Summary
Recent studies describe a network of signaling proteins centered around Goα and Gqα that regulates neurotransmitter secretion in C. elegans by controlling the production and consumption of diacylglycerol (DAG). We sought other components of the Goα–Gqα signaling network by screening for aldicarb-resistant mutants with phenotypes similar to egl-30 (Gqα) mutants. In so doing, we identified ric-8, which encodes a novel protein named RIC-8 (synembryn). Through cDNA analysis, we show that RIC-8 is conserved in vertebrates. Through immunostaining, we show that RIC-8 is concentrated in the cytoplasm of neurons. Exogenous application of phorbol esters or loss of DGK-1 (diacylglycerol kinase) rescues ric-8 mutant phenotypes. A genetic analysis suggests that RIC-8 functions upstream of, or in conjunction with, EGL-30 (Gqα).
Introduction
Chemical synapses play a key role in all nervous systems by controlling the transfer of information between cells. The information transfer, designated synaptic transmission, occurs when synaptic vesicles fuse with the presynaptic membrane and release neurotransmitter, which then diffuses across a narrow extracellular space and activates postsynaptic receptors on another cell. A better understanding of how synaptic transmission is regulated, both by electrical signals and by signal transduction pathways, is likely to yield important insights into how nervous systems establish, maintain, and modify behavior. We seek to understand the nature of the signal transduction pathways that regulate neurotransmitter secretion, which is the presynaptic component of synaptic transmission.
In vertebrates, one common class of signaling pathways in neurons is built around heterotrimeric G proteins of the Gq/G11 class (Watson and Arkinstall, 1994). The binding of neurotransmitter to a Gq-coupled receptor leads to activation of the G protein, which in turn activates phospholipase Cβ (PLCβ) (Singer et al., 1997). PLCβ cleaves phosphatidylinositol 4,5-bisphosphate (PIP2) into the small signaling molecules diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3). The finding that phorbol esters (molecular analogs of DAG) stimulate neurotransmitter secretion suggests that the Gqα path way positively regulates synaptic transmission (Malenka et al., 1986; Shapira et al., 1987; Stevens and Sullivan, 1998). However, genome complexity and the difficulty of mutational approaches have left important questions about the Gqα pathway unanswered in vertebrates. Is the Gqα pathway a crucial regulator of synaptic transmission, or is it used only for modulatory purposes? Are there other downstream effectors of Gqα in addition to PLCβ? What is the relationship of the Gqα pathway to other G protein pathways?
Since synaptic transmission is a highly conserved process, genetic studies in model systems provide a way to address these questions. In C. elegans, the egl-30 gene encodes a highly conserved homolog of Gqα (82% identical to vertebrate forms) that is essential for life (Brundage et al., 1996). Hypomorphic egl-30 mutations result in strongly reduced rates of locomotion and egg laying (Trent et al., 1983; Brundage et al., 1996). More recent studies provide evidence that a network of signaling proteins involving both Goα and Gqα regulates neurotransmitter secretion in C. elegans by controlling the production and consumption of DAG (Figure 1). In brief, these studies suggest that EGL-30 (Gqα) acts through EGL-8 (PLCβ) to produce the small signaling molecule DAG, which in turn positively regulates neurotransmitter secretion, in part via interactions with the DAG binding protein UNC-13 (Lackner et al., 1999; Miller et al., 1999; Nurrish et al., 1999). The EGL-30 (Gqα) pathway is negatively regulated by GOA-1 (Goα) and the RGS protein EAT-16 (Hajdu-Cronin et al., 1999; Miller et al., 1999). Another RGS protein, EGL-10, negatively regulates GOA-1 (Goα) (Koelle and Horvitz, 1996). DGK-1 (aka SAG-1) (DAG kinase) antagonizes the EGL-30 pathway, presumably by converting DAG to phosphatidic acid (Hajdu-Cronin et al., 1999; Miller et al., 1999; Nurrish et al., 1999). The fact that all of these components are conserved in vertebrates suggests that the Goα–Gqα signaling network may be a basic regulator of synaptic transmission in many, if not all, nervous systems.
Figure 1. One Possible Arrangement of the Components of the Goα–Gqα Signaling Network.

Model based on the findings of Koelle and Horvitz (1996), Hajdu-Cronin et al. (1999), Miller et al., (1999), Nurrish et al. (1999), and Lackner et al. (1999). An alternative arrangement, in which GOA-1 acts through EAT-16, is equally consistent with the available data (Hajdu-Cronin et al., 1999). Arrows denote activation. Bars at the end of a line denote inhibition. Proteins that promote or activate synaptic transmission are shown in green blocks. Reducing the function of these proteins results in aldicarb resistance and reduced locomotion rates. Proteins that inhibit synaptic transmission are shown in red blocks. Reducing the function of these proteins results in aldicarb hypersensitivity and hyperactive locomotion. Neurotransmitter inputs into this pathway are not specified.
Studies of the Goα–Gqα signaling network have been facilitated by the drug aldicarb, an inhibitor of acetylcholinesterase that causes a toxic accumulation of secreted acetylcholine (ACh) at synapses (Cambon et al., 1979; Risher et al., 1987). Since toxic accumulations of ACh can be reduced by mutations that decrease neurotransmitter release, aldicarb has been a powerful tool for investigating both the mechanics of synaptic transmission and its regulation by signaling pathways. Many aldicarb resistance genes encode proteins that either function in the synaptic vesicle cycle or are homologous to synaptic vesicle cycle proteins (Miller et al., 1996; Rand and Nonet, 1997; Nonet, 1999). However, a subclass of aldicarb resistance genes encodes proteins that function as regulators of neurotransmitter secretion. This subclass includes egl-30 (Gqα), egl-8 (PLCβ), and egl-10 (RGS). Importantly, mutants in this subclass share unique phenotypes that distinguish them from the synaptic vesicle cycle mutants (Miller et al., 1999).
In a previous study, we began an investigation of the EGL-30 (Gqα) pathway by screening for additional aldicarb-resistant mutants with phenotypes similar to egl-30 mutants (Miller et al., 1999). In this study, we focused on one of the genes identified in that screen: ric-8. We show that ric-8 encodes a novel protein, named RIC-8 (synembryn), that is conserved in vertebrates. Through immunostaining, we show that RIC-8 is concentrated in the cytoplasm of neurons. Exogenous application of phorbol ester or loss of DGK-1 (diacylglycerol kinase) results in a striking rescue of ric-8 mutant phenotypes. A genetic analysis suggests that RIC-8 functions upstream of, or in conjunction with, EGL-30 (Gqα).
Results
Reduction-of-Function Mutations in egl-30 (Gqα) and ric-8 Lead to Similar Phenotypes
Through an extensive series of aldicarb selections that represented ~35-fold coverage of the genome, we identified two ric-8 alleles (md303 and md1909). A large noncomplementation screen (~16-fold coverage of the genome) resulted in only one additional allele (md2230). All three alleles are recessive and exhibit the egg laying–defective and reduced body flexion phenotypes characteristic of egl-30 reduction-of-function mutants (Figure 2).
Figure 2. ric-8 and egl-30 (Gqα) Reduction-of-Function Mutants Share Similar Phenotypes.

Photographs comparing wild-type C. elegans (N2) with strains con taining reduction-of-function mutations in snt-1 (a synaptotagmin homolog) (Nonet et al., 1993), egl-30, or ric-8. Note that snt-1 mutants have a partially coiled posture, are smaller in size, and have fewer eggs in their uteri than do wild type. In contrast, egl-30 and ric-8 mutants, in addition to being aldicarb resistant, are both bloated with eggs and exhibit decreased body flexion phenotypes that are characteristic of the subclass of aldicarb resistance mutants with defects in the Goα–Gqα signaling network.
