Abstract
We accumulated mutations for 1952 generations in 79 initially identical, haploid lines of the fission yeast Schizosaccharomyces pombe, and then performed whole-genome sequencing to determine the mutation rates and spectrum. We captured 696 spontaneous mutations across the 79 mutation accumulation (MA) lines. We compared the mutation spectrum and rate to a recently published equivalent experiment on the same species, and to another model ascomycetous yeast, the budding yeast Saccharomyces cerevisiae. While the two species are approximately 600 million years diverged from each other, they share similar life histories, genome size and genomic G/C content. We found that Sc. pombe and S. cerevisiae have similar mutation rates, but Sc. pombe exhibits a stronger insertion bias. Intriguingly, we observed an increased mutation rate at cytosine nucleotides, specifically CpG nucleotides, which is also seen in S. cerevisiae. However, the absence of methylation in Sc. pombe and the pattern of mutation at these sites, primarily C → A as opposed to C → T, strongly suggest that the increased mutation rate is not caused by deamination of methylated cytosines. This result implies that the high mutability of CpG dinucleotides in other species may be caused in part by a methylation-independent mechanism. Many of our findings mirror those seen in the recent study, despite the use of different passaging conditions, indicating that MA is a reliable method for estimating mutation rates and spectra.
Keywords: mutation accumulation, insertion bias, cytosine mutation
Spontaneous mutation is the fuel for evolution and the ultimate source of all genetic differences within and between species. A complete understanding of the mutational process, both in terms of the rate at which different mutations arise, including kind and location, and the determination of their fitness effects, is thus critical to understanding genetic variation at all levels. However, elucidating the important parameters of mutation is difficult for two reasons. First, spontaneous mutations occur infrequently, making it difficult to obtain large samples of spontaneous mutations to robustly detect patterns. One workaround is to artificially increase the mutation rate using chemical mutagens and X-rays (Muller 1930; Greene et al. 2003; Blumenstiel et al. 2009), or genetic methods such as repair pathway knock-outs (Hoffman et al. 2004; Denver et al. 2006). However, it is clear that such manipulations bias the mutational spectrum in various ways (Koornneeff et al. 1982; Greene et al. 2003). Another way to deal with the rarity of spontaneous mutations is to examine genetic differences between individuals, populations or species that have arisen via mutation. Unfortunately, because many mutations are acted upon by natural selection, this raises the second main difficulty with studying spontaneous mutations: the observed genetic variation has been acted upon by selection and is thus a biased sample of spontaneous mutations (Nachman and Crowell 2000; Ho et al. 2005). Exacerbating this problem is the finding that sites in the genome previously thought to be essentially free of selection, such as those in intronic regions, intergenic regions and fourfold redundant codon positions are, in fact, often constrained by selection, making the study of mutation at these sites biased by selection (Andolfatto 2005; Hershberg and Petrov 2008).
One approach that has been employed to overcome the problems of rarity and selection in studying spontaneous mutations is the mutation accumulation (MA) experiment. MA experiments maintain multiple, initially identical lines at very low effective population sizes for many generations (Halligan and Keightley 2009). Lines accumulate spontaneous mutations at rates proportional to their occurrence since selection is ineffective at enriching for beneficial or decreasing deleterious mutations, unless their fitness effects are large (Sung et al. 2012a). Each line accumulates only a few mutations but, by having numerous lines, several hundred mutations can be captured across the lines. Whole-genome sequencing (WGS) of the MA lines allows mutations to be identified and used to estimate the frequency and spectrum of spontaneous mutation. This approach has been used to examine spontaneous mutations in MA lines in a number of eukaryotic species including Arabidopsis thaliana, Caenorhabditis elegans, Chlamydomonas reinhardtii, Dictyostelium discoideum, Drosophila melanogaster, Paramecium tetraurelia, Saccharomyces cerevisiae and Schizosaccharomyces pombe (Keightley et al. 2009; Denver et al. 2012; Ness et al. 2012; Rutter et al. 2012; Saxer et al. 2012; Sung et al. 2012b; Zhu et al. 2014; Farlow et al. 2015).
The haploid, fission yeast S. pombe was originally isolated from millet beer, and is an important model organism in molecular biology (Leupold 1950; Humphrey 2000; Wood et al. 2002). While both are ascomycete yeasts, Sc. pombe is a distant relative of S. cerevisiae, with a divergence time of 600–1200 million years (Heckman et al. 2001; Douzery et al. 2004). Even so, they exhibit similar life histories as single-celled, sexual yeasts, and are cultured in the lab using essentially identical methods. In addition, they have similar genomic G/C content (36.06% in Sc. pombe vs. 38.29% in S. cerevisiae) and genome size [13.8 Mb in Sc. pombe (Wood et al. 2002) vs. 12.1 Mb in S. cerevisiae (Goffeau et al. 1996)]. While similar, they differ in one feature that is expected to have implications for their mutation rate and spectrum: Sc. pombe is a haploid as opposed to a diploid species in nature. The similarities and difference between these two yeast species allowed us to make several a priori predictions concerning mutation rate and spectrum in Sc. pombe. Before listing these predictions, a brief note on our terminology is useful: G/C and A/T indicate G or C and A or T, and are used when discussing nucleotide composition of regions of the genome; adjacent nucleotides in a single strand of DNA are indicated by the juxtaposition of the base designations with the 5′ nucleotide on the left, e.g., CGA indicates a C nucleotide adjacent to a G nucleotide adjacent to an A nucleotide, with the A in the 3′ position; dinucleotides in the same strand that are Watson-Crick partners when in different strands, are indicated as CpG, GpC, ApT, and TpA to clarify that the nucleotides are in same strand; and base pairs in a DNA double helix are indicated with a colon, e.g., C:G.
Our first prediction was that the genome-wide single-nucleotide mutation (SNM) rate would be higher in Sc. pombe than in S. cerevisiae based on estimates from reporter genes that average 8.2 × 10−10 single nucleotide substitutions per base per generation (summarized in Lynch 2010), which is 4.8-fold higher than the most recent estimate in S. cerevisiae (Zhu et al. 2014).
Our second prediction was that most of the SNM biases, which are the relative rates of mutations among the different base pairs, in Sc. pombe would generally be the same as in S. cerevisiae. This prediction comes from the fact that the genomic G/C content of Sc. pombe (36.06%) is similar to that of S. cerevisiae (38.29%). Interestingly, the SNM biases observed in S. cerevisiae predict a lower G/C content (32%) than observed (Zhu et al. 2014), suggesting another force acts on G/C content. One possibility is selection. If weak selection drives G/C content to a higher equilibrium than predicted based on SNM bias in both species, then we expect Sc. pombe to be closer to the mutation-bias equilibrium than S. cerevisiae, i.e., lower, as observed, because of its smaller effective population size (calculated using previous estimates of diversity and mutation rate), which reduces the efficacy of selection. The SNM bias in S. cerevisiae shows an elevated mutation rate of C:G base pairs (Zhu et al. 2014). A major surprise concerning the elevated mutation rate at C:G base pairs was the finding that their mutagenicity is affected by trinucleotide context. Specifically, when C:G is the middle base pair in CCG (equivalent to CGG) and TCG (equivalent to CGA) trinucleotides, the mutation rate is elevated even more than for other C:G base pairs. Both of these trinucleotides include a CpG dinucleotide, which is a well-characterized target for methyl-transferases (Bestor and Verdine 1994). The bias at CpG dinucleotides in S. cerevisiae was in the C → T direction. For this reason, the finding of elevated C:G mutation at CCG and TCG trinucleotides was interpreted as indicating a very low occurrence of DNA methylation in S. cerevisiae (Zhu et al. 2014), in agreement with a previous study (Tang et al. 2012). Sc. pombe is believed to lack DNA methylation so we did not expect to see an elevation of the G:C mutation rate at CpG sites (Antequera et al. 1984).
Our third prediction was that among small insertions and deletions (indels) there would be an insertion bias. Small indels occur primarily at microsatellites, presumably primarily due to slippage of the DNA polymerase during replication (Levinson and Gutman 1987). In S. cerevisiae, indels of less than 50 bp were moderately biased toward deletions (18 deletions vs. eight insertions) across the diploid MA lines (Zhu et al. 2014). However, analysis of indels in haploid MA lines of S. cerevisiae showed a bias toward insertions (34 insertions and eight deletions at microsatellite loci) (Lynch et al. 2008). Attributing this insertion bias to haploidy is complicated by the fact that many of the haploid MA lines reverted to diploidy over the course of the MA experiment (Lynch et al. 2008). Nevertheless, Sc. pombe is a natural haploid and was passaged as such in our MA experiment, and so we hypothesized that we would see a bias toward insertions for small indels in this species.
