Abstract
FG nucleoporins (Nups) are the class of proteins that both generate the permeability barrier and mediate selective transport through the nuclear pore complex (NPC). The FG Nup family has 11 members in Saccharomyces cerevisiae, and the study of mutants lacking different FG domains has been instrumental in testing transport models. To continue analyzing the distinct functional roles of FG Nups in vivo, additional robust genetic tools are required. Here, we describe a novel collection of S. cerevisiae mutant strains in which the FG domains of different groups of Nups are absent (Δ) in the greatest number documented to date. Using this plasmid-based ΔFG strategy, we find that a GLFG domain-only pore is sufficient for viability. The resulting extensive plasmid and strain resources are available to the scientific community for future in-depth in vivo studies of NPC transport.
Keywords: nuclear pore complex, FG nucleoporin, S. cerevisiae
The nuclear pore complex (NPC) is the essential, conserved, selective portal for nucleocytoplasmic transport in eukaryotic cells. By controlling transport across the NPC and maintaining the separation of transcription and translation machinery, intricate levels of gene regulation are supported in both single and multicellular eukaryotic organisms (reviewed in Raices and D’Angelo 2012). The 60–120 MDa NPC complex is built from multiple copies of a conserved set of ∼30 nuclear pore proteins (nucleoporins, Nups; reviewed in Field et al. 2014). Nups are organized into subcomplexes that assemble to generate a transport channel across the nuclear envelope (NE) with nuclear basket and cytoplasmic filament structures extending from the NE (Figure 1). Diverse technologies have been used to enhance our understanding of how structural Nups interact to build the NPC scaffold (Alber et al. 2007; Field et al. 2014; Chug et al. 2015; Stuwe et al. 2015). However, despite extensive study using a variety of approaches, questions remain regarding how the NPC forms a barrier to nonspecific transport of large macromolecules (>40 kDa) while at the same time facilitating specific import and export of molecules against concentration gradients (Rout et al. 2000; Yamada et al. 2010; Hulsmann et al. 2012; Lim et al. 2015). Importantly, the combined use of in vivo and in vitro experimental approaches is critical to fully unravel the mechanisms for nuclear transport and to define discrete Nup functions in a cell.
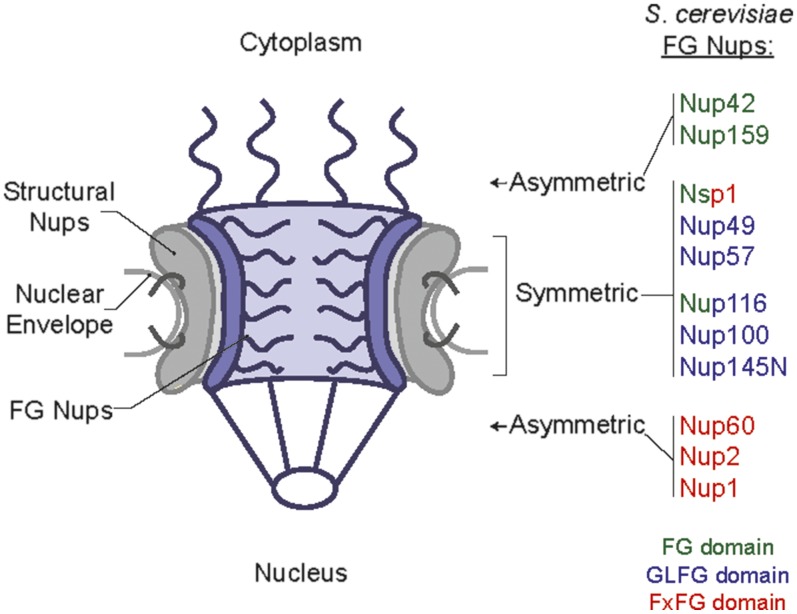
Figure 1.
Schematic of NPC depicting relative structural location of FG Nups, based on Rout et al. (2000) with the image adapted from Adams and Wente (2013). FG Nups are color-coded based on the type of FG repeats enriched in their FG domains: Green, FG; Blue, GLFG; Red, FxFG. Nsp1 contains both FG and FxFG domains, and Nup116 contains both FG and GLFG domains.
The FG Nups (11 members in Saccharomyces cerevisiae and humans) are the class of NPC proteins that both generate the NPC permeability barrier and provide binding sites for facilitated transport (Hulsmann et al. 2012; Lord et al. 2015; reviewed in Terry and Wente 2009). Each FG Nup contains an unstructured domain with multiple phenylalanine-glycine (FG) repeat motifs separated by ∼10–20 spacer residues comprised mostly of polar amino acids. The 11 FG Nups are characterized by different types of FG repeat motifs (classified into FG; glycine-leucine-phenylalanine-glycine, GLFG; or phenylalanine-any-phenylalanine-glycine, FxFG domains; reviewed in Rout and Wente 1994). Unless specified, in this report, we use the terminology “FG” generically to refer to the entire family of FG, FxFG, and GLFG Nups or when referring to multiple FG domains in a subcomplex. In each FG Nup, structured region(s) flank the FG domain to allow interaction with scaffold Nups. These structural domains effectively anchor FG Nups at discrete NPC sites, either symmetrically in the channel or asymmetrically at the cytoplasmic or nuclear face (Figure 1; Rout et al. 2000). Furthermore, a trio of highly conserved symmetric FG Nups (in S. cerevisiae: Nsp1, Nup49, and Nup57; in vertebrates: Nup62, Nup58/Nup45, and Nup54) forms a subcomplex through interactions of their coiled-coil structural domains (Chug et al. 2015; Stuwe et al. 2015).
