tube growth in Arabidopsis by modulating F-actin organization. Plant Cell 25: 851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC (1999) The Arabidopsis thaliana TAG1 mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J 19: 645–653 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.