Hair color differences are among the most obvious examples of phenotypic variation in humans. Although genome-wide association studies (GWAS) have implicated multiple loci in human pigment variation, the causative base pair changes are still largely unknown1. Here we dissect a regulatory region of the KIT ligand (KITLG) gene that is significantly associated with common blond hair color in northern Europeans2. Functional tests demonstrate that the region contains a regulatory enhancer that drives expression in developing hair follicles. This enhancer contains a common single nucleotide polymorphism (SNP rs12821256) that alters the binding site for the lymphoid enhancer-binding factor (LEF) transcription factor, reducing LEF responsiveness and enhancer activity in cultured human keratinocytes. Mice carrying ancestral or derived variants of the human KITLG enhancer exhibit significant differences in hair pigmentation, confirming that altered regulation of an essential growth factor contributes to the classic blond hair phenotype found in northern Europeans.
Humans show striking differences in hair pigmentation within and among populations. While dark hair color predominates in many African, Asian, and southern European populations, lighter hair colors are common in northern Europe. Blond hair, which occurs naturally in a small fraction of humans, has had a notable range of both positive and negative associations in human history. In some cultures, light skin and hair is stigmatized as a ghost-like abnormality, sign of promiscuity, or signature of unusual ancestry3,4. In contrast, fair hair was associated with youth and beauty in the earliest written works of ancient Greece5, and has been frequently imitated (or masked) using bleaches, dyes, and wigs in both ancient and modern populations4. Despite thousands of years of interest in hair color variation, the molecular basis of common human hair color phenotypes are still incompletely understood. Recent genome-wide surveys have shown that genetic variants linked to eight genes are significantly associated with blond hair color in Europeans2,6–8. Some human blond-associated variants alter the coding regions of known pigmentation genes1,9. However, many GWAS signals for pigmentation and other human traits map outside protein-coding regions of genes1,10, are enriched in likely regulatory sequences11, and have been difficult to trace to particular DNA base-pair mutations12. Knowing the causative nucleotides underlying human traits may improve genetic predictions compared to common linked markers13, and facilitate comparison of traits and mutations among both past and present populations14.
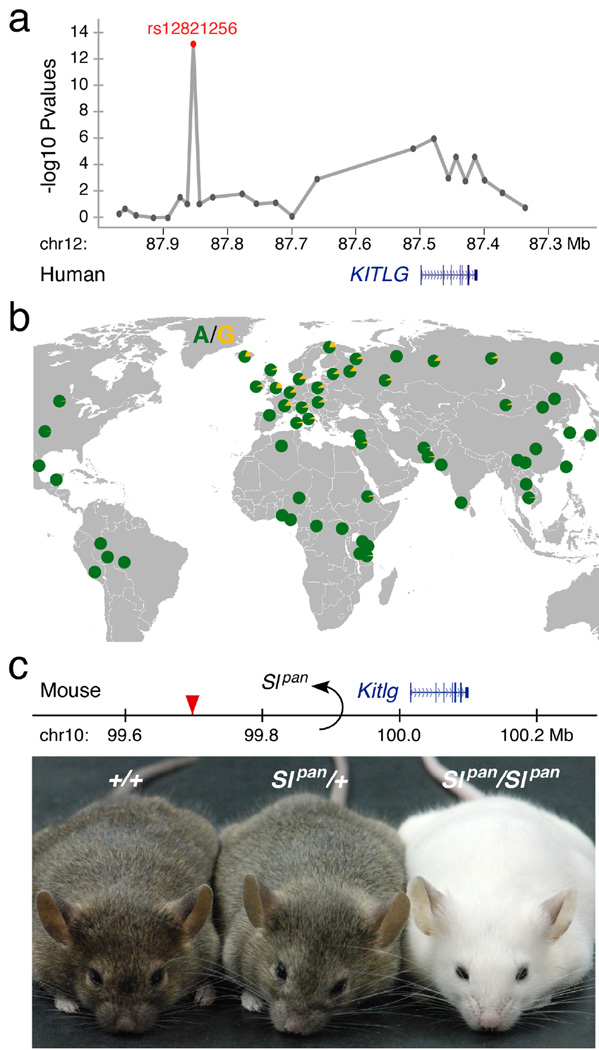
KITLG encodes a secreted ligand for the receptor tyrosine kinase, KIT and plays an essential role in development, migration, and differentiation of many different cell types in the body, including melanocytes, blood cells, and germ cells15. Null mutations affecting Kitlg or Kit are lethal in mice, and hypomorphic alleles cause white hair, mast cell defects, anemia, and sterility16–18. A non-coding SNP (rs12821256) located in a large intergenic region over 350 kb upstream of the KITLG transcription start site is significantly associated with blond hair color in Iceland and the Netherlands2 (Fig. 1a). This SNP shows relatively large odds ratios of 1.9 – 2.4 per risk allele for blond vs. brown hair in northern Europeans (multiplicative model2). Together with other genes, rs12821256 helps explain 3 to 6% of variance in categorical hair color scores2, and is now one of several markers used for predictive testing of human hair color19. The blondassociated A to G substitution at this position is prevalent in northern European populations but virtually absent in Africa and Asia2,20,21 (Fig. 1b), suggesting that regulatory changes associated with an essential signaling gene may contribute to common blond hair color in Europe.
Figure 1. A distant regulatory region upstream of the KITLG gene controls hair pigmentation in humans and mice.
(a) SNPs on human chromosome 12 are associated with blond hair in Europeans (modified from2). The peak association is found at rs12821256 (red), which is 355 kb upstream of the KITLG transcription start site. (b) The frequency distribution of rs12821256 in different populations. The allele associated with blond hair, G (yellow), is most prevalent in northern Europe. (c) The Slpan allele at the mouse Kitlg locus consists of a 65.6 megabase chromosome inversion (GRCm38/mm10 chr10 breakpoints: 34.301 Mb - 99.916 Mb) that displaces upstream sequences orthologous to rs12821256 (red triangle). Heterozygous (Slpan/+) and homozygous (Slpan/Slpan) mice have lighter coats than control (+/+) animals, demonstrating that alteration of even a single copy of the region upstream of Kitlg can reduce hair pigmentation.
