Using the established 5i/L/FA system, fibroblasts from a β-thalassemia patient were directly reprogrammed into transgene-free naïve induced pluripotent stem cells (iPSCs) with ground-state pluripotency. Human urinary cells were also reprogrammed into a ground state using a similar approach. These findings demonstrate the feasibility and superiority of using patient-specific iPSCs in the naïve state for disease modeling, gene editing, and future clinical therapy.
Keywords: Reprogramming, Naïve state, β-Thalassemia patient, Gene correction, CRISPR/Cas9
Abstract
Conventional primed human embryonic stem cells and induced pluripotent stem cells (iPSCs) exhibit molecular and biological characteristics distinct from pluripotent stem cells in the naïve state. Although naïve pluripotent stem cells show much higher levels of self-renewal ability and multidifferentiation capacity, it is unknown whether naïve iPSCs can be generated directly from patient somatic cells and will be superior to primed iPSCs. In the present study, we used an established 5i/L/FA system to directly reprogram fibroblasts of a patient with β-thalassemia into transgene-free naïve iPSCs with molecular signatures of ground-state pluripotency. Furthermore, these naïve iPSCs can efficiently produce cross-species chimeras. Importantly, using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 nuclease genome editing system, these naïve iPSCs exhibit significantly improved gene-correction efficiencies compared with the corresponding primed iPSCs. Furthermore, human naïve iPSCs could be directly generated from noninvasively collected urinary cells, which are easily acquired and thus represent an excellent cell resource for further clinical trials. Therefore, our findings demonstrate the feasibility and superiority of using patient-specific iPSCs in the naïve state for disease modeling, gene editing, and future clinical therapy.
Significance
In the present study, transgene-free naïve induced pluripotent stem cells (iPSCs) directly converted from the fibroblasts of a patient with β-thalassemia in a defined culture system were generated. These naïve iPSCs, which show ground-state pluripotency, exhibited significantly improved single-cell cloning ability, recovery capacity, and gene-targeting efficiency compared with conventional primed iPSCs. These results provide an improved strategy for personalized treatment of genetic diseases such as β-thalassemia.
Introduction
Human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) derived from embryos or via somatic cell reprogramming [1–4], respectively, represent a primed state of pluripotency similar to mouse epiblast stem cells (EpiSCs) [5, 6] but distinct from the naïve pluripotent state of mouse ESCs and iPSCs [7]. Several studies comparing naïve and primed pluripotent stem cells have indicated that the two types of cells exhibit different molecular and functional properties, including colony morphologies, single-cell passage abilities [7, 8], dependent signaling pathways [6, 9, 10], pluripotent gene expression profiles, and, most important, mouse embryo integration capacity to generate cross-species chimeras [7, 11, 12]. Previous studies have also suggested that human primed iPSCs and mouse EpiSCs can be converted to the naïve ground state via chemical manipulation or the overexpression of specific transcription factors [12–17]. Recently, several groups have further modified culture conditions to maintain human ESCs or iPSCs in the naïve ground state [12, 13, 15, 18]. In particular, the 5i/L/FA system, developed by Jaenisch’s group, contains a combination of inhibitors and growth factors sufficient for the induction and maintenance of the naïve ground state [15].
β-Thalassemia, an inherited blood disorder characterized by reduced or absent synthesis of hemoglobin (HB) subunit β (HB β chain), is one of the most common genetic diseases worldwide. The genetic mutations of β-thalassemia are diverse. The most common molecular defects are either point mutations or small fragment deletions in the HBB gene that affect mRNA assembly or translation. Individuals with β-thalassemia major (also called Cooley’s anemia) develop severe microcytic and hypochromic anemia, which always causes hepatosplenomegaly, skeletal abnormalities during infancy, and a shortened life expectancy if untreated. Although the pathogenesis of β-thalassemia has been extensively studied, no effective treatments are available thus far.
The emergence of iPSC technologies and the development of gene targeting strategies bring new hope for the treatment of genetic diseases, including β-thalassemia [19, 20]. Recent studies have shown that personalized iPSCs can be derived from β-thalassemia patient fibroblasts via the induction of transcription factors, and the mutations can be corrected using a transcription activator-like effector nuclease or clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 nuclease (CRISPR/Cas9) system [21–23]. However, these primed-state iPSCs derived from β-thalassemia patient fibroblasts have shown extremely low levels of single cell cloning efficiencies, thus impairing the subsequent targeting efficiencies. Moreover, the extremely low efficiencies in the recovery process also impede future clinical manipulations of such patient-specific iPSCs.
In the present study, we successfully derived human transgene-free naïve iPSCs directly from β-thalassemia patient fibroblasts with transcription profiles and epigenetic signatures similar to those of mouse ESCs or iPSCs. Significant improvements in mutation correction efficiencies were achieved using a CRISPR/Cas9 editing system in these naïve iPSCs, which are capable of hematopoietic differentiation. In addition, human naïve iPSCs could also be directly generated from noninvasively collected urinary cells [24], which are easily acquired and thus represent an excellent cell resource for further clinical trials. Thus, our study offers an improved strategy for personalized treatment of β-thalassemia and has important implications for the clinical use of human naïve iPSCs in the future.
Materials and Methods
Animal Maintenance
All mice had free access to food and water. All experiments were performed in accordance with the University of Health Guide for the Care and Use of Laboratory Animals and were approved by the Biological Research Ethics Committee of Tongji University.
Human Skin Tissue Acquisition
Human skin specimens were obtained from the Third Affiliated Hospital, Guangzhou Medical College. The patients provided informed consent for tissue donations, and the Biological Research Ethics Committee of Tongji University approved the study.
Generation of β-Thalassemia Patient Naïve iPSCs
Fibroblasts were isolated from β-thalassemia patients carrying the β-41/42 mutation. episomal vectors, including pCXLE-hOCT3/4-shp53, pCXLE-hSOX2-KLF4, and pCXLE-hc-Myc-Lin28-NANOG (Addgene, Cambridge, MA, http://www.addgene.org) were transfected into 2 × 105 fibroblasts through electroporation and were then cultured in conventional human embryonic stem cell medium (hESM) containing knockout Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) supplemented with 20% Knockout Serum Replacement (Invitrogen), 10 ng/ml basic fibroblast growth factor (bFGF; PeproTech, Rocky Hill, NJ, http://www.peprotech.com), 10−4 M nonessential amino acids (EMD Millipore, Billerica, MA, http://www.emdmillipore.com), 10−4 M β-mercaptoethanol (EMD Millipore), 2 mM l-glutamine (Invitrogen), and 50 µg/ml penicillin/streptomycin (EMD Millipore) for 6 days. Then, the medium was replaced with human naïve medium (5i/L/FA medium) and cultured for 14–20 days. Dome-shaped colonies similar to mouse ESCs were selected and expanded by single cell passaging.