Our fourth prediction was that mutations resulting from nonrecombinational repair of double-strand breaks would be more common in haploid Sc. pombe than in diploid S. cerevisiae. Nonrecombinational repair is substantially more mutagenic than recombinational repair (Daley et al. 2005). In a diploid cell, there is always a nonsister homolog present that can be used for recombinational repair. However, in a haploid cell, recombinational repair is only possible during the S or G2 phase of the cell cycle, when a sister chromatid is present. Repair of double-strand breaks is thus expected to be more error-prone in a haploid because of the higher likelihood of using nonrecombinational repair. While double-strand breaks cannot be directly observed or always inferred with certainty in the MA framework, their occurrence can be indicated, if they are inaccurately repaired, by the presence of multiple mutations in close proximity (Strathern et al. 1995; Holbeck and Strathern 1997). In diploid S. cerevisiae, three double mutations, which consist of two SNMs adjacent to one another, were observed across the MA lines (Zhu et al. 2014). The relative occurrence of double SNMs was thus ∼0.35% compared to SNMs. In addition, there were five “complex mutations”, in which multiple SNMs, and often small indels, were in close proximity, giving a relative occurrence of ∼0.58% compared to SNMs. If both double and complex mutations were due to error-prone double-strand break repair, then the observed number of these mutations accounted for ∼1% of SNMs. The haploid MA experiment in S. cerevisiae was too small to detect such rare events (Lynch et al. 2008). However, our MA experiment is large enough that we predicted we would see an increase in the rate of double mutations and complex mutations in Sc. pombe compared to S. cerevisiae, assuming these events are indeed caused by repair of double-stranded breaks. Our prediction was somewhat tempered by the fact that Sc. pombe spends only ∼10% of its cell cycle in the G1 phase when growing exponentially (Knutsen et al. 2011), and is thus haploid for a minority of the time, though this proportion can increase substantially when growth slows (Nasmyth 1979). An average, exponentially growing, Sc. pombe strain, is thus expected to have two copies of its genome in each cell 90% of the time, and a single copy 10% of the time, implying an average ploidy of 1.9 genomes. S. cerevisiae, in contrast, is in G1 (with two copies of each DNA region) about 25–35% of the time when growing exponentially (Slater et al. 1977; Brewer et al. 1984), implying an average ploidy of 3.4 genomes. These data imply that a double-strand break in Sc. pombe will be in a cell containing one homologous copy for possible recombinational repair about 90% of the time, and will have no template for repair about 10% of the time. In contrast, a double-strand break in S. cerevisiae will always have at least one copy, and will have three copies ∼70% of the time. So, while Sc. pombe may have a copy available for repair up to 90% of the time, the average number of copies available for repair is about 2.5 times less (0.9 vs. 2.4 extra copies) than in S. cerevisiae.
Our final a priori prediction was that we would not observe aneuploidy in Sc. pombe. Since Sc. pombe is haploid, loss of a single chromosome (nullisomy) results in loss of all copies of the genes on that chromosome, which would likely be lethal and thus unobservable. In addition, gain of a single chromosome (disomy) would result in a doubling of gene dose for all genes on the chromosome, which is a larger increase than the 1.5-fold increase that occurs with trisomy in diploid S. cerevisiae. If deleterious effects due to gene dosage were increased with larger differences in dosage across genes, this would further reduce the likelihood of observing aneuploidy. In addition, Sc. pombe has only three chromosomes, implying that many more genes would be affected by an aneuploidy event than in S. cerevisiae, which has a similar genome size but 16 chromosomes. The only instance of aneuploidy reported in previous work in Sc. pombe is disomy of chromosome III, which was highly deleterious (Niwa et al. 2006). For these reasons, we predicted we would see no nullisomy, and likely no disomy in any of our MA lines.
One month prior to submission of our results, Farlow and colleagues (Farlow et al. 2015) published the results of an MA experiment on Sc. pombe of essentially identical scope. While the inadvertent duplication of effort is unfortunate, their results allow us to investigate the repeatability of the MA framework for determining estimates of parameters of mutation. Their study utilized a different starting strain, different media, different growth temperatures, and longer time between transfers, allowing us to determine whether these factors together alter parameter estimates.
This study presents genome-wide estimates of mutational parameters for Sc. pombe. We examined 79 MA lines, cultivated as haploids for an average of 1952 generations. We were able to identify a total of 696 mutations. These mutations allowed us to calculate precise estimates of the mutation rate and spectrum for Sc. pombe, test our a priori predictions and determine the repeatability of MA for parameter estimation.
Materials and Methods
Mutation accumulation lines
Sc. pombe MA lines were passaged in the same manner as described for S. cerevisiae (Joseph and Hall 2004). Briefly, the haploid ancestral line, 972 h– (ATCC 26189), was streaked onto rich, solid YPD medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar) and incubated at 30°. From the streaked ancestor, 96 random isolated colonies were selected after 48 hr and used to found 96 MA lines. Lines were cultured six to a YPD plate, and bottlenecked by randomly selecting one isolated colony per line every 48 hr and transferring to a new plate. Lines were passaged for a total of 100 transfers (200 days). Every 10 transfers, a random colony from each line was frozen and stored in 15% glycerol at –80°.
Every 10 transfers, photographs were taken of all 96 MA lines. From these photographs, colony size was measured for five random colonies per line using ImageJ (Schneider et al. 2012). In addition, the numbers of cells per colony for colonies of various sizes were recorded at transfer 10 (T10) and 100 (T100) by suspending a single colony in 1 ml of water, and counting individual cells using a hemocytometer. These measurements were used to determine standard curves of colony size vs. cell number at these two transfers. Measurements of average colony sizes at every 10th transfer were then used to calculate the average number of cells at those transfers. From the average number of cells at every 10th transfer, the number of cell generations per transfer was estimated, using the fact that, with no mortality, the number of cells in a colony is equal to 2g, where g is the number of generations. The number of generations per transfer was then multiplied by the number of transfers to estimate the number of generations across the entire experiment. The effective population size across the experiment was determined by calculating the harmonic mean of the number of cells present each generation, assuming cell number doubles each generation between transfers, at which point it bottlenecks to one cell.
Sequencing
MA lines were cultured from frozen T100 stock on solid YPD medium at 30° for 48 hr. A single colony from each line was selected, inoculated into 3 ml liquid YPD, and incubated on a rotator at 30° for 48 hr. Cells were then pelleted and DNA was extracted using the YeaSTAR kit (Zymo Research) protocol with chloroform, and an extended digestion time with zymolase of 2.5 hr at 37°. Whole genome shotgun libraries were prepared by the Georgia Genomics Facility using the Kapa Library Low-Throughput Library Preparation Kit with Standard PCR Amp Module KK8232 with dual SPRI size selection cleanup to generate 100 bp paired-end fragments with ∼300 bp inserts. After seven cycles of PCR, the libraries (96 haploid MA lines and two haploid ancestor samples) were pooled across two lanes of Illumina HiSequation 2000 machines (raw sequences are deposited at NCBI, BioProject SRP065886).
Quality control (QC), mapping, and identification of mutations
Sequence reads from each library were quality controlled with the ea-utils and fastx toolkit in order to remove low quality reads and residual adaptor sequence (Gordon and Hannon 2010; Aronesty 2011)(Workflow deposited at https://github.com/behrimg/Scripts/blob/master/Hall_Projects/Pombe/MA_Pipeline.txt). Based on the workflow outlined in Zhu et al. (2014) and adjusted for a haploid dataset, quality controlled (QCed) reads were then mapped to the Sc. pombe reference genome ASM294v2.24 with BWA v1.1.2, sorted and indexed with SAMtools v1.0, and assigned line identification numbers with Picard Tools v1.87 (Wood et al. 2002; Li and Durbin 2009). Duplicated reads were marked with Picard Tools and removed, and then the remaining sequence reads were locally realigned with GATK v3.2.2 (McKenna et al. 2010). SNM and indel variants for each line and the ancestor were identified simultaneously using GATK’s Unified Genotyper tool with parameter settings for haploid organisms. The resulting VCF files were converted to tab delimited text using VCFtools v0.1.12a vcf-to-tab function (Danecek et al. 2011). All sequence differences between the MA ancestor, which was sequenced twice, and the reference were identified to determine the sequence of the ancestor. The differences between each MA line and the reference were determined, and those that were present in the ancestor were ignored. In order to call a variant, a minimum of four reads with ≥75% of the reads favoring the variant allele was needed. Regions of the genome that corresponded to centromeres, telomeres, and mating type loci (approximately 472 kbp) were excluded from the analysis to avoid inaccurate mapping. This was in addition to the two tandem rDNA repeat arrays on chromosome III accounting for 1,465 kbp, which are excluded from the reference genome. Identified SNMs and small indels were annotated using Ensembl’s variant effect predictor (VEP) while flanking regions were determined using the fill-fs program from the VCFtools package (Li and Durbin 2009; McLaren et al. 2010).
Presence of medium and large structural variants were investigated using the Delly software package (Rausch et al. 2012), and variants that passed Delly’s QC were investigated further using the integrated genome viewer (IGV) v2.1.23 (Robinson et al. 2011). When IGV supported a structural variant call, the variant was tested with PCR.
Sequencing also allowed the detection of across-line and other microbial contamination. Across-line contamination was deemed to have occurred if any two lines shared an identical new mutation. When this happened, one of the lines (chosen by coin flip) was discarded from the remainder of the analysis.
