Abstract
L. pneumophila is the causative agent of Legionnaires’ disease, a human illness characterized by severe pneumonia. In contrast to those derived from humans, macrophages derived from most mouse strains restrict L. pneumophila replication. The restriction of L. pneumophila replication has been shown to require bacterial flagellin, a component of the type IV secretion system as well as the cytosolic NOD-like receptor (NLR) Nlrc4/ Ipaf. These events lead to caspase-1 activation which, in turn, activates caspase-7. Following caspase-7 activation, the phagosome-containing L. pneumophila fuses with the lysosome, resulting in the restriction of L. pneumophila growth. The LegS2 effector is injected by the type IV secretion system and functions as a sphingosine 1-phosphate lyase. It is homologous to the eukaryotic sphingosine lyase (SPL), an enzyme required in the terminal steps of sphingolipid metabolism. Herein, we show that mice Bone Marrow-Derived Macrophages (BMDMs) and human Monocyte-Derived Macrophages (hMDMs) are more permissive to L. pneumophila legS2 mutants than wild-type (WT) strains. This permissiveness to L. pneumophila legS2 is neither attributed to abolished caspase-1, caspase-7 or caspase-3 activation, nor due to the impairment of phagosome-lysosome fusion. Instead, an infection with the legS2 mutant resulted in the reduction of some inflammatory cytokines and their corresponding mRNA; this effect is mediated by the inhibition of the nuclear transcription factor kappa-B (NF-κB). Moreover, BMDMs infected with L. pneumophila legS2 mutant showed elongated mitochondria that resembles mitochondrial fusion. Therefore, the absence of LegS2 effector is associated with reduced NF-κB activation and atypical morphology of mitochondria.
Introduction
The facultative intracellular pathogen L. pneumophila multiplies within human alveolar macrophages and modulates host cell signaling. Following internalization by macrophages or amoeba, L. pneumophila form a unique compartment called the Legionella containing vacuole (LCV) that evades fusion with lysosomes [1–7]. The LCV provides the bacteria with a protected environment in which L. pneumophila can secure replication. The Dot/Icm type IV secretion system, which translocates effector proteins into the host cell cytoplasm and manipulates host cell signaling, coordinates the formation of the LCV [7–9]. Several injected effectors have been previously identified as substrates of the Dot/Icm secretion system [10–14]. Some effectors are involved in the recruitment of the endoplasmic reticulum vesicles to the LCV and thus disrupting host trafficking [15–20]. Others are modulators of the NF-κB pathway [20,21]. Some proteins are homologous to eukaryotic proteins and have domains with enzymatic activity required in various post-translational modifications, including phosphorylation, glycosylation, methylation, prenylation, AMPylation and ubiquitination, of host cell proteins [14,20,22–26].
The subcellular compartments in eukaryotic cells are designated by their lipids and proteins content. Trafficking is tightly controlled to guarantee the correct delivery of cargo to the correct compartment [27]. For example, phosphatidylinositol 4- phosphate PtdIns (4)P and phosphatidylinositol 3- phosphate PtdIns (3)P have been shown to regulate phagolysosome biogenesis [28]. Some L. pneumophila secreted effectors anchor to the cytoplasmic face of the LCV membrane by binding to phosphoinositide (PI) lipids [29]. This process is achieved through the modulation of the vacuole membrane PI pattern such as the accumulation of PtdIns (4)P, which is catalyzed by L. pneumophila effector proteins that directly manipulate PIs or indirectly control them through effectors that recruit host PI-metabolizing enzymes [27,29]. Moreover, L. pneumophila largely controls the localization of secreted bacterial effectors and the recruitment of host factors by modulating the PI patterns at the LCV.
The legS2 (lpg2176) was identified in a bioinformatics screen of the L. pneumophila Philadelphia-1 genome and encodes for a protein that is highly homologous to the eukaryotic sphingosine 1-phosphate lyase [30,31]. LegS2 exhibits 36% identity and 52% similarity to Tetrahymena thermophila SPL and functions as a sphingosine 1-phosphate lyase [31]. The L. pneumophila SPL harbors a C-terminal domain that is required for translocation to eukaryotic cells via the Icm/Dot system [31]. This domain is absent in the eukaryotic homologues.
SPL is required for the degradation of Sphingosine-1-Phosphate S1P to phosphoethanolamine and hexadecanal in eukaryotic cells [31]; Sphingosine-1-Phosphate S1P is a sphingolipid metabolite that regulates cell migration, angiogenesis, and development [32]. The intracellular pool of S1P is regulated by three highly conserved enzymes: sphingosine kinase (SPHK) that catalyzes the phosphorylation of sphingosine producing S1P, S1P phosphatase (S1PP) that reverses the former reaction, and S1P lyase (SPL) that catalyzes the irreversible cleavage of S1P to ethanolamine phosphate and a long chain aldehyde [32].
In this study, we show that L. pneumophila lacking the sphingosine-1-phosphate lyase, legS2 replicated in higher numbers compared to WT strain in WT BMDMs. The increase in bacterial multiplication is not attributed to the compromised activation of caspase-1, caspase-7, or caspase-3 or phagosome lysosome fusion. Instead, the disruption of L. pneumophila SPL is associated with a reduction in the level of inflammatory cytokines. This reduction results from the inhibition of the NF-κB pathway. Moreover, infection with the legS2 mutant results in an elongation of the mitochondria that is a characteristic of mitochondrial fusion.
Materials and Methods
Ethics statements
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of health and The Ohio State University. The Institutional Animal Care and Use Committee (IACUC) has approved our Animal Use Protocol (AUP), 2010A00000066-R1 and approved this study. Approved by Donna McCarthy Beckett, Ph.D., R.N., F.A.A.N. Human cells were obtained from anonymous samples from the Red Cross, http://www.redcrossblood.org/centralohio. Blood from the red cross does not have any information or link to the donor and therefore does not require IRB or consent. Primary macrophages derived from mice bone marrow were used.
Bacterial strains
L. pneumophila JR32 [33] and the legS2 mutant [30] were kindly provided by Dr. Amal Amer, at The Ohio State University. L. pneumophila strains were grown on buffered charcoal yeast extract solid media (BCYE) plates at 37°C, or in BYE broth containing L-cysteine, thymidine and ferric nitrate supplements at 37°C with shaking [2,34].