Karyotype Analysis
The iPSCs were incubated in culture medium containing 0.25 mg/ml colcemid (Invitrogen) for 4 hours, and the cells were harvested by Accutase (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). After incubation in hypotonic solution containing 0.4% sodium citrate and 0.4% potassium chloride (1:1, vol/vol) at 37°C for 5 minutes, the cells were fixed with a methanol/acetic acid mixture (3:1, vol/vol) for 6 hours. The slides were digested with 0.8% trypsin and stained with Giemsa for 10 minutes. At least 20 metaphase chromosome karyoschisis images were examined, and G-band images were analyzed.
Quantitative Polymerase Chain Reaction Analysis
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. cDNA synthesis was performed with the M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, http://www.promega.com) in accordance with the manufacturer’s instructions. Quantitative reverse transcription-polymerase chain reaction (PCR) was performed using SYBR Green PCR Master Mix (Takara, Otsu, Japan, http://www.takara.co.jp), and signals were detected with ABI7500 Real-Time PCR System (Applied BioSystems, Foster City, CA, http://www.appliedbiosystems.com).
Immunofluorescent Staining
For immunofluorescent staining, the cells were fixed with phosphate-buffered saline (PBS) containing 4% paraformaldehyde (Sigma-Aldrich) overnight at 4°C and permeabilized for 15 minutes in PBS containing 0.5% Triton X-100. Next, the cells were incubated with PBS containing 4% bovine serum albumin for 30 minutes at room temperature. The following primary antibodies were used: OCT3/4 (1:500; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com), SOX2 (1:500; Santa Cruz Biotechnology Inc.), NANOG (1:500; Abcam, Cambridge, U.K., http://www.abcam.com), stage-specific embryonic antigen (SSEA)-3 (1:50; ES Cell Marker Sample Kit; EMD Millipore), SSEA4 (1:50; ES Cell Marker Sample Kit; EMD Millipore), TRA-1-60 (1:50; ES Cell Marker Sample Kit; EMD Millipore), and SSEA1 (1:50; ES Cell Marker Sample Kit; EMD Millipore). The following secondary antibodies were used in the present study: Alexa Fluor 594-conjugated donkey anti-mouse IgG (1:500; Invitrogen), fluorescein isothiocyanate (FITC) 488-conjugated donkey anti-rabbit IgG (1:500; Invitrogen), and FITC 488-conjugated donkey anti-goat IgG (1:500; Invitrogen). Nuclei were stained with 4′,6-diamidino-2-phenylindole (1:10,000; Sigma-Aldrich). Stained cells mounted on slides were observed on a confocal microscope (A1 Nikon; Nikon, Tokyo, Japan, http://www.nikon.com).
Teratoma Formation
The iPSCs were harvested by Accutase (Sigma-Aldrich) dissociation and centrifuged. The pellets were resuspended in DMEM/F12 (Invitrogen). Cells from a 60-mm dish were injected subcutaneously into dorsal flank of a severe combined immunodeficiency (SCID) mouse. At 6–8 weeks after injection, the tumors were dissected and fixed with PBS containing 10% paraformaldehyde (Sigma-Aldrich). Analysis of the hematoxylin and eosin-stained tissue sections was performed.
Hematopoietic Differentiation
For naïve and primed iPSCs hematopoietic differentiation, OP9 stromal cells were plated onto gelatinized 35-mm dishes in α-minimal essential medium (α-MEM; Invitrogen) containing 20% FBS (HyClone, Logan, UT, http://www.hyclone.com), 2 mM l-glutamine (Invitrogen), and 50 µg/ml penicillin/streptomycin (EMD Millipore). After formation of confluent cultures on days 4 and 5, one half of the medium was changed, and the cells were cultured for an additional 3–4 days. Naïve iPSCs were digested into single cells by Accutase (Sigma-Aldrich), and primed iPSCs were harvested by mechanical dissociation into small clumps. Both iPSCs were seeded onto OP9 cultures at a density of 3 × 106 per 4 ml per 35-mm dish in α-MEM (Invitrogen) supplemented with 10% FBS (HyClone) and 100 µM monothioglycerol (Sigma-Aldrich). The cocultured cells were incubated for up to 10 days with a one half-medium change every other day and then were harvested for flow cytometry analysis.
Flow Cytometry Analysis
To analyze the phenotype of hematopoietic progenitor cells, both iPSC/OP9 cocultures were collected and washed with fluorescence-activated cell sorting (FACS) buffer (PBS with 2% FBS). Cells were stained with anti-human CD43-APC monoclonal antibody (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com). The cells were then washed and resuspended in FACS buffer. All analyses were performed on a MoFlo XDP cell sorter (Beckman Coulter, Fullerton, CA, http://www.beckmancoulter.com) running Summit software.
Gene Targeting of Naïve and Primed Patient-Specific iPSCs
Three specific single-guide RNAs (sgRNAs) were designed according to a previously established protocol [25], cloned into pX330 vector (Addgene) and tested by transfection into HEK293T cells. The cleavage efficiencies at the HBB mutation site were tested, and the most efficient sgRNA2 was selected by the T7E1 assay. The targeting donor DNA was amplified from wild-type human genomic DNA with ∼250-base pair (bp) 5′ and 3′ homologous arms. The primers used for the donor DNA amplification were as follows: HBB, forward: 5′-TGACACAACTGTGTTCACTAGC-3′; and HBB, reverse: 5′-TGAGACTTCCACACTGATGC-3′. The donor DNA fragment was then constructed into T-vectors using the pEASYTM-T5 Zero cloning Kit (TransGen Biotech, Beijing, China, http://www.transgenbiotech.com) for DNA sequencing validation and digested by NotI/PstI for linearization.