As an initial comparison of the amount of ric-8 function remaining in these alleles, we measured their locomotion rates on an agar surface. Wild-type worms exhibit a stereotyped, spontaneous locomotion behavior that can be quantified by counting body bends. Table 1 compares the mean locomotion rates of wild-type worms and ric-8 mutants. The locomotion rates of the md1909 and md2230 alleles were about 15% that of wild type, while the locomotion rate of the md303 allele was 2.6% that of wild type, a level that is similar to that seen in strong reduction-of-function alleles of egl-30 (Miller et al., 1999). The aldicarb resistance, as well as the reduced locomotion rate, body flexion, and egg laying of ric-8(md303), is corrected to approximately wild-type levels by the intragenic suppressor mutation md1712, which we identified in a screen for suppressors of ric-8(md303) (Table 1; data not shown). This shows that the ric-8(md303) mutant phenotypes result from defects at a single genetic locus. The spectrum of phenotypes shared by ric-8 and egl-30 mutants suggests that RIC-8 and EGL-30 have overlapping functions. Unlike egl-30 mutants, however, ric-8 mutants exhibit partial embryonic lethality that appears to result from defects in the regulation of a subset of centrosome movements during early embryogenesis (K. G. Miller and J. B. Rand, submitted).
Table 1.
Locomotion Rates of ric-8 Mutants
| Allele | Locomotion Rate (Body Bends/min)a |
|---|---|
| N2 (wild type) | 15.6 ± 1.7 |
| ric-8 (md1909) | 2.53 ± 0.85 |
| ric-8 (md2230) | 2.02 ± 1.0 |
| ric-8 (md303) | 0.4 ± 0.32 |
| ric-8 (md1712 md303) | 20.8 ± 1.3 |
mean ± standard error, n = 5 for ric-8 (md303) and ric-8 (md2230); n = 10 for all other strains.
ric-8 Encodes a Novel Protein that Is Related to Vertebrate Proteins of Unknown Function
To investigate the molecular basis of the similarities between ric-8 and egl-30 mutants, we cloned a portion of the ric-8 gene by transposon tagging and then used that sequence as a probe to isolate full-length cDNAs. The open reading frame of the full-length cDNA predicts a protein of 566 amino acids with a molecular mass of 63 kDa. Sequence analysis programs did not identify any of the known subcellular localization signals in RIC-8. This suggests that RIC-8 is a cytoplasmic protein. To reflect the dual roles of RIC-8 in synaptic transmission and early embryogenesis, we also refer to RIC-8 as synembryn.
The Drosophila and human genome sequencing projects have each identified one synembryn-related gene (GenBank accesssion numbers AAF46477 and BAA91717, respectively). In addition, there is at least one synembryn-related gene in both the mouse and human expressed sequence tag (EST) databases. To investigate the evolutionary conservation of RIC-8, we isolated and sequenced mouse synembryn cDNAs. The open reading frame of mouse synembryn cDNAs predicts a 530 amino acid peptide that is 30% identical and 43% similar to C. elegans RIC-8, while the predicted Drosophila protein is 578 amino acids, and 29% identical and 40% similar (Figure 3A). The highest homology occurs in a 190 amino acid region near the amino terminus that is 37% identical between the C. elegans and mouse proteins. Of the more than 30 synembryn EST sequences in the mouse EST database, the representation is highest in databases of brain, embryo, and mammary gland ESTs (seven cDNAs each); however, synembryn cDNAs were also found in other tissues, such as blastocyst and tissues of the immune system (three cDNAs each).
Figure 3. ric-8 Encodes a Novel Protein that Is Conserved in Vertebrates.

(A) Amino acid alignment of mouse (top), Drosophila (middle), and C. elegans (bottom) synembryn. Gray and black backgrounds represent similar and identical residues, respectively.
(B) The ric-8 gene, transcript, and mutations. Schematic of ric-8 gene structure and mutations. Boxed regions are exons, black boxed regions indicate coding sequence, and lines indicate introns. The break in the intron near the middle of the gene indicates that this intron has not been completely sequenced. The positions and descriptions of the three ric-8 mutations are indicated. The md1909 mutant has a 1.6 kb Tc1 transposon insertion following amino acid 221. The md303 mutant has a C-to-A change at nucleotide 832 of the cDNA that converts alanine 275 to glutamic acid. md1712, isolated as an intragenic suppressor of md303, converts leucine 267 to phenylalanine.
ric-8 Mutations Reduce the Function of RIC-8
To investigate how ric-8 mutations affect the RIC-8 protein, we analyzed genomic DNA from ric-8 mutants. We found that the md1909 allele contains a Tc1 transposon insertion in one of the coding exons near the middle of the gene (Figure 3B). Although C. elegans can sometimes remove Tc1 sequences during mRNA splicing (Rushforth et al., 1993), this mutation should reduce the production of RIC-8 protein. The md303 allele contains a nonconservative amino acid substitution. Interestingly, the ric-8(md1712 md303) strain, in which the phenotypes of md303 are suppressed to nearly wild-type levels, contains a second missense mutation eight amino acids upstream from the md303 lesion (Figure 3B).
Since the rate at which we obtained ric-8 mutations was ~17-fold lower than the frequency expected for loss-of-function mutations, we think it is most likely that the mutations do not completely eliminate RIC-8 function. Since ric-8 mutants exhibit 19%–29% embryonic lethality (K. G. Miller and J. B. Rand, submitted), it is possible that complete elimination of RIC-8 would result in 100% embryonic lethality. To further explore the ric-8 loss-of-function phenotype, we injected double stranded RNA derived from a ric-8 cDNA into the gonads of wild-type animals and examined their progeny for the effects of RNA interference (RNAi). RNAi is an effective method of blocking the production of specific proteins in the C. elegans embryo and in many adult cell types, as well (Fire et al., 1998). We observed that a high percentage of the progeny of the injected animals exhibited larval phenotypes similar to those seen in ric-8(md303) and ric-8(md1909) (Table 2). These animals moved little and exhibited the straight posture that is characteristic of ric-8 and egl-30 mutants. The remaining progeny of the injected animals exhibited embryonic lethality (0%–32%) or resembled wild type (4%–22%) (Table 2). Since RIC-8 protein is likely to be contributed maternally (K. G. Miller and J. B. Rand, submitted), ric-8 RNAi injections into young adults may not eliminate RIC-8 protein. However, we found that even larval injections of double stranded ric-8 RNA resulted in similar or lower levels of embryonic lethality (data not shown). In summary, although this method does not conclusively determine the null phenotype of ric-8, our finding that ric-8 RNAi phenotypes in larvae mimic the mutant phenotypes confirms that the mutant phenotypes are caused by reduction-of-function mutations in ric-8.
Table 2.
ric-8 RNA Interference Mimics ric-8 Larval Phenotypes
| Animal Number | Unhatched Eggs | Ric-8-like Larvae | Wild-type Larvae |
|---|---|---|---|
| 1 | 0 (0%) | 53 (91%) | 5 (9%) |
| 2 | 3 (8.3%) | 25 (69%) | 8 (22%) |
| 3 | 2 (4.9%) | 30 (73%) | 9 (22%) |
| 4 | 10 (14%) | 57 (80%) | 4 (5.6%) |
| 5 | 24 (32%) | 48 (64%) | 3 (4%) |
Animals were transferred to individual plates 2–6 hr after injection and allowed to lay eggs for 18 hr, after which the injected animals were killed or transferred to fresh plates. The brood laid during the 18 hr period, which showed the greatest effect, was observed at 24 hr intervals for 4 days. Unhatched eggs, Ric-8-like larvae, and wild-type larva were scored at the first 24 hr interval. Control injections using egl-30 dsRNA yielded 2.8% embryonic lethality (n = 176 progeny).
We also observed that as the Ric-8-like progeny of the RNAi-injected animals developed to adulthood, they gradually lost their Ric-8 phenotypes and eventually resembled wild-type animals. This indicates that the Ric-8-like larvae are not significantly impaired by developmental defects. Since some cell types, including adult neurons, are strongly resistant to RNAi (Nonet, 1999), this loss of phenotype in the developing animals is likely to reflect a gradual accumulation of ric-8 mRNA and protein.