Regulatory mutations in mice confirm the importance of distant upstream sequences in the control of hair color. The Steel panda (Slpan) mutation, a viable X-ray induced allele that reduces pigmentation in both heterozygotes and homozygotes22, results from a large chromosome inversion upstream of Kitlg23. We sequenced the precise molecular breakpoints of this inversion for the first time (Methods), showed that the inversion spans 65.6 megabases, and confirmed that the position of the breakpoints would displace mouse sequences orthologous to the blond-associated GWAS peak. Mice homozygous for the mutation are white, and heterozygous animals carrying only a single copy of the Slpan mutation are significantly lighter than control mice (Fig. 1c). Quantitative RT-PCR assays show that heterozygote animals express 61±9.1% of wild type Kitlg RNA levels in skin (mean ± s.e.m., P=0.0022), showing that displacement of a single copy of distant upstream Kitlg regulatory sequences is sufficient to reduce Kitlg expression and lighten hair color.
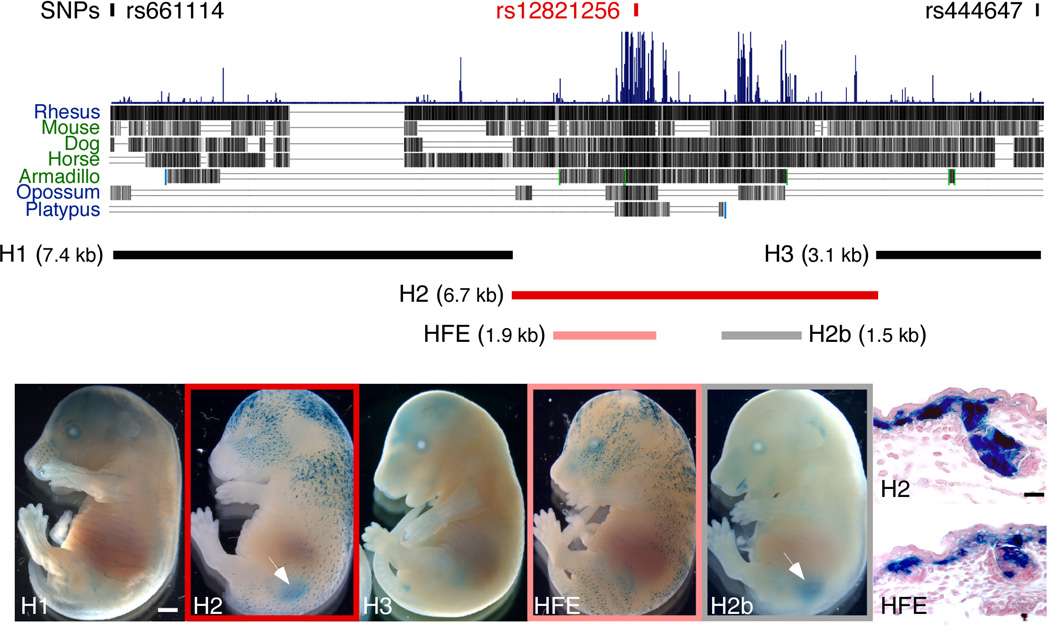
To identify the base pair changes responsible for the blond hair association in humans, we used a transgenic approach to search for functional enhancers throughout the GWAS candidate interval surrounding rs128212562. Three segments of human DNA completely spanning the 17.1 kb blond-associated region (as defined by the nearest flanking non-significant markers in the GWAS study) were separately cloned upstream of a minimal promoter and lacZ reporter gene (Fig. 2a). Only the 6.7 kb region, H2, drove consistent reporter expression in transgenic mouse embryos (Fig. 2c and Supplementary Fig. 1). lacZ expression was visible in the kidney (N=14/15) and in developing hair follicles (N=13/15). We subsequently tested two smaller clones from the H2 region, each of which overlapped major peaks of mammalian sequence conservation. One of these fragments, H2b, drove consistent expression in kidney (Fig. 2f and Supplementary Fig. 2; N=12/13) while the other fragment, HFE (for hair follicle enhancer), drove consistent expression in developing hair follicles (Fig. 2e and Supplementary Fig. 3; N=8/11). Histological analysis demonstrated that the latter expression occurred in the epithelial cells of the developing hair and skin (Fig. 2g,h), corresponding to a known site of endogenous Kitlg expression24 that plays an important role in attracting melanocytes to developing epidermis and hair follicles25.
Figure 2. The human blond-associated region contains a functional hair enhancer.
(a) A 17.1 kb region bounded by SNPs rs444647 and rs661114 defines the candidate interval for blondness2. Within this region, a large block of mammalian sequence conservation overlaps peak marker rs12821256. Five human fragments were cloned upstream of a lacZ reporter gene and tested for in vivo enhancer activity in transgenic mice. (b–f) Representative transgenic embryos generated by pronuclear microinjection of different lacZ constructs, processed at E16.5 to reveal lacZ gene activity (blue staining). Scale bar, 1 mm. (b) H1. (c) H2. (d) H3. (e) HFE (for hair follicle enhancer). (f) H2b. Of the three clones spanning the entire interval, only the 6.7 kb clone, H2, produced consistent lacZ expression in skin and kidney (arrow). Analysis of two subclones from H2 separated HFE skin (e) and H2b kidney (f, arrow) enhancers. (g,h) 6 µm crosssections through E16.5 dorsal skin from (g) H2 and (h) HFE transgenic embryos counterstained with nuclear fast red. Strong lacZ expression is visible in the basal epithelium and developing hair follicles. Scale bar, 30 µm.