For gene targeting, 106 naïve and primed iPSCs were digested by Accutase (Sigma-Aldrich) and electroporated with 2.5 µg sgRNA-pX330 plasmid and 2.5 µg linearized donor DNA in 100 µl primary buffer 3 using nucleofactor II (Lonza, Walkersville, MD, http://www.lonza.com). The transfected cells were subsequently plated onto the feeder layers and cultured in naïve and conventional human embryonic stem medium, respectively. Next, 10 mM ROCK inhibitor (ROCKi) was supplemented for the first 24 hours. Individual colonies were picked 7 days after transfection, which were then expanded and verified by DNA sequencing (Sangon Biotech, Co., Ltd., Shanghai, China, http://www.sangon-biotech.com).
Mouse Embryo Micromanipulation, Whole-Mount Staining, and Imaging
For naïve and primed iPSC injection, the cells were digested, and 10–15 cells were microinjected into 8-cell embryos or blastocysts of ICR (Crl:CD1) diploid mouse embryo. Approximately 15–20 injected embryos were transplanted into embryonic day (E) 2.5 post coitum pseudopregnant mice by uterine transfer. The embryos were harvested and dissected at the E10.5 developmental stage for analysis.
Western Blot Analysis
For Western blot analysis, naïve iPSCs and primed iPSCs were collected and lysed by RIPA buffer supplemented with protease inhibitor cocktail (Roche, Indianapolis, IN, http://www.roche.com) for 30 minutes at 4°C, followed by centrifugation at 20,000g for 10 minutes at 4°C. The supernatants were loaded onto SDS polyacrylamide gel electrophoresis, and Western blot analysis was performed using specific OCT3/4 (1:2,000; Santa Cruz Biotechnology), NANOG (1:2,000; Abcam), KLF4 (1:2,000; Abcam), and REX1 (1:2,000; Abcam) antibodies.
Results
Reprogramming β-Thalassemia Fibroblasts Directly Into Ground State-Naïve iPSCs
First, β-41/42 fibroblasts with a TCTT deletion between the 41st and 42nd amino acids of the HBB gene were used to generate naïve iPSCs in accordance with the procedures summarized in Figure 1A (supplemental online Fig. 1A). Six days after electroporation, morphological changes occurred in the transfected cells, and small aggregates could be observed in the culture dishes. A dome shape could be observed as early as 7 days after culturing in naïve hESM. These colonies were then selected 20 days after electroporation and further expanded by single cell passage with Accutase every 4–5 days (Fig. 1B). When cultured on mitomycin C-treated ICR murine embryonic fibroblasts or on Matrigel (BD Biosciences), all the naïve iPSC lines that we established showed typical morphologies similar to those of mouse ESCs/iPSCs (Fig. 1C, 1D; supplemental online Fig. 1B). Karyotype examinations showed that most of these ground-state cells were normal (46,XY) and could be maintained after additional extended passaging (Fig. 1E, 1F). In addition, the ground-state cell lines that we derived could be propagated for more than 20 passages without showing differences in morphology and doubling time (supplemental online Fig. 1C).
Figure 1.
Generation of naïve iPSCs from β-thalassemia patient fibroblasts. (A): Scheme of the strategy for the generation of naïve iPSCs from human β-thalassemia patient fibroblasts. (B): Representative images of morphological changes on reprogramming of human fibroblasts into naïve iPSCs on days 1, 7, and 15 and P1. Scale bar = 50 μm. (C): Phase images of naïve iPSC lines established from human β-thalassemia fibroblasts cultured on feeders. Scale bar = 200 μm. (D): Phase images of two naïve iPSCs cell lines derived from human β-thalassemia fibroblasts cultured on Matrigel. Scale bar = 200 μm. (E): Karyotype analysis of n2-iPSCs generated from human β-thalassemia fibroblasts. (F): Karyotype analysis of n5-iPSCs generated from human β-thalassemia fibroblasts. (G): Single-cell cloning efficiencies of several primed and naïve pluripotent stem cell lines cultured in medium with or without ROCKi. Primed cell lines included pr2-iPSCs, pr9-iPSCs, and by1-ESCs and naïve cell lines included n2-iPSCs and n5-iPSCs. Data are shown as the mean ± SEM; t test; ∗∗, p < .01; n = 3 individual experiments. (H): Recovery efficiencies of several primed and naïve pluripotent stem cell lines. Primed cell lines included pr2-iPSCs, pr9-iPSCs, and by1-ESCs and naïve cell lines included n2-iPSCs and n5-iPSCs. Data are shown as mean ± SEM; t test; ∗∗, p < .01; n = 3 individual experiments. Abbreviations: ESC, embryonic stem cell; d, day; HDF, human dermal fibroblast; hESM, human embryonic stem cell medium; iPSC, induced pluripotent stem cell; P1, passage 1; Rocki, ROCK inhibitor.
For further analysis, we adopted two primed iPSC (pr2-iPSC and pr9-iPSC) lines and one ESC (by1-ESC) line as controls. The primed iPSC lines were derived from the fibroblasts of the same patient by electroporation of the same episomal vectors followed by hESM culture, and the by1-ESC line was established from a donated human blastocyst. Calculation of the colony numbers revealed that the efficiencies of naïve reprogramming and primed reprogramming were approximately 0.135% and 0.155%, respectively, without significant differences between each other (supplemental online Fig. 1D).
When passaging these cell lines, the single-cell cloning efficiency of the naïve iPSCs reached up to 88%, far higher than the efficiency of the primed iPSCs derived from the same fibroblasts or the ESCs that we previously established, both of which always barely survived after single cell digestion. Even in the presence of ROCKi, the single-cell cloning efficiency of the primed iPSCs or ESCs was still less than 30% (Fig. 1G). Moreover, the recovery efficiencies of the naïve iPSCs could reach up to 95%, significantly higher than those in primed iPSCs or ESCs (Fig. 1H). These results indicated that transgene-free naïve iPSC lines can be successfully generated from β-thalassemia fibroblasts under defined culture conditions.
Characterization of Ground-State Pluripotency in β-Thalassemia Patient-Specific Naïve iPSCs
Next, we examined the pluripotency of the naïve iPSCs derived from patient fibroblasts. Quantitative PCR analysis showed that several pluripotent genes were expressed in naïve iPSCs and in primed iPSCs. Importantly, multiple transcription factors typically associated with naïve pluripotency were significantly upregulated in naïve iPSCs compared with those in primed human ESCs or iPSCs, including NANOG, KLF4, REX1, STELLA, KLF2, and TFCP2L1 (Fig. 2A; supplemental online Fig. 2A). Western blot analysis also revealed higher expression levels of NANOG, KLF4, and REX1 in the naïve iPSCs compared with those in the primed iPSCs (Fig. 2B). We also detected marker gene expression in naïve iPSCs via immunofluorescence. Consistent with primed iPSCs, naïve iPSC lines were positive for pluripotency-associated markers such as OCT4, SOX2, and NANOG and surface markers such as SSEA3, SSEA4, and TRA1-60, but not SSEA1 (Fig. 2C; supplemental online Fig. 2B). These results indicate that these naïve iPSCs exhibit the characteristics of human pluripotent stem cells with ground-state features.
