Abstract
Antibiotic resistance has increased in recent years, raising the concern of public health authorities. We conducted a study of Escherichia coli isolates obtained from human and food samples to assess the prevalence of antimicrobial resistance and to determine the genotype and clonal relationship of 84 E. coli isolates (48 from humans and 36 from foods). An antimicrobial susceptibility test was performed using the disk diffusion method. Virulence factors were evaluated by multiplex PCR, and the clonal relationship among the resistant isolates was studied by Pulsed Field Gel Electrophoresis (PFGE). All isolates were susceptible to ceftriaxone. Overall, 26%, 20.2%, 15.4% and 6% of the isolates were resistant to tetracycline, ampicillin, sulfamethoxazole/trimethoprim and cephalotin, respectively. Twenty two percent of the isolates exhibited resistance to more than one antimicrobial agent. Multiple-drug resistance was mostly observed in the human isolates and involved the antibiotics ampicillin and tetracycline. None of the six virulence genes were identified among the isolates. Analysis of genetic diversity by PFGE of 31 resistant isolates, revealed 29 distinct restriction patterns. In conclusion, E. coli from humans and foods are resistant to commonly used antibiotics and are highly genetically diverse. In this setting, inappropriate use of antibiotics may be a cause of high resistance rate instead of clonal spread.
Keywords: Escherichia coli, antimicrobial resistance, food contamination, genetic diversity
Introduction
Escherichia coli are a common inhabitant of the intestinal tract of humans and animals and the most common cause of nosocomial- and community-acquired infections (von Baum and Marre, 2005). Within the gastrointestinal tract, commensal E.coli can transfer its antibiotic-resistant genes to diverse microorganisms, such as pathogenic bacteria, especially when exposed to antimicrobials (Smith et al., 2007). The transfer of antibiotic-resistant genes was first described by Smith (1969) who isolated E.coli strains from the digestive tract of humans and animals. Over the years, this finding has been confirmed by numerous studies (Aaerestrup and Wegener, 1999; Winokur et al., 2001; Angulo et al., 2004; Wang et al., 2006).
Humans become colonized and/or infected through physical contact, occupational exposure or food consumption. Food, especially of animal origin, is an important vehicle of antibiotic-resistant pathogens (von Baum and Marre, 2005; Riaño et al., 2006). Some studies have shown epidemiological relationships among different E.coli strains isolated from humans and foods and an increased number of resistant isolates (Voltattoni et al., 2002 Ramchandani et al., 2005; Manges et al., 2007; Johnson et al., 2007; Thorsteinsdottir et al., 2010).
The aims of this study were to evaluate the antimicrobial susceptibility of E.coli strains isolated from humans and foods, to identify potential pathogenic strains and to determine the possible epidemiological relationship among them by pulsed-field gel electrophoresis (PFGE).
Materials and Methods
Escherichia coli Strains
A total of 84 E.coli isolates from human (48) and food (36) samples were evaluated. Human isolates were obtained from stool (39) and urine (9) samples of outpatients at two major hospitals in the city of Salvador, Brazil. At the Food Microbiology Research Laboratory of the Federal University of Bahia (UFBA) and at the Central Laboratory of Public Health Prof. Gonçalo Moniz (LACEN/BA), food isolates were obtained from meat (8), chicken meat (1), milk (2), dairy products (6) and ready-made meals (19) according to Downes and Ito (2001).
Antimicrobial susceptibility profile
Susceptibility testing was carried out by a disc diffusion method according to the Clinical and Laboratory Standard Institute guidelines (CLSI, 2010). The tested antibiotics (Laborclin, Paraná, Brazil) included ampicillin (10 μg), cephalothin (30 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), tetracycline (30 μg), chloramphenicol (30 μg), trimethoprim/sulfamethoxazole (1.25/23.75 μg), amoxicillin/clavulanic acid (20/10 μg), levofloxacin (5 μg) and ceftriaxone (30 μg). E. coli ATCC 25922 was used as a quality control. Multidrug resistance was defined as resistance to at least two classes of antimicrobial drugs (Knezevic and Petrovic, 2008).