RIC-8 Positively Regulates Synaptic Transmission
We quantified the aldicarb resistance of the strong reduction-of-function ric-8 mutant md303 by measuring its population growth rate on various concentrations of aldicarb. We observed that ric-8(md303) mutants exhibit an aldicarb dose-response curve that is similar to that of the strong reduction-of-function egl-30 mutant ad805 (Figure 4A). Both mutants are able to grow up to concentrations of aldicarb that are about 4-fold higher than the concentration that stops the growth of wild type (Figure 4A). Although aldicarb resistance could theoretically arise through defects in the reception of ACh, a previous study showed that ric-8 and egl-30 mutants have a normal muscle response to the ACh receptor agonist levamisole, and, in addition, mutants in both genes have defects consistent with the impaired release of multiple neurotransmitters (Miller et al., 1996). Taken together, these studies suggest that the aldicarb resistance of ric-8 and egl-30 reduction-of-function mutants is due to decreased neurotransmitter secretion and that the normal function of RIC-8 and EGL-30 is to promote, or positively regulate, neurotransmitter secretion.
Figure 4. RIC-8 Appears to Function Upstream of EGL-30 (Gqα) or in a Parallel Intersecting Pathway.

(A) egl-30(ad805) and ric-8(md303) exhibit a similar degree of aldicarb resistance. Shown are the aldicarb dose-response curves for wild type, egl-30(ad805), and ric-8(md303). One hundred percent represents the number of progeny produced from a starting population of L1 larvae over a 96 hr period in the absence of aldicarb. Curves are representative of duplicate experiments.
(B) The aldicarb resistance of ric-8(md303) is suppressed to near wild-type levels by overexpression of egl-10+. Shown are aldicarb dose-response curves of ric-8(md303); egl-10+[nls51] animals and control strains. Note that the aldicarb sensitivity of ric-8(md303); egl-10+[nls51] animals is close to that of wild type. Curves are representative of duplicate experiments.
(C) The locomotion rate of md303 is rescued to near wild-type levels by eliminating or blocking GOA-1 function. Shown are the mean locomotion rates of wild type, ric-8(md303), egl-30(ad805), goa-1(n363), egl-10+ [nIs51], and two double mutant combinations. Note that the locomotion rate of goa-1(n363); ric-8(md303) is close to that of wild type. Error bars represent the standard error of the mean in a population of five to ten young adults.
In addition to their similar aldicarb dose-response curves, ric-8(md303) and egl-30(ad805) also exhibit similar locomotion rates, which are 2.6% and 0.8% that of the wild-type rate, respectively (Figure 4C). Strongly reducing the function of either protein, therefore, has similar consequences with respect to aldicarb resistance and locomotion rate.
RIC-8 Appears to Function Upstream of EGL-30 (Gqα) or in a Parallel Intersecting Pathway
To investigate the relationship of RIC-8 to the Goα–Gqα signaling network, we first investigated the relationship of RIC-8 to GOA-1 (Goα). Loss-of-function goa-1 mutants exhibit hyperactive locomotion and hypersensitivity to aldicarb (Mendel et al., 1995; Ségalat et al., 1995; Miller et al., 1999; Nurrish et al., 1999). Previous studies suggest that these phenotypes result from excess EGL-30 pathway activity (Hajdu-Cronin et al., 1999; Miller et al., 1999).
To investigate the relationship of RIC-8 to GOA-1, we first analyzed the phenotype of double mutant strains containing a loss-of-function mutation in goa-1 and a strong reduction-of-function mutation in ric-8. In a separate investigation, we found that GOA-1 and RIC-8 exhibited a strong maternal effect interaction that resulted in 95%–100% embryonic lethality for the progeny of goa-1/+; ric-8/ric-8 animals (K. G. Miller and J. B. Rand, submitted). In this study, we found that we were able to bypass this embryonic lethality by selecting goa-1; ric-8 double mutants from the progeny of goa-1/+; ric-8/+ animals. goa-1; ric-8 double mutants produced no eggs and thus are completely sterile. This suggests that goa-1 and ric-8 act together in some aspect of germ cell production or maturation. Although the sterility of the goa-1; ric-8 double mutants prevented us from assaying their aldicarb resistance by our standard assay, we observed that the locomotion rates of the goa-1(n363); ric-8(md303) double mutants were similar to, or greater than, the wild-type locomotion rate (Figure 4C). This result is in striking contrast to the egl-30(ad805); goa-1(n363) double mutant, which has a locomotion rate not significantly different from the egl-30 single mutant (Miller et al., 1999).
We also analyzed the aldicarb sensitivity and locomotion rate of ric-8(md303) in a background containing the egl-10+ [nls51] integrated transgenic array, which overexpresses wild-type EGL-10. EGL-10 is an RGS protein that negatively regulates GOA-1 (Koelle and Horvitz, 1996). Analogous studies in vertebrates have found that RGS proteins inhibit G protein signaling by functioning as GTPase-activating proteins (Berman et al., 1996; Hunt et al., 1996; Watson et al., 1996). egl-10+ [nls51] animals exhibit hyperactive locomotion and hypersensitivity to aldicarb, presumably because the excess EGL-10 RGS protein inhibits GOA-1 signaling. We found that ric-8(md303); egl-10+ [nIs51] animals have aldicarb sensitivities that are indistinguishable from those of wild type and locomotion rates that are in between those of ric-8 single mutants and wild type (Figures 4B and 4C). Once again, this result is in striking contrast to egl-30(ad805); egl-10+ [nls51] animals, whose locomotion rates and levels of aldicarb resistance are not significantly different from those of the egl-30 single mutant (Miller et al., 1999).
These results provide key insights into the relationship of RIC-8 to GOA-1 and EGL-30. First, RIC-8 does not appear to function as a negative regulator of GOA-1 signaling because, if it did, the phenotypes of the goa-1; ric-8 double mutant would resemble goa-1 single mutants, and they clearly do not. Second, although strong reduction-of-function egl-30 and ric-8 mutants exhibit similar degrees of impairment, egl-30 mutants are not suppressed by goa-1 (loss of function) or egl-10 (overexpression) (Miller et al., 1999). Since ric-8(md303) mutants are strongly suppressed by excess EGL-30 pathway activity (produced through goa-1 loss of function or egl-10 overexpression), the results suggest that RIC-8 functions upstream of, or in conjunction with, EGL-30 (Gqα).
Loss of DGK-1 (Diacylglycerol Kinase) or Exogenous Application of Phorbol Ester Results in a Striking Rescue of ric-8 Mutant Phenotypes
DAG is a major end product of the Gqα–PLCβ pathway, and previous studies suggest that phorbol esters (DAG analogs) promote synaptic transmission in C. elegans (see references in Introduction). If RIC-8 functions up stream of, or in conjunction with, EGL-30 (Gqα), then one should be able to partially or completely bypass the function of RIC-8 by supplying DAG or phorbol esters. We therefore examined the effect of increased DAG levels on ric-8 reduction-of-function mutants. We first analyzed the phenotype of ric-8; dgk-1 double mutants. dgk-1 encodes a diacyglycerol kinase (Nurrish et al., 1999) and is a component of the Goα–Gqα signaling network in the C. elegans nervous system (Miller et al., 1999; Nurrish et al., 1999). Diacylglycerol kinases function to reduce DAG levels by converting DAG to phosphatidic acid (Sakane and Kanoh, 1997). Loss-of-function mutations in dgk-1 result in strong hypersensitivity to aldicarb and hyperactive locomotion, presumably as a result of DAG accumulation in neurons.