Figure 2.
Pluripotency validation of the naïve iPSCs derived from β-thalassemia fibroblasts. (A): Quantitative PCR analysis of genes associated with ground-state self-renewal and pluripotency in naïve iPSCs and primed iPSCs and ESCs. Data are shown as the mean ± SEM; t test; ∗, p < .05; ∗∗, p < .01; n = 3 individual experiments. (B): Western blot analysis of ground-state pluripotency-associated transcription factors such as NANOG, KLF4, and REX1 in naïve iPSC lines and primed iPSC lines. β-Actin was used as an endogenous control. (C): Immunostaining images of pluripotency-associated markers OCT4, SOX2, NANOG, SSEA3/4, and TRA-1-60. Scale bars = 20 μm. (D): Representative images of n2-iPSC morphologies after withdrawal of individual inhibitors and growth factors from 5i/L/FA culture system. Scale bars = 100 μm. (E): Quantitative PCR analysis of pluripotency-associated gene expressions in naïve iPSCs after withdrawal of individual inhibitors from 5i/L/FA culture system. Data are shown as mean ± SEM; n = 3 individual experiments. (F): Quantitative PCR analysis of pluripotency-associated gene expression in naïve iPSCs after withdrawal of growth factors from 5i/L/FA culture system. Data are shown as mean ± SEM; n = 3 individual experiments. (G): Representative images of morphological changes from naïve state to primed state as the culture system changed. Scale bar = 100 μm. Abbreviations: bFGF, basic fibroblast growth factor; Ctrl, control; ESCs, embryonal stem cells; DAPI, 4′,6-diamidino-2-phenylindole; GSK3βi, GSK3β inhibitor; hESM, human embryonic stem cell medium; iPSCs, induced pluripotent stem cells; LIF, leukemia inhibitory factor; MEKi, MEK inhibitor; PCR, polymerase chain reaction; RAFi, BRAF inhibitor; SRCi, SRC inhibitor; SSEA3, stage-specific embryonic antigen 3; SSEA4, stage-specific embryonic antigen 4; ROCKi, ROCK inhibitor.
To investigate whether the individual components of the 5i/L/FA medium were essential for the maintenance of the naïve iPSCs, we checked the changes in colony morphologies and pluripotency gene expression on removal of individual components from the culture system. Withdrawal of the MEK inhibitor and BRAF inhibitor significantly impaired the colony morphologies and pluripotency-associated gene expression. In contrast, withdrawal of the SRC inhibitor and GSK3β inhibitors affected colony morphologies but not pluripotency gene expression. In addition, withdrawal of the ROCKi resulted in significantly reduced proliferation and increased cell death. Although withdrawal of growth factors bFGF or activin A, individually, had no effect on the expression of pluripotency genes, we observed a downregulation of pluripotency genes when bFGF and activin A were both depleted (Fig. 2D–2F). Moreover, when transferred to a conventional hESC medium, the naïve iPSCs were unable to maintain their ground-state morphologies; instead, they exhibited primed iPSC phenotypes (Fig. 2G).
Characterization of Differentiation Capacities of β-Thalassemia Patient-Specific Naïve iPSCs
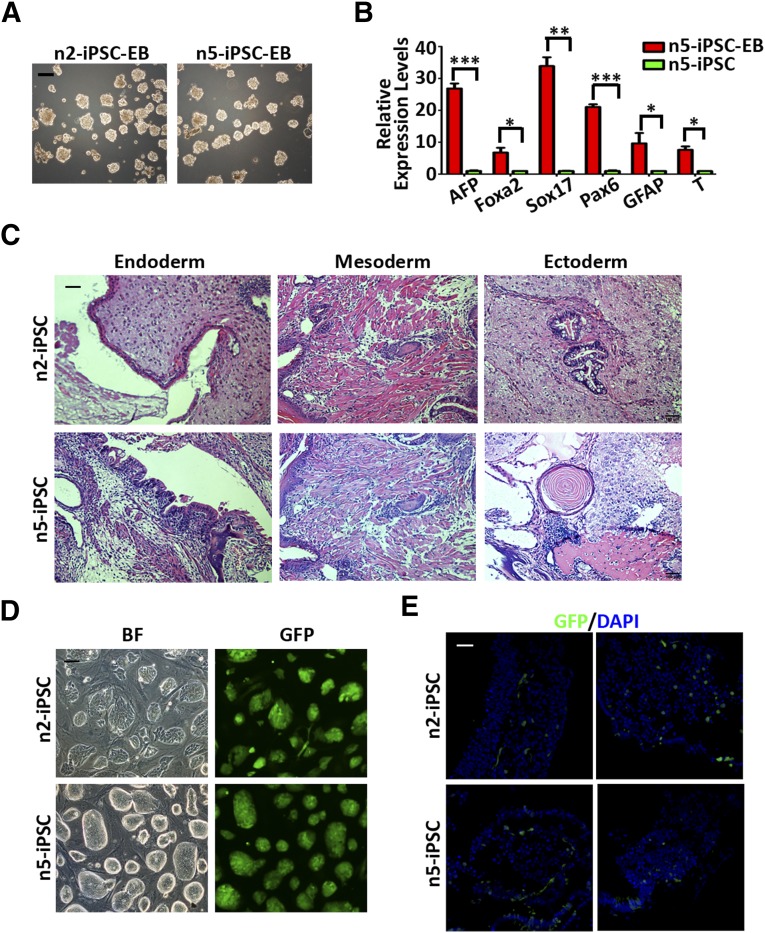
Next, to assess the in vitro and in vivo differentiation capacity of the naïve iPSCs that we derived, embryoid bodies were generated from different naïve iPSC lines by single-cell suspension followed by culturing in differentiation medium (Fig. 3A). On differentiation of the naïve iPSCs, quantitative PCR analysis showed significant upregulation in marker genes associated with the three germ layers (Fig. 3B). For differentiation in vivo, two naïve iPSC lines were injected subcutaneously into NOD/SCID mice for teratoma formation. Similar to primed iPSCs, as previously reported [26], both naïve iPSC lines could form teratomas 6 weeks after injection, which contained derivatives from all three embryonic germ layers (Fig. 3C).
