Summary
While transcription factors are prevalent among yeast prion proteins, the role of prion-mediated transcriptional regulation remains elusive. We show here that the yeast prion [SWI+] abolishes flocculin (FLO) gene expression and results in a complete loss of multicellularity. Further investigation demonstrates that besides Swi1, multiple other proteins essential for FLO expression, including Mss11, Sap30, and Msn1 also undergo conformational changes, and become inactivated in [SWI+] cells. Moreover, the asparagine-rich region of Mss11 can exist as prion-like aggregates specifically in [SWI+] cells, which are SDS-resistant, heritable, and curable, but become metastable after separation from [SWI+]. Our findings thus reveal a prion-mediated mechanism through which multiple regulators in a biological pathway can be inactivated. In combination with the partial loss-of-function phenotypes of [SWI+] cells on non-glucose sugar utilization, our data therefore demonstrate that a prion can influence differently on distinct traits through multi-level regulations, providing insights into the biological roles of prions.
Keywords: protein-aggregation, amyloids, prion, protein conformation change, Saccharomyces cerevisiae, yeast, Swi1, SWI/SNF, flocculin, filamentous growth, multicellularity
Introduction
Prions are self-perpetuating protein conformations that are often associated with protein misfolding, aggregation, and amyloidogenesis (Prusiner, 1998). Although the term prion was originally used to describe PrPSc, a causative agent of the infectious neurodegenerative disease known as transmissible spongiform encephalopathy or prion disease, it has now been extended to include a large group of proteins in fungi and mammals that can also be transmitted as altered and self-propagating conformations (Cascarina and Ross, 2014; Crow and Li, 2011; Prusiner, 2012; Soto, 2012). In the budding yeast Saccharomyces cerevisiae, at least nine prion proteins have been identified (Crow and Li, 2011; Garcia and Jarosz, 2014; Suzuki et al., 2012). Yeast prions can be manifested as epigenetic modifiers of important cellular processes, including translation and transcription, resulting in distinct and heritable phenotypic changes (Crow and Li, 2011; Sugiyama and Tanaka, 2014; Tuite, 2015).
There are at least six transcription factors among the identified yeast prions (Crow and Li, 2011). This prevalence of transcription factors suggests that the prion-based conformational switch may play a role in transcriptional regulation. Although a number of studies have shed light on how prions modify transcription of yeast (Holmes et al., 2013; True et al., 2004), the impact on transcriptional regulation by prion remains to be fully understood. Comparing to gene mutations, prion-mediated regulation has the advantage of not only being heritable but also reversible, as well as responsive to extreme environmental changes (Halfmann and Lindquist, 2010; Tuite, 2013). In particular, prion conformational switch of a global transcriptional regulator like Swi1 or Cyc8 may robustly alter the yeast transcriptome by turning on or off the expression of many target genes simultaneously (Du et al., 2008; Patel et al., 2009). However, we are just at the beginning of this line of research and definitive evidence, particularly the mechanisms through which yeast prions impose their impacts on transcription, remain to be elucidated.
S. cerevisiae can undergo a reversible change from a single-cell form to multiple distinct multicellular forms and it is believed that such a dimorphic switch is important for yeast to survive extreme environmental conditions (Gimeno et al., 1992). Yeast multicellular features include flocculation, biofilm formation, invasive growth of haploid cells, and pseudohyphal development of diploid cells. Flocculins or adhesins, a group of lectin-like cell wall proteins, are shown to be important for yeast to exhibit the described multicellular growth features (De Las Penas et al., 2003; Dranginis et al., 2007). In S. cerevisiae, flocculins are encoded by the FLO gene family, which includes the genes of FLO1, FLO5, FLO9, FLO10, and FLO11 (Guo et al., 2000; Hahn et al., 2005). These genes may have been evolved via gene duplication and they often undergo genomic silencing, noncoding RNA insertion and rearrangement, thus their expression and effect on multicellular growth are strain specific (Halme et al., 2004; Octavio et al., 2009). For instance, FLO11 is the only active FLO gene identified in Σ1278b, a common strain used for this line of research (Guo et al., 2000; Halme et al., 2004), whereas FLO1 and FLO11 are shown to be the two active genes of S288C (Kobayashi et al., 1999). In S288C derived strains, Flo1 is responsible for flocculation and adhesive growth on minimal agar plates and plastic surfaces, whereas Flo11 is the major flocculin that determines haploid invasive growth and diploid pseudohyphal growth (Fichtner et al., 2007).
At least five prion proteins, Ure2, Swi1, Cyc8, Mot3, and Sfp1, the protein determinants of [URE3], [SWI+], [OCT+], [MOT3+], and [ISP+] (Alberti et al., 2009; Du et al., 2008; Patel et al., 2009; Rogoza et al., 2010; Wickner, 1994), respectively, are potential transcriptional regulators of the FLO genes (Barrales et al., 2012). Recently, [MOT3+], the prion form of Mot3, a transcriptional repressor, has been shown to promote yeast multicellular growth, possibly through de-repression of FLO11 (Holmes et al., 2013). In this study, we examined how Swi1 and its prion form ([SWI+]) affect yeast multicellular growth. Swi1 is a key component of SWI/SNF, an ATP-dependent chromatin-remodeling complex, which functions as a global transcriptional regulator modulating the expression of ~6% of yeast genes (Sudarsanam et al., 2000). Upon prion conformational switch, [SWI+] causes a partial loss-of-function in utilizing non-glucose sugars (Du et al., 2008). We report here that [SWI+] also leads to a complete abolishment of multicellular growth in S288C cells. We further show that the lack of multicellularity of [SWI+] cells is caused by not only an insufficiency of Swi1 function but also by the functional sequestration of multiple additional co-activators that are essential for FLO gene expression. Our results demonstrate a prion-mediated mechanism through which the conformational switch of a prion protein can trigger the conformational changes of multiple proteins in the same biological pathway resulting in heritable changes in phenotypes.
Results
Adhesive growth, flocculation, and pseudohyphal growth are absent in swi1Δ and [SWI+] cells
Due to a nonsense mutation in FLO8, a gene encoding a key transcriptional activator of FLO genes, the most commonly used laboratory strain S288C completely lacks multicellular features (Liu et al., 1996). Upon FLO8 repair, the transcription of FLO1 and FLO11 in S288C derivative strains can be activated and all multicellular features except biofilm formation can be restored (Kobayashi et al., 1999). Although earlier research indicated that Swi1 is essential for flocculin synthesis in a couple of S. cerevisiae strains commonly used for studies on multicellularity (Barrales et al., 2008; Barrales et al., 2012), the requirement of Swi1 for FLO gene expression has not yet been shown for S288C. To investigate the effects of swi1Δ and [SWI+] on FLO gene expression and multicellularity, we repaired the chromosomal FLO8 mutation in isogenic S288C strains of [swi−] (wild-type, non-prion), swi1Δ, and [SWI+], and examined their multicellular phenotypes.
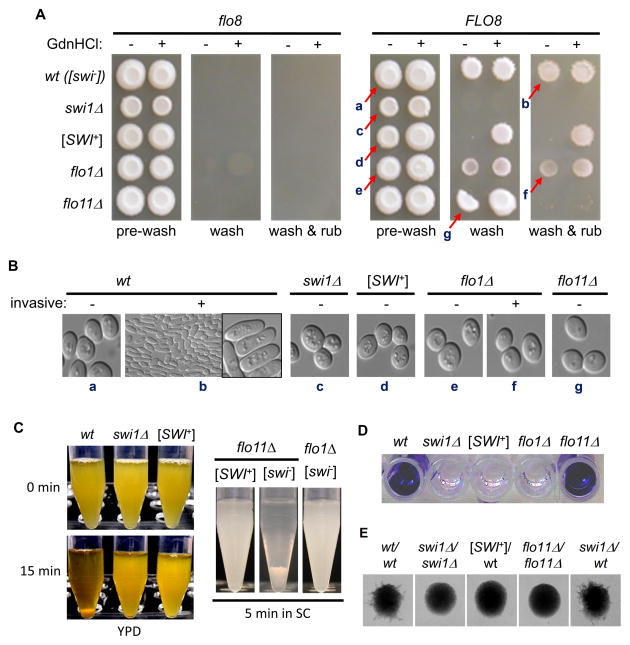
We first examined invasive growth of haploids on YPD plates. As shown in Figure 1A (left), in the absence of Flo8, none of the tested strains were able to adhere to YPD plates. After FLO8 repair, [swi−] cells could no longer be washed off by a mild wash, and a layer of cells remained on the plate even after wash with rubbing (Figure 1A, right). This suggests that cells regain the invasive growth ability after FLO8 repair. For flo11Δ cells, although their top layers could not be easily removed by a mild wash, all cells were completely washed off as big clumps upon wash with rubbing. In contrast, the top layers of flo1Δ cells could be easily washed off, but a layer of cells still remained on the agar plate even after a wash with rubbing. We observed that swi1Δ cells were completely removed by a mild wash, indicating that Swi1 function is required for invasive growth (Figure 1A). Surprisingly, like swi1Δ cells, [SWI+] cells exhibited no adhesive growth at all (Figure 1A). This result suggests that swi1Δ and [SWI+] might be similarly defective in flocculin synthesis. As expected, [SWI+] cells regained invasive growth after being treated with 5 mM guanidine hydrochloride (GdnHCl), which inactivates the molecular chaperone Hsp104 essential for [SWI+] propagation (Du et al., 2008) (Figure 1A). This result demonstrates that the reversible defect in adhesive growth of [SWI+] cells is conferred by [SWI+].
