Abstract
The cytochrome P450 2D6 (CYP2D6) gene is perhaps the most well characterized gene involved in drug metabolism and is known to have both gene duplication and deletion variants that are inheritable and stable. In a set of over 30 000 deidentified clinical samples we found that 12.6% of all patients tested had zero, one, or three or more copies of the CYP2D6 gene. On the basis of the combined frequency and impact of these variants, we believe that CYP2D6 copy number variation may account for the single most impactful genetic anomaly as it relates to pharmacogenetic directed therapies.
Keywords: codeine, cytochrome P450 CYP2D6, gene deletion, gene duplication, pharmacogenetics
The cytochrome P450 superfamily of genes are well understood for their role in drug metabolism, with CYP2D6 being the most extensively studied 1–4. In clinical practice, CYP2D6 has been referenced in the FDA approved labeling of over 40 different drugs citing issues of safety and/or efficacy in individuals with an altered metabolizer status (http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm). In addition, CYP2D6 has been the focus of three separate professional guidelines for the utilization of codeine, tricyclic antidepressants, and selective serotonin reuptake inhibitors making it arguably one of the most valuable pharmacogenetic tools in guiding meaningful drug selection 5–7. Although many CYP2D6 variants have been identified that contribute to variability in enzymatic function, the gene is also known to have stably inherited gene deletion and duplication events 1–4,8. A deletion of CYP2D6 is referred to in the literature as CYP2D6*5 and, when in a compound heterozygous state with another loss-of-function allele (e.g. CYP2D6*4), has been shown to result in a poor metabolizer (PM) phenotype. CYP2D6 gene duplication events are given the nomenclature of CYP2D6*xN, with ‘N’ signifying the presence of two or more copies of the gene on the same chromosome, often giving rise to an ultrarapid metabolizer (UM) phenotype (http://www.cypalleles.ki.se/cyp2d6.htm) 7. These PM and UM phenotypes are used as the basis for translating complex genotype information into clinically actionable results for pharmacologists and physicians. The prevalence of CYP2D6 copy number variation (CNV) has been reported in the literature previously, but often with modest sample sizes or as a meta-analysis combining the data from several dipartite studies 7. In this report, we provide CYP2D6 CNV data on the single most comprehensive US population data set reported to date. The cohort consisted of 31 563 male (40.5%) and female (59.5%) patients with an average age of 61.2±20.7 years (±1 SD), referred for pharmacogenetic testing by an attending physician. CYP2D6 copy number results were generated using gDNA isolated from buccal epithelial cells using QiaSymphony DNA extraction chemistry (cat. no. 937255; Qiagen, Germantown, Maryland, USA) and a commercially available copy number TaqMan assay for CYP2D6 CNV determination targeting Exon 9 (cat. no. 4400291; Thermo Scientific, Waltham, Massachusetts, USA) with an RNase P internal reference TaqMan assay (cat. no. 4403328; Thermo Scientific) coamplified in a duplex PCR reaction using the ViiA 7 Real Time PCR System (Thermo Scientific). Although this biplexed TaqMan assay combination is commonly used in clinical practice, it should be noted that in rare instances, an assay targeting Exon 9 could produce a three copy result from a two copy sample when a sample has a nonfunctional CYP2D7/2D6 hybrid gene structure (which includes the CYP2D6 Exon 9 region) in tandem with a functional copy of a CYP2D6 (e.g. *76+*1, *77+*2, and *78+*2). Samples were run in quadruplicates and a custom algorithm used to average replicate data and assign a predicted copy number based on sample ΔΔCt values calibrated against a CYP2D6 two copy control 2. On the basis of these calculations, samples were classified as containing zero, one, two, or three or more copies of the CYP2D6 gene (Fig. 1a). The overall distribution of CNVs demonstrated that only 87.4% (27 589) of the patients possessed two copies of the CYP2D6 gene, whereas 12.6% (n=3974) of the patients were classified as having zero, one, or three or more copies of the gene. Approximately 7.2% (2288) of the population had only one copy of CYP2D6 and 5.2% (1643) of the population had three or more copies. Roughly two-thirds of the samples (n=21 472) in the cohort included data on self-reported ethnicity with selections including; African American, Asian, Caucasians, Hispanic, or other. The distribution of ethnicity among the self-reported samples closely mirrors the results of the 2010 US census (http://www.census.gov/prod/cen2010/briefs/c2010br-02.pdf), suggesting the CYP2D6 CNV frequency data obtained in this study is reflective of the relative diversity of the US population (Fig. 1b).
Fig. 1.