The unstructured FG domains are modeled to extend into the NPC transport channel (reviewed in Terry and Wente 2009; Kabachinski and Schwartz 2015). For facilitated movement through the NPC, specialized transport receptors bind both the cargo and the FG repeats of FG domains, allowing entry into and through the FG domain network (reviewed in Field et al. 2014; Kabachinski and Schwartz 2015). Directionality of transport is mediated by additional soluble factors found at the NPC faces, or in the nucleus or cytoplasm (reviewed in Kabachinski and Schwartz 2015). With regard to roles in inhibiting the diffusion of macromolecules, the vertebrate GLFG Nup98 is critically important (Hulsmann et al. 2012) and the S. cerevisiae orthologs Nup116 and Nup100 also contribute to the permeability barrier (Lord et al. 2015). Thus, due to their bifunctional role in inhibiting diffusion of molecules and providing binding sites for transport receptors, FG domains constitute the fundamental basis for selective nucleocytoplasmic trafficking.
To date, S. cerevisiae is an important model system for investigating FG domain function in vivo, and diverse mutant construction approaches have been developed over the nearly three decades of study. Analyses of strains generated with entire genes deleted found that some FG Nup encoding genes are individually essential (Hurt 1988; Davis and Fink 1990; Wente et al. 1992; Del Priore et al. 1997). Thus, for functional studies with full gene deletions, analysis is limited to nonessential genes. Early studies also used plasmid-based expression of nup FG domain deletion (ΔFG) alleles to complement lethal chromosomal nup null mutants, and demonstrated that most individual FG domains can be removed with no loss in cell viability (Nehrbass et al. 1990; Grandi et al. 1995; Iovine et al. 1995; Del Priore et al. 1997). Indeed, most plasmid-based individual ΔFG strains with only the FG domain absent have minimal growth and transport defects (reviewed in Terry and Wente 2009). Given such functional redundancy within the NPC, to analyze FG domain function, multiple combined deletions of sequences encoding different FG domains must be included within a given strain. However, with 11 FG Nups, the availability of auxotrophic markers to maintain multiple plasmids, each encoding individual NUP genes, has limited analysis using such a strategy to only a few Nups within one strain.
To overcome these limitations, we originally developed a collection of S. cerevisiae mutants wherein NUP genes, with only the sequence encoding the respective FG domain deleted (ΔFG), are expressed from the endogenous chromosomal locus (Strawn et al. 2004; Terry and Wente 2007). In this approach, ΔFG alleles were generated by replacement of the FG domain-encoding region of the NUP gene with a floxed SpHIS5 “replacement” cassette, selection on media lacking histidine, and subsequent looping out of the SpHIS5 sequence by expression of Cre recombinase (Strawn et al. 2004). The replacement cassette also included sequence encoding one of four small epitope tags (FLAG, myc, T7, or HA) that was retained with the remaining loxP sequence after SpHIS5 was looped out. The resulting in-frame ΔFG gene expressed a protein with both the respective epitope tag and the translated loxP sequence, “TTLNITSYNVCYTKLL”, in place of the FG domain. By classic yeast genetic strategies, ΔFG alleles were then combined to generate higher-order, multiple ΔFG mutant strains (Strawn et al. 2004). Deletion of all five asymmetric FG domains results in a mutant strain with minimal growth and transport defects. Subsequent analysis went further to remove one or two symmetric FG domains from the background where all asymmetrical FG domains were deleted from Nup1, Nup2, Nup60, Nup42, and Nup159 together (Terry and Wente 2007). Functional analysis of such multiple, higher order ΔFG mutants for perturbations in the transport of different import and export cargos revealed that the absence of specific FG domains leads to unique transport defects (Terry and Wente 2007). Overall, FG domains serve specialized roles during transport, but it is unknown what attributes (FG type, spacer sequence, location within the NPC) lead to these particular functions.
Although the chromosomal ΔFG mutant strains have been instrumental in NPC functional analysis, they have several important caveats. First, chromosomal deletions preclude easy modification of genes in comparison to plasmid-based expression. Second, the remaining epitope and loxP tags result in nonspecific defects in some of the higher-order multiple ΔFG mutant strains. For instance, we previously reported that the lethality of T7-loxP-nup1ΔFxFG myc-loxP-nup2ΔFxFG myc-loxP-nup60ΔFG HA-loxP-nup42ΔFG myc-loxP-nup159ΔFG T7-loxP-nup49ΔFG is rescued by plasmid-based expression of untagged nup49ΔFG (Terry and Wente 2007). Therefore, our goal in this study was to generate a new collection of ΔFG mutants which (1) avoid indirect effects from epitope or loxP tagging during strain construction, (2) allow straightforward future mutational analysis of the sequences encoding individual domains, and (3) enable functional analysis of the resulting mutants.