Figure 1. Yeast adhesion, flocculation and pseudohyphal growth were similarly impaired in [SWI+] and swi1Δ BY4741 cells.
(A) The indicated strains prior to and after repair of FLO8 were treated with (+) or without (−) 5 mM guanidine hydrochloride (GdnHCl), and spotted onto YPD plates. After 6 days of growth (pre-wash), yeast colonies were subjected to a washing assay. Wash, washing in a water bowl; wash & rub, washing with rubbing. (B) The arrowed cells in panel A were sampled and morphologically analyzed under a microscope, (+): invasive; (−): non-invasive. (C) Stationary-phase cultures of the indicated FLO8-repaired strains in YPD (left) or SC (right) were tested for flocculation. Cultures were vortexed and kept still for the indicated minutes before imaging. (D) The indicated strains were cultured in YPD in micro-titer plates and assayed for adhesion on polystyrene surface.. (E) Indicated FLO8-repaired diploids were examined for their pseudohyphal growth. The medium used was SLAD plates containing 4% glucose. See also Figure S1.
We found that the invasive [swi−] (non-prion) cells (retained on the plate after a vigorous wash) had an elongated cellular shape, which was not seen for the top-layer cells of the same [swi−] colony or any other tested [SWI+], swi1Δ, flo1Δ, or flo11Δ strains (Figure 1B), indicating that this unique morphology requires the functions of Swi1, Flo1, and Flo11. It is interesting to note that the Flo8-restored flo1Δ cells could undergo invasive growth but did not show an elongated cellular morphology, suggesting that the elongated cell-morphology and invasive-growth can be decoupled. We also found that the invasive growth was minimal and difficult to detect on SC plates and the elongated cell shape was not seen for all tested strains (data not shown). These results suggest that the elongated cell morphology is tightly associated with invasive growth and triggered by particular nutrient conditions that can be only achieved in rich media.
We next examined flocculation, a multicellular feature of cell-cell aggregation (Kobayashi et al., 1996), in FLO8-repaired [SWI+], [swi−], flo1Δ, and flo11Δ strains. We observed that flocculation can occur in both YPD and SC media, and it requires the function of Flo1 but not Flo11 (Figure 1C). Flocculation is absent for both [SWI+] and swi1Δ strains (Figure 1C). We also examined another multicellular feature – adhesive growth onto plastic surfaces. As shown in Figure 1D and S1A, Flo1, but not Flo11, was the major determinant of this feature and this adhesion was completely eliminated from both swi1Δ and [SWI+] cells. In addition, we found that another multicellular feature, pseudohyphae development of diploid cells, was absent in flo11Δ cells, and similarly abolished in swi1Δ and [SWI+] strains (Figure 1E). Noticeably, deletion of one copy of SWI1 did not significantly affect the pseudohyphal growth of the heterozygous diploid of SWI1/swi1Δ. However, diploids of [SWI+]/wt showed no pseudohyphal growth due to the prion dominance (Figure 1E). We also confirmed a previous report (Fichtner et al., 2007) that biofilm formation was absent in S288C-derivative strains after Flo8 restoration (data not shown). Taken together, our results show that Swi1 is essential for the tested multicellular phenotypes in S288C-derived strains and the formation of [SWI+] can result in a complete loss of these multicellular features.
Transcription of FLO1 and FLO11 is inactivated in both [SWI+] and swi1Δ cells
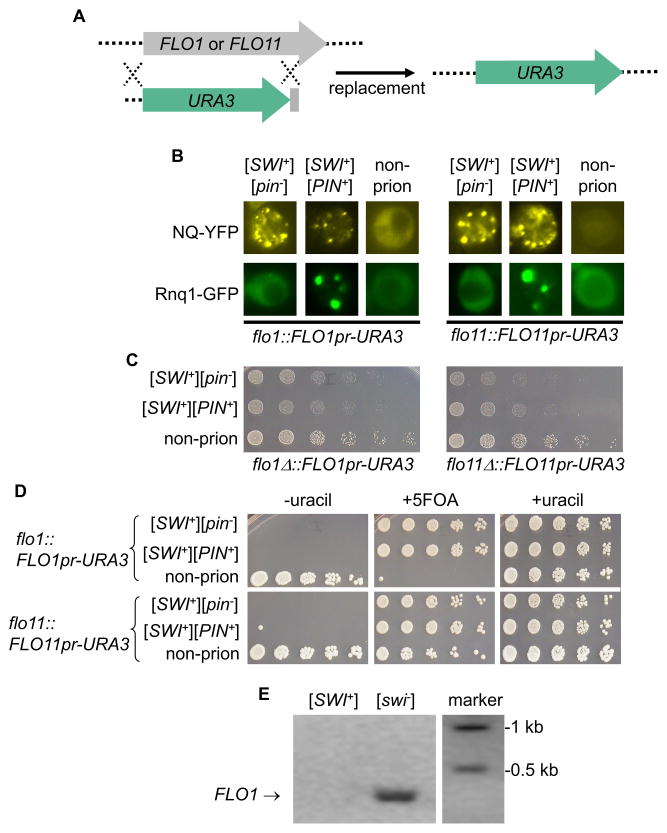
Since FLO1 and FLO11 are the only active FLO genes of S288C upon FLO8 repair (Kobayashi et al., 1999), we next investigate whether the abolishment of multicellularity in [SWI+] cells was due to repression of FLO1 and/or FLO11 expression. To do so, the ORF of FLO1 or FLO11 was replaced with the URA3 ORF to make a transcriptional fusion of FLO1 promoter-URA3 (FLO1pr-URA3) or FLO11 promoter-URA3 (FLO11pr-URA3) (Figure 2A). With the engineered strains, the promoter activity of FLO1 or FLO11 can be measured by a uracil/5-FOA (5-fluoroorotic acid) growth assay. We transferred the [SWI+] and/or [PIN+] prions into the engineered FLO1pr-URA3 or FLO11pr-URA3 cells. We found that Swi1-NQYFP and Rnq1-GFP formed fluorescence foci in these engineered [SWI+] and [PIN+] cells, respectively (Figure 2B). In addition, the engineered [SWI+] cells also exhibited partial deficiency of raffinose usage (Raf±) (Figure 2C). These results indicate that the prion features of [SWI+] and [PIN+] were not changed by the described genetic manipulations. [PIN+] was included in this study because it can be often spontaneously induced by [SWI+] (Du and Li, 2014). Since the normal biological function of Rnq1, the protein determinant of [PIN+], is currently unknown, we were not certain whether the presence of [PIN+] would influence FLO gene expression and the associated multicellular phenotypes.
Figure 2. FLO1 and FLO11 were not transcribed in [SWI+] and swi1Δ cells.
(A) A diagram showing the gene-replacement strategy of FLO1 or FLO11 ORF with the URA3 ORF, resulting in a transcriptional fusion of FLO1pr- or FLO11pr-URA3. (B) The FLO8 repair and FLO1 or FLO11 replacement did not alter aggregation patterns of Swi1 and Rnq1. The engineered strains were transformed with p415TEF-NQYFP or pCUP1-RNQ1GFP. NQ-YFP and Rnq1-GFP (induced by 50 μM CuSO4 overnight) signals were examined. (C) YPD cultures of the indicated FLO8-repaired strains were spotted onto raffinose plates, and images were taken after 3 days of growth. (D) YPD cultures of the indicated FLO8-restored strains harboring FLO1pr-URA3 or FLO11pr-URA3 were spotted onto SC plates without uracil (−uracil), with 1% 5 FOA (+5FOA), or with uracil (+uracil). Images were taken after a 3 days of growth. See also Figure S1. (E) RT-PCR to detect the mRNA level of FLO1 gene.