Investigation of pathogenic serotypes
The E. coli isolates were screened by multiplex-PCR (Tornieporth et al., 1995) with primers (IDT, USA) specific to the virulence genetic markers ipaH, eae, stx, bfp, LT and ST for E. coli enteroinvasive (EIEC), enteroaggregative (EAEC), enteropathogenic (EPEC), enterohemorrhagic (EHEC) and enterotoxigenic (ETEC), respectively, as described by Tornieporth et al. (1995) and Meng et al. (1998). The primers, the size of the amplification products (i.e., amplicons) and the references used in the detection of the six virulence gene markers are shown in Table 1. The non-pathogenic E. coli K12 HB101 was used as a PCR negative control, and EPEC O111: H2, H34x ST ETEC, ETEC LT 52593, EIEC O144: H25, and EHEC EDL933 were used as positive controls.
Table 1. Primers and amplicons size of the virulence genes searched in the 84 E.coli isolates studied.
| Virulence Gene | Serotype | Primers | Sequence (5′-3′) | Size of product PCR | Reference |
|---|---|---|---|---|---|
| BFP gene | EPECa | EP1 | CAA-TGG-TGC-TTG-CGC-TTG-CT | 324 | Tornieporth et al., 1995 |
| EP2 | GCC-GCT-TTA-TCC-AAC-CTG-GT | ||||
| LT gene | ETEC (LT)b | ETLT1 | GCG-ACA-AAT-TAT-ACC-GTG-CT | 708 | Tornieporth et al., 1995 |
| ETLT2 | CCG-AAT-TCT-GTT-ATA-TAT-GT | ||||
| Sta gene | ETEC (ST)c | ETST1 | CTG-TAT-TGT-CTT-TTT-CAC-CT | 182 | Tornieporth et al., 1995 |
| ETST2 | GCA-CCC-GGT-ACA-AGC-AGG-AT | ||||
| ipaH | EIECd | EI-1 | GCT-GGA-AAA-ACT-CAG-TGC-CT | 424 | Tornieporth et al., 1995 |
| EI-2 | CCA-GTC-CGT-AAA-TTC-ATT-CT | ||||
| eae | EHECe | EHeae1 | GTG-GCG-AAT-ACT-GGC-GAG-ACT-A | 435 | Meng et al., 1998 |
| EHeae2 | GAT-CGT-AAC-GGC-TGC-CTG-ATA-TAA | ||||
| hly | EHECe | EHhly1 | AGC-CGG-AAC-AGT-TCT-CTC-AG | 526 | Meng et al., 1998 |
| EHhly2 | CCA-GCA-TAA-CAG-CCG-ATG-T |
enteropathogenic;
enterotoxigenic the heat-labile enterotoxin (LT)-producing;
enterotoxigenic the heat-stable enterotoxin(STa)-producing;
enteroinvasive;
enterohemorrhagic.
Pulsed-Field Gel Electrophoresis (PFGE)
The Center for Disease Control and Prevention protocol for molecular subtyping of E. coli O157:H7 by PFGE was used (Bender et al., 1997). PFGE of XbaI-digested DNA (Sigma-Aldrich, USA) was performed on the isolates that were resistant to an antibiotic. Isolates exhibiting identical PFGE patterns were considered to be genetically indistinguishable, those exhibiting 1-3 band differences were considered to be closely related, and those exhibiting 4-6 band differences were considered to be possibly related.
Data Analysis
The XbaI (Sigma-Aldrich, USA) fingerprints were analyzed using GelCompar II software (Applied Maths, Kortrijk, Belgium). A similarity dendrogram was constructed by the unweighted pair group (UPGMA) method using the Dice similarity coefficient; the relatedness between the isolates was interpreted according to the method described by Tenover (1995).