We report here a new approach based on chromosomal null alleles complemented by plasmid-based expression of ΔFG nups, wherein each plasmid encodes multiple FG Nups that are colocated in specific NPC substructures. Using this strategy, we find that the FG domains of the Nsp1-Nup49-Nup57 subcomplex and those located exclusively at the nuclear (Nup1, Nup2, and Nup60) and cytoplasmic faces (Nup42, and Nup159) of the NPC can all be deleted without loss of viability. Although harboring severe growth defects, these deletions result in a new GLFG domain-only NPC. This collection will be of use to the community and set the stage for future experiments further probing of FG domain function in vivo.
Results and Discussion
Our efforts focused on deleting the FG domains of the conserved Nsp1-Nup49-Nup57 subcomplex in combination with deletion of FG domains from the nuclear (Nup1, Nup2, Nup60) and cytoplasmic (Nup159, Nup42) faces of the NPC. Analysis of such an octameric (eight) ΔFG mutant was not technically possible via former approaches. The basic strategy underlying the generation of a new collection of haploid S. cerevisiae ΔFG mutants included: (1) deletion of the entire endogenous FG NUP gene in the presence of a plasmid expressing the corresponding wild type (WT) FG NUP, and (2) shuffling the WT FG NUP plasmid for respective ΔFG nup constructs. Plasmids were engineered to allow expression of multiple FG NUP genes with their respective endogenous 5′ and 3′ UTRs (Figure 2A). Importantly, this plasmid-based expression strategy should not alter Nup stoichiometry within the NPC, because sequence encoding the anchoring structured domains is still present in ΔFG nup constructs.
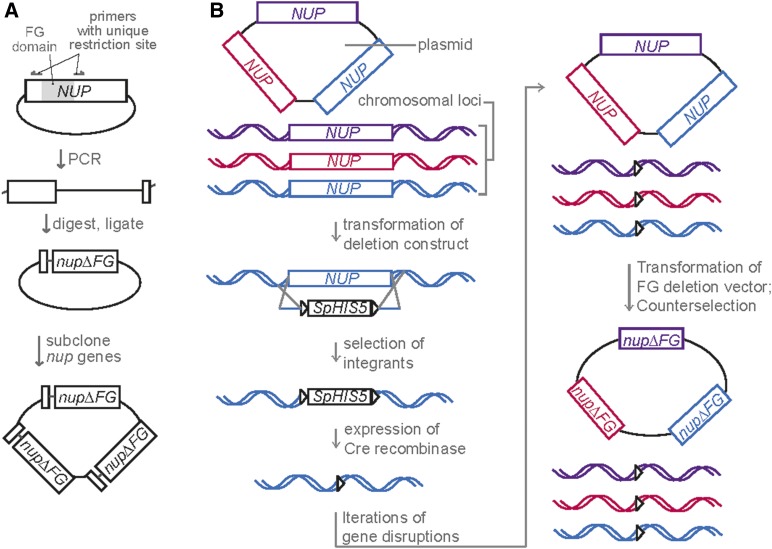
Figure 2.
(A) Schematic of ΔFG plasmid construction. Centromeric plasmids encoding a WT NUP gene with its endogenous 5′ and 3′ UTR were PCR amplified with primers that annealed outside of the FG domain and generated a unique in-frame restriction site. PCR products were cut and ligated back together to generate the ΔFG plasmid. ΔFG nups or WT NUPS were subcloned into one plasmid encoding multiple genes (Table 2). (B) Schematic depicting ΔFG strain construction. Plasmids harboring multiple NUP genes were transformed into parent strains followed by disruption of the chromosomal ORF with sequence encoding floxed Schizosaccharomyces pombe HIS5 (SpHIS5). SpHIS5 was then looped out by transformation with a plasmid for inducible expression of Cre recombinase. Iterative transformation, disruption, and SpHIS5 recycling cycles were used to generate indicated strains. Strains were subsequently transformed with ΔFG nup plasmids and counterselected.
The specific combinations of FG NUP or ΔFG nup genes cloned into a single expression plasmid was chosen based on the Nups, common physical association in NPC subcomplexes and/or NPC substructural localization (Figure 1 and Figure 2A). One set of plasmids harbored the three genes encoding the three FG Nups of the symmetric Nsp1 subcomplex: Nsp1, Nup49, and Nup57 (Grandi et al. 1995) (designated as NSP1/NUP49/NUP57, or nsp1/nup49/nup57ΔFG when lacking the FG domains). A second set contained genes encoding the two cytoplasmic-oriented FG Nups: Nup159 and Nup42 (designated as C-WT, or CΔFG when lacking the FG domains), and a third, the three nuclear-oriented FG Nups: Nup1, Nup2, and Nup60 (designated as N-WT, or NΔFG when lacking the FG domains).