Mice, cell culture and infections
Wild-type mice were purchased from Jackson laboratory. Mice were housed in AALAC-accredited facilities and kept in ventilated cages with automatic water valves (lixit). Mice were fed Harlan 7912 as needed and kept in 12hr light/dark cycles 6am-6pm light, 6pm-6am—dark for their circadian rhythms. Plastic houses/huts were provided for enrichment if one mouse per cage or mice needed to feel more secure. Mice were euthanized by CO2 then cervical dislocation. Anesthesias for in vivo experiments were done by exposing the mice to isoflurane gas for 30 seconds until they are asleep. Mice were monitored daily for signs of pain or distress. All of the animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals published by National Institutes of Health and The Ohio State University. C57BL/6 BMDMs were prepared from the femurs of five to eight-week-old mice as previously described [2,35] and grown in IMDM (Iscove’s modified Dulbecco’s medium supplemented with the L929 fibroblast cell line (ATCC)-cultured supernatant and 10% heat inactivated fetal bovine serum (HIFBS) (Gibco). Infection was done in IMDM supplemented with 10% HIFBS. The isolation and preparation of the hMDMs from peripheral blood (obtained from the Red Cross) was carried out as previously described [34,36,37].
In vitro and in vivo infection
The in vitro BMDM infections were carried with a multiplicity of infection (MOI) of 0.2–0.5 unless otherwise specified. For in vivo experiments, C57BL/6 mice were infected with 1x106 bacteria intratracheally. The lungs were harvested 4 or 48 h post-infection, homogenized, and plated on BCYE with streptomycin. Colony forming units (CFUs) were then quantified.
Western blot
Cell extracts were prepared and immunoblotted with antibodies that recognize caspase-1, caspase-7 and caspase-3 (Cell Signaling), followed by the appropriate secondary antibody. Equal concentration of protein was loaded on a 12% SDS polyacrylamide gel.
Macrophage cytotoxicity assay
In vitro quantification of cytoplasmic histone-associated-DNA-fragments (apoptosis) was performed using the Cell Death Detection ELISA and the photometric enzyme immunoassay (Roche Applied Science) according to the manufacturer’s specifications. The fold change of macrophage necrosis was determined by measuring the release of host cell cytoplasmic lactate dehydrogenase (LDH) using the cytotoxicity detection kit (Roche Applied Science) according to the manufacturer’s specifications. BMDMs (0.5 x 106) were plated in 12-well plates and infected with either JR32 or the legS2 mutant at an MOI of 0.5, 1, or 5. The supernatants were then collected 24 h post-infection and assayed.
NF-κB DNA binding activity assay
Nuclear extracts of WT BMDMs untreated or treated with the JR32 or legS2 mutant were prepared as described [2]. EMSA was used to measure NF-κB DNA binding activity as noted [38].
Lyso tracker red colocalization
Wild-type mouse macrophages were plated at a density of 0.5 x 106 cells per well in a 24 well plate containing sterile coverslips for 48 h. The lyso tracker red was added to the monolayer for 30 min at 37°C. The monolayer was then washed with the infection medium 3 times and kept for 30 min at 37°C in infection media alone. Next, it was infected with JR32, dotA, or legS2 at an MOI of 0.2 for 1 h. Then, the monolayer was washed with the infection medium, fixed with paraformaldehyde, and finally stained with the anti-L. pneumophila antibody, followed by the secondary antibody Alexa flour 488 goat anti-mouse. The coverslips were mounted and viewed by the Olympus FV1000 spectral Confocal Laser Scanning Microscopy (CLSM).
Enzyme-linked immuno sorbent assay (ELISA)
Macrophages were infected with L. pneumophila JR32 or the legS2 mutant for 24 h. Then, culture supernatants were collected, centrifuged and stored at -20°C until assayed for cytokine content. The amount of IL-1β, IL-6, IL10 and TNFα in the supernatants was determined by specific sandwich ELISA following the manufacturer’s protocol (R&D system Inc.,
Quantitative PCR
Total RNA was extracted from mouse macrophages using Trizol (Invitrogen Life Technologies). Then, 1–2 μg of the RNA were converted to cDNA by ThermoScript RNase H- Reverse Transcriptase (Invitrogen, Life Technologies). 20–60 ng of the converted cDNA was used for quantitative PCR with SYBR Green I PCR Master Mix in the Step One Plus Real Time PCR System (Applied Biosystems). The target gene Ct values were normalized to the Ct values of two housekeeping genes (human GAPDH and CAP-1, accordingly to the cell origin) and expressed as relative copy number (RCN) (34).
Transmission Electron Microscopy
BMDMs were plated at a density of 4x106 per well on coverslips in Permanox chamber slides, with 2 chambers and 8 units per tray (lab tech) for 48 h; they were then infected with an MOI of 1 for 4 h or MOI of 0.5 for 24 h. The coverslips were fixed with 2.5% glutaraldehyde in 0.1M phosphate buffer and 0.1M sucrose at a pH of 7.4. The cells were post-fixed with 1% osmium tetroxide in the phosphate buffer; then, they were en bloc stained with 2% uranyl acetate in 10% ethanol, dehydrated in a graded series of ethanol, and embedded in Eponate 12 epoxy resin (Ted Pella Inc., 18012). Ultrathin sections were cut on a Leica EM UC6 ultramicrotome (Leica microsystems, EM FC7), collected on copper grids and stained. Images were acquired with an FEI Technai G2 Spirit transmission electron microscope (FEI), a Macrofire (Optronics) digital camera, and AMT image capture software.
Statistical analysis
The comparisons of groups for statistical difference were done using a Student’s two-tailed t-test. P values less than 0.05 were considered significant.
Results
L. pneumophila legS2 strain replicates in WT BMDMs and hMDMs
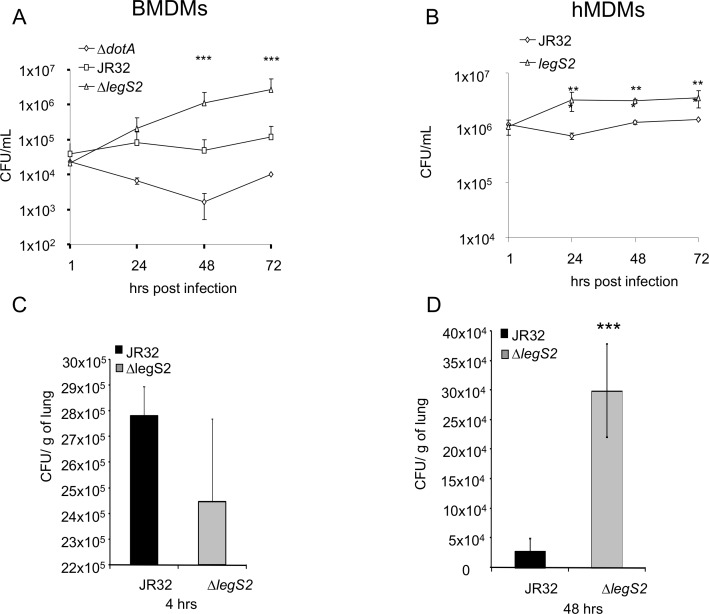
To delineate the role of LegS2 in L. pneumophila infection, we tested the replication of the JR32 and legS2 mutant in WT BMDMs. The BMDMs restricted the growth of the JR32 strain, while the legS2 mutant showed a 10- fold increase in the colony forming units (CFUs) over time (48–72 h) compared to the JR32 strain (Fig 1A). Since humans are permissive to L. pneumophila replication (39), we also examined the intracellular growth of legS2 mutant in hMDMs (Fig 1B). Both JR32 and legS2 mutant replicated in hMDMs; however, the legS2 mutant replicated more than the JR32 strain.