We found that the presence of the dgk-1 mutation in the ric-8 background caused a striking increase in the aldicarb sensitivity of ric-8 mutants, to the point that they were nearly as hypersensitive as the dgk-1 single mutants were (Figure 5A). This result suggests that dgk-1 is epistatic to ric-8 with respect to aldicarb resistance; however, we observed that, with respect to other phenotypes, ric-8; dgk-1 double mutants did not resemble dgk-1 mutants but instead showed intermediate phenotypes that were close to wild-type. For example, the locomotion rate of the ric-8; dgk-1 double mutants, though strikingly improved relative to ric-8 single mutants, was close to that of wild-type worms (Figure 5B). In addition, the size, egg laying, movement, and body flexion characteristics of the double mutants were closer to those of wild type than to those of either single mutant. (For supplementary video, go to http://www.neuron.org.cgi/content/full/27/2/289/DC1).
Figure 5. Loss of DGK-1 (Diacylglycerol Kinase) or Exogenous Application of Phorbol Ester Results in a Striking Rescue of ric-8 Mutant Phenotypes.
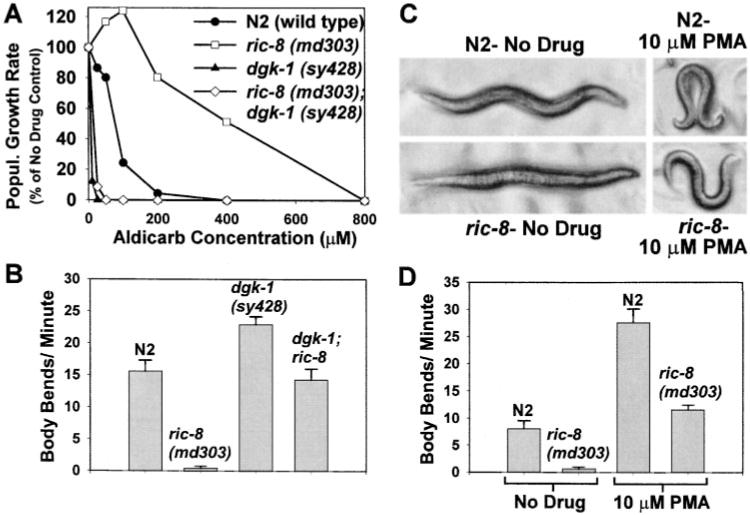
(A) ric-8; dgk-1 double mutants are hypersensitive to aldicarb. Aldicarb sensitivity is quantified by measuring population growth rates of wild-type and mutant strains on various concentrations of aldicarb. Shown are aldicarb dose-response curves for wild type, ric-8(md303), dgk-1(sy428), and ric-8(md303); dgk-1(sy428). Note that ric-8(md303) is strongly resistant to aldicarb, while dgk-1(sy428) is strongly hypersensitive to aldicarb. The aldicarb sensitivity of the ric-8; dgk-1 double mutant is similar to that of the dgk-1 single mutant. Curves are representative of duplicate experiments.
(B) The mean locomotion rate of ric-8; dgk-1 double mutants is close to wild-type. The mean locomotion rate of ric-8(md303) is 2.6% that of the wild-type rate. dgk-1(sy428) mutants, on the other hand, exhibit hyperactive locomotion. ric-8; dgk-1 double mutants have intermediate locomotion rates that are close to that of wild type. Error bars represent the standard error of the mean in a population of ten young adult animals. A supplementary video (http://www.neuron.org/cgi/content/full/27/2/289/DC1) shows that ric-8; dgk-1 double mutants also exhibit normal body flexion and coordinated locomotion.
(C) Effect of phorbol esters on wild type and ric-8. Shown are photographs of N2 (wild type) and ric-8(md303) on agar plates containing carrier (no drug) or 10 mM phorbol myristate acetate. Note that the phorbol ester induces a hyperflexive posture in both wild type and the ric-8 mutant. In addition, note that ric-8 mutants on phorbol esters are no longer bloated with eggs.
(D) Phorbol esters induce hyperactive locomotion in wild type and rescue ric-8 locomotion rates to near wild-type levels. Shown are the mean locomotion rates of N2 (wild type) and ric-8(md303) on agar plates containing carrier (no drug) or 10 μM phorbol myristate acetate. Error bars represent the standard error of the mean in a population of eight young adult animals.
To further investigate the effects of the DAG signal on ric-8 mutants, we examined wild type and ric-8 mutants in the presence of phorbol esters. Previous studies have shown that, in the presence of phorbol esters, wild-type worms become strikingly hypersensitive to aldicarb (Lackner et al., 1999; Miller et al., 1999; Nurrish et al., 1999). In this study, we found that, in the presence of phorbol myristate acetate, wild-type worms also exhibit increased body flexion and hyperactive locomotion (Figures 5C and 5D). The phenotypes of ric-8 mutants were also corrected to greater than wild-type levels by the exogenous application of the phorbol ester. The body posture of ric-8 mutants, which is nearly straight in the absence of the drug, became hyperflexive in the presence of the drug (Figure 5C). The phorbol ester also improved the locomotion rate of ric-8 mutants from ~2% that of the wild-type rate to levels slightly greater than those of wild type (Figure 5D).
In summary, the striking rescue of strong reduction-of-function ric-8 mutant phenotypes by DAG and phorbol esters is consistent with RIC-8 functioning upstream of, or in conjunction with, EGL-30 (Gqα).
RIC-8 Is Present throughout the Nervous System in Juveniles and Adults
A key assumption of our genetic analysis is that RIC-8 functions in the same cells as the other components of the Goα–Gqα signaling network. Previous studies have found that the proteins of the Goα–Gqα signaling network are expressed and/or localized broadly, though not in all cases exclusively, throughout the nervous system (Mendel et al., 1995; Ségalat et al., 1995; Koelle and Horvitz, 1996; Zwaal et al., 1996; Hajdu-Cronin et al., 1999; Lackner et al., 1999; Miller et al., 1999; Nurrish et al., 1999). To investigate the cellular and subcellular distribution of RIC-8, we produced a polyclonal antibody against purified recombinant RIC-8 and used it to visualize RIC-8 protein. In immunoblots of total worm protein, the RIC-8 antibody recognizes a major band of 62 kDa and a minor band of 64 kDa (Figure 6A). Although we have not investigated the molecular basis of the difference in mass between the two forms, both forms are close to the predicted weight of 63 kDa.
Figure 6. Localization of RIC-8 in Juvenile C. elegans.

(A) An affinity-purified RIC-8 antibody recognizes proteins of 62 kDa (major band) and 64 kDa (minor band) in a Western blot of total C. elegans protein. A parallel control blot of total C. elegans protein (probed with a RIC-8 antibody that had been preadsorbed to purified, recombinant RIC-8) was devoid of immunoreactivity (data not shown).
(B) Wild-type C. elegans juvenile stained an antibody to RIC-8. Staining (green) is seen in the head and tail ganglia, as well as near the nuclear membrane of nonneuronal cells, including germ cell nuclei.
(C) Control immunostaining experiment: a wild-type C. elegans juvenile stained with a RIC-8 antibody that was preadsorbed to purified, recombinant RIC-8. To confirm nervous system permeabilization, this animal was double stained with antibodies to CHA-1 (choline acetyltransferase; data not shown).
By immunofluorescence staining, we observed RIC-8 immunoreactivity throughout the nervous system in both juvenile and adult worms (Figures 5B and 6A). This suggests that RIC-8 has the opportunity to interact with other components of the Goα–Gqα signaling network. In juveniles, but not adults, however, we observed immunoreactivity around the nuclei of many nonneuronal cells, including germ cell nuclei (Figure 6B). Both neuronal and nonneuronal RIC-8 staining were absent in animals stained with a RIC-8 preadsorbed antibody preparation (Figure 6C). Neither the neuronal nor the nonneuronal staining was substantially altered in the missense mutant ric-8(md303) (data not shown). In ric-8(md1909) mutants, we observed a significant reduction in neuronal staining in adults (Figure 7E); however, the apparent nuclear membrane staining in juveniles does not diminish in this mutant (data not shown). Similarly, nuclear membrane staining in embryos also appears unchanged in ric-8(md1909) (K. G. Miller and J. B. Rand, submitted). We therefore regard the nuclear membrane staining pattern as a possible artifact; however, it is also possible that the md1909 mutant protein is simply less stable in neurons than in other cells.