By classic mating and sporulation, we first generated a triple deletion strain in which the endogenous chromosomal locus encoding each of the Nsp1-Nup49-Nup57 complex members was deleted in the presence of single WT NUP plasmids. The individual plasmids were then exchanged for a NSP1/NUP49/NUP57 plasmid in the nsp1Δ nup49Δ nup57Δ triple mutant, which was subsequently exchanged for an nsp1/nup49/nup57ΔFG plasmid (Figure 3 and Table 1). We analyzed growth of the resulting nsp1/nup49/nup57ΔFG mutant by serially diluting equal numbers of cells onto rich media and growing the cells at the indicated temperatures (Figure 4A). The nsp1/nup49/nup57ΔFG mutant with the simultaneous deletion of all three of the FG domains in the Nsp1 complex was viable with no noted growth defects at the temperatures tested. This result was consistent with previous genetic analysis of the genes encoding this complex (Fabre et al. 1995), indicating that the reported lethality with the Cre-loxP approach was likely due to tag-specific effects (Strawn et al. 2004).
Figure 3.
Construction history of ΔFG deletion strains. Beginning with a WT strain, NSP1, NUP49, and NUP57 were individually deleted in the presence of a WT NUP vector. Strains were mated and sporulated to generate a triple null, and pSW3643 was transformed with counterselection of single gene-encoding plasmids to generate SWY4684. SWY4684 was transformed with pSW3547, and pSW3643 was counterselected on with the TRP1 counterselective drug 5-FAA to generate SWY4683. SWY4684 was transformed with pSW3641, and NUP1, NUP2, and NUP60 were deleted iteratively to generate SWY4688. SWY4688 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY4690. LYS2 was deleted from SWY4688 with a floxed SpHIS5 cassette, which was recombined. This strain was then transformed with pSW3646, and NUP42 and NUP159 were deleted iteratively to generate SWY4779. SWY4779 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY6359. SWY4779 was transformed with pSW3642, and colonies with spontaneous loss of LEU2 were selected to generate SWY6360. SWY6360 was transformed with pSW3547, and pSW3643 was counterselected on 5-FAA to generate SWY6361. Additional strain and plasmid information is described in Table 1 and Table 2.
Table 1. Strain table.
| Strain | Description | Source | |
|---|---|---|---|
| SWY2284 | MATα trp1-1, ura3-1 his3-11,15, LYS2, leu2-3,112 | (Strawn et al. 2004) | |
| SWY4684 | nsp1::KANR nup49::loxP nup57::loxP MATα trp1-1, ura3-1 his3-11,15, LYS2, leu2-3,112 pSW3554 | This study | |
| SWY4683 | nsp1::KANR nup49::loxP nup57::loxP MATα trp1-1, ura3-1 his3-11,15, LYS2, leu2-3,112 pSW3547 | This study | |
| SWY4688 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP MATα trp1-1, ura3-1 his3-11,15, LYS2, leu2-3,112 pSW3643 pSW3641 | This study | |
| SWY4690 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP MATα trp1-1, ura3-1 his3-11,15, LYS2, leu2-3,112 pSW3547 pSW3641 | This study | |
| SWY4779 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP nup42::loxP nup159::loxP MATα trp1-1, ura3-1 his3-11,15, lys2::loxP, leu2-3,112 pSW3643 pSW3641 pSW3636 | This study | |
| SWY6359 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP nup42::loxP nup159::loxP MATα trp1-1, ura3-1 his3-11,15, lys2::loxP, leu2-3,112 pSW3547 pSW3641 pSW3636 | This study | |