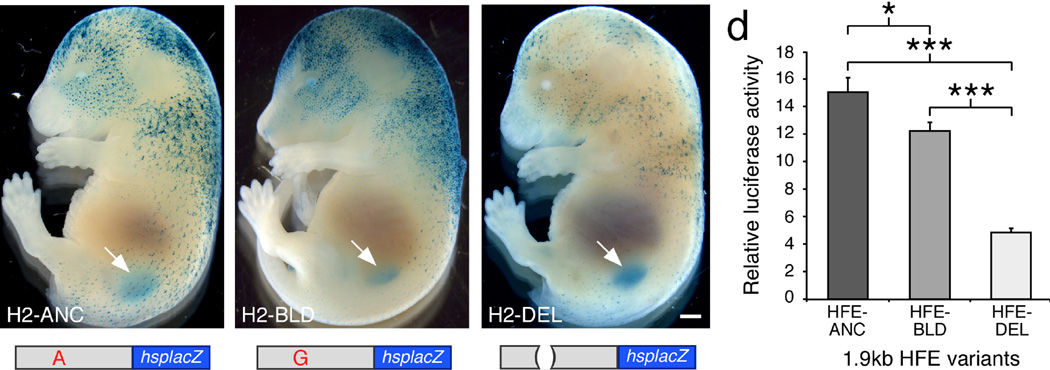
The location of HFE overlaps SNP rs12821256 (Fig 2a), which shows the strongest association with blond hair color2. Complete genome sequences of 270 individuals reveal no additional linked sequence changes in the HFE that are common in northern Europeans and missing in Africans (Methods). To test the effect of the rs12821256 polymorphism, we constructed derivatives of the H2 clone containing the ancestral allele (A, H2-ANC), the blond-associated allele (G, H2-BLD), or an 11 bp deletion overlapping the rs12821256 SNP position (H2-DEL) (Fig. 3). No obvious qualitative differences in lacZ expression patterns were noted between H2-ANC and H2-BLD transgenics (Fig. 3a,b and Supplementary Figs. 1 and 4). However, for the H2-DEL construct, relatively weak hair expression was observed even in transgenic embryos with strong kidney expression, as expected if sequences surrounding rs12821256 modulate hair, but not kidney expression (Fig. 3c and Supplementary Fig. 5).
Figure 3. Variant hair follicle enhancers produce altered levels of gene expression.
(a–c) Representative E16.5 transgenic embryos, generated by pronuclear injection of different 6.7 kb H2-lacZ constructs (shown below), processed for lacZ gene activity (blue). The full H2 region was used for these experiments, as expression in kidney provided a control for successful integration and expression of constructs even if expression in hair was disrupted. The clones tested were: (a) H2-ANC, "A" at rs12821256, (b) H2-BLD, "G" at rs12821256, or (c) H2-DEL, an 11 bp deletion that removes the rs12821256 position. lacZ gene activity was observed in developing hair follicles and kidneys (arrows) in all transgenic embryos. Although no consistent difference was noted between H2-ANC (N=15) and H2-BLD (N=9) embryos, H2-DEL embryos (N=8) (c) showed reduced activity in skin but normal kidney expression (arrow). Scale bar, 1 mm. (d) Expression analysis of different 1.9 kb HFE-luciferase reporters in the human HaCaT keratinocyte cell line. Bars represent the mean increase in luciferase gene activity over an empty vector control measured 48 hours after transfection from a typical experiment. The enhancers tested differed only by the following: HFEANC (A at rs12821256), HFE-BLD (G at rs12821256), and HFE-DEL (11 bp deletion removing rs12821256). Both the HFE-BLD and HFE-DEL constructs exhibited significantly reduced activity in HaCaT keratinocytes compared to the HFE-ANC plasmid. Error bars indicate s.e.m. Unpaired t test P-values; * P<0.05, *** P<5×10−4.
Although gross differences in lacZ staining are readily observed in mouse transgenics, quantitative changes can be difficult to score due to random variation in copy number and integration site from embryo to embryo. We therefore cloned the ancestral (HFE-ANC), blond-associated (HFE-BLD), or 11 bp deleted (HFE-DEL) version of the hair enhancer upstream of a luciferase reporter gene, and examined quantitative levels of expression in a human keratinocyte cell line, HaCaT26 (Fig. 3d). The HFE-ANC construct produced a 17-fold increase over background activity, confirming that the HFE is active in human keratinocytes (Stouffer’s Z method, P=1E-24, N=8). HFE-DEL showed a marked 73% reduction in luciferase activity compared to HFE-ANC (Stouffer’s Z method, P=8E-16, N=5). The HFE-BLD construct with the rs12821256 base pair change also showed a small but consistent reduction in activity, 22% less than HFE-ANC (Stouffer’s Z method, P=1E-8, N=8).
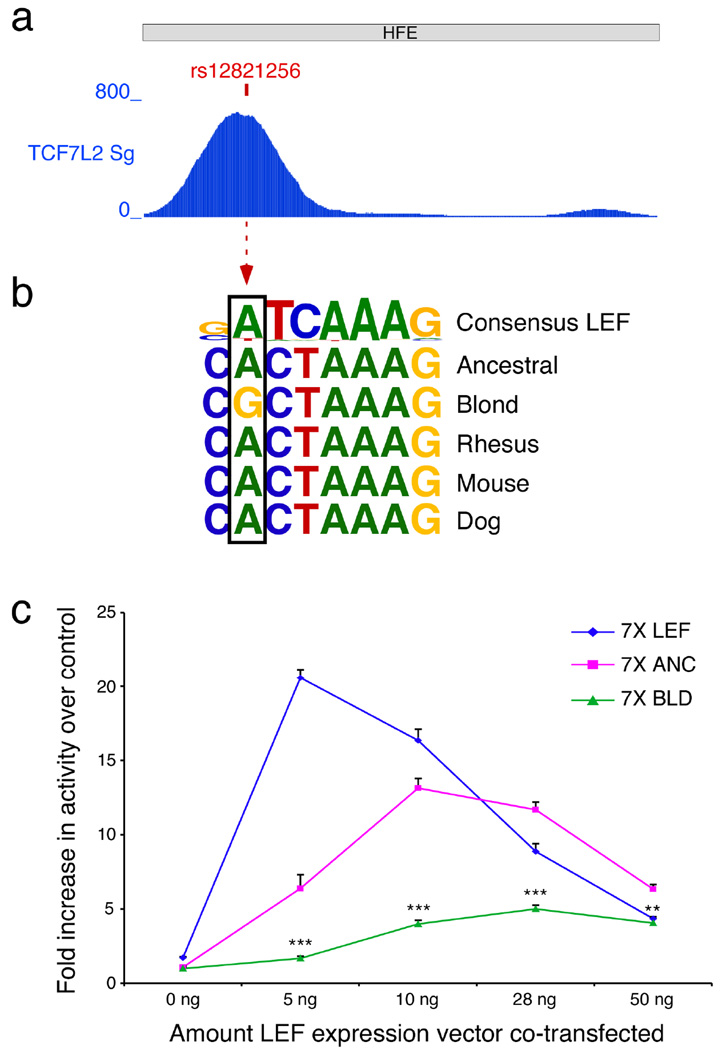