Figure 7. RIC-8 Is Concentrated in the Nervous System of Adult Animals.
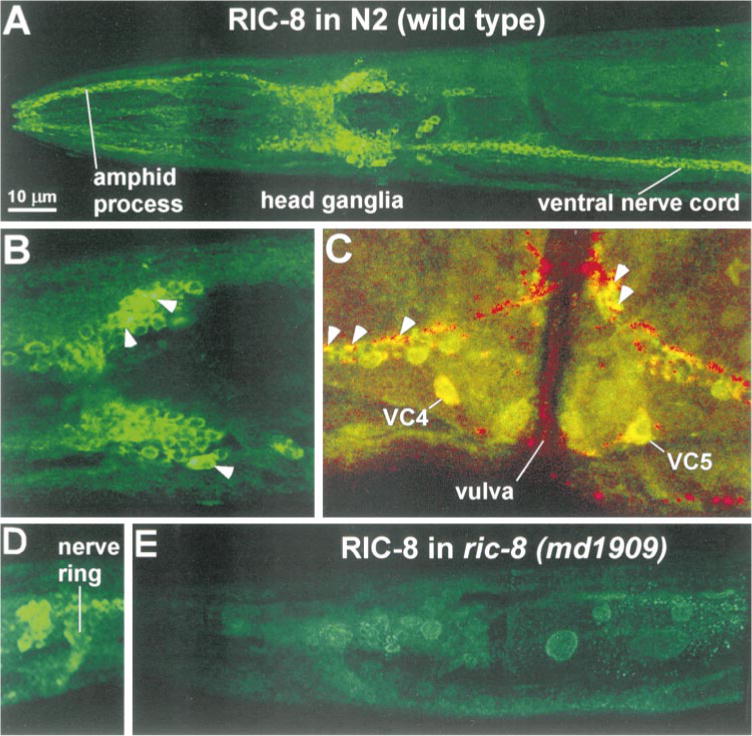
(A) Head region of a wild-type C. elegans adult stained with an antibody to RIC-8. Note the prominent staining of the head ganglia, amphid process, and ventral nerve cord.
(B) Close-up view of a section of the head ganglia. White arrowheads indicate examples of several strongly stained neurons in which RIC-8 immunoreactivity is present throughout the cytoplasm of the cell. Note the variability in RIC-8 staining intensity between different neurons.
(C) RIC-8 is also present at some cholinergic synapses in the ventral cord. Close-up view of a region near the vulva that was double stained with RIC-8 (green staining) and CHA-1 (red staining), a marker that is localized to cholinergic synapses (represented by bright puncta) (Janet Duerr, personal communication). Regions of overlap show up as yellow. Arrowheads indicate a subset of synapses along the ventral nerve cord that are positive for both RIC-8 and CHA-1.
(D) Expanded view of a region of head ganglia showing RIC-8 staining in the nerve ring, which contains axonal processes.
(E) Head region of a ric-8(md1909) adult stained with an antibody to RIC-8. Note that the nervous system staining is decreased, though not absent, in this transposon insertion mutant.
Within the nervous system, RIC-8 appears to be present in most or all neurons, including those comprising the ventral nerve cord, although the amount of staining varied greatly between individual neurons (compare brightly and weakly stained neurons in Figures 7A and 7B). This is interesting, because it suggests that RIC-8 production or stability is regulated over a wide range in a neuron-specific manner. Although RIC-8 was most heavily concentrated in neuronal somas, we also observed RIC-8 staining in neuronal processes, including strong staining of amphid dendritic processes and weaker staining at some cholinergic synapses in the ventral nerve cord, as well as the axonal processes of the nerve ring (Figures 7A–7D). RIC-8 staining in neuronal cell somas did not appear localized but instead appeared to fill the cytoplasm (Figures 7A–7D).
Discussion
RIC-8 Encodes a Novel Protein that Functions in the Adult Nervous System to Positively Regulate Synaptic Transmission
To investigate the EGL-30 pathway in C. elegans, we screened for aldicarb-resistant mutants with phenotypes similar to egl-30 and identified, among other genes, alleles of ric-8. Our molecular analysis of C. elegans RIC-8 (synembryn) demonstrates that it is a novel protein that has been conserved during evolution. We did not, however, identify a homolog of RIC-8 in yeast, which uses a heterotrimeric G protein to mediate the pheromone response (Leberer et al., 1997). This could indicate that RIC-8 acts in conjunction with one or more specific G proteins (such as Gqα) but is not necessary for signaling mediated by all classes of G proteins. Alternatively, a yeast ortholog of RIC-8 may be too diverged to recognize by amino acid sequence alignment.
Although we can’t rule out that subtle developmental defects contribute to the adult ric-8 mutant phenotypes, several of our findings suggest that developmental defects cannot be a major contributor to ric-8 mutant phenotypes. First, we observed that the Ric-8-like progeny of the RNAi-injected animals gradually lost their Ric-8-like phenotypes as they progressed to adulthood. This could not occur if the phenotypes of the Ric-8-like larvae were due to developmental defects, which would be permanent. Second, we found that exposure of adult ric-8 mutants to phorbol esters essentially rescued their phenotypes. Third, ric-8; dgk-1 mutants were rescued to approximately wild-type with respect to adult behaviors (this study) without affecting the level of embryonic lethality (K. G. Miller and J. B. Rand, submitted).
By immunostaining, we found that RIC-8 is concentrated in the nervous systems of both juveniles and adults. The localization of RIC-8 within neurons suggests that RIC-8 has the opportunity to interact with other components of the Goα–Gqα signaling network. Like RIC-8, the distributions of GOA-1 (Goα), GPB-1 (Gβ), EGL-10 (RGS), and EGL-8 (PLCβ) are divided between neuronal cell somas and axonal processes throughout the nervous system (Koelle and Horvitz, 1996; Zwaal et al., 1996; Miller et al., 1999; K. G. Miller and J. B. Rand, submitted; K. G. M., unpublished data); however, in contrast to these other proteins, RIC-8 appears to be more concentrated in cell somas than in axonal processes. The subcellular localizations of EGL-30 (Gqα), DGK-1 (diacylglycerol kinase), and EAT-16 (RGS) have not yet been reported.
The Role of RIC-8 in the Goα–Gqα Signaling Network
The shared phenotypes of ric-8 and egl-30 mutants suggest that RIC-8 and EGL-30 function in the same path way to positively regulate synaptic transmission. Our genetic epistasis analysis further narrowed the potential roles of RIC-8 by showing that RIC-8 is unlikely to function as a negative regulator of GOA-1 or as a downstream effector of EGL-30. Genetic epistasis analyses generally rely on the use of null alleles (Huang and Sternberg, 1995). The nonnullness of the ric-8(md303) mutation must, therefore, be taken into account. However, our comparison of ric-8(md303) and egl-30(ad805) phenotypes shows that ric-8(md303) reduces pathway activity to essentially the same extent as egl-30(ad805), which is the strongest known allele of egl-30 that does not result in larval lethality (Brundage et al., 1996; K. G. M., unpublished data). Our findings that egl-30(ad805) is epistatic to goa-1 loss of function (Miller et al., 1999) and that ric-8(md303) is strongly suppressed by goa-1 loss of function (this study) therefore suggest that RIC-8 functions upstream of, or in conjunction with, EGL-30 (Gqα); however, we have not yet determined whether RIC-8 is actively required to maintain proper activity of the Goα–Gqα signaling network or whether RIC-8 indirectly affects the signaling network by ensuring proper production or localization of a network component.
Consistent with RIC-8 functioning upstream of, or in conjunction with, EGL-30, we found that loss-of-function mutations in DGK-1 (diacylglycerol kinase) or exogenous application of phorbol esters results in a striking suppression of the synaptic transmission phenotypes of ric-8 mutants. However, our results also suggest that DAG cannot bypass the entire function of RIC-8. Further studies will be needed to determine if IP3, which is produced along with DAG upon PLCβ-mediated hydrolysis of PIP2, also plays a role. Previous studies suggest that EGL-8 (PLCβ) is not the only effector for EGL-30 (Gqα) (Lackner et al., 1999; Miller et al., 1999). Therefore, the component of RIC-8’s function that is not bypassed by DAG might represent an unidentified effector of EGL-30 signaling.