| SWY6360 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP nup42::loxP nup159::loxP MATα trp1-1, ura3-1 his3-11,15, lys2::loxP, leu2-3,112 pSW3643 pSW3642 pSW3636 | This study | |
| SWY6361 | nsp1::KANR nup49::loxP nup57::loxP nup1::loxP nup2::loxP nup60::loxP nup42::loxP nup159::loxP MATα trp1-1, ura3-1 his3-11,15, lys2::loxP, leu2-3,112 pSW3647 pSW3642 pSW3636 | This study |
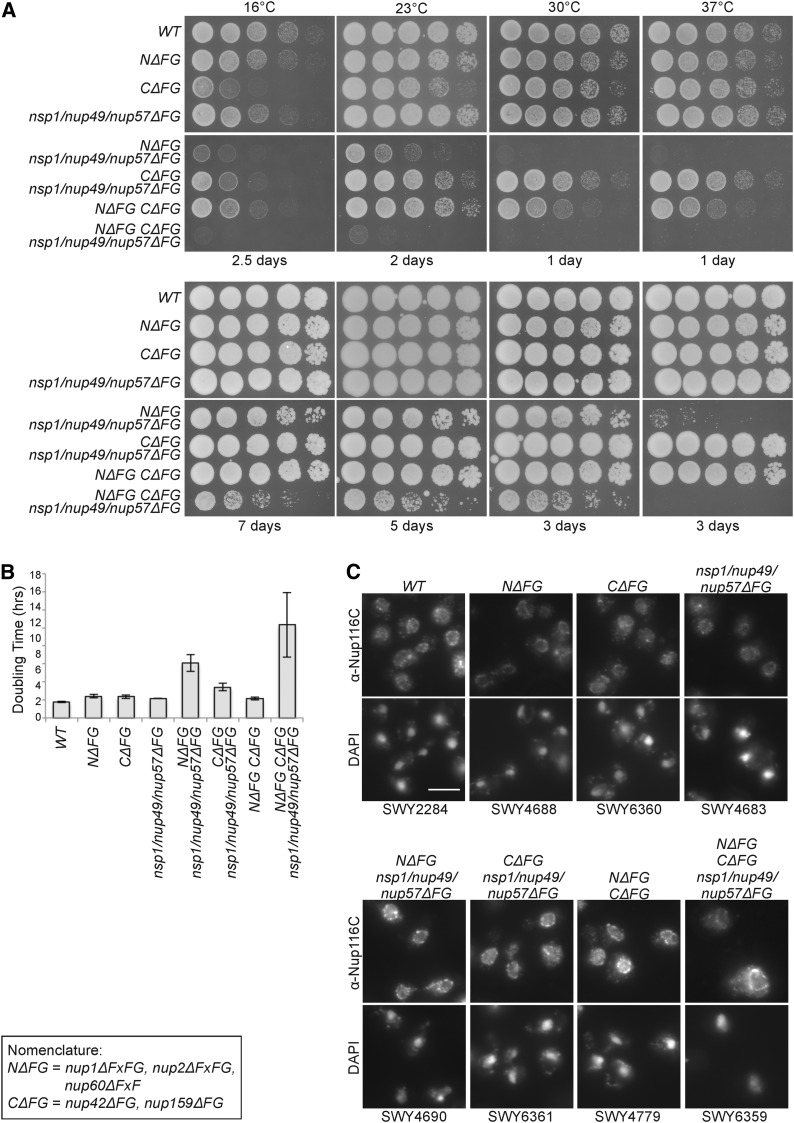
Figure 4.
(A) Growth analysis of ΔFG strains at different temperatures. Yeast strains were grown at 23° to midlog phase and five-fold serially diluted on YPD plates for growth at the indicated temperature for 1–7 days. (B) Liquid growth analysis of ΔFG strains. Yeast strains were grown at 23° to early log phase, and OD600 was analyzed to determine doubling times. Error bars indicate standard deviation from three replicates. (C) Nup116 is properly assembled into NPCs of ΔFG strains. Indicated strains were grown at 23° to midlog phase and processed for indirect immunofluorescence microscopy using the anti-(α)-Nup116-CTD antibodies. DAPI staining marks the nucleus. NΔFG CΔFG nsp1/nup49/nup57ΔFG was scaled independently due to increased cellular autofluorescence. Scale bar, 5 μm.
Using the nsp1Δ nup49Δ nup57Δ triple mutant as a starting point, the sequences encoding the FG domains on the nuclear and cytoplasmic face of the NPC were subsequently deleted (Figure 3 and Table 1). Because higher order gene deletions are difficult to generate and track by mating and sporulation, we adopted an iterative approach in haploid strains where the endogenous FG NUP gene was deleted by a floxed SpHIS5 cassette followed by recombination of the SpHIS5 sequence by expression of Cre recombinase (Figure 2B). This approach permitted deletion of multiple genes within one strain without losing availability of auxotrophic markers. In order to accommodate available markers, the asymmetric NUP genes were deleted in the presence of NΔFG and CΔFG plasmids with NSP1/NUP49/NUP57 covering the nsp1Δ nup49Δ nup57Δ deletions (Figure 3). We reasoned that this approach would not select for off-target effects because we previously observed that absence of all five asymmetric FG domains results in minimal growth defects (Terry and Wente 2007). Once the nuclear and cytoplasmic FG Nups were deleted, the WT NSP1/NUP49/NUP57 plasmid was exchanged for the nsp1/nup49/nup57ΔFG plasmid.
By serial dilution and analysis of growth on YPD, we observed that absence of FG domains from both the Nsp1-Nup49-Nup57 subcomplex and the nuclear face had growth defects at all temperatures tested (NΔFG nsp1/nup49/nup57ΔFG). In contrast, the absence of the FG domains both the Nsp1-Nup49-Nup57 subcomplex and the cytoplasmic face resulted in only mildly impacted growth (CΔFG nsp1/nup49/nup57ΔFG) (Figure 4A). Deletion of all asymmetric FG domains in combination with nsp1/nup49/nup57ΔFG (NΔFG CΔFG nsp1/nup49/nup57ΔFG) resulted in a viable strain with drastic growth defects (Figure 4A). To quantitatively analyze growth of all strains, liquid culture growth analysis was conducted at 23° (Figure 4B). Whereas most strains had doubling times of ∼2 to 3 hr, NΔFG nsp1/nup49/nup57ΔFG had a doubling time of 6.1 hr; CΔFG nsp1/nup49/nup57ΔFG, 3.4 hr; and NΔFG CΔFG nsp1/nup49/nup57ΔFG 10.3 hr.