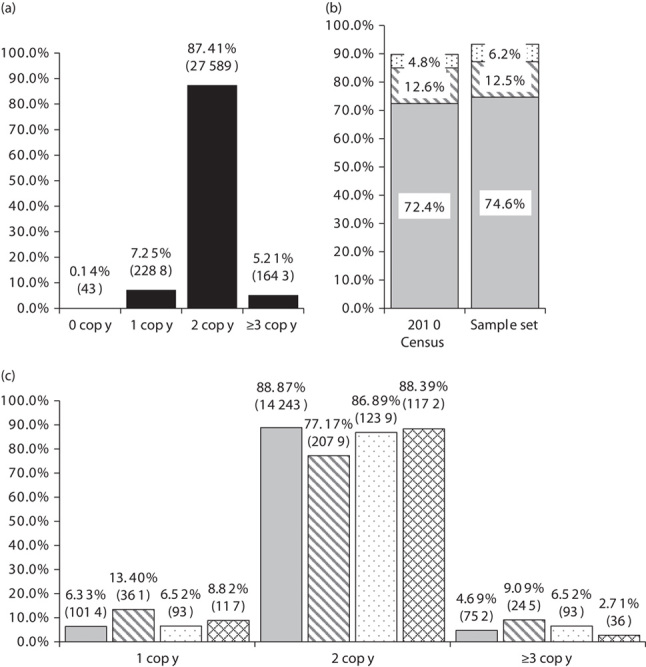
Distribution of samples containing zero, one, two, and three or more copies of the CYP2D6 gene. (a) Percentages of samples containing zero, one, two, and three or more copies of the CYP2D6 gene throughout the entire sample set (n=31 563), actual sample numbers in parentheses. (b) Percentages of self-reported ethnicity in the US 2010 Census as compared with the selected sample set, Caucasians (light grey), African Americans (diagonal lines), and Asians (dots). (c) Percentages of samples containing one, two, and three or more copies of the CYP2D6 gene stratified based on self-reported ethnicity (n=21 472), actual sample numbers in parentheses, Caucasians (light grey), African Americans (diagonal lines), Asians (dots), and Hispanics (diamonds).
For the sample set in which ethnicity was provided, data was stratified accordingly and the percentages of CYP2D6 one, two, and three or more copy patients were determined. Although the frequency of CYP2D6 ‘two copy’ patients was indistinguishable among Caucasians, Asians, and Hispanics; patients of African American ethnicity demonstrated statistically significant differences when compared with other ethnicities (Z-test for two proportions, P<0.01). Consistent with this observation, the percentage of one copy samples was 1.5- to 2.1-fold higher and the percentage of three or more copy samples was 1.4- to 3.4-fold higher in African Americans, as compared with other ethnicities (Fig. 1c). In addition, we identified 43 patients in our set of 31 563 who had no detectable copies of CYP2D6 (i.e. CYP2D6*5 homozygotes or ‘zero copy’ samples), translating to ∼0.14% of the US population. Although stratification by ethnicity resulted in single sample representation for Asians and Hispanics, African American patients demonstrated the highest probability of being CYP2D6*5 homozygotes at 0.33% (n=9/2694 or ∼1/300), followed next by Caucasians at 0.11% (17/16 026 or ∼1/940) (Table 1). Of particular note, 38 of the 43 patients identified as having ‘zero copies’ of the gene had a list of the medications the patient was currently taking. For those 38 patients, 68% (n=26) were taking at least one medication metabolized by CYP2D6.
Table 1.
Frequency of CYP2D6*5 homozygotes
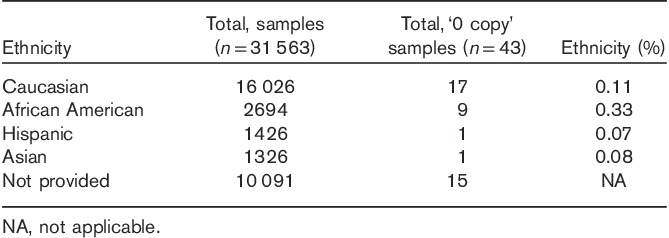
We believe this report represents the single largest evaluation of CYP2D6 CNV distribution in a clinically relevant US population. Recently, a comprehensive meta-analysis of CYP2D6 CNV frequencies was published in 2015 CPIC guidelines 7. Although the maximum and minimum frequencies reported in these guidelines were broad, the average allele frequencies calculated in these guidelines are generally supported by our findings (Table 2). However, it is of particular interest that the CYP2D6*5 allele frequencies for patients of African American ethnicity appear higher (+0.93%) in this study as compared with the percentage calculated from the CPIC meta-analysis (Table 2). This could have resulted from using a population that had been referred for pharmacogenetic testing or this may simply be a reflection of a large singular data set, obtained under rigorous clinical conditions, revealing a more statistically meaningful set of outcomes. Regardless, the relevance of this data set is in looking at a significant cross-section of the US population deemed eligible for pharmacogenetic testing by physicians and understanding the relative impact of CYP2D6 CNV determination.