Gene expression
To estimate mRNA concentrations, as a surrogate for gene expression levels for our ancestor strain, we sequenced mRNA from 10 biological replicates. We selected 10 colonies, inoculated each into 3 ml liquid YPD medium, and incubated on a rotator at 30° for 48 hr. After 48 hr, mRNA was extracted using the MasterPure Yeast RNA Purification kit (Epicentre). mRNA libraries were constructed using the Illumina Truseq mRNA Stranded Kit, amplified using 13 cycles of PCR and sequenced on an Illumina HiSequation 2500. Libraries were sequenced as 100 bp single-end reads (NCBI SRA BioProject: SRP065886). Sequenced reads were QCed in the same manner as genomic sequencing reads, reference-mapped with TopHat v.2.0.13 (Trapnell et al. 2009), and assembled with Cufflinks (Trapnell et al. 2010). The log-median fragments per kilobase of exon per million fragments mapped (FPKM) for each site across ancestor replicates was chosen to represent the level of expression at that site.
Verification of identified mutations to determine false positive rate
Five lines were randomly selected to verify the mutations that were identified bioinformatically with Sanger sequencing (Supporting Information, Table S1). Primers were designed using Primer3 (Rozen and Skaletsky 1999) and PCR products destined for sequencing were cleaned using a standard Exo-SAP protocol (Bell 2008), and sequenced with an ABI BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Completed sequencing reactions were submitted to the Georgia Genomics Facility, and analyzed using an Applied Biosystems 3730xl 96-capillary DNA Analyzer.
Identification of ancestor mutations across lines to determine false negative rate
Mutations present in the ancestor relative to the reference should be present in every MA line. This implies that an ancestral mutation that is not found bioinformatically in an MA line is a false negative. To estimate the false negative error rate, the total number of ancestral mutations that should have been scored (equal to the product of the number of mutations in the ancestor and the number of MA lines) was compared to the number actually scored, and the probability of missing a mutation was calculated.
Relative mutation rate analysis
To investigate the effects of G/C content, replication time, and transcription rate, relative mutation rates were calculated across groups differing in these variables. G/C content was determined in nonoverlapping 10 kb windows across the entire genome. The number of SNMs was determined for each window and separated into 14 bins representing G/C content rounded down to the nearest 1%. Relative mutation rates were determined for each bin by determining the per base pair mutation rate for each bin and dividing by the observed genome-wide per base pair mutation rate. Bins were then separated into groups so that each group contained roughly equal numbers of SNMs, but not necessarily equal numbers of bins. Weighted averages and standard errors for each group were determined by weighting each bin by the amount of the genome covered by the bin. The same bins were used to determine relative mutation rates at A/T and G/C bases by calculating the per base pair mutation rate for just A/T and G/C bases.
Replication time was determined genome wide by assigning each SNM a replication time based on the closest origin of replication (Heichinger et al. 2006). SNMs were separated into 17 bins, each representing 1 min of S phase of the cell cycle. Relative mutation rates were then determined in an analogous manner as for G/C content.
Transcript abundance was determined genome wide and transcripts were separated into 15 bins based on abundance increasing from 0 to 3.5 in intervals of 0.25 log10FPKM. SNMs within genes were assigned a bin based on the transcript in which they reside; SNMs not mapping to a transcript were assigned a gene expression of 0. Relative mutation rate was then determined in an analogous manner as G/C content and replication time.
Data availability
Scripts written to process and analyze the data are available on GitHub (https://github.com/behrimg/Scripts/blob/master/Hall_Projects/Pombe/MA_Pipeline.txt) and raw sequences are available on the Sequence Read Archive at NCBI (BioProject SRP065886). All other data is available upon request.
Results
MA lines
The relationship between colony area and the number of cells in the colony did not differ between transfers 10 and 100 (data not shown). The regression between colony size and transfer number indicated that average colony size declined over the course of the experiment such that the number of cells at T100 was approximately half the number at T10, indicating an average reduction in per-generation growth rate of approximately 5%. A reduction in growth rate is expected as haploid MA lines accumulate deleterious mutations. In diploid S. cerevisiae, a trend toward reduced colony size was not observed (Joseph and Hall 2004), presumably because new mutations accumulated in the heterozygous state and thus had a smaller effect on fitness due to masking. The average number of cells in a colony was used to estimate the average effective population size in a MA line as 10.26 cells. This small effective population size implies that deleterious (beneficial) mutations with a fitness effect greater than ∼0.097 would be underrepresented (overrepresented) due to the action of selection (Hall et al. 2008; Lynch et al. 2008). Mutations with smaller fitness effects are expected to accumulate approximately at random in the MA lines. The average number of cells in a colony was also used to estimate that the MA experiment lasted 1952 cell generations.
Mapping of sequencing reads to the Sc. pombe reference genome revealed that coverage was uniform across all three chromosomes except for telomeric regions in those lines with average sequencing depth below 43x, where telomeric sequences appeared to be affected by amplification bias (Figure S1). After sequencing and identification of nucleotide variants, we discarded 17 of the 96 MA lines due either to contamination by a different microbial species (six lines) or to across MA line contamination (11 lines), which left 79 lines for further analysis. We note that for most of the contaminated lines (10 of 11), all nucleotide variants were shared, indicating that contamination likely occurred either at one of the last transfers, or when the lines were removed from the freezer for DNA sequencing.
Approximately 472 kbp, which is ∼3.8% of the sequenced genome, was excluded from the analysis to ensure precise identification of new mutations. This included the low-complexity sequence, comprising centromeres, telomeres, the mating type region, and the representative rDNA repeat belonging to the two tandem rDNA repeat arrays on chromosome III, which had been previously excluded from the 12.47 Mb reference genome (Wood et al. 2011). The exact coordinates of the regions removed from the analysis are listed in Table S2.
Differences between the MA ancestor and Sc. pombe reference genome
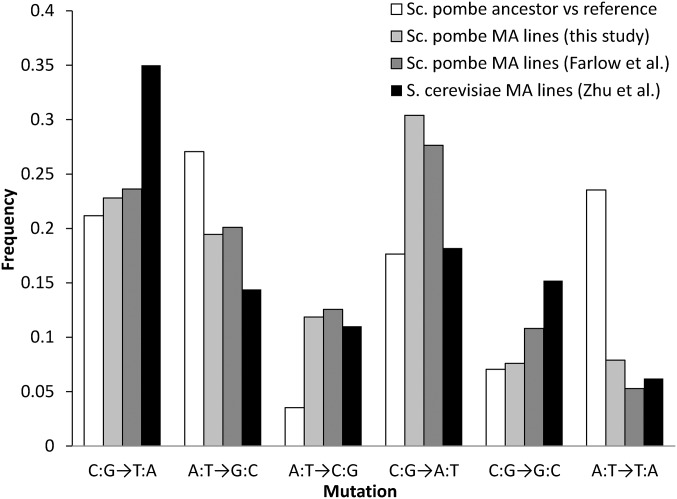
Sequencing of the ancestor revealed a total of 262 differences between it and the reference genome. These differences included 80 SNMs, 120 small insertions (< 50 bp), 42 small deletions (< 50 bp), six double SNMs, and 14 complex mutations (Figure S2). We define double SNMs as two SNMs that occur within 50 nucleotides of one another. Complex mutations are three or more SNMs and/or indels occurring within 50 nucleotides of one another. Both complex mutations and double SNMs were deemed to be nonindependent events and were thus analyzed separately (Schrider et al. 2011). Of the 80 SNMs, 39 were transitions and 41 were transversions, giving a transition to transversion ratio of 0.95. Within transitions, after correcting for genomic G/C content, and assuming that all changes were mutations from the reference strain sequence to the ancestral strain sequence, G:C → A:T or T:A mutations were 1.38 times more frequent than A:T → G:C or C:G mutations (Figure 1).
Figure 1.
Summary of mutations for each of six possible nucleotide changes for the ancestor (ATCC 26189) vs. the reference, the 79 mutation accumulation (MA) lines in this study, and the 96 Farlow et al. (2015) MA lines. Also shown are the mutation frequencies observed in the 145 diploid Saccharomyces cerevisiae MA lines (Zhu et al. 2014).
The distribution of SNMs across chromosomes was not significantly different from the expectation based on chromosome length (X2 = 5.45, P = 0.06). This result held whether we tested all three chromosomes, or just chromosomes I and II (X2 = 3.05, P = 0.06), which together represent 81% of the genome. The distribution of small insertions was nonrandom, with chromosome I (5.58 Mb) containing 68, which is greater than the 53 expected based on chromosome length (X2 = 7.603, P = 0.006). Other mutations, including small deletions, complex mutations, and double SNMs, did not show bias across chromosomes, after accounting for length (Figure S3).