Figure 3.
Differentiation properties of the naïve iPSCs derived from β-thalassemia fibroblasts. (A): Morphologies of embryoid bodies differentiated from two naïve iPSC lines. Scale bar = 200 μm. (B): Quantitative PCR analysis of lineage-related markers in embryoid bodies generated from n5-iPSCs. Data are shown as the mean ± SEM; t test; ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; n = 3 individual experiments. (C): Hematoxylin and eosin staining of teratomas containing tissues of all three germ layers derived from n2-iPSCs and n5-iPSCs. Scale bar = 100 μm. (D): Morphologies of GFP-labeled n2-iPSCs and n5-iPSCs. Scale bar = 200 μm. (E): Representative confocal images showing integration of GFP+ cells differentiated from n2- and n5-iPSCs into different sites of a mouse embryo at E10.5 stage. Scale bar = 20 μm. Abbreviations: AFP, α-fetoprotein; BF, bright field; DAPI, 4′,6-diamidino-2-phenylindole; EB, embryoid body; Foxa2; forkhead box protein A2; GFAP, glial fibrillary acidic protein; GFP, green fluorescent protein; iPSCs, induced pluripotent stem cells; PCR, polymerase chain reaction.
Finally, to test whether the human naïve iPSCs that we derived could be integrated into interspecies chimeras in vivo, naïve and primed iPSCs were labeled with green fluorescent protein (GFP; Fig. 3D; supplemental online Fig. 2C), followed by microinjection into ICR mouse blastocysts (E3.5) in parallel, and allowed to develop in vivo for a specific amount of time. The transplanted embryos were then dissected and observed under confocal microscopy to detect the presence of human iPSC-derived cells. Notably, in contrast to human primed iPSCs, we were able to obtain chimeric embryos with human naïve iPSCs corresponding to the E10.5 developmental stage, consistent with previous reports [12]. Moreover, these chimeric embryos showed a widespread integration of human naïve iPSCs at the organogenesis stages of embryonic development, which could be observed in different tissues, including the heart, brain, and neck (Fig. 3E). Collectively, these data indicate that the naïve iPSCs that we derived possess differentiation capacity both in vitro and in vivo. Most importantly, the naïve iPSC-derived cells could generate interspecies chimeras.
Genome-Wide Expression Analysis of Patient-Specific Naïve iPSCs
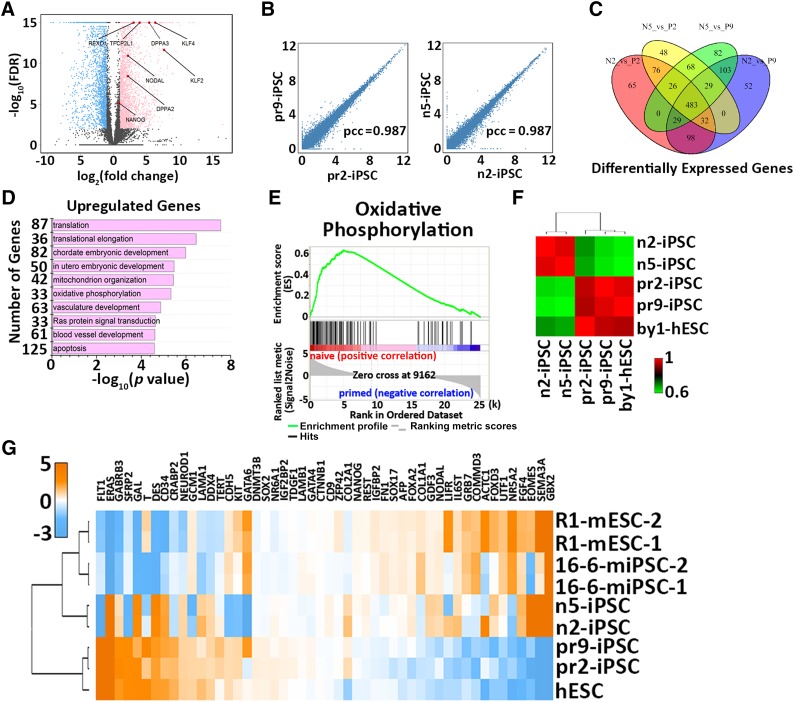
To validate the global gene expression patterns of naïve iPSCs derived from patient fibroblasts, RNAs were collected from both naïve iPSC lines (n2-iPSC and n5-iPSC) and primed cell lines (pr2-iPSC, pr9-iPSC, and by1-ESC) and subjected to RNA-sequence analysis. A comparison of gene expression profiles revealed significant differences between naïve iPSCs and primed iPSCs, although they share the same genetic origins (Fig. 4A). In contrast, the cell lines within the same categories exhibited high similarities (Fig. 4B, 4C). More importantly, a number of transcription factors associated with ground-state self-renewal and pluripotency were upregulated in naïve iPSCs, including DPPA2, DPPA3 (also known as STELLA), REX1, KLF2, KLF4, TFCP2L1, NANOG, and NODAL (Fig. 4A). Gene ontology [18] enrichment analysis showed that the upregulated genes in naïve iPSCs were mainly involved in translation, embryonic development, and oxidative phosphorylation (Fig. 4D). These results were further validated by KEGG pathway analysis (Fig. 4E). This is consistent with previous observations in mice in which mouse ESCs used oxidative phosphorylation, and EpiSCs showed low mitochondrial respiratory capacity [27].
Figure 4.
Transcriptional profiling of naïve iPSCs derived from β-thalassemia fibroblasts. (A): Volcano plot showing gene expression changes between naïve and primed iPSCs. The light blue dots represent the genes that are significantly downregulated in the naïve state [defined by log2(FC) <−1 and FDR <0.01]; pink dots represent the genes significantly upregulated in the naïve state [defined by log2(FC) >1 and FDR <0.01]. Highlighted in red are the genes of interest related to ground-state self-renewal and pluripotency. (B): Scatter plots showing gene expressions between the two cell lines of primed (left) or naïve state (right). (C): Venn diagram illustrating the overlapped and differentially expressed gene numbers identified among naïve iPSCs and primed iPSCs. (D): Gene ontology analysis of the upregulated genes in naïve iPSCs. (E): Upregulated oxidative phosphorylation by KEGG pathway analysis in naïve iPSCs compared with primed iPSCs. (F): Hierarchical clustering of naïve and primed iPSCs derived from patient fibroblasts. (G): Cross-species hierarchical clustering of naïve and primed pluripotent cells from mouse and human tissue. Abbreviations: FC, fold change; FDR, false discovery rate; hESC, human embryonal stem cell; iPSCs, induced pluripotent stem cells; mESC, mouse embryonal stem cell; miPSCs, murine iPSCs; N5, n5-iPSC; P2, pr2-iPSC; P9, pr9-iPSC; pcc, Pearson correlation coefficient.