Upon FLO8 repair, [swi−] cells of the flo1::FLO1pr-URA3 strain could grow on SC-uracil but not on SC media supplemented with 5-FOA, indicating that the FLO1 promoter was activated (Figure 2D). Interestingly, the non-prion, wild-type flo11::FLO1pr-URA3 strain could grow on both SC-uracil and SC+5FOA plates (Figure 2D), suggesting that this promoter was partially active. Remarkably, the transcription of FLO1 and FLO11 was completely inactivated in [SWI+] or swi1Δ cells (Figure 2D and data not shown), which is consistent with the observed absence of multicellularity (Figure 1 and S1A). Results from a RT-PCR experiment confirmed that FLO1 transcription was present in [swi−] cells but absence in cells harboring [SWI+] (Figure 2E). Further experiments indicated that the transcription of FLO1 and FLO11 was not affected by [PIN+] as both isogenic [pin−] and [PIN+] strains showed similar multicellular features of adhesion and flocculation (data not shown). We also show that changes in carbon sources do not alter the tested multicellular patterns (Figure S1A and S1B). Taken together our results indicate that the lack of multicellularity of the FLO8-repaired swi1Δ and [SWI+] strains is caused by transcriptional inactivation of FLO1 and FLO11.
The lack of FLO gene expression and multicellularity in [SWI+] cells is not solely caused by insufficient Swi1 function
The complete loss of FLO gene expression and multicellularity is in a sharp contrast to the partial loss-of-function in raffinose usage exhibited by [SWI+] cells (compare Figure 2C and 2D). Our results suggest that different mechanisms are likely involved in regulating these two phenotypes: SWI/SNF-targeted gene(s) required for raffinose usage, such as SUC2 (Carlson and Botstein, 1982), may be partially repressed whereas the FLO1 and FLO11 genes required for multicellular features are completely inactivated in [SWI+] cells.
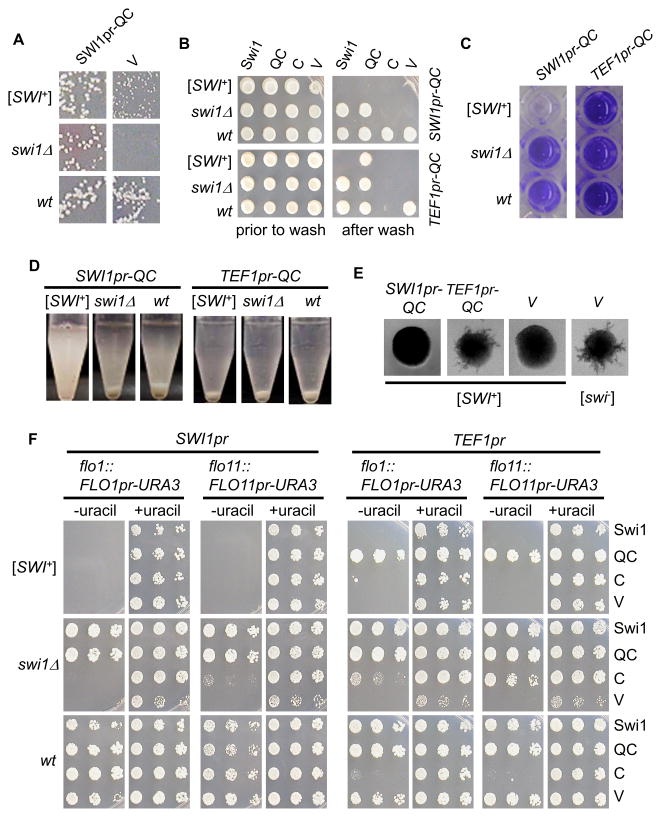
The Swi1 protein is comprised of three regions, an NH2-terminal asparagine-rich region (N), a middle glutamine-rich region (Q), and a COOH-terminal region (C) (Du et al., 2010). The N-region contains the prion domain (PrD) and contributes to aggregation and the [SWI+] prion features but is dispensable for the Swi1 function. The C- and QC-regions cannot join the Swi1 prion aggregates due to lack of the Swi1-PrD but provide normal chromatin-remodeling function of Swi1. Previously, we reported that QC or C was able to restore raffinose-usage of [SWI+] and swi1Δ cells (Du et al., 2010). To determine if this was also the case for FLO gene expression, we expressed the full-length Swi1, QC and C from a single-copy (CEN) plasmid using the SWI1 promoter (SWI1pr) or TEF1 promoter (TEF1pr) in FLO8-repaired strains. As expected, expression of Swi1, QC or C under either promoter fully suppressed the Raf− deficiency of the swi1Δ strain, and only QC or C, but not Swi1, was able to rescue the Raf± phenotype of [SWI+] cells (Figure 3A and data not shown). However, Swi1 or QC but not C driven by either promoter could fully restore the invasive growth of the swi1Δ strain (Figure 3B). An earlier study suggested that the Swi1-Q region might contain binding sites for several transcriptional regulators that recruit SWI/SNF to their target promoters (Prochasson et al., 2003). Our results suggest that such a transcription factor-interacting activity of Swi1-Q might be essential for invasive growth. The fact that C expression under TEF1pr greatly reduced invasive growth of the wild-type non-prion strain but C expression under SWI1pr, a much weaker promoter, had no detectable effect also supports the notion that the Q-region of Swi1 is essential for multicellularity. The overproduced nonfunctional Swi1 C domain may compete with the endogenous Swi1 to interact with other interacting partners and lead to a dominant negative effect.
Figure 3. The defect of [SWI+] cells in FLO gene expression and multicellularity was not solely due to insufficient Swi1 function.
As shown, full-length Swi1 (Swi1) and Swi1 functional domain with (QC) or without (C) the Q-rich region were expressed in the indicated FLO8-repaired strains driven by the SWI1 promoter (SWI1pr) or TEF1 promoter (TEF1pr). An empty vector (V) was used as a control. Transformants were analyzed for (A) non-glucose utilization assay on raffinose plates; (B) Wash assay on SC-ura plates; (C) Adhesion assay on polystyrene surface in SC-uracil medium (see also Figure S2); (D) Flocculation assay in SC-uracil medium (see also Figure S3); and (E) Pseudohyphal growth assay on SLAD (without uracil, 4% glucose) plates (see also Figure S4). (F) Transcription of FLO1 or FLO11 on SC plates without uracil (−uracil), with 5-FOA (+5FOA), or with uracil (+uracil). Strains are diploids (see materials and methods for information) for (E) but haploids for other panels. See also Figure S5.
As expected, expression of SWI1 or C could not restore any multicellular phenotypes of [SWI+] cells in which FLO8 was repaired (Figure 3B and S2–S4). Unexpectedly, the SWI1pr-QC could not rescue the multicellular phenotypes of [SWI+] cells but it could fully rescue the same defects of swi1Δ cells (Figure 3B–3E and S2–S4). Since SWI1pr-QC could fully restore the Raf± phenotype of [SWI+] (Figure 3A), these results demonstrate that insufficient Swi1 function is not the only cause of the multicellular defects observed in [SWI+] cells. Intriguingly, TEF1pr-QC was able to restore all tested multicellular growth phenotypes (Figure 3B–3E and S2–S4). The mechanism underling this dosage effect of Swi1 was further investigated and will be discussed in a later section of this paper.
To gain a better understanding of the observed phenotypes, we further examined the effect of SWI1, QC, and C expression on FLO1 and FLO11 transcription. To do so, SWI1 and its two sub-regions were expressed from the SWI1pr or TEF1pr in a CEN-plasmid in a FLO8-repaired flo1::FLO1pr-URA3 or flo11::FLO11pr-URA3 strain that was either [swi−], [SWI+], or swi1Δ (Figure 3F and data not shown). Results from SC-uracil growth assays showed that the effect of Swi1 and its sub-regions on multicellularity acted through modulation of the expression of the two FLO genes (compare Figures 3F, 3B–3E and S2–S5). Taken together, we conclude that the QC region of Swi1 is required for activating FLO gene expression and the lack of FLO expression and multicellularity in [SWI+] cells was not solely attributable to insufficient Swi1 function.
Other non-Swi1 FLO up-regulators have comprised functions in [SWI+] cells
We next examined if the observed lack of multicellularity was attributed to the repression of FLO8 expression in [SWI+] cells. We observed that Flo8 was expressed in similar levels in [SWI+] and [swi−] cells, suggesting that the observed multicellular defects of [SWI+] cells was not due to repression of FLO8 expression (Figure S6A). Since the multicellular deficiency of [SWI+] cells was reversible upon the GdnCl treatment, it is unlikely caused by a gene mutation(s). We hypothesized that in addition to Swi1, other non-Swi1 FLO gene up-regulators may also have compromised functions due to possible functional sequestrations by [SWI+]. To test this hypothesis, we chose six proteins - Mss11, Sap30, Gts1, Msn1, Snf5, and Flo8 - for examination. Our choice was based on two considerations: 1) Except for Flo8, they all contain a Q/N-rich region; and 2) All function as FLO gene up-regulators (Barrales et al., 2008; Bossier et al., 1997; Gagiano et al., 2003; van Dyk et al., 2005).