Dual Roles for GOA-1 (Goα) and RIC-8 (Synembryn) in Regulating Synaptic Transmission and Early Embryogenesis
In a separate study, we found that, in addition to their roles in the adult nervous system, RIC-8 and GOA-1 are required to regulate a subset of centrosome movements in the early embryo (K. G. Miller and J. B. Rand, submitted). At two different points in the animal’s life, therefore, the functions of RIC-8 and GOA-1 are closely associated. A closer analysis, however, reveals important and potentially informative differences between the two pathways. First, EGL-30 (Gqα) appears not to play a role in the embryonic pathway (K. G. Miller and J. B. Rand, submitted). Second, in the embryonic pathway, reduction-of-function mutations in goa-1 and ric-8 lead to similar phenotypes that are enhanced in goa-1; ric-8 double mutants, whereas in the adult neuronal pathway, the same goa-1 and ric-8 mutants have opposite phenotypes and suppress each other.
One possible explanation for the apparently different relationship of RIC-8 to GOA-1 in the embryo versus the nervous system is that RIC-8 positively regulates both GOA-1 and EGL-30 signaling in the nervous system; however, because EGL-30 acts downstream of GOA-1 in the nervous system, reducing RIC-8’s function results in an egl-30 reduction-of-function phenotype rather than a goa-1 reduction-of-function phenotype. In the embryo, on the other hand, where EGL-30 apparently does not play a role, reducing RIC-8’s function results in a goa-1 reduction-of-function phenotype. GPB-1 (Gβ) is one candidate for a molecule that is likely to be required for both EGL-30 (Gqα) and GOA-1 (Goα) function and whose regulation by or of RIC-8 could account for our findings. GPB-1’s role in centrosome positioning during early embryogenesis, as well as locomotion and egg laying in adults, is consistent with this possibility (Zwaal et al., 1996).
Experimental Procedures
Strains
Worms were cultured using standard methods (Brenner, 1974). Wild-type worms were C. elegans variety Bristol, strain N2. Variety Bergerac, strain EM1002, was used for STS mapping. TR638 and RM25, derived from TR403, were used as starting strains for isolating spontaneous aldicarb-resistant mutants. The following C. elegans mutant strains were used in this work. Single mutants: RM2224 dgk-1(sy428)X (gift of Paul Sternberg), DA823 egl-30(ad805)I, RM2226 goa-1(n363)I, MT8190 lin-15(n765) egl-10+ [nls51]X, TR638 mut-3(r456), RM1702 ric-8(md303)IV, RM2209 ric-8(md1909)IV, and RM2235 ric-8(md1712 md303)IV. Double mutants: RM2291 egl-30(ad805)I; ric-8(md303)IV, goa-1(n363)I; ric-8(md303)IV, RM35 lin-1(e1275) unc-33(e204)IV, RM2218 ric-8(md303)IV; dgk-1(sy428)X, RM1797 ric-8(md303)IV; dpy-11(e224)V, and RM2165 ric-8(md303)IV; lin-15(n765) egl-10+ [nls51]X.
Genetic Screens and Mapping
The isolation of ric-8(md303) as a spontaneous aldicarb-resistant mutant was described previously (Miller et al., 1996). ric-8(md1909) was isolated in a similar screen for transposon insertion aldicarb-resistant mutants using the mutator strain TR638. In this screen, 400 independent lines were analyzed (~20,000 animals per line). By noting the frequency of loss-of-function mutations in genes such as snt-1 and unc-13, we estimate that the two screens together represented at least 35-fold coverage of the genome. ric-8(md1712 md303) was obtained in a screen for ethyl methanesulfonate-(EMS-) induced suppressors of ric-8(md303) that covered 15,000 genomes (~12-fold coverage of the genome). The md1712 mutation is tightly linked to ric-8(md303) and has not been separated from the md303 background. ric-8(md2230) was identified in a noncomplementation screen (~20,000 genomes, or ~16-fold genome coverage) in which EMS-mutagenized N2 males were crossed to ric-8(md1909); dpy-11(e224). Plates were allowed to produce F1 cross progeny for 2 days, at which point progeny were rinsed off and transferred to 0.4 mM aldicarb plates for selection of noncomplementing cross progeny (the dpy-11 mutation causes self progeny to die under these conditions). We do not know if loss-of-function ric-8 alleles are viable in trans to md1909, since deficiencies that overlap with ric-8 are not available. Therefore, we do not know if this noncomplementation screen could have identified loss-of-function ric-8 alleles.
md303 was previously mapped to the interval between lin-1 and dyf-3 on linkage group IV (Miller et al., 1996). In this study, we further mapped ric-8 to a region close to lin-1, ~35% of the distance between lin-1 and unc-33. The ric-8(md1909) allele also mapped to the same interval.
Mutants identified in this study were outcrossed at least twice before use in assays or double mutant construction.
Double Mutant Strain Construction and Verification
For double mutant constructions, we chose strong reduction-of-function or loss-of-function alleles. The ric-8 reduction-of-function alleles are described in this study. dgk-1(sy428) and goa-1(n363) are loss-of-function or null alleles (this study; Ségalat et al., 1995; Hajdu-Cronin et al., 1999). dgk-1(sy428) contains an early stop codon in the coding sequence of dgk-1 (S. Nurrish and J. Kaplan, personal communication).
Double mutants were constructed using standard genetic methods, without additional marker mutations. Homozygosity of ric-8(md303) in double mutants was confirmed by sequencing amplified genomic DNA from double mutant strains. Homozygosity of dgk-1 alleles (X-linked) was confirmed by complementation tests. goa-1(n363); ric-8(md303) double mutants were selected from the progeny of goa-1(n363)/+; ric-8(md303)/+. The genotype of individual animals, which were sterile, was confirmed by PCR (for n363) and sequencing amplified genomic DNA (for md303).
Cloning and Analysis of C. elegans and Mouse ric-8 cDNA Sequences
The ric-8(md1909) mutant, obtained from the Tc1 transposon mutator strain TR638, was outcrossed 11 times to wild type. As part of this outcrossing, the closely flanking markers lin-1 and unc-33 were crossed on and then off the md1909 chromosome to remove linked Tc1 transposons. Genomic sequence corresponding to C. elegans ric-8 was then identified using a transposon display method developed by Henri van Luenen and Ronald Plasterk and previously described for the cloning of the egl-8 gene (Miller et al., 1999). The genomic fragment was then sequenced, labeled, and used to screen a C. elegans mixed stage oligo dT-primed cDNA library and obtain cDNA clones. The sequence of two cDNA clones began with a portion (9 bp) of the trans-spliced leader SL1, which is found on the 5′ end of some C. elegans transcripts (Krause and Hirsh, 1987). Limited sequence and restriction analysis of 11 other clones revealed no evidence of alternative splicing. A single full-length clone was sequenced on both strands. The sequence was used to search the database of C. elegans genomic sequence, and intron-exon boundaries were determined. The predicted peptide sequence was used to search the GenBank nonredundant and EST databases using BLAST 2.0 with default search parameters (Altschul et al., 1997).
To obtain the full-length mouse synembryn cDNA sequences, the non full-length mouse cDNA clone (GenBank number AA209749) was used to screen a mouse embryonic day 12.5 cDNA library (Stratagene) and isolate additional clones; 11 clones, all of which contained ~2.4 kb inserts, were analyzed by restriction analysis and end sequencing. A single clone (RM545) was then sequenced in entirety on both strands. The presence of a Kozak consensus sequence (GCCATGG) (Kozak, 1991) at the first methionine residue (72 nucleotides from the 5′ end) suggests that translation begins at that methionine. When we screened the mouse EST database with the RM545 sequence, we identified >30 additional synembryn EST sequences, none of which extended significantly beyond the 59 end of RM545. The predicted peptide sequence was analyzed using PSORT II (K. Nakai) to look for subcellular localization signals. The mouse, C. elegans, and Drosophila synembryn sequences were aligned using the ClustalW computer program.