Ethics
This project was approved by the Research Ethical Committee of Department of Health of the State of Bahia (No. 0028.0053.000-09).
Results and Discussion
Susceptibility profile
The prevalence of antimicrobial resistance among the E. coli strains isolated from humans and foods is shown in Table 2. Overall, 29 (34.6%) of the isolates were resistant to at least one of the 11 antibiotics tested, two (2.4%) had an intermediate susceptibility, and 53 (63%) were pan-sensitive. Among those isolates recovered from humans (n = 48), 30 (62%) were susceptible to all the tested antibiotics, 13 (27%) were resistant to ampicillin, 13 (27%) were resistant to tetracycline, and 11 (23%) were resistant to trimethoprim/sulfamethoxazole. Among those isolates recovered from food, 23 (64%) were susceptible to all the antibiotics tested, 9 (25%) were resistant to tetracycline, 4 (11%) were resistant to ampicillin, and 2 (5%) had an intermediate resistance to cephalotin (Table 2).
Table 2. Prevalence of antimicrobial resistance among Escherichia coli isolates from human and food source.
| Antibiotics | % Resistance | ||
|---|---|---|---|
| Human (N = 48) | Food (N = 36) | Overall (N = 84) | |
| Ampicillin | 27.0 | 11.0 | 20.2 |
| Amoxicillin/Clavulanic acid | 0 | 5.5 | 2.3 |
| Cephalotin | 4.0 | 8.3 | 6.0 |
| Cefotaxime | 2.0 | 0 | 1.2 |
| Ceftriaxone | 0 | 0 | 0 |
| Ciprofloxacin | 6.2 | 2.7 | 4.7 |
| Chloramphenicol | 6.2 | 2.7 | 4.7 |
| Gentamicin | 2.0 | 2.7 | 2.3 |
| Levofloxacin | 6.2 | 0 | 3.5 |
| Tetracycline | 27.0 | 25.0 | 26.0 |
| Trimethoprim/sulfamethoxazole | 23.0 | 5.5 | 15.4 |
These results are consistent with those reported by Jakobsen et al. (2010), who found that human isolates had a higher resistance percentage to ampicillin, trimethoprim/sulfamethoxazole and tetracycline than to other drugs. Furthermore, Pires et al. (2007) reported that the highest resistance percentage was to ampicillin, followed by trimethoprim/sulfamethoxazole.
In this study, the prevalence of tetracycline resistance was similar in E. coli isolates obtained from humans and foods, contrary to the results reported by Meyer et al. (2008), who investigated E. coli isolates obtained from foods, animals and humans in Germany. The authors found a higher rate of tetracycline resistance in E. coli isolated from animals and foods than in E. coli isolated from humans. However, in both studies, the rate of resistance to ampicillin was the second highest in the strains isolated from dietary sources. Ampicillin resistance has been widely observed even when the use antimicrobial agents is restricted, as reported by Hoyle et al. (2006), who analyzed E. coli isolates obtained from animal feces living in organic farms.
In a similar study conducted by Jakobsen et al. (2010) in Denmark, the rate of tetracycline resistance was higher in E. coli isolates obtained from food than in those obtained from humans. In Denmark, tetracycline is used mainly in swine farms (Vieira et al., 2009). Meyer et al. (2008) attributed this high rate of tetracycline resistance in E. coli isolates from food to the widespread use of antimicrobials during food production. According to the Brazilian Ministry of Agriculture, Livestock and Supply, the use of antimicrobials has not been allowed since 1998 (Ordinance No. 193 of 12.05.1998 repealed by the Normative Instruction number 26, 07.09.2009) (Brazil, 2009).
The rate of antimicrobial resistance observed in this study among the E. coli isolates obtained from food was lower than that reported by Van et al. (2007), Van et al. (2008) and Altalhi et al. (2000), who reported resistance rates close to 100%. Because of the use of antimicrobial agents in animal feed and the selective pressure that such drugs exert on the microorganisms, it is assumed that isolates obtained from foods of animal origin have a resistance profile that is superior to those from other food sources.