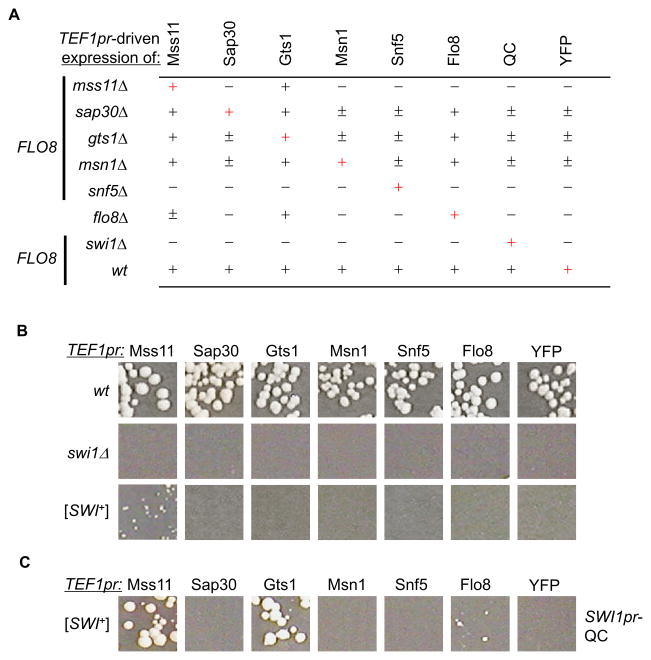
We first analyzed the functional relationship of the chosen regulators. We ectopically expressed each gene as a YFP fusion driven by TEF1pr in individual deletions. As shown in Figure 4A, for YFP only (controls), invasive growth was absent for mss11Δ, flo8Δ, snf5Δ, and swi1Δ cells, but partially seen for sap30Δ, gts1Δ, and msn1Δ cells. As expected, the deficiency of invasive growth of each mutant could be complemented by ectopic expression of its corresponding gene. We also found that MSS11- or GTS1-YFP expression under TEF1pr could suppress the deficiency of invasive growth shown by all tested mutants except snf5Δ and swi1Δ and TEF1pr FLO8-YFP showed similar effects except that it did not complement mss11Δ. These results confirmed earlier reports that Mss11, Gts1, and Flo8 are versatile regulators whose overexpression has a dominant suppressive effect over multiple non-SWI/SNF deletion mutants (Bester et al., 2006; Jin et al., 2008; Shen et al., 2006). Interestingly, overproduction of Swi1-QC or Snf5-YFP was only able to complement its own deletion (Figure 4 and data not shown) and overproduction of neither Swi1 nor QC could restore the multicellularity of the flo1Δ, flo11Δ, or flo8Δ mutants (Figure S4 and data not shown). Thus, FLO gene expression requires both the SWI/SNF complex and a group of additional transcriptional activators to work concertedly, and they are not functionally substitutable.
Figure 4. Additional FLO gene up-regulators besides Swi1 were functionally compromised in [SWI+] cells.
(A) Except for flo8Δ, CEN-FLO8 was ectopically expressed in all indicated strains. The listed 8 YFP fusions were expressed in the indicated strains driven by TEF1pr. Washing assays were done on selective SC-his-leu plates. Results from at least five experiments were summarized. +, adhesive; −, non-adhesive; ±, partial adhesive. Self-complementation is indicated by red +. (B) The FLO8-repaired wild-type (wt), swi1Δ, and [SWI+] strains of flo1::FLO1pr-URA3 were transformed with a CEN-plasmid expressing one of the indicated YFP fusions from TEF1pr. Cells were spread onto SC-uracil to examine FLO1pr-URA3 expression. Images were taken after 3 days of growth. (C) Co-expression of the indicated YFP fusions from TEF1pr and QC from SWI1pr in the FLO8-repaired [SWI+], flo1::FLO1pr-URA3 strain. Cells were spread onto SC-uracil plates and images were taken after 3 days of growth. Shown in (B) and (C) are representative data from at least five independent experiments.
We next tested whether overproduction of any of the FLO regulators, particularly Mss11, Gts1, and Flo8 could reactivate FLO gene expression in [SWI+] cells. We found that MSS11 was the only gene whose overexpression partly reactivated FLO1 expression in [SWI+] cells (Figure 4B). Interestingly, by providing Swi1 function at endogenous level, expression of Mss11-YFP fully activated the FLO1 transcription, and under this condition, Gts1-YFP and Flo8-YFP also had partially rescued the FLO1 transcription (Figure 4C). As the three transcription factors can suppress diverse mutations in the flocculin synthetic pathways under overproduction (Bossier et al., 1997; Lorenz and Heitman, 1998) (Figure 4A), one can postulate that additional FLO up-regulators besides Swi1 might be also functionally sequestered in [SWI+] cells.
Multiple non-Swi1 FLO gene activators undergo conformational changes in [SWI+] cells
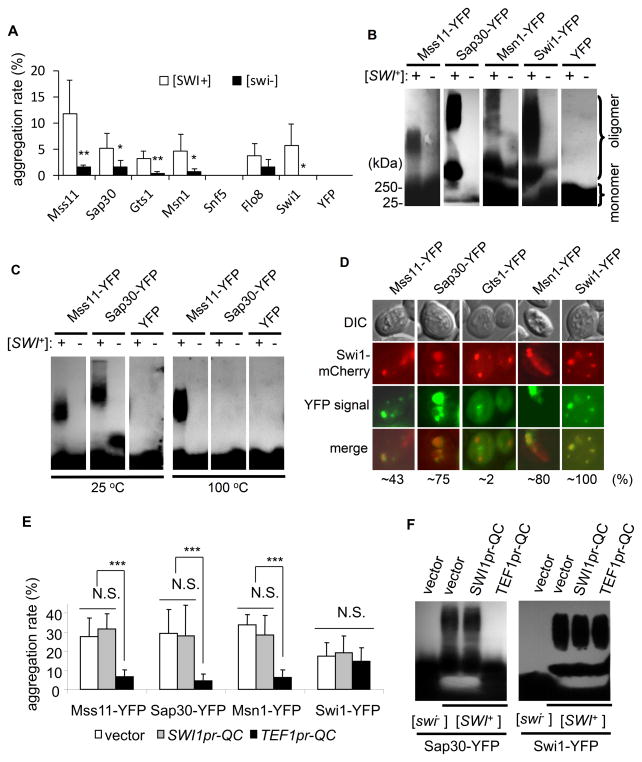
We next investigated if any of the six proteins, Mss11, Sap30, Gts1, Msn1, Flo8, and Snf5 had undergone conformational changes in [SWI+] cells. To do so, we expressed them in a pair of [SWI+] and [swi−] strains as YFP fusions driven by a TEF1pr, or GAL1pr in a CEN- or 2μ plasmid. With TEF1pr, Mss11-, Sap30-, and Gts1-YFP aggregated in [SWI+] cells; however, YFP foci were also seen in [swi−] cells (data not shown). It was challenging to tell if these foci in [swi−] cells represented aggregation or simply nuclear localization. We thus used GAL1pr to express and analyze the aggregation propensity of these proteins (Figure 5A, S6B and S7A). The frequency of foci appearance in isogenic [SWI+] and [swi−] cells was examined in a time course after adding galactose as an inducer to a final concentration of 0.5%. While Flo8-YFP, Snf5-YFP, and YFP did not show significant differences in aggregation between the [SWI+] and [swi−] strains, we found that Mss11-YFP, Sap30-YFP, Gts1-YFP, Msn1-YFP, and Swi1-YFP displayed significantly higher aggregation frequencies in [SWI+] cells than in [swi−] cells (Figure 5A and S6B).
Figure 5. A few Q/N-rich FLO up-regulators underwent conformational changes in [SWI+] cells.
(A) Individual YFP fusions were ectopically expressed from GAL1 promoter in a 2μ-plasmid in FLO8-repaired [SWI+] and [swi−] cells harboring flo1::FLO1pr-URA3. Galactose was added to 0.5% in log-phase culture of the transformants in SC-leucine + 2% sucrose to induce the expression. Aggregation frequency of each YFP fusion was assayed in a time course (see complete results in Figure S6B). Shown is a summary of five repeats upon 4 h of galactose addition. (B) The indicated YFP fusions were produced in isogenic [SWI+] and [swi−] strains from a CEN-GAL1pr plasmid. Log-phase cultures (in SC selective medium with sucrose as carbon source) were induced in the presence of 0.5% galactose before SDD-AGE assay. An anti-GFP monoclonal antibody was used to probe the blot. Shown is a representative result of five independent experiments. +, [SWI+]; −, [swi−]. (C) Experiments were done similarly to that shown in the panel B except that proteins were expressed from TEF1pr. Lysates were treated with 2% SDS at the indicated temperatures for 30 min before SDS-AGE assay. (D) The indicated YFP fusions were co-expressed with SWI1-mCherry driven by TEF1pr from a CEN-plasmid in isogenic [SWI+] and [swi−] strains. The overlapping frequency of a YFP fusion with Swi1mCherry is shown at the bottom. Results for the [swi−] strain were not shown as Swi1 did not aggregate. (E) The indicated YFP-fusions were expressed from GAL1pr in a 2μ-plasmid in [SWI+] cells. Swi1-QC (QC) was also co-expressed under SWI1pr or TEF1pr from a CEN-plasmid (with empty vector included as a control). Galactose was added to 2% in log-phase cultures to induce the expression of the indicated YFP fusions, and cells with YFP aggregation was examined after 4 h of induction. (F) SDD-AGE assay to verify the results shown in the panel E. Cultures in panel E co-expressing a YFP-fusion and Swi1-QC were prepared for SDD-AGE. The error bars represent standard deviation in panel A and E, and significance was analyzed by single-tail t-test (***, P<0.001; **, P <0.01; *, P < 0.05; N.S., not significant). See also Figures S6 and S7.