Differences between the MA lines and the MA ancestor
Single nucleotide mutations:
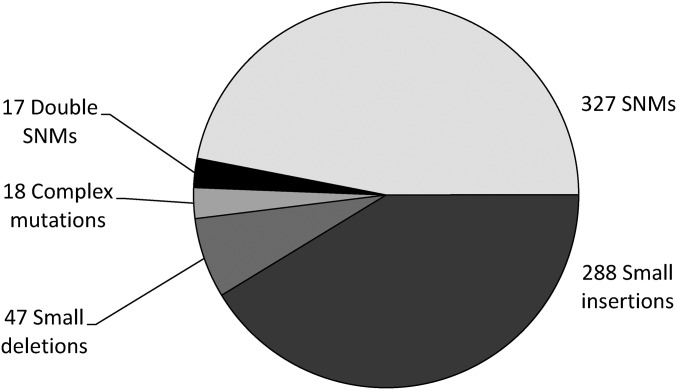
Across the 79 MA lines, 326 SNMs arose during MA, which gives the SNM rate (± 1 SE) for Sc. pombe as 1.70 ± 0.13 × 10−10 per base per generation (Figure 2 and Table S3). The SE of the estimate is based on the variance across MA lines. This mutation rate estimate does not include the 123 SNMs that were in double or complex mutations. Including SNMs in double and complex mutations increases the genome-wide mutation rate estimate to 2.34 ± 0.23 × 10−10 per base per generation.
Figure 2.
Summary of types of mutations identified across 79 MA lines. SNM, Single nucleotide mutation.
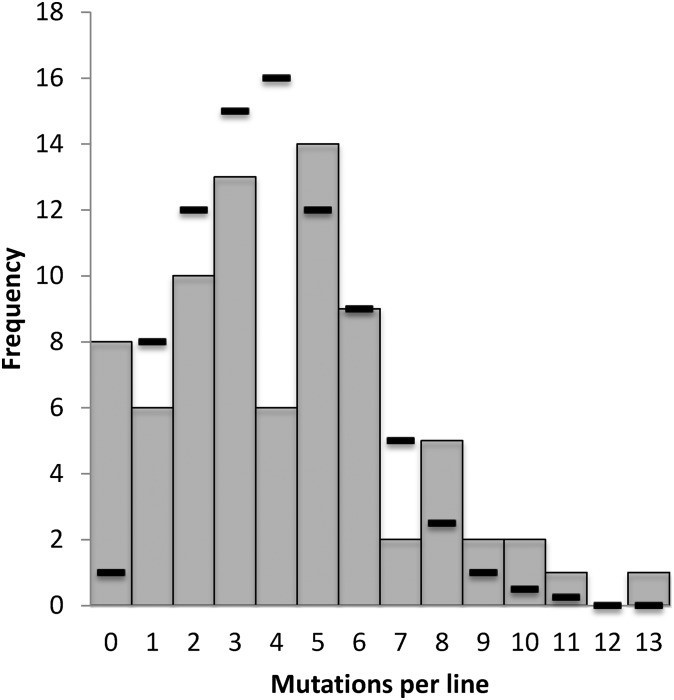
The number of SNMs varied across the 79 MA lines from zero (eight lines) to 13 (one line), with an average of 4.13 SNMs per line (ignoring doubles and complex mutations). Surprisingly, the distribution of mutations across the MA lines was not consistent with a Poisson distribution (χ2; P < 0.001). A negative binomial distribution (Gamma-Poisson), with mean = 4.13 and overdispersion parameter = 2.06, could not be rejected (χ2; P = 0.954, Figure 3). Using Akaike’s information criterion (AIC), the negative binomial was a substantially better fit to our data than the Poisson (Poisson AIC = 15.58, negative binomial AIC = 4.45; Poisson is 0.0034 as likely to explain the data). SNMs occurred at random with respect to protein-encoding genes: 52.4% of SNMs were in the 57% of the genome that is protein coding (Fisher’s exact, P = 0.27), and 3.6% of SNMs were in the 3% of the genome that is intronic sequence (Fisher’s exact, P = 0.83), suggesting that selection was inefficient during MA.
Figure 3.
Histogram of SNM numbers per line across the 79 MA lines. The distribution is consistent with a negative binomial (P = 0.95, χ2 test). Dashes represent the expected numbers for the best-fit negative binomial distribution (mean = 4.13, overdispersion = 2.06). The total number of SNMs across all 79 MA lines is 326.
The distribution of SNMs and all other mutation classes (small insertions, deletions, complex, double SNMs) across the three chromosomes were not significantly different from the expectation based on chromosome length.
Single nucleotide mutation biases:
Ignoring SNMs in complex and double mutations, we observed a transition to transversion (Ts/Tv) bias of 0.72 (Figure 1). Within transitions, G:C → A:T mutations were 2.02 times more frequent than A:T → G:C. Within transversions, G:C → T:A mutations were 4.55 times more frequent than A:T → C:G. Across transitions and transversions, G/C → A/T mutations were 2.97 times more frequent than A/T → G/C mutations. If the mutational process were the sole determinant of the G/C content in Sc. pombe, the equilibrium genome G/C content would be 25.14%, far less than the 36.06% observed in the reference genome.
G/C content has been shown to affect local mutation rate in rodents (Hurst and Williams 2000). However, in this study, we found no effect of G/C content on the local mutation rate. We determined the G/C content in 10-kb windows across the genome, and then separated the data into groups such that each contained a similar number of SNMs. Regardless of whether we split the data into two or three groups, we found no significant difference in mutation rate across them, suggesting that local G/C content has no effect on local mutation rate. Data for the two-group case where the division is for a GC content of 36% is shown in Figure S4A.
In humans, late replicating regions have higher mutation rates (Stamatoyannopoulos et al. 2009). We tested for a similar relationship and found that replication time did not affect mutation rate. We assigned each nucleotide in the genome a replication time based on the time at which the closest origin of replication (ori) initiates replication during mitosis (Heichinger et al. 2006). There is little variation in the replication initiation times of different oris in Sc. pombe, with all oris firing between 68 and 85 min after release from G2 arrest (Heichinger et al. 2006). Further, published initiation times are reported in 1-min intervals, and so we could not split times into smaller intervals. We separated replication times into three groups and found no significant differences in mutation rate for different replication times (Figure S4B).
Mutation rate has been shown to correlate with gene expression in S. cerevisiae and humans (Park et al. 2012), but see Zhu et al. (2014). We found that mRNA levels were not associated with mutation rate. Using RNA-seq data collected from our ancestor line, we examined whether gene expression, measured as transcript abundance, influenced local mutation rate. Transcription rates were split into 15 bins that were then separated into two groups. These two groups did not differ in mutation rate. Adding rRNA and tRNA genes, and assuming they are expressed in the highest bin, did not alter this conclusion (Figure S5).
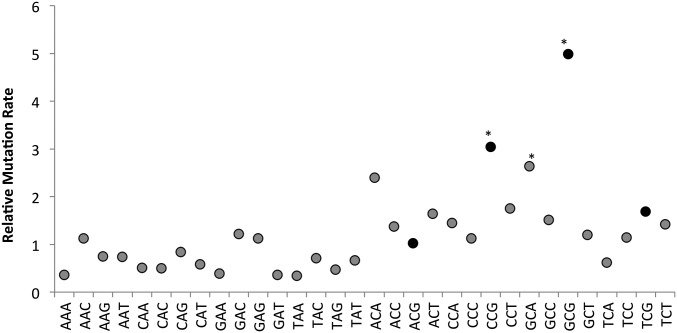
We did find an effect of trinucleotide context, which is the identity of the nucleotides immediately before and after the mutated nucleotide, on mutation rate (Zhu et al. 2014). Each nucleotide position within the genome, along with its neighboring bases, was assigned to one of the 64 trinucleotide possibilities. Ignoring strand orientation allowed us to reduce the 64 possibilities to 32 groups, defined such that complementary trinucleotides belong to the same group (e.g., GCA and TGC are in the same group). Mutation rates at the center position across the 32 groups were quite variable (Figure 4). We analyzed the 16 groups with A (or T) at the center position separately from those with G (or C) because C:G base pairs have a higher mutation rate than A:T base pairs, as is clearly apparent in Figure 4. After correcting for multiple comparisons, we found context did not affect mutation rate at A:T base pairs. In contrast, at C:G base pairs, mutation rates were increased in the CCG (equivalent to CGG), GCG (equivalent to CGC), and GCA (equivalent to TGC) trinucleotide groups (t-test; P = 0.0003, P = 0.0001, and P = 0.0009 respectively; Bonferroni corrected critical P-value = 0.0031). Interestingly, two of these trinucleotides are two of the four trinucleotide groups that contain a CpG dinucleotide, where the C is the at the center position. If increased CpG mutation rate was due to deamination of methylated cytosine (giving thymine), then we would expect CpGs to mutate to TpGs. However, of the 44 mutations occurring at CpG sites, 23 were C → A mutations (52.3%), 18 were C → T (40.1%) and three were C → G (6.6%); which is no different than expected based on the mutational spectrum of Sc. pombe (X2 = 1.76, P = 0.41). For the GCA trinucleotide group, C → A mutations were overrepresented relative to the mutational spectrum (18 of 22 mutations were C → A, 11 were expected; X2 = 8.91, P = 0.011).
Figure 4.