Examination of human ENCODE regulatory tracks11 identified a robust TCF/LEF signal that overlaps the position of rs12821256 in ChIP-seq experiments with TCF7L2 in HCT116 colorectal carcinoma epithelial cells (Fig. 4a). LEF1 is a major WNT-activated transcription factor that binds DNA through a high mobility group (HMG) domain27–29. LEF1 is known to be expressed during hair follicle development and regeneration30,31, and Lef1 knockout mice have defects in hair and whisker formation32. Interestingly, Lef1 knockout mice appear light-colored due to local failure of melanogenesis in mutant hair follicles32. Conversely, hyperactivation of WNT signal transduction in epithelial cells leads to increased Kitlg expression and hyperpigmentation in vivo33. Finally, LEF1 is known to induce sharp DNA bends27,34 and stimulate DNA looping interactions35,36. Thus, LEF1 is an excellent candidate for an upstream factor that could act on distant regulatory enhancers of KITLG and other melanogenic genes37. The rs12821256 SNP alters a well-conserved base within a sequence that resembles a consensus LEF binding motif (Fig. 4b). Examination of the UniPROBE collection of protein binding microarray data confirms that oligonucleotides matching the HFE ChIP-seq motif bind directly to TCF/LEF in vitro38,39 (Supplementary Table 1). Oligonucleotides containing the rs12821256 A to G change show significantly less experimental binding, consistent with the observed differences in activity of the two HFEs in our assays. To further quantify the LEF responsiveness of the KITLG HFE sequences, we co-transfected different minipromoter constructs containing canonical, human ancestral, or human blond-associated binding motifs along with a LEF-1 expression plasmid in HaCaT keratinocytes (Fig. 4c). As expected the canonical LEF1 response sequence showed the highest level of activation. Similarly, the ancestral binding motif found in the human KITLG enhancer also mediated strong LEF1 responsiveness. In contrast, the blond-associated motif with the rs12821256 base pair change reduced but did not eliminate LEF1 responsiveness in vitro (Fig. 4c).
Figure 4. The blond allele at rs12821256 alters a TCF/LEF binding site and reduces LEF responsiveness in keratinocytes.
(a) A 4 kb window centered on the blond SNP shows that TCF7L2 ChIP-seq reads from the ENCODE project11 accumulate over rs12821256 in the 1.9 kb HFE (NCBI36/hg18 chr12:87,852,100-87,853,992; HCT116 cells, TCF7L2 Sg data set). (b) The sequence surrounding rs12821256 resembles a consensus LEF binding motif39. The blond-associated allele in humans changes a highly conserved, consensus-matching A nucleotide to a non-consensus G within the predicted LEF binding motif. (c) Response of mini-promoters to increasing levels of LEF protein 48 hours following co-transfection into HaCaT keratinocytes. The three luciferase reporter constructs tested contained 7 tandem copies of an artificial consensus LEF binding site (7X LEF) (SuperTOPFlash43), seven copies of the human ancestral binding site (7X ANC), or seven copies of the blond-associated sequence variant (7X BLD). All three mini promoters demonstrated elevated activity in response to increased LEF protein. The magnitude of the response to moderate LEF levels corresponds with the predicted binding capabilities of the variant LEF sites, with 7X LEF >> 7X ANC >> 7X BLD. Note that the 7X BLD human variant shows significantly lower activation than the 7X ANC human sequence at every level of LEF1 tested (5 ng, P<0.0001; 10 ng, P<0.0001; 28ng P<0.0001; 50 ng, P<0.001; Mann-Whitney test). Representative experiment shown of N=2 replicates. Error bars indicate s.e.m.
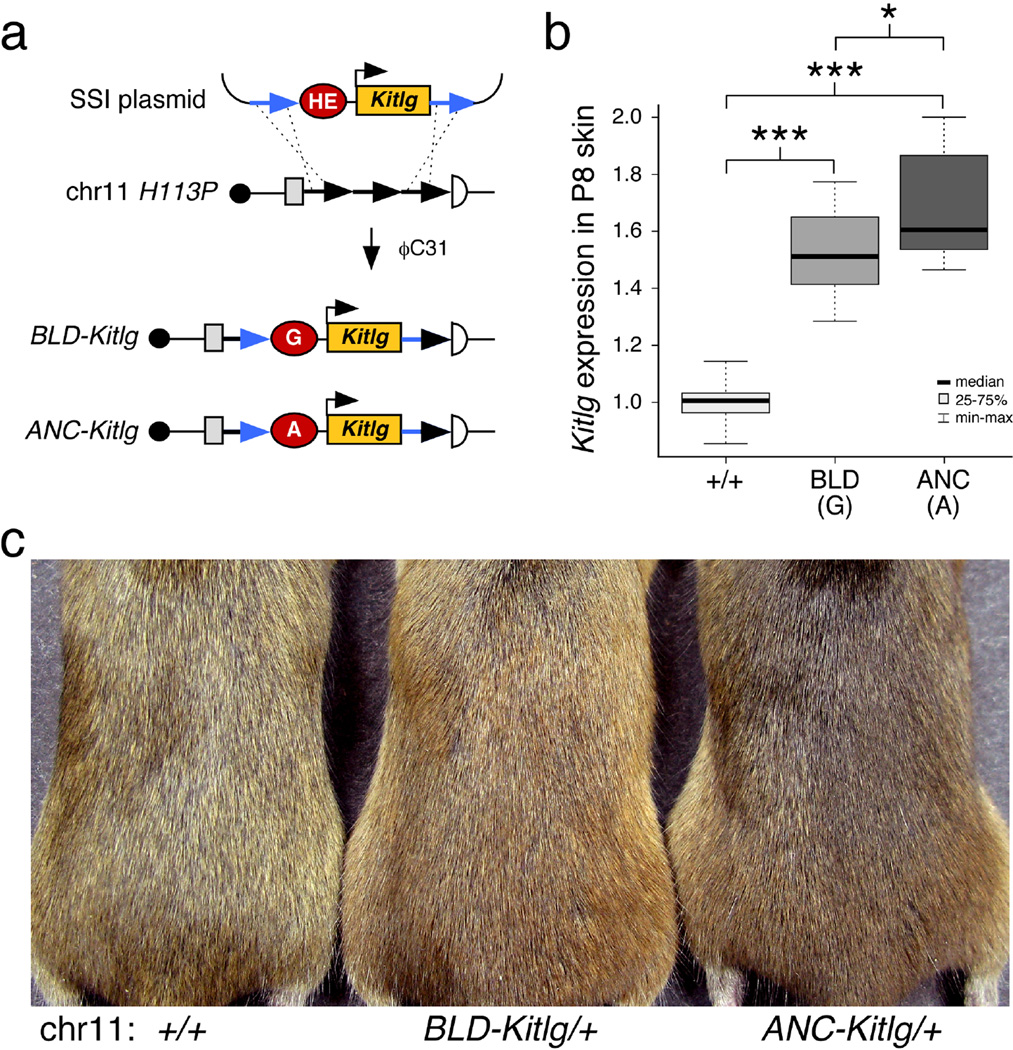
To determine whether small quantitative changes in enhancer activity are sufficient to alter hair color in vivo, we generated matched lines of transgenic mice that express Kitlg cDNA under the control of either the ANC or BLD hair enhancer (Fig. 5a). To eliminate possible differences due to transgene copy number, orientation, or integration location, we used the ϕC31 integrase system to produce single-copy integrants at the H11P3 locus on mouse chromosome 1140. We verified that integration occurred at the same position in both transgenic lines. Therefore, any phenotypic differences can be attributed to the status of a single base pair present in the hair enhancer, the presence of an A (ANC-Kitlg) or a G (BLD-Kitlg) corresponding to rs12821256.