Next, we examined the biophysical properties of these YFP fusion aggregates in [SWI+] and [swi−] cells using semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) assay (Fan et al., 2007; Halfmann and Lindquist, 2008). We observed that under TEF1pr, Mss11, Sap30, and Msn1 formed SDS-resistant, high molecular-weight polymers in [SWI+] cells but not in isogenic [swi−] cells (Figure S7B). Similar results were obtained with the CEN-GAL1pr and 2μ-GAL1pr constructs (Figure 5B and S7C). Frequently Sap30 and Msn1 formed separate sized polymers on SDD-AGE blot, suggesting that they may have formed different SDS-tolerant aggregates with distinct conformations (Figure 5B and S7C). Interestingly, Mss11 aggregates from [SWI+] cells showed an extreme thermal stability and SDS resistance as high molecular-weight polymers were still detectable after a 30-minute boiling in the presence of 2% SDS (Figure 5C). For Gts1, Flo8, and Snf5, their SDD-AGE patterns were similar for [SWI+] and [swi−] cells (data not shown). Taken together, our results suggest that Mss11, Msn1, Sap30, and Gts1 have a higher aggregation propensity in [SWI+] cells, however, only the first three can form SDS-resistant aggregates specifically in [SWI+] cells. Thus, the lack of multicellularity of [SWI+] cells are likely a combined effect of conformational changes of Swi1 and additional FLO up-regulators including, but not necessarily limited to Mss11, Msn1, Sap30, and Gts1. To see if the aggregation of these proteins were caused by direct recruitment of Swi1 aggregates, Swi1-mCherry and one of these YFP fusions were co-produced in [SWI+] cells. We observed that Swi1-mCherry aggregates partially overlapped with that of Mss11-YFP, and a higher overlapping signal was seen for Sap30-YFP and Msn1-YFP aggregates (Figure 5D). These results suggest that Swi1 aggregates may directly interact with Mss11, Msn1, and Sap30 and such physical interactions are likely the cause of the aggregation of the three proteins in [SWI+] cells. However, the overlapping signals of Swi1-mCherry and the examined YFP fusions were not always seen, suggesting that their interactions may be dynamic in nature or these non-Swi1 proteins may also aggregate autonomously without direct interactions with Swi1 prion aggregates.
We then asked the question why TEF1pr-QC, but not SWI1pr-QC, could restore FLO gene expression and multicellular phenotypes in [SWI+] cells (Figure 3 and S3–S5). We hypothesized that Swi1-QC, which does not contain Swi1-PrD and thus cannot join Swi1 aggregates, can interact with diverse FLO gene up-regulators and recruit them to the FLO gene promoters. To test this hypothesis, we examined whether QC overproduction would reduce the aggregation of Mss11, Sap30, and Msn1 in [SWI+] cells. We co-transformed a [SWI+] strain with a GAL1pr-based plasmid expressing MSS11-, SAP30-, MSN1-, or SWI1-YFP and a plasmid expressing QC from SWI1pr or TEF1pr. The aggregation frequency of each YFP fusion was assayed upon galactose induction. As shown in Figure 5E, SWI1pr-QC had no detectable effect on the aggregation frequency of all tested proteins. However, overproduction by TEFpr-QC significantly reduced the aggregation of Mss11-, Sap30-, and Msn1-YFP, but not Swi1-YFP. This finding was confirmed by SDD-AGE analysis. As shown in Figure 5F, Swi1-QC overproduction significantly reduced the amount of SDS-tolerant Sap30 aggregation but had no effect on Swi1 aggregation. These results suggest that excessive QC can interact with these FLO up-regulators, thereby release them from aggregation in [SWI+] cells.
Mss11 prion-like domain can exist as prion-like aggregates in [SWI+] cells
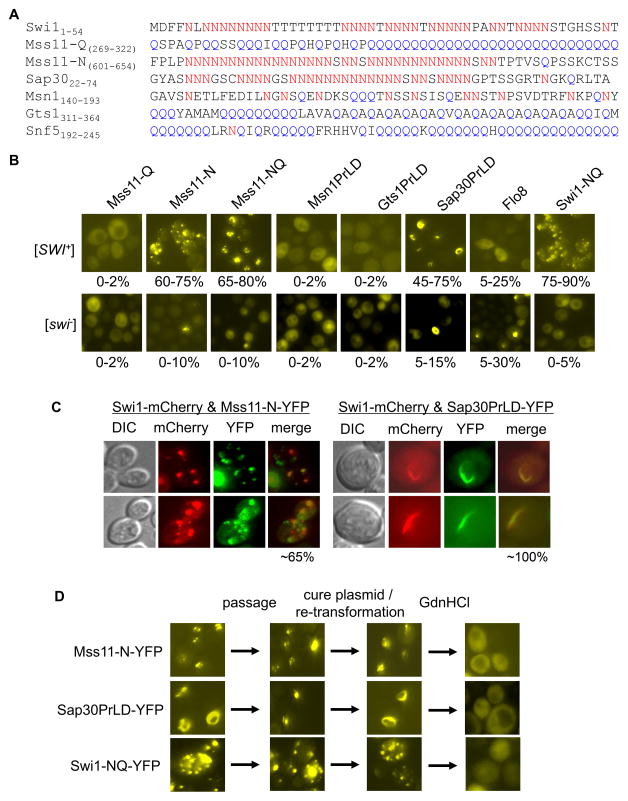
Our finding that the full-length Mss11, Sap30, and Msn1 could form SDS-resistant aggregates in [SWI+] cells raised another question: Was such aggregation mediated by the Q/N-rich prion-like domain (PrLD)? Based on a previous prediction (Alberti et al., 2009), Mss11 has two possible PrLDs: a Q-rich region (Mss11-Q, aa 254-456) and an N-rich region (Mss11-N, aa 487-654) (Figure 6A). Sap30 also has an N-rich PrLD (aa 10-74) resembling that of the Swi1-PrD, and the PrLDs of Gts1 (aa 296-396) and Snf5 (aa 1-294) are highly enriched in Q but not N residues (Figure 6A). When these putative PrLDs and the full-length Flo8 were fused to YFP and expressed in isogenic [SWI+] and [swi−] strains from TEF1pr in a CEN-plasmid, different YFP fluorescence patterns were observed. Mss11-N and Mss11-NQ (containing both N and Q regions) but not Mss11-Q aggregated in [SWI+] but not in [swi−] cells (Figure 6B), indicating that the N-rich but not the Q-rich PrLD contributed to the Mss11 aggregation in [SWI+] cells. Sap30-PrLD also had a significantly higher frequency of fluorescent foci in [SWI+] than in [swi−] cells. Other tested proteins did not show aggregation in either [SWI+] or [swi−] cells (Figure 6B). The Mss11-N-YFP aggregates were dot-like and partially overlapped with Swi1-mCherry aggregates in [SWI+] cells (Figure 6C). Sap30PrLD-YFP formed ring/rod-like aggregates that almost perfectly overlapped with the Swi1-mCherry aggregates that were, remarkably, changed from dot-shaped to ring/rod-shaped (Figure 6C). This prion aggregate remodeling is similar to the remodeling of [RNQ+] and [SWI+] prion aggregates from multiple dots to ring/rod/ribbons which significantly co-localize with the newly formed Sup35 ribbon-like aggregates upon Sup35 overexpression (Du and Li, 2014). Noticeably, the diffuse pattern of PrLD-YFP fusions of Msn1 and Gts1 (Figure 6B) is in contrast with the substantial aggregation of their full-length proteins in [SWI+] cells under similar conditions (Figure 5), suggesting that their PrLDs do not contribute to the aggregation of the full-length proteins.
Figure 6. Aggregation of the PrD-like domain (PrLD) of Mss11 and Sap30 in [SWI+] cells.