Mutation rate is affected by context. Relative SNM rate adjusted by its trinucleotide context. Trinucleotide classes represent mutation rates of both strand orientations. For example, aCa trinucleotide class includes overall mutation rate at aCa and tGt sites. Mutation rate is shown relative to the average SNM rate across all sites (= 1.7 × 10−10 per base, per generation). The average, relative mutation rate of 1.8 at G:C bases shows clear overall elevation over the corresponding rate of 0.66 at A:T bases. The four black-filled points are those trinucleotides with a C in the central position with a G as the 3′ neighbor. * P ≤ 0.003 Bonferroni corrected t-test.
Small insertion and deletion mutations:
Across the MA lines, we identified 335 small indels of less than 50 bp, including 288 insertions and 47 deletions. A complete list of indels is in Table S4. Average insertion size was 1.6 bp, while average deletion size was 3.1 bp, with insertions occurring six times more frequently than deletions, resulting in a net gain in DNA sequence across all lines of 340 bp. Small indels occurred as frequently as SNMs across the MA lines, and the resulting spontaneous indel rate, 0.174 × 10−9 indels/base/generation, is essentially identical to the mutation rate calculated for SNMs, ignoring double SNMs and complex mutations. Indels, however, were not randomly distributed with respect to genomic features. They were substantially underrepresented in protein coding sequence (observed: 33, expected: 191; Fisher’s exact test: P < 0.001), occurred as frequently as expected in introns (observed 21, expected 10; P = 0.064), and were over represented in noncoding regions (observed 288, expected 134; P < 0.001).
Effects of SNMs and indels:
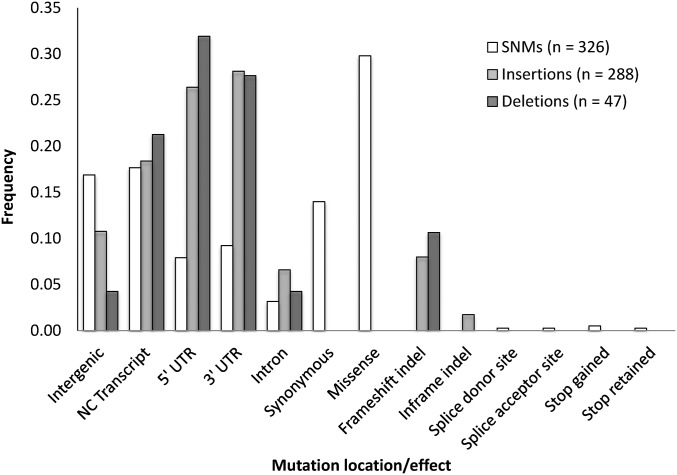
We annotated the expected functional effects of our SNM and small indel mutations using Ensembl’s VEP software (McLaren et al. 2010). For the 183 SNMs that occurred within coding regions, 53 were synonymous, 113 were missense, 12 were within introns, one was within a splice donor site, one was within a splice acceptor site, two were nonsense, and one changed a termination codon (UAA) into another termination codon (UAG) (Figure 5). The ratio of synonymous to missense changes (0.398) was higher but not statistically different from the expected ratio (0.323), computed using randomly generated protein coding sequences (Z-test for proportions: 1.104, P = 0.271) (Figure S6) (Graur 2003). This suggests selection did not substantially reduce the number of missense mutations captured during MA.
Figure 5.
Summary of the locations and predicted functional consequences of the 326 SNMs (excluding SNMs in double and complex mutations), 288 small insertions and 47 small deletions accumulated in the MA lines. To facilitate comparison across the three mutational classes, frequencies are shown. NC, noncoding; UTR transcribed, untranslated region.
Among the 54 small indels that occurred within protein and intronic coding sequence, seven were deletions, of which five were frameshift deletions and two were within introns. For the remaining 47 insertions, 23 were frameshift insertions, five were in-frame insertions ranging from 6 to 12 bp in length, and 19 were within introns (Figure 5). The proportion of indels in protein coding sequence that were in-frame was not significantly different from 1/3 (Fisher’s exact test P > 0.2). Again, these results suggest that selection did not substantially reduce the number of frameshift mutations captured during MA.
Double mutations, complex mutations, and structural variants:
As expected, no aneuploids were detected among the haploid MA lines. We specifically searched for other structural variants including medium-sized deletions (50–1000 bp), inversions, translocations and duplications using the DELLY software that predicts structural variants from short insert paired-end sequences (Rausch et al. 2012). We detected three medium-sized deletions, two of which were flanked by a number of SNMs (Table S5). In addition to multiple SNMs in the region of the three medium deletions, 16 other complex mutations and 17 separate double SNMs were also detected (Table S6 and Table S7). Complex mutations thus occurred at a rate of 8.32 × 10−12 per base per generation and double SNMs occurred at a rate of 8.84 × 10−12 per base per generation.
Error in identification of mutations
Our Sc. pombe lines were haploids and our sequencing coverage exceeded 25x in every line (average = 44x). Given previous work in diploid S. cerevisiae, we predicted we would have a very low likelihood of incorrectly calling a SNM or indel. To confirm a low false positive error rate, we randomly chose five lines and checked all predicted mutations using Sanger sequencing. Together, these lines contained 15 SNMs, 20 small insertions, three small deletions, one double SNM and one complex mutation. Sanger sequencing confirmed all of the mutations identified from next-generation sequencing, suggesting that the false positive error rate is likely is no larger than 0.025, and could be close to zero. We did find that, in the region of the complex mutation (MA Line 58, Chromosome II, positions 4,273,519–4,274,319), additional mutations not identified in next-generation sequencing data were present (i.e., false negatives). These mutations were probably not detected due to the difficulty in reference mapping sequencing reads that are too divergent from the reference genome. This conjecture is supported by the extreme loss of coverage that was observed in this 800-bp region. Just 80 bp on either side of the region coverage was 48x, while across the complex mutation coverage averaged 9x, with three nucleotides in the center only being represented by a single read. Our results for this complex mutation suggest that the numbers of detected changes present in the five complex mutations we identified between the ancestor and reference strain and the 18 in the MA lines, are underestimates of the number actually present.
We also examined the probability of false negatives for SNMs. This was done by asking how many of the 272 mutations that differ between the reference and the ancestor were not found in each of the 79 MA lines. Any mutation present in the ancestor that is not identified in an MA line is a false negative, assuming the probability that a mutation converting the ancestor base back to the reference base at any of these sites is negligible. Given our estimate of the mutation rate, the probability of a mutation occurring at any of these sites in any line during the experiment is ∼0.007, indicating that failure to detect a site that differed in the ancestor is almost certainly a false negative. For SNMs, the false negative rate was 0.0001, while for small insertions and small deletions it was 0.011 and 0.017, respectively. Thus, we may have underestimated the base substitution, small insertion and small deletion mutation rates by 0.01, 1.1 and 1.7%, respectively. No mutations were found in three or more MA lines that were not also found in the ancestor, indicating that we likely detected all of the mutations in the ancestor.
Discussion
Rate of mutation at single nucleotides
We expected to observe a higher SNM rate in Sc. pombe than in S. cerevisiae, based on data using reporter genes (Lynch 2010). Our estimate of the base pair mutation rate in Sc. pombe, from the 326 accumulated SNMs, is 1.70 ± 0.13 × 10−10 per base per generation, which is almost identical to the mutation rate of 1.67 × 10−10 per base per generation, based on 867 accumulated SNM mutations, in S. cerevisiae (Zhu et al. 2014). Our estimate is slightly, but significantly, lower than the 2.00 ± 0.10 × 10−10 mutation rate per base per generation estimate reported by Farlow et al. (2015), but that estimate did not remove SNMs in double and complex mutations, except for in flocculation-related genes. Including the 123 SNMS that we observed in double and complex mutations raises the genome-wide mutation rate estimate to 2.34 ± 0.23 × 10−10 per base per generation, which is not significantly different from the Farlow et al. (2015) estimate. In both studies, there was no effect of either local G/C content, transcript abundance, used as a proxy for gene expression, or timing of replication on the local mutation rate.
With our estimates of the mutation rate and the genome-wide estimate of π from Jeffares et al. (2015), we can calculate the effective population size for Sc. pombe as either 6.52 × 106 (including SNMs in double and complex mutations) or 8.80 × 106 (ignoring SNMs in double and complex mutations), which span the estimate of effective population size for S. cerevisiae, 8.53 × 106 (Liti et al. 2009). Thus, Sc. pombe and S. cerevisiae do not seem to differ very much in their effective population size, suggesting that selection will encounter the drift barrier (Sung et al. 2012a), the point at which the fitness advantage of new mutation is the same magnitude as the strength of genetic drift, at similar selection coefficients in both species. We note that our population size estimates are smaller than the estimate of 12 million reported in Farlow et al. (2015), the reason being that they utilize π calculated from intergenic regions, which is larger (0.0048) than the genome-wide π (0.0030) that we use here.