To assess whether NPCs are assembled in these ΔFG strains, indirect immunofluorescence microscopy was performed using an antibody raised against the carboxy-terminal (non FG) domain of Nup116. Nup116 is an FG Nup that localizes to cytoplasmic foci when NPC assembly is perturbed (Ryan and Wente 2002), and the vertebrate ortholog, Nup98, associates with the nuclear envelope only after scaffold Nups are recruited following mitosis (Dultz et al. 2008). Therefore, Nup116 localization to the NE rim is a marker for proper NPC assembly. In all the ΔFG strains tested, anti-Nup116 signal was located at the nuclear rim surrounding the nuclear DAPI signal, suggesting that NPC assembly was not notably altered in the mutants (Figure 4C).
The NΔFG CΔFG nsp1/nup49/nup57ΔFG strain results in a GLFG-only NPC: the GLFG domains of Nup100, Nup116, and Nup145 (paralogous to each other and orthologous to vertebrate Nup98; Ryan and Wente 2000) are the only FG domains remaining. The other two GLFG domains in Nup49 and Nup57 are absent. Considering previous reports that GLFG domains are required for the formation of the NPC permeability barrier (Hulsmann et al. 2012; Lord et al. 2015), and that modification of GLFG Nups relaxes the barrier in vitro (Labokha et al. 2013), this strain will be of interest for subsequent studies of NPC transport capacity and nuclear permeability. We have generated plasmids encoding Nup100, Nup116, and Nup145 and FG deletions (Table 2) for use in such analysis.
Table 2. Plasmid table.
| Vector | Name in Text | Description | Residues Deleted | Plasmid Backbonea | Auxotrophic Marker | ΔFG Restriction Site | Residues Added | Source |
|---|---|---|---|---|---|---|---|---|
| pSW222 | NSP1 | pRS315 | LEU2 | This study | ||||
| pSW3428 | nsp1ΔFxFG | 179–591 | pRS315 | LEU2 | NheI | Ala Ser | This study | |
| pSW3524 | nsp1ΔFGΔ-FxFG | 3–591 | pRS314 | TRP1 | SpeI | Thr Ser | This study | |
| pSW3444 | NUP49 | pRS315 | LEU2 | This study | ||||
| pSW3513 | NUP49 | pRS314 | TRP1 | This study | ||||
| pSW3548 | NUP49 | pRS313 | HIS3 | This study | ||||
| pSW3549 | nup49ΔGLFG | 2–223 | pRS314 | TRP1 | SpeI | Thr Ser | This study | |
| pSW3431 | NUP57 | pRS314 | TRP1 | This study | ||||
| pSW3512 | NUP57 | pRS316 | URA3 | This study | ||||
| pSW3550 | nup57ΔGLFG | 2–236 | pRS314 | TRP1 | NheI | Ala Ser | This study | |
| pSW3521 | NSP1, NUP57 | pRS316 | URA3 | This study | ||||
| pSW3554 | NSP1, NUP49, NUP57 | pRS316 | URA3 | This study | ||||
| pSW3555 | NSP1, NUP49, NUP57 | pRS313 | HIS3 | This study | ||||
| pSW3643 | NSP1/NUP-49/NUP57 | NSP1, NUP49, NUP57 | pRS314 | TRP1 | This study | |||
| pSW3551 | nup49ΔGLFG, nup57ΔGLFG | pRS313 | HIS3 | This study | ||||
| pSW3552 | nsp1ΔFGΔ-FxFG, nup57ΔGLFG | pRS313 | HIS3 | This study | ||||
| pSW3553 | nsp1ΔFGΔ-FxFG, nup49ΔGLFG | pRS313 | HIS3 | This study | ||||
| pSW3644 | nsp1ΔFGΔ-FxFG, nup49ΔGLFG, nup57ΔGLFG | pRS315 | LEU2 | This study | ||||
| pSW3547 | nsp1/nup49/ nup57ΔFG | nsp1ΔFGΔ-FxFG, nup49ΔGLFG, nup57ΔGLFG | pRS313 | HIS3 | This study | |||
| pSW812 | NUP1 | pRS315 | LEU2 | This study | ||||
| pSW3634 | NUP1 | pRS314 | TRP1 | This study | ||||
| pSW3637 | nup1ΔFxFG | 384–888 | pRS315 | LEU2 | AvrII | Pro Arg | This study | |
| pSW3635 | NUP2 | pRS314 | TRP1 | This study | ||||
| pSW3638 | nup2ΔFxFG | 189–527 | pRS314 | TRP1 | AvrII | Pro Arg | This study | |
| pSW3636 | NUP60 | pRS314 | TRP1 | This study | ||||
| pSW3639 | nup60ΔFxF | 397–512 | pRS314 | TRP1 | AvrII | Pro Arg | This study | |
| pSW3640 | NUP1 NUP2 NUP60 | pRS314 | TRP1 | This study | ||||
| pSW3642 | N-WT | NUP1 NUP2 NUP60 | pRS316 | URA3 | This study | |||
| pSW3641 | NΔFG | nup1ΔFxFG nup2ΔFxFG nup60ΔFxF | pRS315 | LEU2 | This study | |||
| pSW3801 | NUP42 | pRS315 | LEU2 | This study | ||||
| pSW3802 | NUP42 | pRS314 | TRP1 | (Adams et al. 2014) | ||||
| pSW3645 | nup42ΔFG | 4–364 | pRS315 | LEU2 | XhoI | Leu Glu | This study | |
| pSW3448 | nup42ΔFG | 4–364 | pRS317 | LYS2 | XhoI | Leu Glu | This study | |
| pSW3657 | nup42ΔFG | 4–364 | pRS314 | TRP1 | XhoI | Leu Glu | (Adams et al. 2014) | |
| pSW3647 | NUP159 | pRS314 | TRP1 | (Adams et al. 2014) | ||||