Genome-wide expression profile comparisons of the five cell lines showed that the two naïve iPSC lines clustered together closely and were clearly separate from the three primed cell lines, which were clustered into another group (Fig. 4F). In addition, cross-species gene expression analysis demonstrated that naïve iPSCs clustered with naïve mouse ESCs in the upregulation of ground-state pluripotency genes and the downregulation of lineage-specific markers (Fig. 4G). Collectively, these data suggest that the gene expression pattern of naïve iPSCs is more similar to that of mouse ESCs and distinct from that of primed iPSCs, which is consistent with previous studies in mice and humans [14–16].
Epigenetic Property Analysis of Patient-Specific Naïve iPSCs
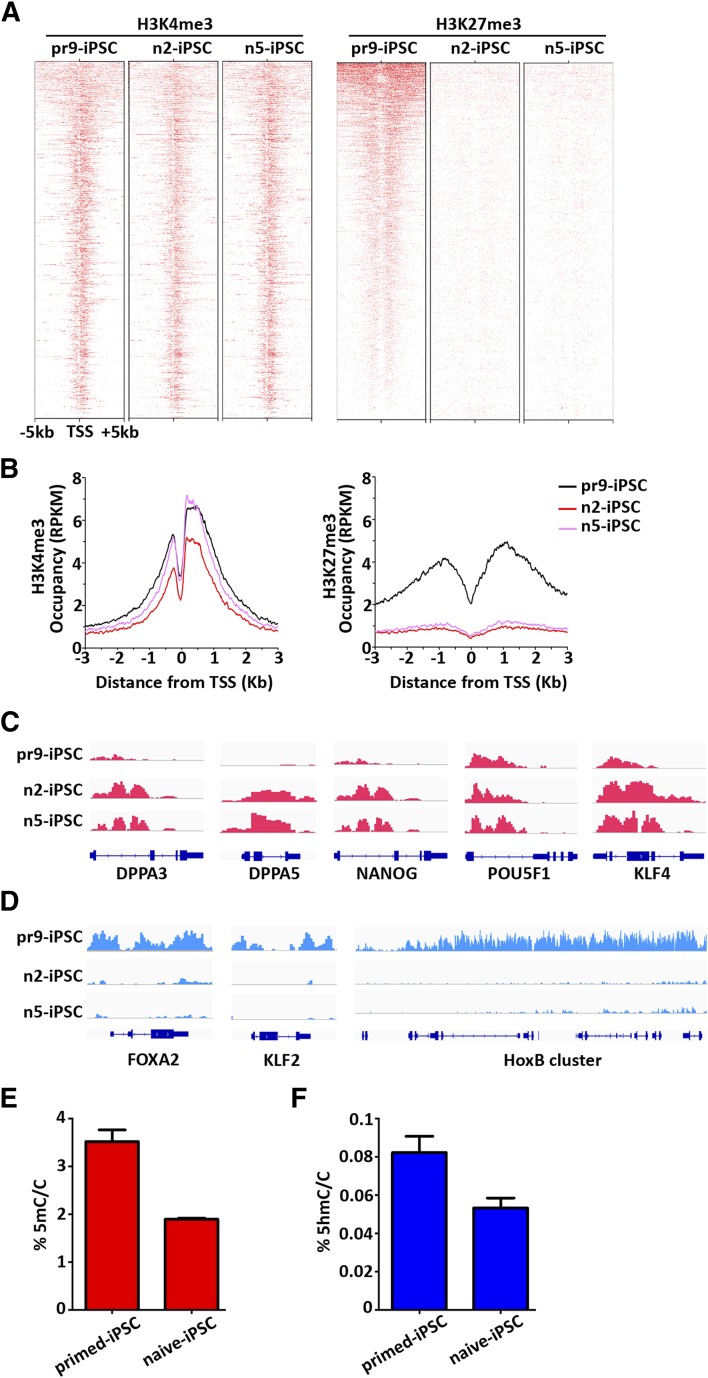
Next, to investigate the genome-wide distribution of H3K4me3 and H3K27me3, which indicate activation and repression of transcription, respectively, we performed chromatin immunoprecipitation-sequence analysis of the n2-iPSCs, n5-iPSCs, and pr9-iPSCs. Consistent with previous studies, the H3K27me3 signal was dramatically reduced at the transcription start site of polycomb group target genes in naïve iPSCs compared with that in primed iPSCs, whereas H3K4me3 showed no significant changes among the three iPSC lines (Fig. 5A, 5B). On further examination of the H3K4me3 profile at different gene loci, genes related to ground-state pluripotency, such as DPPA3, DPPA5, and KLF4, showed enhanced signal in naïve iPSCs, and genes associated with core pluripotency, such as POU5F1 and NANOG, exhibited similar or slightly higher signals in naïve iPSCs compared with that in primed iPSCs (Fig. 5C). However, the H3K27me3 profile in our naïve iPSCs was almost nonexistent for genes associated with naïve state pluripotency, such as KLF2, KLF4, and KLF5, and genes involved in development, such as FOXA2, GATA6, and the HOXA and HOXB clusters (Fig. 5D; supplemental online Fig. 3).
Figure 5.