The percentage of human isolates that exhibited multidrug resistance was 77.7% (14/18); the percentage of food isolates with multidrug resistance was 45.5% (5/11) (Table 3). Similar results were described by Thorsteinsdottir et al. (2010), who found a higher proportion of multidrug resistant E. coli in human isolates than in food isolates.
Table 3. Multiple antimicrobial drug resistance among Escherichia coli isolates by source.
| Antimicrobial | % resistant | ||
|---|---|---|---|
| Human (N = 48) | Food (N = 36) | Total (N = 84) | |
| AMP | 4.2 (2) | 2.7 (1) | 3.5 (3) |
| TET | 4.2 (2) | 13.8 (5) | 8.3 (7) |
| CFL | 0 | 0 | 0 |
| AMP TET | 2.0 (1) | 0 | 1.2 (1) |
| AMP, SUT | 6.2 (3) | 0 | 3.5 (3) |
| AMC, CFL | 0 | 2.7 (1) | 1.2 (1) |
| TET, CLO | 0 | 2.7 (1) | 1.2 (1) |
| TET, SUT | 4.2 (2) | 0 | 2.4 (2) |
| AMP, TET, SUT | 4.2 (2) | 2.7 (1) | 3.5 (3) |
| AMP, TET, SUT, CLO | 6.2 (3) | 0 | 0 |
| AMP, AMC, CFL, TET | 0 | 2.7 (1) | 1.2 (1) |
| GEN, TET, CIP, LVX | 2.0 (1) | 0 | 1.2 (1) |
| AMP, CFL, TET CIP, LVX | 2.0 (1) | 0 | 1.2 (1) |
| AMP, CFL, GEN, TET, CIP, SUT | 0 | 2.7 (1) | 1.2 (1) |
AMP-ampicillin; TET-tetracycline. SUT- trimethoprim/sulfamethoxazole; CLO-chloramphenicol. CFL-cephalothin. AMC-amoxicillin/clavulanic acid; LVX-levofloxacin. CIP-ciprofloxacin;GEN-gentamicin. CRO-ceftriaxone. CTX-cefotaxime.
Multiplex PCR
The analysis of the genes encoding important virulence markers of E. coli revealed that none of the isolates carried the ipaH, eae, hly, bfp, LT and ST genes. Paneto et al., 2007, and Gonzalez et al., 2000, reported low prevalence rates of pathogenic E. coli in food samples.
Genetic diversity of E. coli isolates using pulsed-field gel electrophoresis
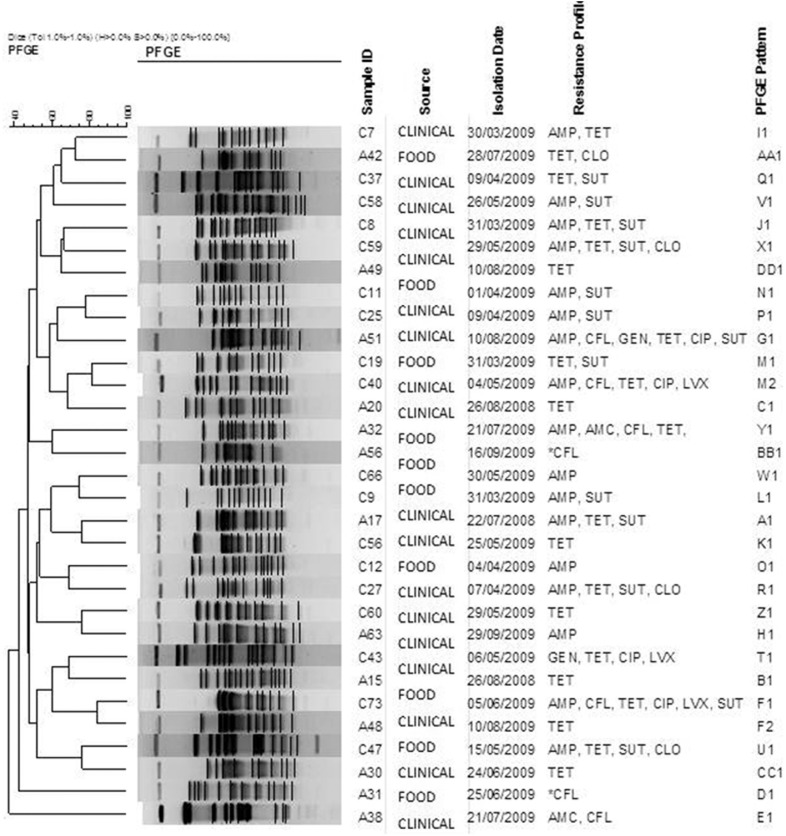
A genetic diversity analysis by PFGE of 31 resistant isolates revealed 29 distinct restriction patterns (Figure 1).