Fig 1. L. pneumophila legS2 mutant replicates in human MDMs and wild-type mice and their derived macrophages.
A) BMDMs were infected with wild-type L. pneumophila, JR32, or ΔlegS2 for 1, 24, 48 and 72h. The colony forming units (CFUs) were scored at the indicated time points. B) hMDMs were infected with wild-type L. pneumophila, JR32, or ΔlegS2 for 1, 24, 48 and 72h. Data are shown as mean + SD of n = 3. Asterisks indicate significant differences, and a two tailed t-test was used to calculate the P value (*P<0.05, **P<0.01, ***P<0.001). (C and D) Four female C57BL/6 mice per group received 1x106 of JR32 or legs2 bacteria intratracheally. Lungs were homogenized and plated for CFUs, counting at (C) 4 or (D) 48 h post infection. (C and D) Data are shown as mean + SD of four mice. Asterisks indicate significant differences (***P<0.001); a two tailed t-test was used to calculate the P-value.
Legionnaires’ disease stems from the successful replication of L. pneumophila in human alveolar macrophages [39,40]. Therefore, we tested legS2 mutant replication in vivo by examining the bacterial CFUs within the lungs of infected mice. C57BL/6 mice were infected with either 1x106 JR32 or legS2 mutant intratracheally. The initial bacterial load was not significantly different 4 h post-infection (Fig 1C). However, 48 h post-infection, the legS2 mutant-infected mice harbored 4 fold more CFUs compared to the JR32 strain (Fig 1D). Our data showed that the legS2 mutant replicates in WT mice and their derived macrophages.
The growth of legs2 mutant is independent of caspase-1 or caspase-7 activation
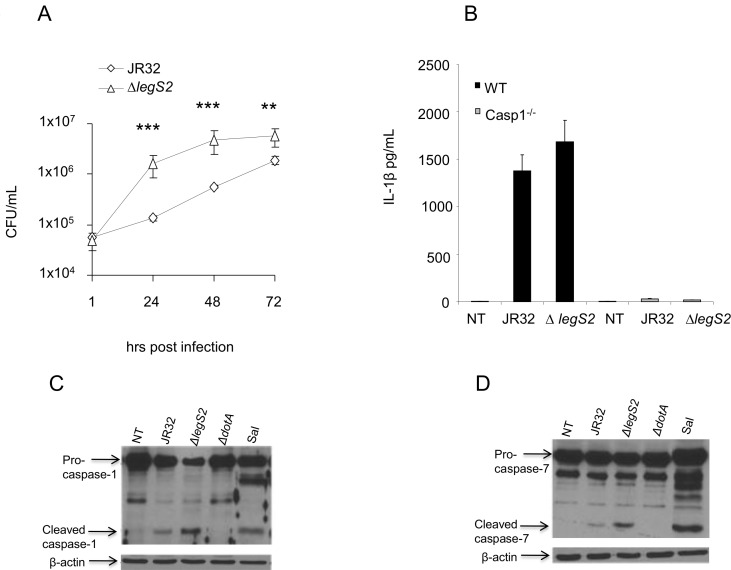
Caspase-1 activates caspase-7, resulting in the restriction of L. pneumophila growth by mediating the fusion of the LCV to the lysosome [41–43]. Casp-1-/- BMDMs are permissive to L. pneumophila replication [2]. Therefore, impaired caspase-1 activation could result in the increased replication of the legS2 mutant. Therefore, we examined the replication of legS2 mutant bacteria in Casp-1-/- BMDMs. The legS2 mutant showed higher replication than JR32 24, 48, and 72 h post-infection in casp-1-/-BMDMs (Fig 2A). Since the cleavage and maturation of IL-1β is mediated by caspase-1 activation, we examined the level of IL-1β 24 h post-infection with the JR32 or the legS2 mutant by ELISA. No significant differences were seen in the level of IL-1β 24 h post-infection with either the JR32 or the legS2 mutant (Fig 2B). Moreover, we tested the possibility of differential activation of caspase-1 by the WT JR32 or legS2 mutant. Indeed, both JR32 and legS2 infected BMDMs showed capsase-1 cleavage as detected by the mature subunits of casp-1 (Fig 2C). Taken together, the replication of the legS2 mutant in WT macrophages is independent of caspase-1 activation.
Fig 2. The permissiveness of mice BMDMs to legS2 mutant bacteria is independent of caspase-1 or caspase-7 activation.
(A) Caspase-1 KO (casp-1-/-) macrophages were infected with L. pneumophila, JR32, or legS2 with an MOI of 0.5 for 1, 24, 48 and 72 h; then, CFUs were measured at the indicated time points. (B) Levels of IL-1β were detected in supernatants of WT or casp-1-/- BMDMs were infected with JR32 or the legS2 mutant after 24 hr, while WT BMDMs were either not treated (NT) or infected with L. pneumophila, JR32, or legS2 mutant bacteria for 2 h. Salmonella infection (Sal) was used as a positive control for caspase-1 or caspase-7 activation. (C) Casp-1 or (D) casp-7 antibodies were used to detect casp-1 and casp-7 activation, respectively, in cell extracts.
The growth restriction of L. pneumophila is also attributed to caspase-7 mediated bacterial delivery to the lysosome and the early death of murine macrophages [2,44]. Hence, impaired caspase-7 activation could result in increased replication of the legS2 mutant. To that end, we assessed the activation of caspase-7 by Western blot. Both the JR32 and the legS2 mutant cleaved caspase-7 in WT macrophages (Fig 2D). Thus, the replication of the legS2 mutant is independent of caspase-1 or caspase-7 activation.