Figure 5. Mouse lines differing at single base pair position in the KITLG hair follicle enhancer show obvious differences in hair color.
(a) Schematic of the site-specific integration (SSI) strategy used to create matched BLD-Kitlg and ANC-Kitlg insertions in mice. Blond or ancestral hair enhancers (HE) were cloned upstream of an Hsp68 minimal promoter-Kitlg transgene (Methods). Blue arrows denote flanking attB sites that recombine with tandem attP sites (black arrows) in the murine chromosome 11 H11P3 locus upon pronuclear injection of a mix containing each SSI plasmid with ϕC31 mRNA. (b) Box plots representing quantitative RT-PCR analysis of Kitlg RNA expression in P8 dorsal skin. Both BLD-Kitlg/+ and ANC-Kitlg/+ heterozygotes exhibit significantly elevated levels of epidermal Kitlg compared to control animals. However, mice carrying the blond-variant transgene produce 21% less Kitlg than the matched ancestral transgene. Mann-Whitney P-values: BLD vs. +/+ = 5e-9; ANC vs. +/+ = 7e-10; BLD vs. ANC = 0.03146. (c) Representative 2-month old mice exhibiting the hair color phenotypes associated with a single copy of each SSI transgene. The mice pictured from left to right are: wild type (FVB/C57Bl/6J F1 hybrid), BLD-Kitlg/+, and ANC-Kitlg/+ heterozygotes. Mice carrying the blond-associated allele at rs12821256 are notably lighter than mice carrying the ancestral allele at the KITLG hair follicle enhancer (also see Supplementary Fig. 7).
Quantitative RT-PCR analysis of Kitlg mRNA expression from postnatal day 8 dorsal skin samples confirmed elevated Kitlg mRNA expression from both site-specific integrants compared to controls (Fig. 5b). The BLD-Kitlg insertion increased Kitlg expression only 79% as high as the ANC-Kitlg insertion (Fig. 5b), consistent with the quantitative differences also observed between the two enhancers in cell culture. Importantly, this quantitative difference was also sufficient to produce obvious visual differences in hair color. Both the ANC-Kitlg and the BLD-Kitlg transgene altered hair pigmentation compared to control animals (Fig. 5c; Supplementary Fig. 6). However, the coats of BLD-Kitlg/+ mice appeared significantly lighter than the coats of ANC-Kitlg/+ mice, and showed lower pigmentation density in hair shafts (P<0.028, Supplementary Fig. 7). Taken together, these data demonstrate that a single base change in a KITLG regulatory sequence is sufficient to significantly alter the activity of a functional hair follicle enhancer. Modulated KITLG expression produces different hair pigmentation in vivo, and provides a molecular link between a regulatory change in an essential developmental signaling gene and a classic morphological phenotype in humans. Additional studies will be required to determine whether the rs12821256 polymorphism alters the number, size, dendricity, or differentiation and pigment synthesis of KIT receptor positive melanocytes, all features that are known to respond to different levels of KITLG signaling in human skin grafts41. However, our results with the rs12821256 polymorphism already illustrate how changes in tissue-specific enhancers can localize phenotypes to particular body regions. Some human pigmentation variants alter general aspects of pigment biosynthesis, producing changes in all melanocytes, and therefore pleiotropic effects on hair, skin, and eye color. However, it is well known that hair and eye colors can also vary independently, producing common human phenotypes such as light-haired individuals with brown eyes, or brown-haired individuals with blue eyes. The rs12821256 variant alters an enhancer that is active specifically in the hair follicle environment, providing a simple genetic explanation for previous observations that this SNP is associated with changes in hair pigmentation but not eye pigmentation in northern Europeans2.
The current data highlight why it is still so difficult to identify the causal basis of human trait associations. The rs12821256 SNP maps more than 350 kb from KITLG, acts at a specific anatomical-site whose active enhancers have not yet been characterized in large-scale studies of human chromatin marks11, alters a sequence that does not perfectly match a LEF1 consensus binding site, and only causes an approximately 20% reduction in activity of a previously unrecognized hair follicle enhancer. However, our results also illustrate how these difficulties can now be overcome using information from human population surveys, large-scale genome annotation projects, and transcription factor interaction databases; when combined with detailed functional tests of enhancer activity in cell lines and in mice. Well-matched animal models may ultimately be necessary to confirm the specific nucleotides that underlie many other human trait associations42, making it possible to link phenotypic variation to particular base pair changes in either the protein coding or the complex regulatory sequences that surround many mammalian genes.
Methods
Primers
All primers used in construct design, genotyping, and RT-QPCR are listed in Supplementary Table 2.