Figure 1. Dendrogram based on XbaI PFGE types identified among 31 E. coli strains. The data were sorted by the UPGMA method. AMP-ampicillin, TET-tetracycline, SUT- trimethoprim/sulfamethoxazole, CLO-chloramphenicol, CFL-cephalothin, AMC-amoxicillin/clavulanic acid, LVX- levofloxacin, CIP-ciprofloxacin, GEN-gentamicin, CRO-ceftriaxone, CTX-cefotaxime. *Isolates that show intermediate susceptibility to cephalotin.
There were no strains with an indistinguishable electrophoretic pattern that could characterize a clone. The highest similarity percentage was 84.2% between the human and food isolates. Two groups (F and M) contained one human isolate and one food isolate that differed by 3 bands (i.e., close related isolates). Despite the limitations, we cannot exclude the hypothesis that a food reservoir exists and that food-borne transmission of E.coli is common.
The genetic diversity among the E. coli isolates in this study can be explained by the fact that the isolates were not epidemiologically related; they had been isolated from different locations. The isolates were obtained in the same period and had some resistance characteristics. Moreover, the human isolates were obtained from outpatients who were epidemiologically unrelated. Even when studying geographically related strains, an absence of clones is possible, as reported by Campos et al. (2009), who observed an absence of an endemic clone and a high diversity of E.coli strains isolated from humans and foods.
In practice, the typing of microorganisms is more effective as an aid during the investigation of outbreaks when applied to a low number of epidemiologically related isolates (Tenover et al., 1995). However, it is necessary to study strains of different origin, especially those not involved in the outbreaks with antimicrobial resistance to assess possible modifications of their genome. Such an analysis is crucial for understanding their origin and their impact on public health and for establishing possible control strategies.
Conclusion
This study, which was performed in the third largest city of Brazil, found that 34.6% of the E. coli isolates were antimicrobial resistant. The most frequent antibiotic resistance observed was tetracycline in both human and food isolates. In addition, we found a high percentage of multidrug resistance among these isolates, particularly in isolates obtained from humans and foods of animal origin. None of the strains was considered to be pathogenic, and the molecular typing by PFGE found a high degree of genetic diversity among the isolates.
Acknowledgments
The authors thank the Ministry of Health of Brazil and the Foundation for Research Support of the State of Bahia - Fapesb for financing the research and the Central Laboratory of Public Health Prof. Gonçalo Moniz (LACEN-BA) for its support.