The growth of legs2 mutant is independent of host cell death
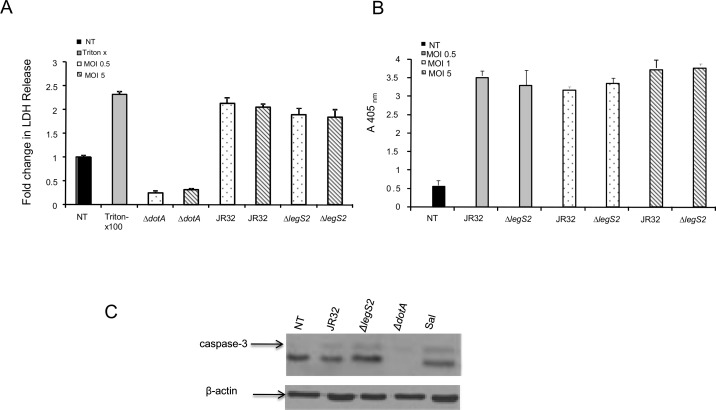
Interference with the host’s survival is a key determinant in the L. pneumophila replication cycle. Early cell death aborts L. pneumophila replication and terminates the infection [45]. It is possible that the legS2 mutant modulates the macrophage’s life span to sustain its replication. To dissect the role of LegS2 in mediating host cell death, we quantified the level of LDH released in the culture supernatants of macrophages infected with either JR32 or the legS2 mutant. Comparable LDH release was observed in the culture supernatants of both the JR32 and the legS2 mutant-infected macrophages at the two different MOIs (Fig 3A). Moreover, we measured apoptosis in the overall population of macrophages by determining the cytoplasmic apoptosis histone-associated-DNA fragments at MOIs of 0.5, 1 and 5. We found that infection by JR32 and the legS2 mutant leads to comparable cell death at both low and high MOIs (Fig 3B). Overall, our data demonstrates that the replication of the legS2 mutant in macrophages is independent of cell death. Further, the activation of caspase-3 is comparable in JR32 and legS2 infected macrophages (Fig 3C). Consequently, the replication of the legS2 mutant is independent of host cell death.
Fig 3. Replication of legS2 mutant is independent of host cell death.
(A) Wild-type BMDMs were not treated (NT) or infected with the type IV secretion mutant dotA, wild-type L. pneumophila, JR32, or legS2 mutant for 24h at MOIs of 0.5 and 5. Then, the fold change in LDH release was measured from the overall population of macrophages. The data represents the mean + SD of n = 3. (B) Wild-type C57BL/6 (B6) were not treated (NT) or infected with wild-type L. pneumophila, JR32, legS2, or dotA mutant for 8 h. Salmonella infection (Sal) was used as a positive control for caspase-3 activation. A Western blot with caspase-3 antibody was used to detect casp-3 activation. β-actin was used as a loading control.
The replication of the legS2 mutant in WT BMDM is not attributed to impaired phagosome-lysosome maturation
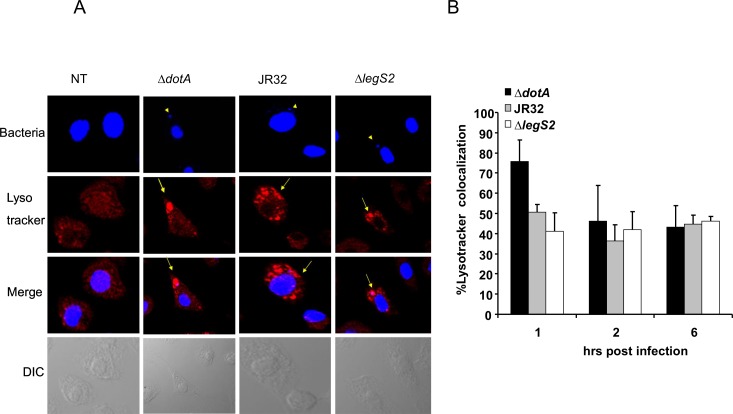
The restriction of L. pneumophila in WT BMDMs is dependent upon the delivery of the LCV to the lysosome, which ultimately results in bacterial degradation. To determine the mechanism by which the legS2 mutant bacteria replicate in macrophages, we examined the colocalization of the JR32, dotA, and legS2 mutants with lyso- tracker Red, an acidic dye used to track the lysosome. (Fig 4A) shows images obtained by the Confocal Laser Scanning Microscopy (CLSM), and (Fig 4B) shows the score of the percentage of bacteria colocalized within the lyso-tracker positive compartment. Approximately 60% of the translocation-defective dotA mutant was delivered to the lyso tracker-labeled compartment within 1 h of infection. In addition, 52% of the JR32 were destined to the lyso tracker labeled compartment. This is due to the fact that WT cells are restrictive to L. pneumophila replication. The trafficking of the legS2 mutant was comparable to that of the JR32 with ~57% lyso tracker labeled compartment. Therefore, the legS2 mutant’s substantial replication in WT macrophages is not attributed to enhanced evasion of LCV-lysosome fusion.
Fig 4. Replication of L. pneumophila legS2 mutant is not due to defective phagosome-lysosome fusion.
(A) Images of wild-type macrophages not infected (NT) or infected with the type IV secretion mutant dotA, wild-type L. pneumophila, JR32, and the legS2 mutant. The first panel presents staining with DAPI, with arrow heads pointing to the bacteria. The second panel shows staining with the lyso tracker. The third panel depicts merged images, with bacteria colocalized with the lysosomal marker. B) The percent of bacteria colocalized with the lyso tracker was scored in 100 infected cells from 3 independent coverslips. The data represents the mean + SD of n = 3.
The increased replication of the legS2 mutant in BMDMs is associated with reduced cytokine production
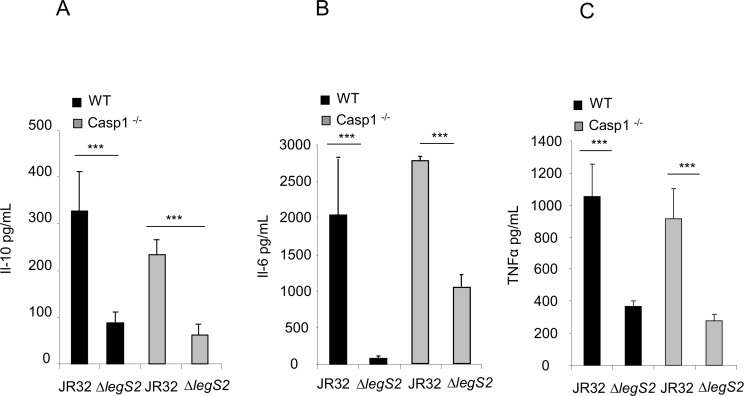
The production and secretion of cytokines and chemokines is concomitant with L. pneumophila intracellular growth [46]. Cytokines are chemical mediators that play fundamental roles in eliciting the necessary immune response for controlling lung infection. We examined the level of several pro-inflammatory and anti-inflammatory cytokines following infection with either the JR32 or the legS2 mutant by ELISA. After 24 h, the levels of the cytokines IL-10, IL-6, and TNFα were significantly less in macrophages infected with the legS2 mutant as compared to those infected with JR32. The level of IL-10 in JR32 infected cells was double the amount in legS2 infected cells, (Fig 5A), the levels of IL-6 and TNFα in JR32 infected cells were triple the amount in legS2 infected cells, (Fig 5B and 5C). Differences in the cytokine levels were also evident in the Casp1-/- BMDMs 24 h post-infection. Our data suggested that cytokine production is associated with increased replication of the legS2 mutant in WT BMDMs.