| pSW3648 | nup159ΔFG | 464–876 | pRS314 | TRP1 | AvrII | Pro Arg | (Adams et al. 2014) | |
| pSW3646 | CΔFG | nup42ΔFG nup159ΔFG | pRS317 | LYS2 | This study | |||
| pSW3500 | NUP100 | pRS313 | HIS3 | This study | ||||
| pSW3501 | NUP100 | pRS314 | TRP1 | This study | ||||
| pSW3502 | nup100ΔGLFG | 2–570 | pRS313 | HIS3 | SpeI | Thr Ser | This study | |
| pSW3503 | nup100ΔGLFG | 2–570 | pRS314 | TRP1 | SpeI | Thr Ser | This study | |
| pSW3504 | NUP116 | pRS313 | HIS3 | This study | ||||
| pSW3506 | NUP145 | pRS314 | TRP1 | This study | ||||
| pSW3656 | nup145ΔGLFG | 10–209 | pRS314 | TRP1 | NheI | Ala Ser | This study |
These plasmids contains bacterial resistance (AMPR) and high copy replication (ori) sequences, yeast centromeric (CEN6) and replication (ARSH4) sequences, and the indicated yeast auxotrophic marker (Siskorski and Hieter 1989).
The plasmid-based expression of ΔFG nups in chromosomal null strains as presented here provides a straightforward way to introduce new sequences, mutations, or deletions into nup genes for analysis of FG Nup function in vivo. We previously assessed FG domain functional complementation using plasmid-based expression of ΔFG nups and swapped FG (SFG) nups (Iovine et al. 1995; Adams et al. 2014; Lord et al. 2015). The “swapped” strategy involves replacing the endogenous FG domain with that of another Nup. These studies revealed that FG domains of different Nups have inherently distinct function in vivo, because only select FG domains could functionally replace those tested. It is likely that sequence differences underlie distinct functionality. Indeed, individual domains from different FG Nups have distinct in vitro biochemical and biophysical characteristics (Lim et al. 2007; Yamada et al. 2010; Labokha et al. 2013). The genetic tools generated in this report will allow future investigations to conduct highly detailed tests of what sequences contribute to specialized function during transport and what biophysical and biochemical properties of FG domains contribute to the NPC permeability barrier and selectivity mechanism.
Materials and Methods
Yeast strains and growth
Table 1 lists the yeast strains generated in this study. Yeast genetic methods were conducted according to standard procedures (Sherman et al. 1986). Yeast strains were grown in either YPD (2% peptone, 2% dextrose, 1% yeast extract) or selective minimal media lacking appropriate amino acids and supplemented with 2% dextrose and 5-fluoroorotic acid (5-FOA; United States Biological) at 1.0 mg/mL or 2-amino-5-fluorobenzoic acid (5-FAA; Sigma-Aldrich) at 0.5 mg/mL as needed. For liquid culture analysis, strains were grown to early log phase (OD600 ∼0.15) at 23°, with OD600 measurements taken every 2 hr and normalized to time = 0.
Plasmid construction
Table 2 lists the plasmid generated in this study. Plasmid cloning was performed according to standard molecular biology strategies, and ΔFG plasmids were generated by amplifying a wild type NUP plasmid to replace the FG domain with a unique restriction site (Figure 2A). Most FG domains were replaced with the restriction sites AvrII, NheI, and SpeI to generate compatible cohesive ends (with the exception of XhoI for nup42ΔFG). FG domain boundaries were based on Strawn et al. (2004), and indicated in Table 2. Immunoblotting confirmed loss of FxFG and GLFG domains in strains transformed with ΔFG plasmids (data not shown).
Immunofluorescence
Yeast strains were grown to midlog phase (OD600 ∼0.5) in YPD medium at 23°, processed and labeled as in Ho et al. (2000). Briefly, samples were incubated with anti-Nup116-CTD rabbit antibodies (WU600, Iovine et al. 1995) overnight at 4°. Bound primary antibodies were detected with Alexa Flour 488-conjugated goat anti-rabbit IgG (1:200, Molecular Probes) and samples were stained with 0.1 mg/mL DAPI. Wide-field images were acquired using a microscope (BX50; Olympus) equipped with a motorized stage (Model 999000, Ludl), Olympus 100× NA1.3 UPlanF1 objective, and digital charge coupled device camera (Orca-R2; Hamamatsu). Images were processed with ImageJ (NIH).