(A) Amino acid composition of the PrLD sequences of the indicated Q/N-rich proteins. The PrLDs were based on the prediction of a published article (Alberti et al, 2009). As indicated, Mss11 has two PrLD regions, Mss11-Q and Mss11-N. (B) YFP fusions of the indicated PrLDs and full-length Flo8 were ectopically expressed in [SWI+] and [swi−] strains from TEF1pr. Aggregation frequency was shown at the bottom of each representative image. (C) The YFP fusion of Mss11-N or Sap30PrLD was co-expressed with Swi1-mCherry, all driven by TEF1pr. Transformants were subjected to fluorescence microscopy assays. Shown is a representative result. The overlapping frequency was shown at bottom of the merged images. (D) YFP fusions of Swi1-NQ, Mss11-N, and Sap30PrLD were analyzed for their prion-like behaviors including aggregation maturation, inheritability and curability in [SWI+] cells.
We next investigated if the Mss11-N-YFP or Sap30-PrLD-YFP aggregates behave like prions in [SWI+] cells by analyzing their stability, maturation, inheritability, and curability (Figure 6D). For Mss11-N-YFP, the dot-shaped aggregates could be stably maintained upon re-streaking, similar to the mature prion aggregates of [PSI+], [RNQ+], and [SWI+] (Du and Li, 2014). The Mss11-N-YFP aggregation frequency and dot-shaped patterns were maintained upon eliminating the URA3-based expression plasmid of Mss11-N-YFP through a 5-FOA treatment then followed by re-introducing the same plasmid back to the system through transformation. When treated with 5mM GdnHCl, the Mss11-N-YFP foci were lost (Figure 6D), suggesting that the aggregation is prion-like. For Sap30-PrLD, the ribbon-shaped aggregates could turn to dots upon further passages. Although GdnHCl-treatment could cure the aggregation, the dot-like pattern cannot be retained after the procedure of eliminating and re-introducing the Sap30-PrLD-YFP plasmid. Under these conditions, Sap30-PrLD aggregation was no longer dot-like but back to ribbon-shaped. These results suggest that Sap30-PrLD aggregates in [SWI+] cells either represent non-prion like aggregation, or a heritable prion that does not cross-seed the full-length endogenous Sap30. Since Sap30 is less likely to be a prion in [SWI+] cells, we focused on the Mss11-PrLD aggregation in our later study.
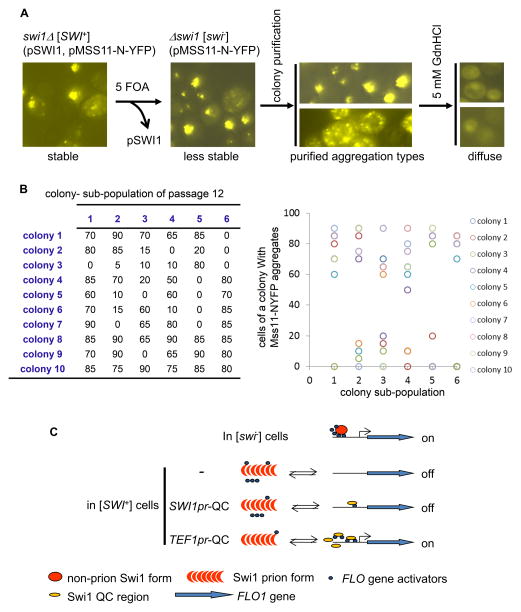
To test if the prion-like properties of Mss11-PrLD aggregation could be maintained and self-propagated in the absence of [SWI+], we introduced [SWI+] into a swi1Δ strain ectopically co-expressing Swi1 and Mss11-N-YFP. As expected, Mss11-N-YFP formed stable, dot-shaped aggregates that were curable by GdnHCl treatment (Figure 7A). Following elimination of the URA3-based SWI1 plasmid to cause the loss of [SWI+], we re-assayed the stability and curability of the Mss11-N-YFP aggregates. We found that the aggregation could be maintained but became unstable in the absence of [SWI+], and it was readily lost upon further passage without a biased selection for aggregate-containing isolates. If we selected and traced the aggregates-containing isolates by colony purification after [SWI+] elimination, Mss11-N-YFP aggregates could be maintained > 20 passages (at the time the experiment stopped) in the swi1Δ strain. Figure 7B represents a typical result of an analysis of one passage. At the 12th passage of this experiment, six individual aggregate-containing colonies were spread onto selective plates to form six subpopulations. Ten colonies were then randomly picked from each subpopulation to assay the Mss11-N-YFP aggregation frequency. We found that a large portion of the tested colonies still retained aggregation with 60–90% of cells containing Mss11-N-YFP aggregates while others had completely lost the aggregation (Figure 7B, left). The large variation in aggregation frequency was shown in Figure 7B, right. Noticeably, similar patterns were observed for each round of passage. We also observed that Mss11-N-YFP exhibited two types of aggregation patterns in [SWI+] cells, dispersed or localized foci. The two aggregation patterns could be isolated and maintained to a great extent after separating from [SWI+] though they were convertible at a low rate (Figure 7A). Taken together, our data suggest that the N-rich Mss11- PrLD can form prion-like aggregates that are associated with [SWI+] and become metastable after separation from [SWI+].
Figure 7. Mss11-N could form prion-like aggregates associated with [SWI+] but became metastable after separation from [SWI+].
(A) A [SWI+] prion strain was created from a swi1Δ strain co-expressing Swi1 and Mss11-N-YFP driven by TEF1pr from CEN-plasmids. Mss11-N-YFP formed stable and inheritable aggregates in [SWI+] cells. The Swi1-expressing plasmid was eliminated by counter-selection on 5 FOA-containing SC plates to eliminate [SWI+]. The Mss11-N-YFP aggregates were then examined for stability, localization, inheritability, and curability after eliminating [SWI+]. Two types of aggregates, dispersed or focused, could be isolated upon colony purification, both of which were eliminable by GdnHCl. (B) An example of the metastable feature of the prion-like Mss11-N aggregates after separating from [SWI+]. Shown is the result at the 12th passage of colony purification. Six aggregate-carrying colonies were spread to generate six sub-populations of colonies. For each sub-population, 10 randomly picked colonies were examined for frequency of cells containing the aggregates (left panel, numbers are percentages). The right panel is a scattered plot showing variations of the aggregation frequency. (C) A diagrammatic model. Briefly, the transcription of FLO genes (blue arrows) requires a coordinated action from the SWI/SNF complex and other up-regulators. In [SWI+] cells, these up-regulators undergo conformational changes and/or become aggregated, and are therefore functionally sequestered from the FLO gene promoters (black lines). Swi1-QC has a full transcriptional activity but does not join Swi1 aggregates in [SWI+] cells due to the lack of Swi1-PrD. This truncated Swi1 might still be able to interact with other SWI/SNF subunits and transcription factors that are essential for FLO gene expression. Therefore overproduction but not endogenous level of Swi1-QC will compete and reduce aggregation of these FLO gene up-regulators, and re-guide them to activate the FLO gene promoters.
Discussion
Yeast prions and their roles in regulating FLO gene expression and multicellular phenotypes
Given the importance of the dimorphic switch of yeast for survival it is not surprising that a significant portion of the yeast genome encodes genes that are involved in the development of multicellularity (Ryan et al., 2012). Among them are a group of genes encoding transcriptional regulators (Barrales et al., 2008; Jin et al., 2008; Ryan et al., 2012; Song et al., 2014), including the prion protein Swi1. In this study, we showed that in a set of FLO8-repaired S288C-derivative strains, the [SWI+] prion completely abolished the expression of FLO genes and multicellularity (Figure 1–2 and S1A). Interestingly, Holmes et al recently showed that the [MOT3+] prion is linked to yeast dimorphic switch - [MOT3+] endows yeast with the ability to grow multicellularly. The observed effects of [MOT3+] on multicellularity are largely opposite from what we observed for [SWI+]. They showed that [MOT3+] promotes FLO11 expression but the effect of [MOT3+] on FLO1 is less clear (Holmes et al., 2013). Some of these observations may reflect strain-specific effects since a wild strain instead of a laboratory strain was used in their study. In contrast to the FLO8-repaired S288C strain, this wild strain does not express FLO11 or show the tested multicellular features in the absence of a prion (Holmes et al., 2013). For [OCT+], one may assume that it imposes an effect that is opposite to [SWI+] on multicellularity because the inhibitory activity of Tup1/Cyc8 co-repressor on the FLO promoters should be abolished or lessened when Cyc8 adopts a prion conformation (Barrales et al., 2012; Fleming and Pennings, 2001; Mao et al., 2008), as proposed earlier (Crow and Li, 2011).