Bias in single nucleotide mutations
There are both similarities and differences between the SNM mutation biases in Sc. pombe and S. cerevisiae. The most obvious difference is that the most common SNM in S. cerevisiae is a C:G → T:A transition, while in Sc. pombe it is a C:G → A:T transversion (Figure 1). In addition, C:G → G:C transversions are more common, and A:T → G:C transitions less common in S. cerevisiae than in Sc. pombe. In contrast, both A:T → C:G and A:T → T:A transversions are similarly frequent in S. cerevisiae and Sc. pombe (Figure 1). Both species show an elevated rate of G/C → A/T, which points to oxidative damage as a major cause of mutagenesis (Cheng et al. 1992; Shen et al. 1994). Within our MA lines, G/C → A/T mutations occurred at a rate that was ∼three times greater than A/T → G/C mutations, which is the same as the bias observed by Farlow et al. (2015), and higher than the ∼two-fold bias seen in S. cerevisiae (Zhu et al. 2014). The equilibrium G/C content calculated from the mutation spectra is 25% and 32% in Sc. pombe and S. cerevisiae, respectively.
In both species the observed G/C content (35% in Sc. pombe and 38% in S. cerevisiae), is higher than predicted from mutation biases, suggesting that there is either selection for lower G/C content or some other mechanism, perhaps biased gene conversion, or selective constraint on protein sequence, which is causing the increase in genomic G/C content. Biased gene conversion is unlikely given the rarity of heterozygote formation in Sc. pombe (Farlow et al. 2015). Suggestive evidence for an additional force is present in the ancestor, whose G/C → A/T bias relative to the reference strain is 24% less than observed in the MA lines, indicating that G/C → A/T mutations are fixing at a lower rate than expected.
Rate and spectrum of indels
In our MA lines, we observed a 6.13-fold insertion bias. Insertions accounted for 86% of small indel events, consistent with previous observations of the GT repeat region in the ade6 gene of Sc. pombe, where 83% of indel events in wild-type were insertions (Mansour et al. 2001). This bias is identical to that observed by Farlow et al. (2015). A similar bias toward insertions has also been observed in C. elegans (3.75 insertions to deletions) (Denver et al. 2004) and haploid, but not diploid, S. cerevisiae (3.55 insertions to deletions) (Lynch et al. 2008). We do not know of any mechanism that would predict more insertion mutations based on ploidy, but if small deletions are more often strongly deleterious than small insertions, they would be underrepresented in MA experiments in haploid species, and in diploid species passaged with strong inbreeding (e.g., C. elegans). In haploid S. cerevisiae, despite their rarity, the size of small deletions was able to negate the effect of small insertions in terms of changes in genome size. This is not the case in Sc. pombe. Even though small deletions were twice as large as small insertions, small insertions occurred six times more frequently resulting in a net gain of 340 bp across the genome across all MA lines. This suggests that Sc. pombe’s current genome size is not at equilibrium with respect to an insertion/deletion mutational balance. Additionally, in the ancestor, small deletions may have been favored by selection: 17.3% of the differences between the ancestor and reference are small deletions compared to only 6.7% in the MA lines, which is a significant difference.
Similar to observations in C. elegans, Sc. pombe experiences small indels as often as SNMs (Denver et al. 2004), resulting in an indel rate that is 10-fold higher than haploid S. cerevisiae and almost 35-fold higher than diploid S. cerevisiae (Sc. pombe: 1.74 × 10−10 vs. haploid S. cerevisiae: 0.2 × 10−10, and diploid S. cerevisiae: 0.05 × 10−10 per base pair per generation). One hypothesis for the differences in indel rate may be due to differences in genome complexity between the two species. Sc. pombe’s genome is 60.2% protein coding (57% excluding introns) while S. cerevisiae’s is 71% protein coding (70.5% excluding introns) indicating a higher prevalence of noncoding DNA in Sc. pombe. Most indels identified in Sc. pombe and S. cerevisiae are within low complexity, intergenic regions such as microsatellites and mononucleotide runs. However, in an analysis of short simple repetitive sequences, there is little observed difference in the amount of repetitive sequence in S. cerevisiae and Sc. pombe (Karaoglu et al. 2005). An alternate hypothesis is that there are differences in mismatch repair pathways between these two species. However, both species contain MSH2 homologs, which are responsible for high fidelity repair of small indels (Rudolph et al. 1999), suggesting that this hypothesis would require differences in fidelity of the mismatch repair pathway, rather than presence vs. absence. A third hypothesis is that there may be differences in polymerase fidelity between Sc. pombe and S. cerevisiae causing differing rates of strand slippage resulting in Sc. pombe’s increased insertion rate. However, given similar effective population sizes, we do not expect fidelity to differ in these two species. Finally, the difference may be due to detection issues, especially the problem of false negatives. Our study yielded an indel rate that is three times as large as that reported by Farlow et al. (2015), which might be explained by false negatives. The methods of sequence mapping and variant calling differ between the two studies. Assuming there is no substantial difference in indel mutation rate between the two strains, the Farlow et al. (2015) study should have accumulated approximately 354 indels. Instead, only 119 were discovered. This difference suggests that our methods reliably identify a greater percentage of the mutations (i.e., we have fewer false negatives). We directly estimated our false negative error rate by determining whether an ancestral mutation, of which there were 272, was detected in all MA lines, which represents a total of 21,488 mutations tested across the 79 MA lines. For small insertions and deletions, the false negative error rates were 1.1% and 1.7%, respectively. A direct estimate of the false negative error rate was not performed in the study of Farlow et al. (2015), though they did introduce simulated SNMs into their reference assemblies to indirectly estimate a false negative error rate for SNMs. If the difference in indel rate estimates between the two studies is due to false negatives, then the Farlow et al. (2015) study missed about two thirds of all indels.
Complex mutations, double mutations, and aneuploidy
We also predicted an increase of mutations associated with double-stranded breaks; particularly double SNMs and complex mutations. Double-stranded breaks are lethal to a cell unless repaired. Repair can involve homologous recombination, which tends to be the preferred mechanism (Raji and Hartsuiker 2006), but can also utilize nonhomologous end-joining (NHEJ). Recombinational repair requires homologous copies of DNA. Compared to S. cerevisiae, Sc. pombe has 2.5 times fewer homologous copies available on average for recombinational repair of a double-strand break, and sometimes has no copies present (see Introduction). In the absence of homologous DNA, double-stranded breaks are repaired through NHEJ. Homologous recombination is considered an error-free method of repair, while nonhomologous end joining is considered to be error-prone, introducing small insertions or deletions when partially degraded ends inhibit precise repair (Cavero et al. 2007; Shrivastav et al. 2008).
The locations of complex mutations were not random. Six of the 18 complex mutations occurred within three of the nine flocculin genes. All of these six mutations occurred within the characteristic tandem repeats found in these genes (Verstrepen et al. 2005). The flocculin tandem repeats are known to cause replication errors and are highly prone to double-stranded breaks and, as a result, recombination (Verstrepen et al. 2005). Their propensity for double-stranded breaks could explain the repeated observation of complex mutations within them if complex mutations are, in fact, caused by mutagenic NHEJ repair of double-stranded breaks. To make sure that inferred mutations in the flocculin genes were not caused by gene conversion/recombination, we checked for and found no evidence of recombination between paralogs. An excess of mutations in flocculin genes was also observed by Farlow et al. (2015). They interpreted this finding as indicating positive selection for some unknown flocculation phenotype on their plates. They noted that only one mutation mapped to flocculation-related genes in S. cerevisiae (Zhu et al. 2014), suggesting that an increased mutation rate was not the underlying cause of the observed overrepresentation of mutations in these genes. However, mapping to flocculation genes in S. cerevisiae is problematic (M. G. Behringer, unpublished data) due to their repetitive nature, so mutations in these genes may have been missed in that study. In addition, given our estimate of the small effective population size within MA lines (10.26 cells), and the location of mutations in the MA lines indicating ineffectual selection, we favor increased mutation rate, caused by a high propensity for double-stranded breaks, as the explanation for more mutations occurring in these genes.
As we predicted given its haploid state, possession of only three chromosomes, and previous work (Niwa et al. 2006), there were no instances of aneuploidy. In Sc. pombe, aneuploidy has only been observed as a disomic haploid of chromosome III. Even if a disomic haploid had been fixed in a line at an intermediate transfer, it would likely have been unstable (Niwa and Yanagida 1985; Niwa et al. 2006) and thus rapidly lost. Interestingly, when S. cerevisiae is passaged as a mitotic haploid it tends to be very unstable (Lynch et al. 2008), and at large population size, where selection is effective, it reverts to a diploid state (Mable and Otto 2001). This instability is in contrast to the relative stability of diploid strains (Nishant et al. 2010; Zhu et al. 2014). This implies that aneuploidy is generally less likely to occur in a natural haploid than in a natural diploid. However, when a natural diploid is kept as a haploid strain artificially, disomies, or other steps toward diploidy, may be tolerated and perhaps even favored. It would be interesting to see if the instability of haploidy in S. cerevisiae, which is a natural diploid, would also be reflected in instability of a diploid Sc. pombe, which is a natural haploid.