Table 2.
CYP2D6 CNV allelic frequencies by ethnicity
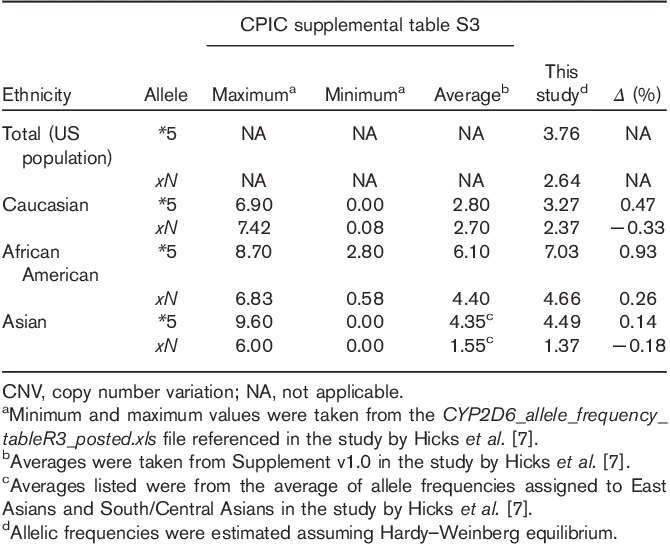
Pharmacogenetics-based therapies reserve the strongest recommendations for therapeutic intervention for individuals classified as PMs and UMs 5–7. The majority of CYP2D6 PM phenotypes arise from diplotypes of four nonfunctional alleles; CYP2D6*3, CYP2D6*4, CYP2D6*5 and CYP2D6*6. Our results establish that the allelic frequency of CYP2D6*5 in the US population is ∼3.76% (assuming Hardy–Weinberg equilibrium) making it the second most frequent ‘nonfunctional’ allele behind CYP2D6*4 7. Although not all patients containing three or more copies of CYP2D6 are UMs, UM phenotypes arise almost entirely from the presence of more than two functional copies of the CYP2D6 gene (CYP2D6*xN alleles). In our data set, we had corresponding CYP2D6 diplotype information for 1448 of the ≥3 copy samples identified. On the basis of the translational tables provided in the study by Hicks and Colleagues 7, ∼52% (750/1448) of the samples could be clearly classified as UMs as only functional variants of CYP2D6 were detected (e.g. *1/*1, *1/*2, and *2/*2). An additional ∼15% (220/1448) were possible UMs based on the presence of a reduced function allele in combination with a functional allele (e.g. *1/*41, *2/*41). This would indicate that the prevalence of functional UM phenotypes in the US population should be estimated between 2.71 and 3.49% (5.21%×0.52 to 5.21%×0.67). It has been recently noted that ‘scientists have largely overlooked the influence of gene duplications or deletions - on drug response’ and that some laboratory-developed tests identify only the single nucleotide polymorphisms of interest and do not interrogate for CYP2D6 CNVs 9. We believe that the combined prevalence of CYP2D6 CNVs, along with their relative contributions to deleterious PM and UM phenotypes, underscores the importance of determining CYP2D6 CNV in the clinical practice of pharmacogenomics.
Acknowledgements
The authors thank Dr Juan-Sebastian Saldivar, Kahuku Oades, and Elaine Sugarman, MS LCGC for their critical review and edits of this communication.
This study was funded internally by AltheaDx.
Conflicts of interest
All of the authors in this manuscript are employed by AltheaDx on either a full time or contract basis.
References
- 1.Ingelman-Sunberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics 2005; 5:6–13. [DOI] [PubMed] [Google Scholar]
- 2.Teh LK, Leif B. Pharmacogenomics of CYP2D6: molecular genetics, interethnic differences and clinical importance. Drug Metab Pharmacokinet 2012; 27:55–67. [DOI] [PubMed] [Google Scholar]
- 3.Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, and biochemistry. Naunyn-Schmiedebergs Arch Pharmacol 2004; 369:23–37. [DOI] [PubMed] [Google Scholar]
- 4.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance. Clin Pharmacokinet 2009; 48:761–804. [DOI] [PubMed] [Google Scholar]
- 5.Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, Caudle KE, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for cytochrome P450 2D6 (CYP2D6) genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014; 95:376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hicks JK, Swen JJ, Thorn CF, Sangkul K, Kharasch ED, Ellingrod VL, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 and CYP2D19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther 2013; 93:402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hicks JK, Bishop JR, Sangkuhl K, Muller DJ, Ji Y, Leckband SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther 2015; 98:127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther 2008; 83:234–242. [DOI] [PubMed] [Google Scholar]
- 9.Willard C. Copy number variations’ effect on drug response still overlooked. Nat Med 2015; 21:206. [DOI] [PubMed] [Google Scholar]