It would be of significance to investigate if the prions mentioned above can coordinate the regulation of FLO gene expression, and how such coordination responds to changes in environmental conditions. Their coordinated regulation of FLO gene expression might be realized through influencing each other’s appearance and/or transmission. Both negative and positive interactions among yeast prions have been described and it is believed that such interactions are largely mediated by the Q/N-rich PrDs (Bradley and Liebman, 2003; Derkatch et al., 2001; Du and Li, 2014; Schwimmer and Masison, 2002). Various prion-prion interactions may have different phenotypic outcomes and thus provide cells a flexibility to alter their surface features epigenetically according to environmental conditions. Intriguingly, formation and propagation of [MOT3+] are influenced by environmental signals such as ethanol and hypoxia, and the formation of [MOT3+] is accompanied by gain of multicellularity and thus considered beneficial (Holmes et al., 2013). We believe that the conversion between [SWI+] and [swi−] would also allow cells to sense and adapt to the environmental changes through the gain or loss of multicellularity, and therefore can be beneficial under certain conditions. For example, multicellularity may help yeast to survive poor nutrient conditions and protect cells from multiple stresses (Bruckner and Mosch, 2012; Granek and Magwene, 2010); however, when environmental conditions improve, yeast cells may favor resuming the unicellular form so that they can freely migrate and quickly multiply.
Mechanistic insight into the [SWI+]-triggered aggregation of multiple FLO gene regulators
Our results demonstrate that in addition to Swi1, there are other transcriptional activators whose functions are also compromised in [SWI+] cells. Although we have only shown that Mss11, Sap30, Msn1, and Gts1 preferentially aggregate in [SWI+] cells (Figure 5A and S6B), there may be more FLO up-regulators whose functions are comprised in [SWI+] cells. Importantly, since the multicellular defects of [SWI+] cells are reversible by a treatment of 5mM GdnHCl, one may conclude that the functional deficiencies of these FLO up-regulators are not due to mutations, but conformational changes are likely the major cause.
How does [SWI+] induce the aggregation of multiple different FLO activators? Swi1 has an N-rich PrD that is similar to the PrLDs of Mss11 and Sap30 but dissimilar to the PrLDs of Gts1, Msn1, and Snf5 in their amino acid composition (Figure 6A). Indeed, we observed co-aggregation of Swi1 with Mss11-N or with the Sap30-PrLD in [SWI+] cells and a partial co-aggregation of Swi1 with the full-length Mss11 and Sap30 (Figure 5D and 6C). These observations suggest that interactions between Swi1 and Mss11 or Sap30 in [SWI+] cells are likely mediated through cross-seeding between PrDs and/or PrLDs. Interestingly, the full-length Msn1 and Gts1 but not their PrLDs co-aggregated with Swi1 in [SWI+] cells (Figure 5D and 6C), suggesting that they may interact with Swi1 aggregates through their non Q/N-rich regions. Considering that many FLO up-regulators are Q/N-rich and some of them are natural interacting partners of SWI/SNF complex (Barrales et al., 2008; Barrales et al., 2012), we suspect that there might be more FLO up-regulators sequestered in [SWI+] cells by such direct interactions. Direct protein-protein interactions may also explain the dosage effect of Swi1-QC in suppressing the multicellular defects of [SWI+] cells (Figure 7C). Overproduction of QC but not the endogenous level is able to prevent the FLO up-regulators from being sequestered. This dichotomy likely results from the sequestration of diverse FLO up-regulators by [SWI+] requiring a large excess amount of QC to compete with the [SWI+] prion aggregates for binding of these FLO up-regulators. In addition to direct interactions, alternative mechanisms might also contribute to the sequestration of FLO up-regulators. The fact that Mss11, Sap30, and Mss11 do not always co-aggregate with Swi1 in [SWI+] cells (Figure 5D) suggests that these heterologous interactions may take place only at the initiation stage of aggregation or they aggregate by a titration mechanism - a preexisting prion can sequester anti-aggregation factors such as chaperones so that another aggregation prone protein has a higher opportunity to aggregate (Derkatch et al., 2001; Osherovich and Weissman, 2001).
We showed that Mss11-N is able to exist as prion-like aggregates in [SWI+] cells but becomes metastable in the absence of [SWI+] (Figure 7). Although it remains to be determined if the full-length Mss11 can be a [SWI+]-associated prion, the fact that it forms SDS-tolerant and reversible aggregates in [SWI+] cells (Figure 5, S6B, S7B and S7C) implicates this possibility. Even though there is no evidence that Sap30 can exist as a prion in [SWI+] cells, our observations that Sap30-PrLD can form ribbon-like aggregates that co-localize with Swi1 in [SWI+] cells and that this Sap30-PrLD aggregation is GdnHCl-curable suggests that the function of Sap30 might be also subjected to a prion-like regulation under certain conditions. Further research to explore under what conditions and to what extent the FLO up-regulators can become a prion(s) will provide mechanistic insights into prion-triggered aggregation of heterologous proteins and aid our understanding of multicellularity regulation.
Implication of prion-triggered protein sequestration in pathology of protein-misfolding disease
Previous studies showed that Sup45 and molecular chaperones, such as Hsp104 and Hsp70-Ssa1, can be sequestered by the Sup35 aggregates in [PSI+] cells (Bagriantsev et al., 2008; Vishveshwara et al., 2009; Yang et al., 2013). Similarly, [PIN+] aggregates are able to sequester Hsp40-Sis1 (Sondheimer et al., 2001).
Importantly, similar aggregation events can also occur in mammals. A number of recent studies have provided firm support to the proposal that a prion-like mechanism may play a role in the etiology of many of these protein misfolding diseases (Soto, 2012). It is notable that many protein misfolding-disease-associated proteins are also Q/N-rich, including huntingtin, TAF15, TDP-43, and FUS, while others such as p53, PrP, Aβ, and α-synuclein are not. Intriguingly, the aggregation of one such a pathogenic protein can often promote the aggregation of other aggregation-prone proteins. For example, Aβ aggregation can promote the aggregation of tau (Gotz et al., 2001), and α-synuclein aggregates can cross-seed and accelerate the aggregation of Aβ and tau (Guo et al., 2013; Ono et al., 2012). The well-known tumor-suppressor p53 can turn into an oncogenic factor when misfolded and aggregated; and in this case, the p53 aggregates possess several prion-like features and can seed and co-aggregate with additional anti-tumor factors and other proteins, including p63, p73, Mdm2, and several molecular chaperones (Ano Bom et al., 2012; Wang and Fersht, 2015). Protein aggregation and prion-mediated functional sequestration of important cellular factors are thus central events contributing not only to features of yeast prions but also pathogenesis of some protein-misfolding diseases. Our finding that [SWI+] can functionally sequester multiple distinct proteins to result in remarkable heritable changes in phenotype provides us an ideal system to study the interactions among prions, prion proteins, and other aggregation prone proteins.
Experimental Procedures
Oligonucleotides, plasmids and yeast strains
Information on oligonucleotides, yeast engineering procedures, and DNA sequences is shown in Table S1. Plasmids used in this study are shown in Table S2. S. cerevisiae strains used in this study are all BY strains derived from S288C except for LY422 and DY902 (Table S3). All yeast cultures were grown at 30°C unless specified. Description of plasmids and strain engineering is available in the Supplemental Experimental Procedures.
Major Experimental Procedures
A protein extract-based transformation method was used to transfer [SWI+] as described previously (Du et al., 2010), using DY902 as extract donor. Methods for yeast mating, sporulation, and mating-type test are described in detail in the supplementary information.
Invasive growth on YPD and SC plates was performed as described by (Braus et al., 2003; Fichtner et al., 2007; Roberts and Fink, 1994) with minor changes. Basically, a washing-based assay was used to examine the binding force of cells to agar plates. Adhesion to plastic surfaces was conducted with flat-bottom 96-well micro-titer polystyrene plates (Reynolds and Fink, 2001) in media with either 2% or 0.1% glucose. Flocculation assays were carried our similarly to a previous report (Kobayashi et al., 1996). Yeast’s mobility associated with biofilm formation was assayed on semi-soft YPD plates with 0.3% and 0.2% glucose according to an earlier study (Reynolds and Fink, 2001). Pseudohyphal growth of diploids was tested on SLAD plates (Gimeno et al., 1992), with 2% or 4% glucose was supplemented as carbon source.
Fluorescence microscopic assay was performed with GFP-, YFP-, and mCherry-tagged proteins, with methods described (Du and Li, 2014). Semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) assay was carried out based on the published protocol (Halfmann and Lindquist, 2008) with modifications. In brief, after clearing cell lysates by centrifugation, both pellet and supernatant fractions were collected and assayed. Lysates were also treated with DNase I if the viscosity was too high. SDS-tolerance was conducted with 2% SDS at 25°C or 100°C for 30 min before sample loading.
For RT-PCR, total mRNA was extracted and purified with RNeasy Mini Kit (QIAGEN, cat# 74104), and cDNA was synthesized with SuperScript First-strand Synthesis SuperMix (Invitrogen, cat# 18080-400) with oligo(dT) according to the manufacturer’s instructions. PCR was performed with primer pair of FLO1-F and FLO1-R to detect the FLO1 gene transcription (50 ng cDNA in a 50 μL system as template, 25 cycles). Detailed methods are described in Supplementary Experimental Procedures.