Fig 5. Increased replication of the legS2 mutant in BMDMs is associated with reduced cytokine production.
WT BMDMs or casp-1-/- macrophages were infected with L. pneumophila, JR32, or the legS2 mutant. 24 h post infection, the supernatants from infected and un-infected cells were assayed by ELISA. (A) IL-10 level, (B) IL-6, (C) TNFα levels. Data represent the mean + SD of n = 3. Asterisks indicate significant differences (***P<0.001), and a two tailed t-test was used to calculate the P-value.
The legS2 mutant infected macrophages exhibited reduced cytokine mRNAs accompanied by inhibition of the NF-κB pathway
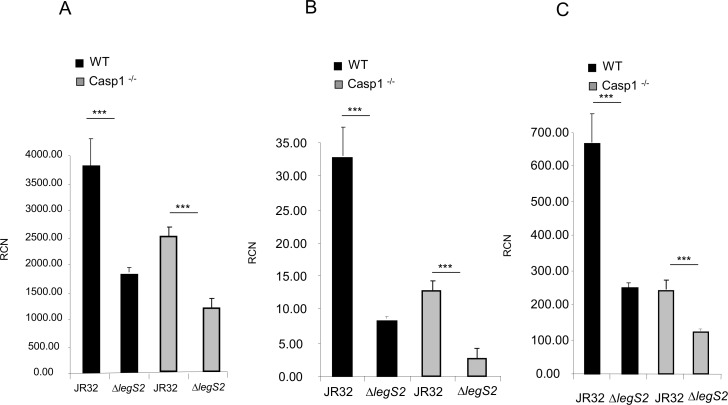
To decipher the mechanism by which legS2 affects cytokine level, we used quantitative RT-PCR to examine the levels of the cytokine mRNA following infection with either the JR32 or the legS2 mutant. At 4 h post-infection, the transcription levels of IL-10, IL-6 and TNFα were reduced in the legS2 infected monolayers as compared to the JR32 infected monolayers (Fig 6A–6C). Our results indicate that the decrease in cytokine level from the legS2 infected cells was due to lower levels of cytokine mRNA.
Fig 6. The legS2 mutant limits the levels of cytokine mRNAs.
WT BMDMs or casp-1-/- macrophages were infected with L. pneumophila, JR32, or the legS2 mutant. 4 h post infection, total RNA was isolated from JR32 or legS2 infected macrophages; then, the converted cDNA was used for quantitative PCR. The target gene Ct values were normalized to the Ct values of two housekeeping genes (human GAPDH and CAP-1, according to the cell origin) and expressed as relative copy numbers (RCN). (A) Represents the IL-10 RCNs, (B) Represents the IL-6 RCNs, (C) Represents the TNFα RCNs. Data represent the mean + SD of n = 3. Asterisks indicate significant differences (***P<0.001); a two tailed t-test was used to calculate the P-value.
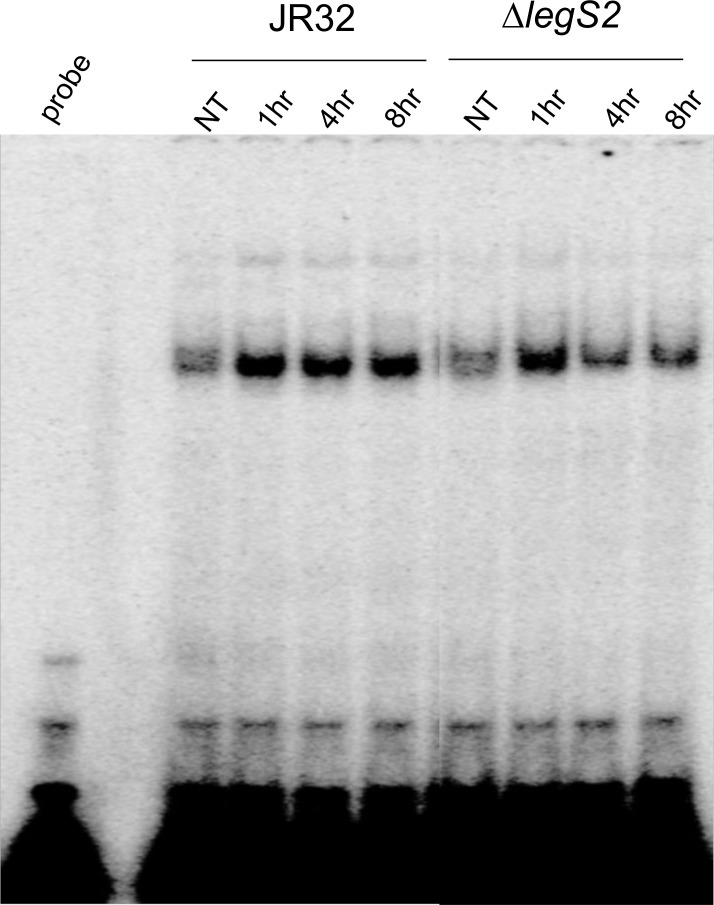
NF-κB is a transcription factor that regulates the mammalian innate immune system by modulating the transcription of pro-inflammatory cytokine and chemokine genes [47]. Indeed, infection with L. pneumophila activates the host NF-κB pathway [1,48]. Therefore, we examined NF-κB activation by an electrophoretic mobility shift assay (EMSA) (Fig 7). The EMSA data showed that, 4–8 h post-infection, NF-κB was inhibited in the legS2 mutant and not in the JR32 strain. This suggests that the reduced cytokine production is associated with the inhibition of the NF-κB pathway.
Fig 7. The legS2 mutant inhibits the NF-κB pathway.
Wild-type macrophages were either not treated (NT) or infected with the JR32, or the legS2 mutant for 1, 4, and 8h. Then, nuclear extracts were processed using electrophoretic mobility shift assay (EMSA) to determine NF-κB activation.
Infection with the legS2 mutant is accompanied by change in mitochondrial morphology
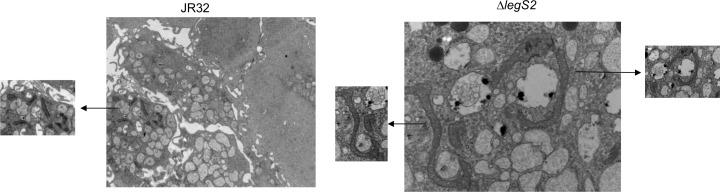
The dynamic nature of mitochondrial morphology is shown by its continuously fusing and interconnecting network [49]. The fusion process is critical for the maintenance of mitochondrial function [50]. However, mitochondrial fission is an early event during apoptosis, either occurring before or coinciding with cytochrome c release [49,51,52]. It is upstream of caspase activation and characterized by mitochondrial fragmentation into multiple small units [49,51,52]. We employed transmission electron microscopy to examine cells infected with either the JR32 or the legS2 mutant at 4 and 24 h post-infection (Fig 8). Our data showed that, 24 h post-infection, BMDMs infected with the legS2 mutant exhibited elongated mitochondria that seemed to be fused with each other. In BMDMs infected with the JR32 strain, however, we observed fragmented mitochondria, indicating fission. These data suggest that the legS2 mutant bacteria manipulate mitochondrial fusion and fission, respectively.