Chromatin landscape of naïve iPSCs derived from β-thalassemia fibroblasts. (A): Heat map showing H3K4me3 (left) and H3K27me3 (right) distribution at polycomb target genes in pr9-iPSCs, n2-iPSCs, and n5-iPSCs. (B): Mean profiles of H3K4me3 (upper) and H3K27me3 (lower) at polycomb target genes in pr9-iPSCs, n2-iPSCs, and n5-iPSCs. (C): ChIP-seq tracks for H3K4me3 in pr9-iPSCs, n2-iPSCs, and n5-iPSCs at genes related to naïve pluripotency and core pluripotency. (D): ChIP-seq tracks for H3K27me3 in pr9-iPSCs, n2-iPSCs, and n5-iPSCs at genes related to naïve pluripotency and development. (E): Quantification by mass spectrometry of global 5mC levels in naïve and primed iPSCs derived from β-thalassemia fibroblasts. (F): Quantification by mass spectrometry of global 5hmC levels in naïve and primed iPSCs derived from β-thalassemia fibroblasts. Abbreviations: 5 hmC, 5-hydroxymethylcytosine; 5 mc/C, 5-methylcytosine per cytosine; ChIP-seq, chromatin immunoprecipitation-sequence; DPPA3, developmental pluripotency associated 3; DPPA5, developmental pluripotency associated 5; HoxB, homeobox B protein; iPSCs, induced pluripotent stem cells; TSS, transcription start site.
Further epigenetic analysis showed that the total 5mC and 5-hydroxymethylcytosine level were significantly reduced in naïve iPSCs compared with primed iPSCs by mass spectrometric quantification (Fig. 5E, 5F). Taken together, these data indicate that the upregulation of specific genes related to ground-state pluripotency is associated with increased H3K4me3 or decreased H3K27me3 signals in a locus-specific manner. Moreover, the reprogramming process from fibroblasts toward the primed or naïve state possesses different epigenetic deconstruction, including DNA methylation and histone modifications.
Genetic Correction of β-41/42 Mutation and Differentiation of Naïve iPSCs
To validate the application of naïve iPSCs in clinical research, a CRISPR/Cas9 genome editing system was adopted to correct the genetic mutation in the naïve iPSCs directly derived from patient fibroblasts (Fig. 6A). Three specific sgRNAs were designed according to a previously established protocol [25], cloned into pX330 vector, and tested by transfection into HEK293T cells. The most efficient cleaving sgRNA2 was selected by the T7E1 assay for the subsequent experiment (supplemental online Fig. 4A). For mutant gene correction, targeting donor DNA amplified from wild-type human genomic DNA with ∼250-bp 5′ and 3′ homologous arms was introduced, together with selected pX330-sgRNA, into n2-iPSCs and two primed iPSCs (pr2- and pr9-iPSCs) by electroporation (Fig. 6A).
Figure 6.
Genetic correction of the naïve iPSCs and derivation of naïve iPSCs directly from human urinary cells. (A): Schematic overview of targeting strategy for correction of the β-thalassemia mutations in both naïve and primed iPSCs using a clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 nuclease editing system. The sgRNA sequence site is shown as a blue dash. The mutation site is indicated and capitalized. The DNA donor contains 200 bp homologies on both sides flanking the DSB, and the corrected sequences are labeled as a green box. (B): Sequencing results of the β-41/42 mutation sites of HBB gene in both naïve and primed iPSCs before and after targeting. (C): Statistical histogram of the targeting efficiencies in naïve and primed iPSCs; t test; ∗, p < .05. (D): Representative images of morphologies of two corrected naïve iPSC lines. Scale bars = 200 μm. (E): Immunostaining images of pluripotent gene expressions in the two corrected naïve iPSC lines. Scale bars = 20 μm. (F): Representative images of morphological changes upon reprogramming from hUCs to naïve iPSCs on days 1, 10, and 14 and P1. Scale bars = 200 μm. (G): Quantitative polymerase chain reaction analysis of genes associated with the ground-state self-renewal and pluripotency in hUC-pr2-iPSC and hUC-n1-iPSC. Data are shown as mean ± SEM, t test, ∗, p < .05; ∗∗, p < .01; ∗∗∗, p < .001; n = 3 individual experiments. (H): Immunostaining images of pluripotency-associated markers OCT4, SOX2, NANOG, SSEA3/4 and TRRA-1-60 in hUC-n1-iPSC. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; hUC, human urinary cell; iPSCs, induced pluripotent stem cells; P1, passage 1; sgRNA, single guide RNA; SSEA3, stage-specific embryonic antigen 3; SSEA4, stage-specific embryonic antigen 4.
Seven days after electroporation, 40 colonies in each cell line were selected and expanded, followed by DNA sequencing validation. In the naïve iPSC group, up to 57% of clones were corrected at one allele of the HBB gene, showing heterozygous peaks at the mutation sites, and only approximately 32% of colonies were corrected in the primed iPSC group (Fig. 6B). We also observed one clone corrected at both alleles in the primed iPSC group. Furthermore, no off targets were detected in the naïve iPSC group, but 2 clones with off targets were detected in the primed iPSC group (data not shown). The statistical analysis of the sequencing results revealed significantly higher targeting efficiency in naïve iPSCs than in primed iPSCs (Fig. 6C). For further characterization, we selected two corrected naïve iPSC lines that showed morphologies similar to those of mouse ESCs (Fig. 6D). Immunostaining analysis confirmed that the corrected iPSCs also expressed pluripotency-associated markers such as OCT3/4, SOX2, and NANOG (Fig. 6E).
To evaluate the hematopoiesis capacity of the naïve iPSCs, we differentiated two corrected and the parental naïve iPSC lines and two primed iPSC lines into the hematopoietic progenitor cells using previously established protocols [26]. On coculture with OP9-GFP stromal cells, the iPSCs expanded with morphological changes (supplemental online Fig. 4B). Eight days after induction, the ratio of CD34+ cells analyzed by flow cytometry was approximately 4.3% in the corrected naïve iPSCs, comparable to that of the parental naïve iPSCs (approximately 3.7%) and the two primed iPSC lines (approximately 4.0%) (supplemental online Fig. 4C–4E). Taken together, our results indicate that the correction of the disease mutations did not compromise the naïve iPSCs’ self-renewal or differentiation abilities, both a prerequisite for further hematopoietic applications.
Generation of Naïve iPSCs From Human Urinary Cells Using a Similar Strategy
As demonstrated in previous reports, human urinary cells (hUCs) can be reprogrammed into primed iPSCs by introducing exogenous genes [24]; thus, we tested whether hUCs could be reprogrammed directly into the naïve iPSCs. Using a similar strategy, 5 × 105 hUCs obtained from a normal male were electroporated with the same episomal vectors and were then cultured in conventional hESM for 9 days. The medium was then replaced with naïve hESM (5i/L/FA medium) for an additional 15–20 days. At 10 days after culturing in the 5i/L/FA medium, dome-shaped colonies emerged (Fig. 6F). The colonies were then picked and expanded further. The hUC-naïve iPSC lines were established, which exhibited typical morphologies similar to those of murine ESC/iPSCs (Fig. 6F).