References
- Aarestrup FM, Wegener HC. The effect of antibiotic usage in food animals on the development of antimicrobial resistance of importance for human in Campylobacter and Escherichia coli. . Microb Infect. 1999;1:639–644. doi: 10.1016/s1286-4579(99)80064-1. [DOI] [PubMed] [Google Scholar]
- Altalhi AD, Gherbawy YA, Hassan SA. Antibiotic Resistance in Escherichia coli Isolated from Retail Raw Chicken Meat in Taif, Saudi Arabia. Foodborne Pathog Dis. 2000;7:281–285. doi: 10.1089/fpd.2009.0365. [DOI] [PubMed] [Google Scholar]
- Angulo FJ, Nargund VN, Chiller TC. Evidence of an Association Between use of Anti-microbial Agents in Food Animals and Anti-microbial Resistance Among Bacteria Isolated from Humans and the Human Health Consequences of Such Resistance. J Vet Med Series B. 2004;51:374–379. doi: 10.1111/j.1439-0450.2004.00789.x. [DOI] [PubMed] [Google Scholar]
- Bender JB, Hedberg CW, Besser JM, et al. Surveillance by molecular subtype for Escherichia coli O157:H7 infections in Minnesota by molecular subtyping. New Eng J Med. 1997;337:388–394. doi: 10.1056/NEJM199708073370604. [DOI] [PubMed] [Google Scholar]
- Brazil Approves the Technical Regulation for manufacturing, quality control, marketing and use of antimicrobial products for veterinary use. 2009. Ministry of Agriculture, Livestock and Supply. Office of the Minister. Normative Instruction No. 26, July 9, 2009. Published in the Official Gazette of July 10, 2009, Section 1.
- Campos MRH, André MCDPB, Borges LJ, et al. Genetic heterogeneity of Escherichia coli strains isolated from raw milk, Minas Frescal cheese, and food handlers. Braz J Vet Anim Sci. 2009;61:1203–1209. [Google Scholar]
- Clinical and Laboratory Standards Institure . Performance standards for antimicrobial disk susceptibility testing: Twentieth informational supplement. CLSI document M100-S20. 1. Vol. 30. Clinical And Laboratory Standards Institute; Wayne, PA: 2010. replaces M100-S19, Vol. 29, nº. 3. January 2010. [Google Scholar]
- Downes FP, Ito K. Compendium of Methods for the Microbiological Examination of Foods. 4th ed. APHA; Washington, DC: 2001. 676 pp [Google Scholar]
- Gonzalez AGM, Rosa ACP, Andrade JRC, et al. Enteropathogenicity markers in Escherichia coli strains isolated from soft white cheese and poultry in Rio de Janeiro, Brazil. Food Microbiol. 2000;17:321–328. [Google Scholar]
- Hoyle DV, Davison HC, Knight HI, et al. Molecular characterisation of bovine faecal Escherichia coli shows persistence of defined ampicillin resistant strains and the presence of class 1 integrons on an organic beef farm. Vet Microbiol. 2006;115:250–257. doi: 10.1016/j.vetmic.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Jakobsen L, Kurbasic A, Skjøt-Rasmussen L, et al. Escherichia coli Isolates from Broiler Chicken Meat, Broiler Chickens, Pork, and Pigs Share Phylogroups and Antimicrobial Resistance with Community-Dwelling Humans and Patients with Urinary Tract Infection. Foodborne Pathog Dis. 2010;7:537–547. doi: 10.1089/fpd.2009.0409. [DOI] [PubMed] [Google Scholar]
- Johnson JR, Sannes MR, Croy C, et al. Antimicrobial Drug-Resistant Escherichia coli from Humans and Poultry Products, Minnesota and Wisconsin, 2002-2004. Emerg Infect Dis. 2007;13:838–846. doi: 10.3201/eid1306.061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knezevic P, Petrovic O. Antibiotic resistance of commensal Escherichia coli of food-producing animals from three Vojvodinian farms, Serbia. Int J Antimicrob Agents. 2008;31:360–363. doi: 10.1016/j.ijantimicag.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Manges AM, Smith SP, Lau BJ, et al. Retail Meat Consumption and the Acquisition of Antimicrobial Resistant Escherichia coli Causing Urinary Tract Infections: A Case-Control Study. Foodborne Pathogs Dis. 2007;4:419–431. doi: 10.1089/fpd.2007.0026. [DOI] [PubMed] [Google Scholar]