Fig 8. Infection with the legS2 mutant is accompanied by change in mitochondrial morphology.
TEM of BMDMs infected with the JR32 or the legS2 mutant. Images were taken from 24 h post-infected cells.
Discussion
In summary, we show that the legS2 mutant bacteria replicate in WT mice and their derived macrophages. The permissiveness of BMDMs to infections with the legS2 mutant, is accompanied by the dampening of pro-inflammatory and anti-inflammatory cytokines, inhibiting the NF-κB pathway resulting in changes in mitochondrial morphology.
Studies with the A/J-derived mouse macrophages, which are permissive to L. pneumophila infection, showed a biphasic activation of NF-κB during infection [1,48,53,54]. The early phase is characterized by strong but transient activation. Even though this phase is beneficial for the host cell, it must be dampened quickly to overcome the toxic effect of inflammatory cytokines [54]. IL-6 and TNFα are major proinflammatory cytokines that are produced in response to NF-κB activation. They regulate inflammatory responses locally and systemically. Moreover, they recruit leukocytes and mediate more cytokine production and increased bactericidal activity [55,56]. The limitation of the above cytokines following infection with the legS2 mutant is likely responsible for the enhanced L. pneumophila growth in vivo. Indeed, patients with Legionnaires’ disease show increased level of IL-6 and TNFα in their lungs [57–60]. Furthermore, the addition of TNFα renders macrophages less permissive for L. pneumophila and renders neutrophils more bactericidal [61] [62]. Also, mice with a mutation that abrogates Toll-like receptors (TLRs) exhibit limited cytokines, which in turn results in increased susceptibility to L. pneumophila lung infection [63–65]. Therefore, the reduction in the levels of IL-10, IL-6 and TNFα could result in the increased replication of the legS2 mutant.
We also observed that infecting BMDMs with legS2 mutant results in the reduction of IL-10 level, and other studies have shown that IL-10 is induced in vivo upon L. pneumophila infection [57,59,66–68]. IL-10 is an anti-inflammatory cytokine that represses the expression of Th1 cytokines [69–71]; therefore, it seems that, by dampening the IL-10, the legS2 mutant reduces L. pneumophila replication. However, it is quite likely that limiting IL-6 and TNFα outweighs the effect of IL-10. Thus, the overall impact of a legS2 mutant infection is promoting growth in BMDMs. Even though we focused our study on IL-6, IL-10, TNFα and IL-1β, it is likely that legS2 affects other cytokines that are produced by macrophages as well.
Based upon our RT-PCR analysis of IL-6, IL-10 and TNFα in macrophages infected with legS2 mutants, we can conclude that infection with L. pneumophila legS2 dampens the mRNA expression of cytokines in infected cells. The reduction in the above cytokines is likely a consequence of the inhibition of the NF-κB pathway seen by EMSA.
Our TEM data show that infection with legS2 mutant bacteria results in changes in mitochondrial fusion or fission. We observed mitochondrial morphologies that resemble fused mitochondria in legS2 infected cells. Mitochondrial fusion has been linked to an increase in the total pool of ATP during starvation [51]. Consistent with our results, Degtyar et. al showed that legS2 colocalizes to the mitochondria in U937 infected cells [31].
Listeria monocytogenes is another pathogen that has been shown to manipulate mitochondrial fusion/fission. The link between mitochondrial dynamics and the efficiency of the L. monocytogenes infection has been demonstrated. Indeed, the inhibition of the mitochondrial fusion has been shown to decrease the efficiency of L. monocytogenes infection [72]. Similarly, inducing mitochondrial fusion the legS2 mutant resulted in a more efficient infection than in the WT. Therefore, modulating mitochondrial fusion/fission by these two pathogens could impact the efficiency of macrophage infection.
We show that legS2 mutant replicated more than the JR32 strain.
It seems contradictory that L. pneumophila would maintain legS2 in the genome, even though it is dispensable for replication in Acanthamoeba castellanii [31]. It is possible that retaining redundant genes in the genome of L. pneumophila is a factor utilized during evolution to prevent chromosomal lesions. Such lesions could affect the ability of the bacteria to multiply intracellularly [30,73]. Moreover, the survival of L. pneumophila in several protozoan hosts could improve the hosts’ tropism. However, it could prevent host specialization by retaining pathways that are essential for growth in macrophages and other hosts. Therefore, growing L. pneumophila in laboratories or in murine macrophages, which are not the natural host for this bacteria, could greatly affect the fitness and host range of L. pneumophila [74]. Furthermore, the restriction of L. pneumophila growth in murine macrophages by the acquisition of legS2 could select for bacteria that has increased adaptability to other hosts.
Acknowledgments
We would like to thank Katherine Ladner for performing the EMSA assays.
Data Availability
All data are in the main manuscript and are available in Dr. Amal Amer's laboratory at the Ohio State University.
Funding Statement
Studies in Dr. Arwa Abu Khweek’s laboratory are supported by Birzeit University. Studies in Dr. Amal Amer’s laboratory are supported by Ohio State University (OSU) Bridge funds, Public Health Preparedness for Infectious Diseases (PHPID), Center for Clinical and Translational Science (CCTS), R21 AI113477, and R01 HL094586). Dr. Amer provided cell lines, bacteria, some kits and working space for some of the conducted work and helped in revising the manuscript [http://cmib.osu.edu/people/faculty/amalamer/]. Dr. Arwa Abu Khweek does not have funding.