We also detected expression of multiple transcription factors associated with ground-state pluripotency, including NANOG, KLF4, REX1, KLF2, and TFCP2L1, which showed significant upregulation in hUC-naïve iPSCs compared with primed iPSCs derived from the same hUCs (Fig. 6G). Moreover, hUC-naïve iPSCs also expressed pluripotency-associated markers, such as OCT4, SOX2, and NANOG, and surface markers such as SSEA3, SSEA4, and TRA1-60 (Fig. 6H). Collectively, our results suggest that hUCs, an easily and noninvasively acquired cell type, could also be directly reprogrammed into naïve iPSCs, providing an excellent cell resource for further clinical trials.
Discussion
β-Thalassemia is one of the most common genetic diseases without an effective treatment. The emergence of conventional iPSCs and the development of targeting techniques have raised hopes for such diseases. However, the properties of primed iPSCs, which closely resemble mouse EpiSCs, with features such as low proliferation ability, poor single-cell cloning, and poor recovery efficiencies, resulting in difficulties in mutant gene targeting or drug screening, strongly restrict the application of iPSCs in disease modeling and regenerative medicine.
In the present study, by using defined culture conditions with a transgene-free strategy, we successfully reprogrammed fibroblasts obtained from a β-thalassemia patient into naïve-state iPSCs. These patient-specific naïve iPSCs exhibited the features of ground-state pluripotency in terms of the proliferation properties, gene expression profiles, dependent-signaling pathways, and the ability to integrate into cross-species chimeric embryos. They also showed significantly higher targeting efficiencies compared with primed iPSCs in the CRISPR/Cas9 system. In addition, the corrected naïve iPSCs, similar to primed iPSCs, possessed the ability to differentiate into hematopoietic progenitor cells. Hence, these results provide strong evidence that the ground-state reprogramming combined with the CRISPR/Cas9 genome editing system could potentially be used to cure genetic diseases, such as β-thalassemia.
To date, multiple human cell types, such as skin fibroblasts, adipose stem cells, peripheral blood cells, and hepatocytes, provide potential cell resources for the generation of induced pluripotent stem cells. However, these cells are all collected invasively. Recently, human urinary cells, an excellent example of a noninvasively collected cell type, have been successfully reprogrammed into primed state iPSCs. In the present study, using similar strategies used in fibroblasts, we successfully generated ground-state iPSCs directly from noninvasively acquired urine cells, which provides a better resource for disease modeling and treatment for future clinical research.
Conclusion
In the present study, using the established 5i/L/FA system, we directly reprogrammed the fibroblasts of a patient with β-thalassemia into transgene-free naïve iPSCs with molecular signatures of ground-state pluripotency, which can efficiently produce cross-species chimeras. Furthermore, using a CRISPR/Cas9 genome editing system, these naïve iPSCs exhibited significantly improved gene-correction efficiencies compared with the corresponding primed iPSCs. Importantly, human urinary cells can also be reprogrammed into a ground state using a similar approach. Therefore, our findings demonstrate the feasibility and superiority of using patient-specific iPSCs in the naïve state for disease modeling, gene editing, and future clinical therapy.
Supplementary Material
Acknowledgments
We thank our colleagues in the laboratory for their assistance with the experiments and preparation of the manuscript. Additionally, we thank Zeyu Xiong for designing the sgRNA targeting the HBB gene locus. This work was supported by the National Natural Science Foundation of China (Grants 31471392, U1132005, and 31325019), the Ministry of Science and Technology of China (Grants 2014CB964601, 2012CBA01300, and 2015CB964503), the Science and Technology Commission of Shanghai Municipality (Grant YF1403900), the Education Commission of Shanghai Municipality (Grant 13CG16), the Program for Young Excellent Talents in Tongji University, and the NSFC-Guangdong Joint Fund (Grant 2011Y1-00038).
Author Contributions
Y.Y.: conception and design, provision of study material or patients, collection and/or assembly of data, data analysis and interpretation, manuscript writing; X.Z.: data analysis and interpretation, manuscript writing; L.Y., Z.H., J.C., X.K., and Y.Z.: collection and/or assembly of data; H.W. and X.-F.S.: provision of study material or patients; C.J.: data analysis and interpretation; Y.W.: conception and design, provision of study material or patients, collection and/or assembly of data, data analysis and interpretation, manuscript writing; S.G.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
References
- 1.Cowan CA, Klimanskaya I, McMahon J, et al. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 2.Suss-Toby E, Gerecht-Nir S, Amit M, et al. Derivation of a diploid human embryonic stem cell line from a mononuclear zygote. Hum Reprod. 2004;19:670–675. doi: 10.1093/humrep/deh135. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 5.Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 6.Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 7.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Boroviak T, Loos R, Bertone P, et al. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat Cell Biol. 2014;16:516–528. doi: 10.1038/ncb2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amit M, Carpenter MK, Inokuma MS, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 11.Bradley A, Evans M, Kaufman MH, et al. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 12.Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 13.Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci USA. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ware CB, Nelson AM, Mecham B, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci USA. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo G, Yang J, Nichols J, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan YS, Göke J, Ng JH, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Ng KM, Tse HF. Modeling hereditary cardiac disease with patient-specific-induced pluripotent stem cells: Opportunities and concerns. J Cardiovasc Pharmacol. 2012;60:406–407. doi: 10.1097/FJC.0b013e31825f3f0f. [DOI] [PubMed] [Google Scholar]
- 20.An MC, Zhang N, Scott G, et al. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma N, Liao B, Zhang H, et al. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J Biol Chem. 2013;288:34671–34679. doi: 10.1074/jbc.M113.496174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111:1048–1053. doi: 10.1002/bit.25018. [DOI] [PubMed] [Google Scholar]
- 23.Xie F, Ye L, Chang JC, et al. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Benda C, Dunzinger S, et al. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012;7:2080–2089. doi: 10.1038/nprot.2012.115. [DOI] [PubMed] [Google Scholar]
- 25.Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Jiang Y, Liu S, et al. Generation of induced pluripotent stem cells from human beta-thalassemia fibroblast cells. Cell Res. 2009;19:1120–1123. doi: 10.1038/cr.2009.100. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, Choi M, Margineantu D, et al. HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.