- Meng J, Zhao S, Doyle MP. Virulence genes of Shiga toxin-producing Escherichia coli isolated from food, animals and humans. Int J Food Microbiol. 1998;45:229–235. doi: 10.1016/s0168-1605(98)00163-9. [DOI] [PubMed] [Google Scholar]
- Meyer E, Lunke C, Kist M, et al. Antimicrobial Resistance in Escherichia coli Strains Isolated from Food, Animals and Humans in Germany. Infect. 2008;36:59–61. doi: 10.1007/s15010-007-7113-9. [DOI] [PubMed] [Google Scholar]
- Paneto BR, Schocken-Iturrino RP, Macedo C, et al. Occurrence of toxigenic Escherichia coli in raw milk cheese in Brazil. Arq Bras Med Vet Zootec. 2007;59:508–512. [Google Scholar]
- Pires MCS, Frota KS, Martins PO, Junior, et al. Prevalence and bacterial susceptibility of community acquired urinary tract infection in University Hospital of Brasília, 2001 to 2005. Rev Soc Bras Med Trop. 2007;40:643–647. doi: 10.1590/s0037-86822007000600009. [DOI] [PubMed] [Google Scholar]
- Ramchandani M, Manges AR, DebRoy C, et al. Possible animal origin of human-associated multidrugresistant, uropathogenic Escherichia coli . Clinic Infect Dis. 2005;40:251–257. doi: 10.1086/426819. [DOI] [PubMed] [Google Scholar]
- Riaño I, Moreno MA, Teshager T, et al. Detection and characterization of extended-spectrum b-lactamases in Salmonella enterica strains of healthy food animals in Spain. J Antimicrob Chemother. 2006;58:844–847. doi: 10.1093/jac/dkl337. [DOI] [PubMed] [Google Scholar]
- Smith DH. Transfer of antibiotic resistance from animal and human strains of Escherichia coli to resident E.coli in the alimentary tract of man. The Lancet. 1969;293:1174–1179. doi: 10.1016/s0140-6736(69)92164-3. [DOI] [PubMed] [Google Scholar]
- Smith JL, Drum DJV, Daí Y, et al. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl Environ Microbiol. 2007;73:1404–1414. doi: 10.1128/AEM.01193-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsteinsdottir TR, Haraldsson G, Fridriksdottir V, et al. Prevalence and genetic relatedness of antimicrobial-resistant Escherichia coli isolated from animals, foods and humans in Iceland. Zoonoses Public Hlth. 2010;57:189–196. doi: 10.1111/j.1863-2378.2009.01256.x. [DOI] [PubMed] [Google Scholar]
- Tornieporth NG, John J, Salgado KJP, et al. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J Clin Microbiol. 1995;33:1371–1374. doi: 10.1128/jcm.33.5.1371-1374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van TTH, Chin J, Chapman T, et al. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Intern J Food Microbiol. 2008;124:217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Van TTH, Moutafis G, Tran LT, et al. Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl Environ Microbiol. 2007;73:7906–7911. doi: 10.1128/AEM.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira AR, Houe H, Wegener HC, et al. Association Between Tetracycline Consumption and Tetracycline Resistance in Escherichia coli from Healthy Danish Slaughter Pigs. Foodborne Pathog Dis. 2009;6:99–109. doi: 10.1089/fpd.2008.0152. [DOI] [PubMed] [Google Scholar]
- Voltattoni P, Hofer C, Redolfi AL, et al. Identificación de virotipos de Escherichia coli aislados de alimentos listos para el consumo. Rev Invest Salud. 2002;5:75–83. [Google Scholar]
- von Baum H, Marre R. Antimicrobial resistance of Escherichia coli and therapeutic implications. Int J Med Microbiol. 2005;295:503–511. doi: 10.1016/j.ijmm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Wang HH, Manuzon M, Lehman M, et al. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol Lett. 2006;254:226–231. doi: 10.1111/j.1574-6968.2005.00030.x. [DOI] [PubMed] [Google Scholar]
- Winokur PL, Vonstein DL, Hoffman LJ, et al. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother. 2001;45:2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]