References
- 1.Losick VP, Isberg RR (2006) NF-kappaB translocation prevents host cell death after low-dose challenge by Legionella pneumophila. J Exp Med 203: 2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhter A, Gavrilin MA, Frantz L, Washington S, Ditty C, et al. (2009) Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog 5: e1000361 10.1371/journal.ppat.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amer AO Modulation of caspases and their non-apoptotic functions by Legionella pneumophila. Cell Microbiol 12: 140–147. 10.1111/j.1462-5822.2009.01401.x [DOI] [PubMed] [Google Scholar]
- 4.Isberg RR, O'Connor TJ, Heidtman M (2009) The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 7: 13–24. 10.1038/nrmicro1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hilbi H, Haas A (2012) Secretive bacterial pathogens and the secretory pathway. Traffic 13: 1187–1197. 10.1111/j.1600-0854.2012.01344.x [DOI] [PubMed] [Google Scholar]
- 6.Urwyler S, Brombacher E, Hilbi H (2009) Endosomal and secretory markers of the Legionella-containing vacuole. Commun Integr Biol 2: 107–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finsel I, Hilbi H (2015) Formation of a pathogen vacuole according to Legionella pneumophila: how to kill one bird with many stones. Cell Microbiol 17: 935–950. 10.1111/cmi.12450 [DOI] [PubMed] [Google Scholar]
- 8.Hubber A, Roy CR (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol 26: 261–283. 10.1146/annurev-cellbio-100109-104034 [DOI] [PubMed] [Google Scholar]
- 9.Rothmeier E, Pfaffinger G, Hoffmann C, Harrison CF, Grabmayr H, et al. Activation of Ran GTPase by a Legionella effector promotes microtubule polymerization, pathogen vacuole motility and infection. PLoS Pathog 9: e1003598 10.1371/journal.ppat.1003598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ninio S, Roy CR (2007) Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol 15: 372–380. [DOI] [PubMed] [Google Scholar]
- 11.Altman E, Segal G (2008) The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol 190: 1985–1996. 10.1128/JB.01493-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Felipe KS, Glover RT, Charpentier X, Anderson OR, Reyes M, et al. (2008) Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4: e1000117 10.1371/journal.ppat.1000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habyarimana F, Al-Khodor S, Kalia A, Graham JE, Price CT, et al. (2008) Role for the Ankyrin eukaryotic-like genes of Legionella pneumophila in parasitism of protozoan hosts and human macrophages. Environ Microbiol 10: 1460–1474. 10.1111/j.1462-2920.2007.01560.x [DOI] [PubMed] [Google Scholar]
- 14.Kubori T, Hyakutake A, Nagai H (2008) Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67: 1307–1319. 10.1111/j.1365-2958.2008.06124.x [DOI] [PubMed] [Google Scholar]
- 15.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR (2002) A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295: 679–682. [DOI] [PubMed] [Google Scholar]
- 16.Shohdy N, Efe JA, Emr SD, Shuman HA (2005) Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A 102: 4866–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machner MP, Isberg RR (2006) Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11: 47–56. [DOI] [PubMed] [Google Scholar]
- 18.Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, et al. (2006) The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8: 971–977. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Luo ZQ (2007) The Legionella pneumophila effector SidJ is required for efficient recruitment of endoplasmic reticulum proteins to the bacterial phagosome. Infect Immun 75: 592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, et al. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun 79: 1936–1950. 10.1128/IAI.00805-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ge J, Xu H, Li T, Zhou Y, Zhang Z, et al. (2009) A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci U S A 106: 13725–13730. 10.1073/pnas.0907200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y (2011) Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334: 1553–1557. 10.1126/science.1212868 [DOI] [PubMed] [Google Scholar]
- 23.Price CT, Jones SC, Amundson KE, Kwaik YA (2010) Host-mediated post-translational prenylation of novel dot/icm-translocated effectors of legionella pneumophila. Front Microbiol 1: 131 10.3389/fmicb.2010.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belyi Y, Tabakova I, Stahl M, Aktories K (2008) Lgt: a family of cytotoxic glucosyltransferases produced by Legionella pneumophila. J Bacteriol 190: 3026–3035. 10.1128/JB.01798-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller MP, Peters H, Blumer J, Blankenfeldt W, Goody RS, et al. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329: 946–949. 10.1126/science.1192276 [DOI] [PubMed] [Google Scholar]
- 26.Al-Quadan T, Price CT, Abu Kwaik Y (2012) Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol 20: 299–306. 10.1016/j.tim.2012.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilbi H (2006) Modulation of phosphoinositide metabolism by pathogenic bacteria. Cell Microbiol 8: 1697–1706. [DOI] [PubMed] [Google Scholar]
- 28.Jeschke A, Zehethofer N, Lindner B, Krupp J, Schwudke D, et al. (2015) Phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate regulate phagolysosome biogenesis. Proc Natl Acad Sci U S A 112: 4636–4641. 10.1073/pnas.1423456112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber S, Wagner M, Hilbi H (2014) Live-cell imaging of phosphoinositide dynamics and membrane architecture during Legionella infection. MBio 5: e00839–00813. 10.1128/mBio.00839-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Felipe KS, Pampou S, Jovanovic OS, Pericone CD, Ye SF, et al. (2005) Evidence for acquisition of Legionella type IV secretion substrates via interdomain horizontal gene transfer. J Bacteriol 187: 7716–7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degtyar E, Zusman T, Ehrlich M, Segal G (2009) A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol 11: 1219–1235. 10.1111/j.1462-5822.2009.01328.x [DOI] [PubMed] [Google Scholar]
- 32.Reiss U, Oskouian B, Zhou J, Gupta V, Sooriyakumaran P, et al. (2004) Sphingosine-phosphate lyase enhances stress-induced ceramide generation and apoptosis. J Biol Chem 279: 1281–1290. [DOI] [PubMed] [Google Scholar]
- 33.Sadosky AB, Wiater LA, Shuman HA (1993) Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun 61: 5361–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y (2008) A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Microbiol 70: 908–923. 10.1111/j.1365-2958.2008.06453.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhter A, Caution K, Abu Khweek A, Tazi M, Abdulrahman BA, et al. (2012) Caspase-11 Promotes the Fusion of Phagosomes Harboring Pathogenic Bacteria with Lysosomes by Modulating Actin Polymerization. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santic M, Molmeret M, Abu Kwaik Y (2005) Maturation of the Legionella pneumophila-containing phagosome into a phagolysosome within gamma interferon-activated macrophages. Infect Immun 73: 3166–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR (2001) How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: implications for conversion of plasma membrane to the ER membrane. J Cell Sci 114: 4637–4650. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar A, Duncan M, Hart J, Hertlein E, Guttridge DC, et al. (2006) ASC directs NF-kappaB activation by regulating receptor interacting protein-2 (RIP2) caspase-1 interactions. J Immunol 176: 4979–4986. [DOI] [PubMed] [Google Scholar]