Molecular Analysis of Mutations
In cases in which genomic regions were amplified from ric-8 mutants, populations of mutant animals were processed for PCR using the method of Williams et al. (1992).
Identification of the transposon insertion in md1909 is described above. PCR amplification and sequencing of the genomic fragment containing the insertion were used to determine the exact site of the insertion. In md303, md1712 md303, and md2230, genomic DNA containing all ric-8 exons and intron-exon boundaries was amplified using PCR, and all exons and intron-exon boundaries were sequenced. The md2230 allele was the only allele that contained no mutations in ric-8 exons or intron-exon boundaries. It is possible that this mutant contains an alteration in the promoter region, since we observed that expression of the RIC-8 protein in this mutant was variable and often restricted to a subset of the nervous system.
RNA Interference
T3 and T7 RNA polymerase were used to synthesize double stranded RNA (dsRNA) from a 1.6 kb ric-8 cDNA clone (RM439) in the pBlue-script SK- vector as described by Fire et al. (1998). The clone includes the 5′ two thirds of the ric-8 cDNA. Control dsRNA was synthesized from a full-length 1.6 kb egl-30 cDNA clone (LB1). dsRNA was checked and quantified by agarose gel electrophoresis, then injected into the gonad syncytia of wild-type worms (Mello et al., 1991). The original DNA template was not removed prior to injection. Worms were transferred to individual plates 2–6 hr after injection and allowed to lay eggs for 18 hr, after which the injected worms were killed or transferred to fresh plates. The brood laid during the 18 hr period, which showed the greatest effect, was observed at 24 hr intervals for 4 days.
RIC-8 Antibodies and Immunostaining
Pfu polymerase (Stratagene) was used to amplify the entire ric-8 coding region, including the natural stop codon. BamHI and XhoI sites were incorporated into the primers to facilitate directional in-frame cloning into the bacterial expression vector pRSETb (In-vitrogen). The construct was transformed into the bacterial expression host BL21(DE3) pLysS, and the HIS6:RIC-8 fusion protein was expressed and then purified under denaturing conditions using Probond nickel resin.
Rabbits were injected with 500 μg of fusion protein and boosted four times with 500 μg each. To affinity purify RIC-8 antibodies, 500 μg of RIC-8 fusion protein was loaded into preparative wells, run out on 8% SDS–PAGE gels, and blotted to nitrocellulose. The blot was stained with Ponceau S, and the RIC-8 fusion protein band was excised and washed for 30 min in blocker (10 mM Tris [pH 8.0], 0.05% Tween 20, 150 mM NaCl, 3% nonfat dry milk, and 0.05% sodium azide). The blocking solution was then removed, and 3 ml of serum was incubated with the RIC-8 protein on the nitrocellulose strips for 1 hr. The serum was removed, and the strips were rinsed with TBST (10 mM Tris [pH 8.0], 0.05% Tween 20, and 150 mM NaCl), followed by 3 × 5 min washes in TBST. Antibodies were eluted from the strips using 4 ml of glycine elution buffer (100 mM glycine, 0.01% bovine serum albumin, 0.05% Tween 20 [pH 2.5]), followed by neutralization in 400 μl 1 of 1 M Tris [pH 8.0], then a second elution with 4 ml 50 mM triethylamine (pH 11.5), followed by neutralization in 400 μl of 1 M Tris (pH 7.0). The eluates were combined and dialyzed against 1× PBS.
Control preparations of this antibody were prepared as follows. His-6-tagged recombinant RIC-8 was purified using the Probond nickel resin and then further purified and immobilized by SDS-PAGE electrophoresis and blotting to nitrocellulose. Nitrocellulose strips containing the RIC-8 protein band were excised, incubated in blocker, and then incubated with the affinity-purified RIC-8 antibody KM1A-5.2. A mock blocked antibody was prepared in an identical manner except that the antibody was incubated with blocked nitrocellulose strips containing no RIC-8 protein. In another control preparation, the affinity-purified antibody was adsorbed to nitrocellulose strips containing a large excess of gel-purified His-6-tagged recombinant EGL-30. This fusion protein contains identical vector-contributed amino acid sequences but otherwise has no homology to RIC-8. The RIC-8 staining pattern was unchanged by this procedure.
Whole mounts of C. elegans for antibody staining were prepared as previously described, using methanol/acetone fixation (Duerr et al., 1999). Affinity-purified anti-RIC-8 antibodies (KM1A-5.2) were used at a 1/150 dilution and were incubated with specimens for 16 hr at room temperature. In double staining experiments, a mixture of anti-CHA-1 mouse monoclonal antibodies was also included (gift of Janet Duerr). Secondary antibodies (adsorbed against 4% formaldehyde-fixed worms to remove antibodies to nematode proteins) were donkey anti-rabbit (Jackson Immunoresearch) coupled to Alexa 488 dye (Molecular Probes) and donkey anti-mouse coupled to Cy3 (to visualize CHA-1). Secondary antibody incubations were for 4 hr at room temperature. Specimens were viewed using a Leica 100× Plan APO 1.4 NA oil immersion lens, and images were collected using the Leica TCS NT confocal system and accompanying software. Images were further processed using Adobe Photoshop 5.0.
Immunoblots
A C. elegans total protein preparation was produced by combining equal volumes of a washed worm pellet and nematode solubilization buffer (0.3% ethanolamine, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, and 1× protease inhibitor mix diluted from 250× stock) in a 1.5 ml screw cap tube. The tube was capped tightly and microwaved for 25 s on high power. An equal volume of 2× Laemmli sample buffer was immediately added, and the tube was placed in a boiling water bath for 7 min. The lysate was then forced through a 26 gauge needle and spun for 1 min to remove insoluble components; 3.5 μl of this preparation was then combined with Laemmli sample buffer, electrophoresed on an 8% SDS–PAGE gel, and blotted to nitrocellulose. Marker proteins were visualized by staining with Ponceau S. Control blots were probed with a 1/150 dilution of the RIC-8 antibody KM1A-5.2 that had been preadsorbed to a gel-purified band of recombinant RIC-8 immobilized on nitrocellulose, as described above. Experimental blots were probed with a mock blocked preparation of KM1A-5.2. Immunoreactive bands were visualized with a horseradish peroxidase-coupled secondary antibody using the LumiGLO Substrate kit (Kirkegaard and Perry) for chemiluminescent detection.
Aldicarb Sensitivity Assays
Aldicarb sensitivity was quantified by placing a fixed number of L1 stage larvae on culture plates containing 0, 10, 25, 50, 100, 200, 400, 800, or 1600 μM aldicarb and allowing them to grow at 20°C for 96 hr. Growth was then stopped by putting the plates at 4°C. The progeny (eggs and larvae) produced during this period were counted on a gridded background, and a percentage of the number of progeny produced on the no drug control plate was calculated for each concentration. The number of L1 larvae plated for the assay was chosen so that at least 300 progeny were produced on the no drug control plate. For strains with wild-type fertility and growth rate, three L1s were placed on each plate in the series; for strains with decreased fertility or growth rate, correspondingly more L1s were placed on each plate.
Locomotion Assays
Standard locomotion assays were performed as previously described (Miller et al., 1999). To assay locomotion rate in the presence of phorbol esters, phorbol myristate acetate (RBI; 5 mg/ml in ethanol) was added to a concentration of 10 μM to molten 55°C media. After cooling, plates were then spread with 35 μl of a fresh overnight culture of OP-50 bacteria and incubated for 40 hr at room temperature to grow a thin lawn of bacteria. Control plates contained the ethanol carrier but no drug. Plates were loaded with young adult worms at staggered intervals and assayed at a fixed time after loading by counting body bends (as described for the standard assay) for a 6 min period. Worms were counted at the point at which they reached maximal activity after loading them on the plates (usually ~75 min, but this time is variable from experiment to experiment).