Mouse Strains and Transgenics
The Slpan allele was maintained on the C3H background23. Additional C3H pups at P7 were obtained from Jackson Laboratories (Bar Harbor, ME). lacZ reporter DNAs were prepared for microinjection as described44. Pronuclear microinjection, harvesting of E16.5 embryos and fixation in cold 4% paraformaldehyde in 1x PBS for 1 hour was carried out at Taconic Inc (USA) or Cyagen Biosciences (China). Fixed embryos were shipped to Stanford where they were hemisected, fixed for an additional 30 minutes and processed for lacZ activity as described45. Skin samples were dehydrated, embedded in paraffin, sectioned, dewaxed and stained with nuclear fast red (Vector Labs).
The HE-Kitlg site-specific insertion plasmids were individually mixed with ϕC31 RNA and injected into pronuclei of H11P3 FVB embryos by the Stanford Transgenic facility as described40. Genomic DNA from putative founder and offspring animals were analyzed with primer pair PR387/PR42540 as well as primer pair PR52240/Kg1576 and Kitlg primers Kg1580/Kg1581 to screen for site-specific and random integrations. HEKitlg/+ mice, with identical insertion sites, on a FVB background were crossed to C57BL/6J mice to examine gene expression and pigmentation phenotypes. Skin samples from 2-month old FVB/C57BL6/J F1 hybrids were fixed with 4% paraformaldehyde in 1x PBS at 4 degrees for 48 hours, washed, dehydrated and embedded in paraffin. 6 µm sections were collected and processed as above. An independent pair of HE-Kitlg/+ mouse lines, which had also inserted single copies of the ANC-Kitlg or BLD-Kitlg transgenes using different combinations of attP sites at the H11P3 locus, were generated to confirm hair phenotypes, Kitlg RNA expression, and to quantify pigmentation density in hair shafts. All procedures were done in accordance with protocols approved by the Stanford University Institutional Animal Care and Use Committee.
Characterization of Slpan breakpoint
Cloning of a genomic fragment containing the Slpan breakpoint was described previously23. The portion of this fragment that contains the distal inversion breakpoint was sequenced using conventional methods and aligned to the mouse genome (Dec 2011 assembly, GRCm38/mm10). The inversion breakpoints occurred between nucleotides 99,915,895 and 99,915,897 (approximately 100 kb upstream of the Kitlg transcriptional start site at 100,015,824), and between nucleotides 34,300,646 and 34,300,649 (consistent with previous genetic and cytological data mapping the inversion to a distant location on proximal chromosome 1023).
Plasmids
The 7X LEF (SuperTOPFlash, originally published as M50 Super 8x TOPFlash43) plasmid was a gift from the Nusse Lab. The LEF-1 expression plasmid XE237 Human LEF-1 pCS2+ was purchased from Addgene (Addgene plasmid 16709). Digesting XE237 with EcoRI and XbaI, blunting the ends with Klenow, and religating, generated the control expression vector pCS2+.
The genomic fragments in H2, H2b, H3, HFE and HFE-ANC were amplified from human genomic DNA (Clontech #636401) and cloned into the NotI site of the Not5’Hsp68lacZ vector44 or the XhoI site of the firefly luciferase reporter plasmid pTAluc (Clontech).
To generate clone H1, 4.7 kb of the 7.4 kb genomic fragment was amplified from human genomic DNA (Clontech) and cloned into the PCR BLUNT II TOPO vector (Invitrogen). An additional 2.7 kb corresponding to NCBI36/hg18 chr12:87,854,729-87,857,439 was synthesized by GenScript and cloned into the PUC19 vector. The GenScript clone was digested with EcoRI and the 4.7 kb EcoRI fragment from the TOPO clone was inserted. The resulting 7.4 kb H1 insert was liberated by NotI digestion and cloned into the Not5’hsp68lacZ vector.
The genomic fragments in H2-BLD and HFE-BLD were amplified from Icelandic human DNA (Coriell #GM15755) and cloned into the Not5’hsp68lacZ or pTA-luc vectors respectively.
The H2-DEL insert was generated by amplification of two PCR products, H2del1 and H2del2, from human genomic DNA (Clontech). These PCR intermediates were joined together in a third round of PCR to generate the 11 bp deletion within the larger 6.7 kb clone. Similarly, the 11 bp deletion within the 1.9 kb HFE-DEL insert was formed by PCR joining of two PCR intermediates, HFEdel1 and HFEdel2.
The 7X ANC and 7X BLD inserts were generated by annealing overlapping oligos, filling in the ends with Klenow and TOPO cloning the resulting products. Clones were sequenced to identify 7X inserts, which were liberated by XhoI digestion and cloned into pTA-luc.
To build the HE-Kitlg plasmids, an attB site was amplified from pBT31640 and cloned into the SacI site of pBSKS+ to make 5’attBpBS. The Hsp68 promoter and SV40 polyadenylation fragments with ends overlapping the Kitlg cDNA were amplified from Not5’Hsp68lacZ. The Kitlg cDNA with ends overlapping the Hsp68 promoter and SV40pA fragments was amplified from Thermo Scientific clone MMM1013_65096. Next, the Kitlg and SV40pA PCR intermediates were joined in a second round of PCR. Finally, the Hsp68 promoter and KitlgSV40pA intermediates were combined in a third round of PCR. A 2.2 kb fragment was gel purified and TOPO cloned. The Hsp68KitlgSV40pA TOPO plasmid was digested with NotI and SalI and the insert was cloned into 5’attBpBS to make attBKitlg. A second attB site was added by digesting attBKitlg with SalI and ligating an attB site liberated from the minicircle vector pBT34640 by SalI digestion to make attBKitlgattB. Minimal 864 bp ANC and BLD hair enhancers were amplified from Clontech or Icelandic human DNA. The PCR products were digested with NotI and cloned into the 5’ NotI site in attBKitlgattB to make the final ANC-Kitlg and BLD-Kitlg constructs.
Sanger sequencing confirmed the sequence validity of all DNA constructs.
Human population genotypes and sequences
Allele frequencies for rs12821256 were compiled from the 1000 Genomes data set21 and ALFRED20. Phased sequences from 93 Finnish, 89 British and 88 Africans (Nigerian Yorubans) showed no additional common base pair changes within the HFE region that were linked with rs12821256.