Supplementary Material
Acknowledgments
The authors thank Dr. G. Braus (Georg August University Göttingen, Institute for Microbiology and Genetics) for the gift of plasmids; and S. Valtierra and D. Goncharoff for critical comments and manuscript editing. This work was supported by grants from the U.S. National Institutes of Health (R01NS056086) and U.S. National Science Foundation (MCB 1122135) to L.L., and from China Scholarship Council (CSC) (File No. [2012]3022-201208110572) and National Natural Science Foundation of China (81100809 and 81271417) to Y.Z.
Footnotes
Document S1. Figure S1–S7, Table S1–S3, Supplemental Experimental Procedures, and Supplemental References.
Author Contributions
Z.D. and L.L. conceived and designed the research. Z.D. conducted most of the experiments. Y.Z. performed some SDD-AGE experiments. Z.D. and L.L. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Bom AP, Rangel LP, Costa DC, de Oliveira GA, Sanches D, Braga CA, Gava LM, Ramos CH, Cepeda AO, Stumbo AC, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: implications for cancer. The Journal of biological chemistry. 2012;287:28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Molecular biology of the cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Jimenez J, Ibeas JI. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics. 2008;178:145–156. doi: 10.1534/genetics.107.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics. 2012;191:791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bester MC, Pretorius IS, Bauer FF. The regulation of Saccharomyces cerevisiae FLO gene expression and Ca2+-dependent flocculation by Flo8p and Mss11p. Current genetics. 2006;49:375–383. doi: 10.1007/s00294-006-0068-z. [DOI] [PubMed] [Google Scholar]
- Bossier P, Goethals P, Rodrigues-Pousada C. Constitutive flocculation in Saccharomyces cerevisiae through overexpression of the GTS1 gene, coding for a ‘Glo’-type Zn-finger-containing protein. Yeast. 1997;13:717–725. doi: 10.1002/(SICI)1097-0061(19970630)13:8<717::AID-YEA132>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braus GH, Grundmann O, Bruckner S, Mosch HU. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Molecular biology of the cell. 2003;14:4272–4284. doi: 10.1091/mbc.E03-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner S, Mosch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS microbiology reviews. 2012;36:25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cascarina SM, Ross ED. Yeast prions and human prion-like proteins: sequence features and prediction methods. Cellular and molecular life sciences: CMLS. 2014;71:2047–2063. doi: 10.1007/s00018-013-1543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow ET, Li L. Newly identified prions in budding yeast, and their possible functions. Seminars in cell & developmental biology. 2011;22:452–459. doi: 10.1016/j.semcdb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Las Penas A, Pan SJ, Castano I, Alder J, Cregg R, Cormack BP. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes & development. 2003;17:2245–2258. doi: 10.1101/gad.1121003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN+] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiology and molecular biology reviews: MMBR. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Crow ET, Kang HS, Li L. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Molecular and cellular biology. 2010;30:4644–4655. doi: 10.1128/MCB.00225-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Li L. Investigating the interactions of yeast prions: [SWI+], [PSI+], and [PIN+] Genetics. 2014;197:685–700. doi: 10.1534/genetics.114.163402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtner L, Schulze F, Braus GH. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Molecular microbiology. 2007;66:1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Pennings S. Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation. The EMBO journal. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano M, Bester M, van Dyk D, Franken J, Bauer FF, Pretorius IS. Mss11p is a transcription factor regulating pseudohyphal differentiation, invasive growth and starch metabolism in Saccharomyces cerevisiae in response to nutrient availability. Molecular microbiology. 2003;47:119–134. doi: 10.1046/j.1365-2958.2003.03247.x. [DOI] [PubMed] [Google Scholar]
- Garcia DM, Jarosz DF. Rebels with a cause: molecular features and physiological consequences of yeast prions. FEMS yeast research. 2014;14:136–147. doi: 10.1111/1567-1364.12116. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Gotz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293:1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- Granek JA, Magwene PM. Environmental and genetic determinants of colony morphology in yeast. PLoS genetics. 2010;6:e1000823. doi: 10.1371/journal.pgen.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Covell DJ, Daniels JP, Iba M, Stieber A, Zhang B, Riddle DM, Kwong LK, Xu Y, Trojanowski JQ, et al. Distinct alpha-synuclein strains differentially promote tau inclusions in neurons. Cell. 2013;154:103–117. doi: 10.1016/j.cell.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, De Bie T, Stajich JE, Nguyen C, Cristianini N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome research. 2005;15:1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. Journal of visualized experiments: JoVE. 2008 doi: 10.3791/838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–632. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116:405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Holmes DL, Lancaster AK, Lindquist S, Halfmann R. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–165. doi: 10.1016/j.cell.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Dobry CJ, McCown PJ, Kumar A. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Molecular biology of the cell. 2008;19:284–296. doi: 10.1091/mbc.E07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi O, Suda H, Ohtani T, Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Molecular & general genetics: MGG. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- Kobayashi O, Yoshimoto H, Sone H. Analysis of the genes activated by the FLO8 gene in Saccharomyces cerevisiae. Current genetics. 1999;36:256–261. doi: 10.1007/s002940050498. [DOI] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics. 1998;150:1443–1457. doi: 10.1093/genetics/150.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Li Y, Wang H, Cao F, Chen J. Antagonistic interplay of Swi1 and Tup1 on filamentous growth of Candida albicans. FEMS microbiology letters. 2008;285:233–241. doi: 10.1111/j.1574-6968.2008.01236.x. [DOI] [PubMed] [Google Scholar]
- Octavio LM, Gedeon K, Maheshri N. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS genetics. 2009;5:e1000673. doi: 10.1371/journal.pgen.1000673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Takahashi R, Ikeda T, Yamada M. Cross-seeding effects of amyloid beta-protein and alpha-synuclein. Journal of neurochemistry. 2012;122:883–890. doi: 10.1111/j.1471-4159.2012.07847.x. [DOI] [PubMed] [Google Scholar]
- Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI+] prion. Cell. 2001;106:183–194. doi: 10.1016/s0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11:344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Molecular cell. 2003;12:983–990. doi: 10.1016/s1097-2765(03)00366-6. [DOI] [PubMed] [Google Scholar]
- Prusiner SB. Prions. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB, Fink GR. Bakers’ yeast, a model for fungal biofilm formation. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes & development. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rogoza T, Goginashvili A, Rodionova S, Ivanov M, Viktorovskaya O, Rubel A, Volkov K, Mironova L. Non-Mendelian determinant [ISP+] in yeast is a nuclear-residing prion form of the global transcriptional regulator Sfp1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10573–10577. doi: 10.1073/pnas.1005949107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin ZY, Cox MJ, Vizeacoumar F, Cheung D, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- Schwimmer C, Masison DC. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Molecular and cellular biology. 2002;22:3590–3598. doi: 10.1128/MCB.22.11.3590-3598.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Iha H, Yaguchi S, Tsurugi K. The mechanism by which overexpression of Gts1p induces flocculation in a FLO8-inactive strain of the yeast Saccharomyces cerevisiae. FEMS yeast research. 2006;6:914–923. doi: 10.1111/j.1567-1364.2006.00086.x. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lopez N, Craig EA, Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. The EMBO journal. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Johnson C, Wilson TE, Kumar A. Pooled segregant sequencing reveals genetic determinants of yeast pseudohyphal growth. PLoS genetics. 2014;10:e1004570. doi: 10.1371/journal.pgen.1004570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Transmissible proteins: expanding the prion heresy. Cell. 2012;149:968–977. doi: 10.1016/j.cell.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S, Tanaka M. Self-propagating amyloid as a critical regulator for diverse cellular functions. Journal of biochemistry. 2014;155:345–351. doi: 10.1093/jb/mvu026. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- Tuite MF. The natural history of yeast prions. Advances in applied microbiology. 2013;84:85–137. doi: 10.1016/B978-0-12-407673-0.00003-5. [DOI] [PubMed] [Google Scholar]
- Tuite MF. Yeast prions: Paramutation at the protein level? Seminars in cell & developmental biology. 2015 doi: 10.1016/j.semcdb.2015.08.016. [DOI] [PubMed] [Google Scholar]
- van Dyk D, Pretorius IS, Bauer FF. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae. Genetics. 2005;169:91–106. doi: 10.1534/genetics.104.033704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishveshwara N, Bradley ME, Liebman SW. Sequestration of essential proteins causes prion associated toxicity in yeast. Molecular microbiology. 2009;73:1101–1114. doi: 10.1111/j.1365-2958.2009.06836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Fersht AR. Propagation of aggregated p53: Cross-reaction and coaggregation vs. seeding. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2443–2448. doi: 10.1073/pnas.1500262112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB. [URE3] as an altered Ure2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Yang Z, Hong JY, Derkatch IL, Liebman SW. Heterologous gln/asn-rich proteins impede the propagation of yeast prions by altering chaperone availability. PLoS genetics. 2013;9:e1003236. doi: 10.1371/journal.pgen.1003236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.