Cytosine mutation in absence of methylation
One of the major surprises of the S. cerevisiae mutation spectrum is the high mutation rate observed at some C:G base pairs, particularly in CpG dinucleotides found in CCG and TCG trinucleotides. We observed a similar, unexpectedly high mutation rate at some C:G base pairs, especially CCG and GCG. In many eukaryotes a major cause of mutation at cytosine nucleotides is spontaneous deamination of methylated cytosines (5-mC) to thymine, which results in a T:G mismatch that can then be repaired, or replicated, to give a C:G → T:A substitution. Deamination of methylated cytosines was hypothesized to in part explain the elevated mutation rate of C:G base pairs in S. cerevisiae (Zhu et al. 2014), in spite of the fact that 5-mC is generally thought to be absent in this species (Antequera et al. 1984; Proffitt et al. 1984). The reason for considering the possibility that CpG mutation is caused by methylation is the low level of DNA methylation reported in S. cerevisiae (Tang et al. 2012). Sc. pombe is thought to lack 5-mC (Antequera et al. 1984), though, unlike S. cerevisiae, it does have a DNA methyltransferase homolog, pmt1 (Wilkinson et al. 1995), so methylation might occur. However, even if cytosines are methylated at a very low rate, our data suggest that methylation cannot explain the mutational bias at these sites. Methylated cytosines cause C:G → T:A transition mutations because of spontaneous deamination, not the observed C:G → A:T transversions. Additionally, the mutation biases at the C:G base pair in a CpG dinucleotide are no different from the biases at a C:G base pair that is not in a CpG context. Together, these observations suggest a mechanism other than deamination of methylated cytosines is driving the increased relative mutation rate at CpG dinucleotides. This suggests that methylation may not be the only cause of high mutation rates at CpG dinucleotides in other species. Surprisingly, the Farlow et al. (2015) study reported that C:G → T:A transitions were significantly more common at CpG sites than in the non-CpG context, which is in sharp contrast to our finding, where both contexts show the same spectrum of mutations.
Spectrum of single nucleotide mutations
If differences between the ancestor and the reference represent both neutral and selected mutations, then the mutation spectrum might differ from that observed among the MA lines. In the MA lines SNMs represent 46.9% of the observed mutations, while in the ancestor they are significantly less common. The lower proportion of SNMs among differences between the ancestor and reference suggests that many of the SNMs that arose in the lineages linking the two strains were deleterious and were thus prevented from fixing. The location of the SNMs in the genome with respect to exon vs. intron, and noncoding regions is similar in both the ancestor and MA lines, suggesting that many of SNMs present in the ancestor relative to the reference are likely neutral (Figure 2 and Figure S6).
Divergence of the ancestor and reference strains
Both the ancestor and the sequenced reference strains have the same strain designation, 972 h–. However, given that they differ in sequence, they must have been separately passaged for some period of time. If all 80 of the observed SNM differences between the ancestor and the reference strains are neutral, that would suggest the two strains are between 26,800 and 36,900 generations diverged, depending on the mutation rate used to estimate the number of generations. Assuming that a strain might be passaged at most 50 times per year (∼1000 cell generations), this would indicate that the strains are almost 32 years separated (16 years of independent culturing in the lab if both were being passaged). The 972 h– strain was brought into the lab in the 1940s (Leupold 1950), so this number of generations is certainly possible. Alternatively, these mutations may have accumulated during refrigerative storage, though we know of no data to test this hypothesis. Another possibility is that the ancestor and reference strains are fewer generations diverged, with many of the sequence differences having been fixed by positive selection. Selection seems to be a less likely explanation because the distribution of effects of mutational differences between the ancestor and reference is strikingly similar to that for the MA lines, where selection is known to be minimal (Figure S6).
Reproducibility of MA experiments
Throughout, we have compared our results to those of a recently published mutation accumulation study in Sc. pombe (Farlow et al. 2015). It is striking how the results from the two studies are so similar, especially given that we used different strains (972 h– vs. ED668, BG_0000H8 h+), and passaged them at different temperatures (30 vs. 32°) on different media (YPD vs. YES) at different intervals (every 2 vs. every 3–4 days). In our study, the specifics of the protocol were chosen in order to be identical to the conditions used in the S. cerevisiae MA study (Zhu et al. 2014) to facilitate a comparison of the results between these two species. Fortuitously, 30° is the optimal growth temperature for our Sc. pombe isolate (http://www.atcc.org/Products/All/26189.aspx).
Comparing the two studies, the SNM mutation rate estimates are essentially identical (when we include SNMs from double and complex mutations), the SNM spectrum (Figure 1) and the indel bias are essentially identical, and mutations in flocculin genes are overrepresented. Further, neither study found an effect of G/C content, transcription levels, scored as transcript abundance, or replication timing on local SNM rate. One discrepancy between the studies, the three-fold difference in indel rate, may be due to a high rate of missed mutations in their study and may thus not be a real difference; these indels could perhaps be discovered upon reanalysis. The long list of similar findings is reassuring given that MA experiments are time-consuming to repeat. However, there is one striking disparity between the two studies. CpG mutations in the Farlow et al. (2015) study were biased in favor of C:G → T:A, even though C:G → A:T mutations were more common when averaged across all C:G sites. In contrast, we found C:G → A:T mutations were more common in CpG dinucleotides, and the spectrum of mutations at CpG sites did not vary from the genome-wide mutation spectrum. This is an important difference because our study indicates that it is not methylation of cytosine that is driving the higher mutation rate at CpG sites, while the results of the Farlow et al. (2015) study suggest that methylation could be the cause. In summary, we find that Sc. pombe and S. cerevisiae have essentially identical base pair mutation rates, which, coupled with genome-wide estimates of within-species polymorphisms, suggests their effective population sizes are similar. We observe considerable differences in the mutation spectrum and indel rate, suggesting that there are species-specific differences in factors affecting mutation bias and indel rate between these two species. Additionally, we note that the rates of small insertions vs. deletions predict a growing genome, suggesting either that the genome size in Sc. pombe is not at equilibrium, or that rare large deletions offset increases caused by small indel bias. The sample size of captured large insertions and deletions was insufficient to test this hypothesis. We also found that CpG sites are highly mutagenic, but the mutation bias at these sites is not caused by deamination of methylated cytosine, suggesting another factor makes these sites prone to mutation. The results of this MA experiment are reassuringly similar to the results reported from a recently published MA experiment using Sc. pombe, suggesting that these experiments can give repeatable estimates despite differences in starting strains and growth conditions.
Acknowledgments
We would like to thank the Associate Editor and two anonymous reviewers for very helpful comments on a previous version of this manuscript. This work was supported by the National Institutes of Health grants R01GM097415 and T32 GM007103.
Author contributions: D.W.H. designed the research. M.G.B. performed the research and did the bioinformatics. D.W.H. and M.G.B. performed the data analysis and wrote the manuscript.
Footnotes
Supporting information is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.022129/-/DC1
Communicating editor: J. C. Fay
Literature Cited
- Andolfatto P., 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437: 1149–1152. [DOI] [PubMed] [Google Scholar]
- Antequera F., Tamame M., Villanueva J., Santos T., 1984. DNA methylation in the fungi. J. Biol. Chem. 259: 8033–8036. [PubMed] [Google Scholar]
- Aronesty, E., 2011 ea-utils: Command-line tools for processing biological sequencing data. Available at: http://code.google.com/p/ea-utils.
- Bell J., 2008. A simple way to treat PCR products prior to sequencing using ExoSAP-IT. Biotechniques 44: 834. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Verdine G. L., 1994. DNA methyltransferases. Curr. Opin. Cell Biol. 6: 380–389. [DOI] [PubMed] [Google Scholar]
- Blumenstiel J. P., Noll A. C., Griffiths J. A., Perera A. G., Walton K. N., et al. , 2009. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics 182: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J., Chlebowicz-Sledziewska E., Fangman W. L., 1984. Cell cycle phases in the unequal mother/daughter cell cycles of Saccharomyces cerevisiae. Mol. Cell. Biol. 4: 2529–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavero S., Chahwan C., Russell P., 2007. Xlf1 is required for DNA repair by nonhomologous end joining in Schizosaccharomyces pombe. Genetics 175: 963–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. C., Cahill D. S., Kasai H., Nishimura S., Loeb L. A., 1992. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G—-T and A—-C substitutions. J. Biol. Chem. 267: 166–172. [PubMed] [Google Scholar]
- Daley J. M., Palmbos P. L., Wu D., Wilson T. E., 2005. Nonhomologous end joining in yeast. Annu. Rev. Genet. 39: 431–451. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Denver D. R., Feinberg S., Steding C., Durbin M., Lynch M., 2006. The relative roles of three DNA repair pathways in preventing Caenorhabditis elegans mutation accumulation. Genetics 174: 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Wilhelm L. J., Howe D. K., Gafner K., Dolan P. C., et al. , 2012. Variation in base-substitution mutation in experimental and natural lineages of Caenorhabditis nematodes. Genome Biol. Evol. 4: 531–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery E. J., Snell E. A., Bapteste E., Delsuc F., Philippe H., 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc. Natl. Acad. Sci. USA 101: 15386–15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farlow A., Long H., Arnoux S., Sung W., Doak T. G., et al. , 2015. The spontaneous mutation rate in the fission yeast Schizosaccharomyces pombe. Genetics 201: 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., et al. , 1996. Life with 6000 genes. Science 274: 546–567. [DOI] [PubMed] [Google Scholar]
- Gordon, A., and G. Hannon, 2010 Fastx-toolkit. FASTQ/A short-reads preprocessing tools (unpublished). Available at: http://hannonlab. cshl. edu/fastx_toolkit.