Luciferase Reporter Assays
The human keratinocyte cell line HaCaT (CLS, Germany) was cultured in DMEM supplemented with 10% fetal bovine serum, 2mM L-glutamine, and 1% Penicillin Streptomycin. Cells were seeded into 24-well plates at a density of 1 × 105 cells/well. 370 ng of pTA-luc or the enhancer clones HFE-ANC, HFE-BLD or HFE-DEL were transfected using Lipofectamine 2000 (Invitrogen) with 30 ng of pRL-tk (Promega). To generate LEF response curves, 0 (220) ng, 5 (215) ng, 10 (210) ng, 28 (192) ng, or 30 (190) ng of XE237 (pCS2+) plasmids were co-transfected using Lipofectamine 2000 with 150 ng pTA-luc, 7X LEF, 7X ANC, or 7X BLD constructs and 30 ng of pRL-tk. 5 hours after transfection, cell culture medium was replaced with standard medium supplemented with 2.8mM calcium chloride (Sigma). Cells lysates were collected 48 hours later and assayed on a Glo/Max Multi+ Detection system (Promega) using the Dual-Luciferase Reporter Assay System (Promega) according to manufacturers instructions.
Quantitative RT-PCR analysis
Dorsal skin samples from P8 or P21 FVB/C57Bl/6J F1 hybrids with +/+, BLD-Kitlg/+ or ANC-Kitlg/+ genotypes and P8 wild type (C3H), Slpan/+, or Slpan/Slpan genotypes were placed in FastPrep Lysing Matrix A vials (MP Biomedicals) with 1 ml TriReagent (Ambion). Tissue was homogenized using a MP FastPrep-24 with 4 × 30 sec pulses at 6.0 M/S, with a 2 min incubation on ice following each 30 sec pulse. Following tissue disruption, total RNA was prepared according to the manufacturers instructions. Contaminating genomic DNA was removed from RNA samples prior to cDNA synthesis using DNase I (Invitrogen). cDNA was transcribed from 500 ng of total RNA using Superscript III First Strand Synthesis Super Mix (Invitrogen). Quantitative RT-PCR was performed with Kitlg primers (Kg1580/Kg1581) and Actb primers (Kg1588/Kg1589). All reactions were done in triplicate using Brilliant Green Low Rox Master mix (Agilent) with a 2-step protocol (40 cycles of 95°C 15 sec, 60°C 1 min) on an Mx3005P PCR system (Stratagene).
Quantification of pigmentation levels in site-specific transgene lines
Dorsal hair samples were collected from FVB/C57Bl/6J F1 hybrids, BLD-Kitlg/+ (line 2), and ANCKitlg/+ (line 2) heterozygotes at P21. Multiple zigzag hairs were separated, attached to a piece of tape and affixed to a slide. A cover slip was applied using Aquamount and photographs were taken of the middle portion of the hair between the distal most bends of 15 hairs per animal using a Zeiss axiophot (32X objective, 3200K lamp) with an exposure time of 16 ms. Images were imported into Adobe Photoshop and were measured to determine average pigment density per hair (defined as the total number of pixels corresponding to pigmented areas, divided by total pixels comprising the hair shaft).
URLs
Statistical analyses
For cell culture experiments, the average activity of a construct in a given experiment was calculated using 6–8 individually transfected wells. Raw firefly luciferase values were normalized to Renilla luciferase values to control for variation in transfection efficiencies. Elevated HFE or mini-promoter expression represents activity over the pTA-luc expression baseline. All experiments were replicated (HFE enhancers N=5–8; 7X mini-promoters N=2). InStat software was used to determine statistical significance of HFE enhancer expression in unpaired, one-tailed t tests following verification of normality. MetaP46 was used to combine P-values from replicate HFE enhancer experiments. The Stouffer’s Z combined P-value, which corrects for sample size, was reported. The Mann-Whitney test, which makes no assumptions regarding data distribution, was used to determine significance in mini-promoter experiments. All graphs and plots show mean ± s.e.m.
For quantitative RT-PCR experiments, Kitlg expression in animals from 3 litters for each transgene were analyzed to give a total of 22 wild type, 11 BLD-Kitlg/+, and 13 ANC-Kitlg/+ samples. To control for variability in experiments run on different days, gene expression in all three genotypic classes was normalized to expression in a single 2-month old wild type dorsal skin sample. The relative Kitlg expression in each skin sample was then normalized to Actb, using the PFAFFL method47. Kitlg expression levels in the two sets of wild type littermates were not significantly different from each other (InStat two-tailed unpaired t test P=0.1666). Elevation of Kitlg expression in BLDKitlg/+ and ANC-Kitlg/+ animals was calculated relative to the expression in their wild type littermates. Plotting and statistical analysis were performed using R. Mann-Whitney rank sum tests were used to determine significance.
Quantitative RT-PCR experiments on dorsal skin samples taken at P21 from the second pair of site-specific lines was performed on animals from 2 litters for a total of 4 wild type, 3 BLD-Kitlg/+, and 3 ANC-Kitlg/+ samples. Samples were treated as above and statistical significance was determined using InStat software to perform unpaired, one-tailed t tests.
For quantitative RT-PCR experiments on Slpan animals, Kitlg expression in 3 litters (2 Slpan litters and 1 C3H litter) was analyzed to give a total of 3 Slpan/Slpan homozygotes, 5 Slpan/+ heterozygotes, and 6 wild type C3H animals. Gene expression of all samples was normalized to expression in a single 2-month old wild type dorsal skin sample. The relative Kitlg expression in each skin sample was then normalized to Actb, using the PFAFFL method47. Mann-Whitney tests were performed using InStat software to determine statistical significance.
For pigment density analysis, 15 individual zigzag hairs were measured from each of 4 wild type, 3 BLD-Kitlg/+, and 3 ANC-Kitlg/+ animals. The reported pigmentation levels for wild type, BLD and ANC genotypic classes represent the average of the pigmentation values for each animal of that genotype. InStat software was used to determine statistical significance using unpaired, one-tailed t tests.
Supplementary Material
Acknowledgements
We thank Randall Moon for the XE237 LEF-1 expression plasmid, Roel Nusse for the SuperTOPFlash plasmid, Craig Lowe for help with statistical and 1000 Genomes analysis, and members of the Kingsley lab for useful comments on the manuscript. This work was supported in part by a Center of Excellence in Genomic Science Grant from the National Institutes of Health (5P50HG2568). LL and DMK are investigators of the Howard Hughes Medical Institute.