- 39.Horwitz MA (1983) Formation of a novel phagosome by the Legionnaires' disease bacterium (Legionella pneumophila) in human monocytes. J Exp Med 158: 1319–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horwitz MA, Silverstein SC (1980) Legionnaires' disease bacterium (Legionella pneumophila) multiples intracellularly in human monocytes. J Clin Invest 66: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, et al. (2006) The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol 7: 318–325. [DOI] [PubMed] [Google Scholar]
- 42.Amer A, Franchi L, Kanneganti TD, Body-Malapel M, Ozoren N, et al. (2006) Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J Biol Chem 281: 35217–35223. [DOI] [PubMed] [Google Scholar]
- 43.Amer AO, Swanson MS (2005) Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol 7: 765–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamkanfi M, Kanneganti TD, Franchi L, Nunez G (2007) Caspase-1 inflammasomes in infection and inflammation. J Leukoc Biol 82: 220–225. [DOI] [PubMed] [Google Scholar]
- 45.Derre I, Isberg RR (2004) Macrophages from mice with the restrictive Lgn1 allele exhibit multifactorial resistance to Legionella pneumophila. Infect Immun 72: 6221–6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newton HJ, Ang DK, van Driel IR, Hartland EL (2010) Molecular pathogenesis of infections caused by Legionella pneumophila. Clin Microbiol Rev 23: 274–298. 10.1128/CMR.00052-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karin M, Lin A (2002) NF-kappaB at the crossroads of life and death. Nat Immunol 3: 221–227. [DOI] [PubMed] [Google Scholar]
- 48.Abu-Zant A, Jones S, Asare R, Suttles J, Price C, et al. (2007) Anti-apoptotic signalling by the Dot/Icm secretion system of L. pneumophila. Cell Microbiol 9: 246–264. [DOI] [PubMed] [Google Scholar]
- 49.Suen DF, Norris KL, Youle RJ (2008) Mitochondrial dynamics and apoptosis. Genes Dev 22: 1577–1590. 10.1101/gad.1658508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, et al. (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomes LC, Scorrano L (2011) Mitochondrial elongation during autophagy: a stereotypical response to survive in difficult times. Autophagy 7: 1251–1253. 10.4161/auto.7.10.16771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomes LC, Di Benedetto G, Scorrano L (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13: 589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bartfeld S, Engels C, Bauer B, Aurass P, Flieger A, et al. (2009) Temporal resolution of two-tracked NF-kappaB activation by Legionella pneumophila. Cell Microbiol 11: 1638–1651. 10.1111/j.1462-5822.2009.01354.x [DOI] [PubMed] [Google Scholar]
- 54.Schmeck B, N'Guessan PD, Ollomang M, Lorenz J, Zahlten J, et al. (2007) Legionella pneumophila-induced NF-kappaB- and MAPK-dependent cytokine release by lung epithelial cells. Eur Respir J 29: 25–33. [DOI] [PubMed] [Google Scholar]
- 55.Medzhitov R (2007) Recognition of microorganisms and activation of the immune response. Nature 449: 819–826. [DOI] [PubMed] [Google Scholar]
- 56.Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, et al. (2008) Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog 4: e1000220 10.1371/journal.ppat.1000220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brieland JK, Remick DG, Freeman PT, Hurley MC, Fantone JC, et al. (1995) In vivo regulation of replicative Legionella pneumophila lung infection by endogenous tumor necrosis factor alpha and nitric oxide. Infect Immun 63: 3253–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y, Tateda K, Fujita K, Ishii T, Ishii Y, et al. (2010) Sequential changes of Legionella antigens and bacterial load in the lungs and urines of a mouse model of pneumonia. Diagn Microbiol Infect Dis 66: 253–260. 10.1016/j.diagmicrobio.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 59.Sporri R, Joller N, Albers U, Hilbi H, Oxenius A (2006) MyD88-dependent IFN-gamma production by NK cells is key for control of Legionella pneumophila infection. J Immunol 176: 6162–6171. [DOI] [PubMed] [Google Scholar]
- 60.Tateda K, Matsumoto T, Ishii Y, Furuya N, Ohno A, et al. (1998) Serum cytokines in patients with Legionella pneumonia: relative predominance of Th1-type cytokines. Clin Diagn Lab Immunol 5: 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanchard DK, Djeu JY, Klein TW, Friedman H, Stewart WE II (1987) Induction of tumor necrosis factor by Legionella pneumophila. Infect Immun 55: 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McHugh SL, Newton CA, Yamamoto Y, Klein TW, Friedman H (2000) Tumor necrosis factor induces resistance of macrophages to Legionella pneumophila infection. Proc Soc Exp Biol Med 224: 191–196. [DOI] [PubMed] [Google Scholar]
- 63.Archer KA, Alexopoulou L, Flavell RA, Roy CR (2009) Multiple MyD88-dependent responses contribute to pulmonary clearance of Legionella pneumophila. Cell Microbiol 11: 21–36. 10.1111/j.1462-5822.2008.01234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Archer KA, Roy CR (2006) MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires' disease. Infect Immun 74: 3325–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuse ET, Tateda K, Kikuchi Y, Matsumoto T, Gondaira F, et al. (2007) Role of Toll-like receptor 2 in recognition of Legionella pneumophila in a murine pneumonia model. J Med Microbiol 56: 305–312. [DOI] [PubMed] [Google Scholar]
- 66.Archer KA, Ader F, Kobayashi KS, Flavell RA, Roy CR (2010) Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect Immun 78: 2477–2487. 10.1128/IAI.00243-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhan U, Trujillo G, Lyn-Kew K, Newstead MW, Zeng X, et al. (2008) Toll-like receptor 9 regulates the lung macrophage phenotype and host immunity in murine pneumonia caused by Legionella pneumophila. Infect Immun 76: 2895–2904. 10.1128/IAI.01489-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fernandez-Serrano S, Dorca J, Coromines M, Carratala J, Gudiol F, et al. (2003) Molecular inflammatory responses measured in blood of patients with severe community-acquired pneumonia. Clin Diagn Lab Immunol 10: 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McPhee JB, Mena P, Bliska JB (2010) Delineation of regions of the Yersinia YopM protein required for interaction with the RSK1 and PRK2 host kinases and their requirement for interleukin-10 production and virulence. Infect Immun 78: 3529–3539. 10.1128/IAI.00269-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park DR, Skerrett SJ (1996) IL-10 enhances the growth of Legionella pneumophila in human mononuclear phagocytes and reverses the protective effect of IFN-gamma: differential responses of blood monocytes and alveolar macrophages. J Immunol 157: 2528–2538. [PubMed] [Google Scholar]
- 71.Yoshizawa S, Tateda K, Matsumoto T, Gondaira F, Miyazaki S, et al. (2005) Legionella pneumophila evades gamma interferon-mediated growth suppression through interleukin-10 induction in bone marrow-derived macrophages. Infect Immun 73: 2709–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stavru F, Palmer AE, Wang C, Youle RJ, Cossart P (2013) Atypical mitochondrial fission upon bacterial infection. Proc Natl Acad Sci U S A 110: 16003–16008. 10.1073/pnas.1315784110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khweek AA, Caution K, Akhter A, Abdulrahman BA, Tazi M, et al. (2013) A bacterial protein promotes the recognition of the Legionella pneumophila vacuole by autophagy. Eur J Immunol 43: 1333–1344. 10.1002/eji.201242835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ensminger AW, Yassin Y, Miron A, Isberg RR (2012) Experimental Evolution of Legionella pneumophila in Mouse Macrophages Leads to Strains with Altered Determinants of Environmental Survival. PLoS Pathog 8: e1002731 10.1371/journal.ppat.1002731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are in the main manuscript and are available in Dr. Amal Amer's laboratory at the Ohio State University.