Video Production
Animals were placed on agar plates containing thin bacterial lawns and videotaped using a Sony CCD-IRIS black-and-white video camera mounted on a Wild MPS 46 dissecting scope. The video was transferred to a computer using the Rainbow Runner G-Series video capture card and edited with Ulead’s Media Studio Pro 5.0 software.
Supplementary Material
Acknowledgments
We thank Henri van Luenen for providing protocols and advice concerning the transposon display method; Paul Sternberg, and Yvonne Hajdu-Cronin for providing dgk-1(sy428) and for communicating results prior to publication; Janet Duerr for assistance with confocal microscopy, cell identification, and CHA-1 monoclonal antibodies; Tony Crowell for technical assistance; and Bob Barstead for providing cDNA libraries. Confocal images were obtained in the Flow and Cytometry Laboratory in the Warren Medical Research Institute, Oklahoma City. Some of the strains used here were provided by the C. elegans Genetics Center. This work was supported by a grant from the National Institute of Neurological Disorders and Stroke to J. B. R. (NS33187).
Footnotes
GenBank Accession Numbers
The GenBank accession numbers for the C. elegans ric-8 and mouse synembryn cDNA sequences reported in this paper are AF288812 and AF288813, respectively.
Note Added in Proof
The data referred to throughout as “K.G. Miller and J.B. Rand, submitted” are now in press: Miller, K.G., and Rand, J.B. (2001). A role for RIC-8 (synembryn) and GOA-1 (Goα) in regulating a subset of centrosome movements during early embryogenesis in C. elegans. Genetics, in press.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;17:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein α subunits. Cell. 1996;86:445–452. doi: 10.1016/s0092-8674(00)80117-8. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of C. elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage L, Avery L, Katz A, Kim U, Mendel JE, Sternberg PW, Simon MI. Mutations in a C. elegans Gqα gene disrupt movement, egg laying, and viability. Neuron. 1996;16:999–1009. doi: 10.1016/s0896-6273(00)80123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambon C, Declume C, Derache R. Effect of the insecticidal carbamate derivatives (carbofuran, pirimicarb, aldicarb) on the activity of acetylcholinesterase in tissues from pregnant rats and fetuses. Toxicol Appl Pharmacol. 1979;49:203–208. doi: 10.1016/0041-008x(79)90242-4. [DOI] [PubMed] [Google Scholar]
- Duerr JS, Frisby DL, Gaskin J, Duke A, Asermely K, Huddleston D, Eiden LE, Rand JB. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–810. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Patikoglou G, Koelle MR, Sternberg PW. Antagonism between Goα and Gqα in C. elegans: the RGS protein EAT-16 is necessary for Goα signaling and regulates Gqα activity. Genes Dev. 1999;13:1780–1793. doi: 10.1101/gad.13.14.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Sternberg PW. Genetic dissection of devel opmental pathways. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegans: Modern Biological Analysis of an Organism. New York: Academic Press; 1995. pp. 97–122. [Google Scholar]
- Hunt TW, Fields TA, Casey PJ, Peralta EG. RGS10 is a selective activator of Gαi GTPase activity. Nature. 1996;383:175–177. doi: 10.1038/383175a0. [DOI] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 1996;84:112–125. doi: 10.1016/s0092-8674(00)80998-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- Krause M, Hirsh D. A trans-spliced leader sequence on actin mRNA in C. elegans. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner MR, Nurrish SJ, Kaplan JM. Facilitation of synaptic transmission by EGL-30 Gqα and EGL-8 PLCβ: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron. 1999;24:335–346. doi: 10.1016/s0896-6273(00)80848-x. [DOI] [PubMed] [Google Scholar]
- Leberer E, Thomas DY, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Madison DV, Nicoll RA. Potentiation of synaptic transmission in the hippocampus by phorbol esters. Nature. 1986;321:175–177. doi: 10.1038/321175a0. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel JE, Korswagen HC, Liu KS, Hajdu-Cronin YM, Simon MI, Plasterk RHA, Sternberg PW. Participation of the protein Go in multiple aspects of behavior in C. elegans. Nature. 1995;267:1652–1655. doi: 10.1126/science.7886455. [DOI] [PubMed] [Google Scholar]
- Miller KG, Alfonso A, Nguyen M, Crowell JA, Johnson CD, Rand JB. A genetic selection for Caenorhabditis ele-gans synaptic transmission mutants. Proc Natl Acad Sci USA. 1996;93:12593–12598. doi: 10.1073/pnas.93.22.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KG, Emerson MD, Rand JB. Goα and diacyl-glycerol kinase negatively regulate the Gqα pathway in C. elegans. Neuron. 1999;24:323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. Studying mutants that affect neurotransmitter release in C elegans. In: Bellen H, editor. Neurotransmitter Release. New York: Oxford University Press; 1999. pp. 265–303. [Google Scholar]
- Nonet ML, Grundahl K, Meyer BJ, Rand JB. Synaptic function is impaired but not eliminated in C. elegans mutants lacking synaptotagmin. Cell. 1993;73:1291–1305. doi: 10.1016/0092-8674(93)90357-v. [DOI] [PubMed] [Google Scholar]
- Nurrish S, Ségalat L, Kaplan JM. Serotonin inhibition of synaptic transmission: Gαo decreases the abundance of UNC-13 at release sites. Neuron. 1999;24:231–242. doi: 10.1016/s0896-6273(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Rand JB, Nonet ML. Synaptic transmission. In: Riddle DL, et al., editors. C elegans II. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 611–643. [PubMed] [Google Scholar]
- Risher JF, Mink FL, Stara JF. The toxicologic effects of the carbamate insecticide aldicarb in mammals: a review. Environ Health Perspect. 1987;72:267–281. doi: 10.1289/ehp.8772267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushforth AM, Saari B, Anderson P. Site-selected insertion of the transposon Tc1 into a Caenorhabditis elegans myosin light chain gene. Mol Cell Biol. 1993;13:902–910. doi: 10.1128/mcb.13.2.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane F, Kanoh H. Molecules in focus: diacylglycerol kinase. Int J Biochem Cell Biol. 1997;29:1139–1143. doi: 10.1016/s1357-2725(97)00037-x. [DOI] [PubMed] [Google Scholar]
- Ségalat L, Elkes DA, Kaplan JM. Modulation of serotonin-controlled behaviors by Go in Caenorhabditis elegans. Nature. 1995;267:1648–1651. doi: 10.1126/science.7886454. [DOI] [PubMed] [Google Scholar]
- Shapira R, Silberberg SD, Ginsburg S, Rahamimoff R. Activation of protein kinase C augments evoked transmitter release. Nature. 1987;325:58–60. doi: 10.1038/325058a0. [DOI] [PubMed] [Google Scholar]
- Singer WD, Brown HA, Sternweis PC. Regulation of eukaryotic phosphatidylinositol-specific phospholipase C and phospholipase D. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- Stevens CF, Sullivan JM. Regulation of the readily releasable vesicle pool by protein kinase C. Neuron. 1998;21:885–893. doi: 10.1016/s0896-6273(00)80603-0. [DOI] [PubMed] [Google Scholar]
- Trent C, Tsung N, Horvitz HR. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics. 1983;124:855–872. doi: 10.1093/genetics/104.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature. 1996;383:172–175. doi: 10.1038/383172a0. [DOI] [PubMed] [Google Scholar]
- Watson S, Arkinstall S. The G-Protein Linked Receptor Factsbook. New York: Academic Press; 1994. [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaal RR, Ahringer J, van Luenen HGAM, Rushforth A, Anderson P, Plasterk RHA. G proteins are required for spatial orientation of early cell cleavages in C. elegans embryos. Cell. 1996;86:619–629. doi: 10.1016/s0092-8674(00)80135-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