Data availability
Strains and plasmids are available upon request. Table 1 contains genotypes for each individual strain. Table 2 contains information for each plasmid.
Acknowledgments
The authors thank the Wente laboratory for discussions and critical reading of the manuscript, and Eric Shows for generation of several plasmids. This work was supported by grants from the National Institutes of Health [R37GM051219 (to S.R.W.) and training position on T32HD007502 (to R.L.A.)].
Footnotes
Communicating editor: S. L. Jaspersen
Literature Cited
- Adams R. L., Wente S. R., 2013. Uncovering nuclear pore complexity with innovation. Cell 152: 1218–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. L., Terry L. J., Wente S. R., 2014. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics 197: 1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., et al. , 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701. [DOI] [PubMed] [Google Scholar]
- Chug H., Trakhanov S., Hulsmann B. B., Pleiner T., Gorlich D., 2015. Crystal structure of the metazoan Nup62•Nup58•Nup54 nucleoporin complex. Science. 350: 106–110 [DOI] [PubMed] [Google Scholar]
- Davis L. I., Fink G. R., 1990. The NUP1 gene encodes an essential component of the yeast nuclear pore complex. Cell 61: 965–978. [DOI] [PubMed] [Google Scholar]
- Del Priore V., Heath C., Snay C., MacMillan A., Gorsch L., et al. , 1997. A structure/function analysis of Rat7p/Nup159p, an essential nucleoporin of Saccharomyces cerevisiae. J. Cell Sci. 110(Pt 23): 2987–2999. [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., et al. , 2008. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180: 857–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E., Schlaich N. L., Hurt E. C., 1995. Nucleocytoplasmic trafficking: what role for repeated motifs in nucleoporins? Cold Spring Harb. Symp. Quant. Biol. 60: 677–685. [DOI] [PubMed] [Google Scholar]
- Field M. C., Koreny L., Rout M. P., 2014. Enriching the pore: splendid complexity from humble origins. Traffic 15: 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Schlaich N., Tekotte H., Hurt E. C., 1995. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 14: 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. K., Shen T. X., Ryan K. J., Kiseleva E., Levy M. A., et al. , 2000. Assembly and preferential localization of Nup116p on the cytoplasmic face of the nuclear pore complex by interaction with Nup82p. Mol. Cell. Biol. 20: 5736–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmann B. B., Labokha A. A., Gorlich D., 2012. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150: 738–751. [DOI] [PubMed] [Google Scholar]
- Hurt E. C., 1988. A novel nucleoskeletal-like protein located at the nuclear periphery is required for the life cycle of Saccharomyces cerevisiae. EMBO J. 7: 4323–4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovine M. K., Watkins J. L., Wente S. R., 1995. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J. Cell Biol. 131: 1699–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabachinski G., Schwartz T. U., 2015. The nuclear pore complex–structure and function at a glance. J. Cell Sci. 128: 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labokha A. A., Gradmann S., Frey S., Hulsmann B. B., Urlaub H., et al. , 2013. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32: 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R. Y., Fahrenkrog B., Koser J., Schwarz-Herion K., Deng J., et al. , 2007. Nanomechanical basis of selective gating by the nuclear pore complex. Science 318: 640–643. [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Huang B., Kapinos L. E., 2015. How to operate a nuclear pore complex by Kap-centric control. Nucleus . 10.1080/19491034.2015.1090061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C. L., Timney B. L., Rout M. P., Wente S. R., 2015. Altering nuclear pore complex function impacts longevity and mitochondrial function in S. cerevisiae. J. Cell Biol. 208: 729–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Kern H., Mutvei A., Horstmann H., Marshallsay B., et al. , 1990. NSP1: a yeast nuclear envelope protein localized at the nuclear pores exerts its essential function by its carboxy-terminal domain. Cell 61: 979–989. [DOI] [PubMed] [Google Scholar]
- Raices M., D’Angelo M. A., 2012. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat. Rev. Mol. Cell Biol. 13: 687–699. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Wente S. R., 1994. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 4: 357–365. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., et al. , 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148: 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R., 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12: 361–371. [DOI] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R., 2002. Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B., Cold Spring Harbor Laboratory , 1986. Laboratory course manual for methods in yeast genetics, Cold Spring Harbor Laboratory, New York, N.Y. [Google Scholar]
- Siskorski R. S., Hieter P. H., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn L. A., Shen T., Shulga N., Goldfarb D. S., Wente S. R., 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat. Cell Biol. 6: 197–206. [DOI] [PubMed] [Google Scholar]
- Stuwe T., Bley C. J., Thierbach K., Petrovic S., Schilbach S., et al. , 2015. Architecture of the fungal nuclear pore inner ring complex. Science 350: 56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J. Cell Biol. 178: 1121–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R., 2009. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 8: 1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente S. R., Rout M. P., Blobel G., 1992. A new family of yeast nuclear pore complex proteins. J. Cell Biol. 119: 705–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J., Phillips J. L., Patel S., Goldfien G., Calestagne-Morelli A., et al. , 2010. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9: 2205–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Table 1 contains genotypes for each individual strain. Table 2 contains information for each plasmid.