Footnotes
Author contributions
CAG and DMK conceived and oversaw the project. MAB isolated and sequenced the Slpan breakpoint. BT and LL provided advice, reagents, and animals for generating site-specific integrants. CAG performed the gene expression analysis in Slpan mutants, carried out the transgenic analysis of the blond GWAS interval, identified the hair follicle enhancer, and performed the in vitro and in vivo tests of the effects of the rs12821256 mutation. CAG and DMK wrote the paper with input from all authors.
References
- 1.Sturm RA. Molecular genetics of human pigmentation diversity. Hum. Mol. Genet. 2009;18:R9–R17. doi: 10.1093/hmg/ddp003. [DOI] [PubMed] [Google Scholar]
- 2.Sulem P, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 3.Cruz-Inigo AE, Ladizinski B, Sethi A. Albinism in Africa: stigma, slaughter and awareness campaigns. Dermatol. Clin. 2011;29:79–87. doi: 10.1016/j.det.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Pitman J. On Blondes. New York: Bloomsbury Publishing; 2003. [Google Scholar]
- 5.The Iliad of Homer. Chicago: The University of Chicago Press; 2011. [Google Scholar]
- 6.Han J, et al. A genome-wide association study identifies novel alleles associated with hair color and skin pigmentation. PLoS Genet. 2008;4:e1000074. doi: 10.1371/journal.pgen.1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sulem P, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat. Genet. 2008;40:835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, et al. Genome-wide association studies identify several new loci associated with pigmentation traits and skin cancer risk in European Americans. Hum. Mol. Genet. 2013;22:2948–2959. doi: 10.1093/hmg/ddt142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kenny EE, et al. Melanesian blond hair is caused by an amino acid change in TYRP1. Science. 2012;336:554. doi: 10.1126/science.1217849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Praetorius C, et al. A polymorphism in IRF4 affects human pigmentation through a tyrosinase-dependent MITF/TFAP2A pathway. Cell. 2013;155:1022–1033. doi: 10.1016/j.cell.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olalde I, et al. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature. 2014;507:225–228. doi: 10.1038/nature12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison-Graham K, Takahashi Y. Steel factor and c-kit receptor: from mutants to a growth factor system. Bioessays. 1993;15:77–83. doi: 10.1002/bies.950150202. [DOI] [PubMed] [Google Scholar]
- 16.Russell ES. Hereditary anemias of the mouse: a review for geneticists. Adv. Genet. 1979;20:357–459. [PubMed] [Google Scholar]
- 17.Nocka K, et al. Molecular bases of dominant negative and loss of function mutations at the murine c-kit/white spotting locus: W37, Wv, W41 and W. EMBO J. 1990;9:1805–1813. doi: 10.1002/j.1460-2075.1990.tb08305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bedell MA, Copeland NG, Jenkins NA. Multiple pathways for Steel regulation suggested by genomic and sequence analysis of the murine Steel gene. Genetics. 1996;142:927–934. doi: 10.1093/genetics/142.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walsh S, et al. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci. Int. Genet. 2013;7:98–115. doi: 10.1016/j.fsigen.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Rajeevan H, et al. ALFRED: the ALelle FREquency Database. Update. Nucleic Acids Res. 2003;31:270–271. doi: 10.1093/nar/gkg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beechey CV, Loutit JF, Searle AG. Panda, a new steel allele. Mouse News Lett. 1986;74:92. [Google Scholar]
- 23.Bedell MA, et al. DNA rearrangements located over 100 kb 5' of the Steel (Sl)-coding region in Steel-panda and Steel-contrasted mice deregulate Sl expression and cause female sterility by disrupting ovarian follicle development. Genes Dev. 1995;9:455–470. doi: 10.1101/gad.9.4.455. [DOI] [PubMed] [Google Scholar]
- 24.Peters EMJ, Tobin DJ, Botchkareva N, Maurer M, Paus R. Migration of melanoblasts into the developing murine hair follicle is accompanied by transient c-Kit expression. J. Histochem. Cytochem. 2002;50:751–766. doi: 10.1177/002215540205000602. [DOI] [PubMed] [Google Scholar]
- 25.Jordan SA, Jackson IJ. MGF (KIT ligand) is a chemokinetic factor for melanoblast migration into hair follicles. Dev. Biol. 2000;225:424–436. doi: 10.1006/dbio.2000.9856. [DOI] [PubMed] [Google Scholar]
- 26.Boukamp P, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 28.Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 29.Waterman ML, Fischer WH, Jones KA. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 30.Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9:700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]
- 31.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126:4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 32.van Genderen C, et al. Development of several organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–2703. doi: 10.1101/gad.8.22.2691. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, et al. Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development. 2008;135:2161–2172. doi: 10.1242/dev.017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love JJ, et al. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 35.Yun K, So JS, Jash A, Im SH. Lymphoid enhancer binding factor 1 regulates transcription through gene looping. J. Immunol. 2009;183:5129–5137. doi: 10.4049/jimmunol.0802744. [DOI] [PubMed] [Google Scholar]
- 36.Jash A, Yun K, Sahoo A, So JS, Im SH. Looping mediated interaction between the promoter and 3' UTR regulates type II collagen expression in chondrocytes. PLoS One. 2012;7:e40828. doi: 10.1371/journal.pone.0040828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visser M, Kayser M, Palstra RJ. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012;22:446–455. doi: 10.1101/gr.128652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berger MF, et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tasic B, et al. Site-specific integrase-mediated transgenesis in mice via pronuclear injection. Proc. Natl. Acad. Sci. USA. 2011;108:7902–7907. doi: 10.1073/pnas.1019507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grichnik JM, Burch JA, Burchette J, Shea CR. The SCF/KIT pathway plays a critical role in the control of normal human melanocyte homeostasis. J. Invest. Dermatol. 1998;111:233–238. doi: 10.1046/j.1523-1747.1998.00272.x. [DOI] [PubMed] [Google Scholar]
- 42.Kamberov YG, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell. 2013;152:691–702. doi: 10.1016/j.cell.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 44.DiLeone RJ, Russell LB, Kingsley DM. An extensive 3' regulatory region controls expression of Bmp5 in specific anatomical structures of the mouse embryo. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mortlock DP, Guenther C, Kingsley DM. A general approach for identifying distant regulatory elements applied to the Gdf6 gene. Genome Res. 2003;13:2069–2081. doi: 10.1101/gr.1306003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 47.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.