- Graur, D., 2003 Single-base mutation, pp. 287–290 in Nature encyclopedia of the human genome, edited by D. Cooper, Macmillan Publishers, Nature Publishing Group, London. [Google Scholar]
- Greene E. A., Codomo C. A., Taylor N. E., Henikoff J. G., Till B. J., et al. , 2003. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics 164: 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. W., Mahmoudizad R., Hurd A. W., Joseph S. B., 2008. Spontaneous mutations in diploid Saccharomyces cerevisiae: another thousand cell generations. Genet. Res. 90: 229–241. [DOI] [PubMed] [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Evol. Syst. 40: 151–172. [Google Scholar]
- Heckman D. S., Geiser D. M., Eidell B. R., Stauffer R. L., Kardos N. L., et al. , 2001. Molecular evidence for the early colonization of land by fungi and plants. Science 293: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Heichinger C., Penkett C. J., Bähler J., Nurse P., 2006. Genome‐wide characterization of fission yeast DNA replication origins. EMBO J. 25: 5171–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R., Petrov D. A., 2008. Selection on codon bias. Annu. Rev. Genet. 42: 287–299. [DOI] [PubMed] [Google Scholar]
- Ho S. Y., Phillips M. J., Cooper A., Drummond A. J., 2005. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol. Biol. Evol. 22: 1561–1568. [DOI] [PubMed] [Google Scholar]
- Hoffman P. D., Leonard J. M., Lindberg G. E., Bollmann S. R., Hays J. B., 2004. Rapid accumulation of mutations during seed-to-seed propagation of mismatch-repair-defective Arabidopsis. Genes Dev. 18: 2676–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbeck S. L., Strathern J. N., 1997. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics 147: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T., 2000. DNA damage and cell cycle control in Schizosaccharomyces pombe. Mutat. Res. Fundam. Mol. Mech. Mutagen. 451: 211–226. [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Williams E. J. B., 2000. Covariation of GC content and the silent site substitution rate in rodents: implications for methodology and for the evolution of isochores. Gene 261: 107–114. [DOI] [PubMed] [Google Scholar]
- Jeffares D. C., Rallis C., Rieux A., Speed D., Převorovský M., et al. , 2015. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nature Genetics 47: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. B., Hall D. W., 2004. Spontaneous mutations in diploid Saccharomyces cerevisiae more beneficial than expected. Genetics 168: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaoglu H., Lee C. M. Y., Meyer W., 2005. Survey of simple sequence repeats in completed fungal genomes. Mol. Biol. Evol. 22: 639–649. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen J. H., Rein I. D., Rothe C., Stokke T., Grallert B., et al. , 2011. Cell-cycle analysis of fission yeast cells by flow cytometry. PLoS One 6: e17175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneeff M., Dellaert L., Van der Veen J., 1982. EMS-and relation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. Fundam. Mol. Mech. Mutagen. 93: 109–123. [DOI] [PubMed] [Google Scholar]
- Leupold U., 1950. Die vererbung von homothallie und heterothallie bei Schizosaccharomyces pombe. C. R. Trav. Lab. Carlsberg., Ser. Physiol. 24: 381–480. [Google Scholar]
- Levinson G., Gutman G. A., 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4: 203–221. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., 2010. Evolution of the mutation rate. Trends Genet. 26: 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable B. K., Otto S. P., 2001. Masking and purging mutations following EMS treatment in haploid, diploid and tetraploid yeast (Saccharomyces cerevisiae). Genet. Res. 77: 9–26. [DOI] [PubMed] [Google Scholar]
- Mansour A. A., Tornier C., Lehmann E., Darmon M., Fleck O., 2001. Control of GT repeat stability in Schizosaccharomyces pombe by mismatch repair factors. Genetics 158: 77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., et al. , 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20: 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., et al. , 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1930. Types of visible variations induced by X-rays in Drosophila. J. Genet. 22: 299–334. [Google Scholar]
- Nachman M. W., Crowell S. L., 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., 1979. A control acting over the initiation of DNA replication in the yeast Schizosaccharomyces pombe. J. Cell Sci. 36: 155–168. [DOI] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant K., Wei W., Mancera E., Argueso J. L., Schlattl A., et al. , 2010. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 6: e1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O., Yanagida M., 1985. Triploid meiosis and aneuploidy in Schizosaccharomyces pombe: an unstable aneuploid disomic for chromosome III. Curr. Genet. 9: 463–470. [Google Scholar]
- Niwa O., Tange Y., Kurabayashi A., 2006. Growth arrest and chromosome instability in aneuploid yeast. Yeast 23: 937–950. [DOI] [PubMed] [Google Scholar]
- Park C., Qian W., Zhang J., 2012. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 13: 1123–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt J. H., Davie J. R., Swinton D., Hattman S., 1984. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol. Cell. Biol. 4: 985–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raji H., Hartsuiker E., 2006. Double and S. Hattman, 1984 5-Methylogous recombination in Schizosaccharomyces pombe. Yeast 23: 963–976. [DOI] [PubMed] [Google Scholar]
- Rausch T., Zichner T., Schlattl A., Stütz A. M., Benes V., et al. , 2012. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28: i333–i339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E. S., et al. , 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 1999 Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Methods in Molecular Biology, Vol. 132: Bioinformatics methods and protocols, edited by S. Misener, and S. A. Krawetz. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- Rudolph C., Kunz C., Parisi S., Lehmann E., Hartsuiker E., et al. , 1999. The msh2 Gene of Schizosaccharomyces pombe Is Involved in Mismatch Repair, Mating-Type Switching, and Meiotic Chromosome Organization. Mol. Cell. Biol. 19: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M. T., Roles A., Conner J. K., Shaw R. G., Shaw F. H., et al. , 2012. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution 66: 2335–2339. [DOI] [PubMed] [Google Scholar]
- Saxer G., Havlak P., Fox S. A., Quance M. A., Gupta S., et al. , 2012. Whole genome sequencing of mutation accumulation lines reveals a low mutation rate in the social amoeba Dictyostelium discoideum. PLoS One 7: e46759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C. A., Rasband W. S., Eliceiri K. W., 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Hourmozdi J. N., Hahn M. W., 2011. Pervasive multinucleotide mutational events in eukaryotes. Curr. Biol. 21: 1051–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J.-C., Rideout W. M., Jones P. A., 1994. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22: 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastav M., De Haro L. P., Nickoloff J. A., 2008. Regulation of DNA double-strand break repair pathway choice. Cell Res. 18: 134–147. [DOI] [PubMed] [Google Scholar]
- Slater M. L., Sharrow S. O., Gart J. J., 1977. Cell cycle of Saccharomyces cerevisiae in populations growing at different rates. Proc. Natl. Acad. Sci. USA 74: 3850–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos J. A., Adzhubei I., Thurman R. E., Kryukov G. V., Mirkin S. M., et al. , 2009. Human mutation rate associated with DNA replication timing. Nat. Genet. 41: 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern J. N., Shafer B. K., McGill C. B., 1995. DNA synthesis errors associated with double-strand-break repair. Genetics 140: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012a Drift-barrier hypothesis and mutation-rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W., Tucker A. E., Doak T. G., Choi E., Thomas W. K., et al. , 2012b Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc. Natl. Acad. Sci. USA 109: 19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Gao X.-D., Wang Y., Yuan B.-F., Feng Y.-Q., 2012. Widespread existence of cytosine methylation in yeast DNA measured by gas chromatography/mass spectrometry. Anal. Chem. 84: 7249–7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L., 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., et al. , 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen K. J., Jansen A., Lewitter F., Fink G. R., 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37: 986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson C. R. M., Bartlett R., Nurse P., Bird A. P., 1995. The fission yeast gene pmt1+ encodes a DNA methyltransferase homologue. Nucleic Acids Res. 23: 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.-A., Lyne M., Lyne R., et al. , 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415: 871–880. [DOI] [PubMed] [Google Scholar]
- Wood V., Harris M. A., McDowall M. D., Rutherford K., Vaughan B. W., et al. , 2011. PomBase: a comprehensive online resource for fission yeast. Nucleic Acids Res. 40: D695–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. O., Siegal M. L., Hall D. W., Petrov D. A., 2014. Precise estimates of mutation rate and spectrum in yeast. Proc. Natl. Acad. Sci. USA 111: E2310–E2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Scripts written to process and analyze the data are available on GitHub (https://github.com/behrimg/Scripts/blob/master/Hall_Projects/Pombe/MA_Pipeline.txt) and raw sequences are available on the Sequence Read Archive at NCBI (BioProject SRP065886). All other data is available upon request.