Abstract
Nature has devised sophisticated cellular machinery to process mRNA transcripts produced by RNA Polymerase II, removing intronic regions and connecting exons together, to produce mature RNAs. This process, known as splicing, is very closely linked to transcription. Alternative splicing, or the ability to produce different combinations of exons that are spliced together from the same genomic template, is a fundamental means of regulating protein complexity. Similar to transcription, both constitutive and alternative splicing can be regulated by chromatin and its associated factors in response to various signal transduction pathways activated by external stimuli. This regulation can vary between different cell types, and interference with these pathways can lead to changes in splicing, often resulting in aberrant cellular states and disease. The epithelial to mesenchymal transition (EMT), which leads to cancer metastasis, is influenced by alternative splicing events of chromatin remodelers and epigenetic factors such as DNA methylation and non-coding RNAs. In this review, we will discuss the role of epigenetic factors including chromatin, chromatin remodelers, DNA methyltransferases and microRNAs in the context of alternative splicing, and discuss their potential involvement in alternative splicing during the EMT process.
Keywords: alternative splicing, EMT, chromatin, cancer, epigenetics
INTRODUCTION
The removal of introns from pre-mRNAs followed by the joining together of exons is an essential step for the generation of mature mRNAs in higher eukaryotes. When the human genome was first sequenced, the number of predicted mRNA coding genes (~25,000) was surprisingly much lower than the number of proteins in a mammalian cell (over a million). Alternative splicing, which joins together different combinations of exons to allow the generation of multiple mRNA isoforms (and in turn proteins) from the same genomic region, is the main mechanism responsible for this disparity. The process of alternative splicing contributes significantly to proteome diversity: 92–94% of protein-coding genes undergo alternative splicing in humans (Pan et al. 2008; Wang et al. 2008). By switching between alternatively spliced isoforms, several functions of the resulting proteins can be affected, including protein or DNA binding, ligand binding, enzyme activity, or localization of the protein.
A macromolecular complex called the spliceosome is responsible for the stepwise catalysis of pre-mRNA splicing, involving four small nuclear ribonucleic proteins (snRNPs or “snurps”), the U1, U2, U4/U6 and U5 snRNPs, and several associated splicing factors. Splicing can be regulated in cis, by RNA sequences: AG/GU(A or G)AGU at the 5′ splice site and YnNCAG/G at the 3′ splice site (the forward slash indicates position of the splice site) (Shapiro and Senapathy 1987), which are important in the pre-mRNA splicing reaction (Jackson 1991; Hall and Padgett 1994). However, splicing can also be regulated in trans, by proteins that can bind to specific RNA sequences. For instance, the serine/arginine rich family of proteins (SR proteins) dictate splice site recognition (Bradley et al. 2015; Zahler et al. 1992) and lead to exon inclusion (Graveley 2000; Shen and Green 2006) by bringing general splicing factors to the splice site to assemble the spliceosome (Roscigno and Garcia-Blanco 1995; Tarn and Steitz 1996; Das et al. 2007). Another family of splicing regulators is the hnRNP group of proteins, which commonly leads to exon exclusion (Smith and Valcarcel 2000) through its ability to wrap the pre-mRNA transcript (Krecic and Swanson 1999). Spliceosome assembly and dynamics is the subject of several excellent reviews (e.g. (Jurica and Moore 2003; Wahl et al. 2009; Will and Luhrmann 2011; Lee and Rio 2015) and will not be discussed here in detail.
Changes in alternative splicing can be triggered by a variety of stimuli, including but not limited to calcium (Razanau and Xie 2013; Sharma et al. 2014), estradiol (Bhat-Nakshatri et al. 2013), stress (Busa and Sette 2010; Lehtinen et al. 2013) and circadian rhythms (Henriques and Mas 2013), to name a few. These environmental stimuli -induced alternative splicing events can in turn regulate a wide variety of important cellular functions, including but not limited to cell cycle, signal transduction, cell proliferation and differentiation, apoptosis, angiogenesis, invasion, motility and metastasis. Therefore, any deviations from the highly controlled alternative splicing events has the potential to severely affect cellular function, leading to disease such as cancer. Splicing in turn can influence transcriptional proteins and the process of transcription in general, which can in turn affect chromatin in the vicinity of the gene that is being transcribed. Both splicing and chromatin have been shown to play important roles in the process of EMT, or epithelial to mesenchymal transition. Research has suggested that the interactions between splicing, chromatin and transcription involve several feedback mechanisms, which regulate gene expression during EMT. In this review, we will discuss the involvement of chromatin remodelers, epigenetic factors and alternative splicing during the cellular program of EMT.
Alternative splicing, transcription and chromatin: closely coupled mechanisms
For many years, pre-mRNA splicing and transcription were thought to occur independently from each other. This concept was challenged in the late 80s, when electron microscopy images of nascent pre-mRNA transcripts from Drosophila embryos revealed that in fact, splicing could occur in concert with transcription (Beyer and Osheim 1988). More recently, several studies have provided compelling evidence that introns can be removed while the nascent transcript is still tethered to the DNA through the RNA polymerase II (Pol II) complex (Dye et al. 2006; Listerman et al. 2006; Pandya-Jones and Black 2009; Ameur et al. 2011; Khodor et al. 2011; Vargas et al. 2011; Khodor et al. 2012; Tilgner et al. 2012).
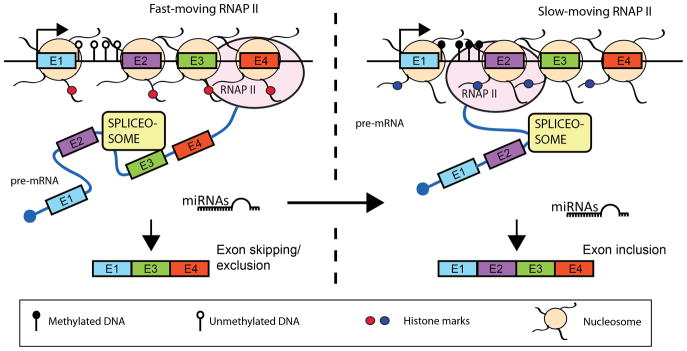
The kinetic model of co-transcriptional splicing was proposed to explain the keen “eyesight” of the spliceosome complex that allowed it to recognize the short, often ~100 nt or less sized exons, the proverbial needles in the haystack of long, several 1000 Kb introns. This model proposed that the rate of Pol II elongation directly affected splice site recognition and spliceosome assembly. In other words, if Pol II transcribed at a rapid rate (either due to it being hyperphosphorylated, or if there was fairly “open” chromatin along the gene), then the spliceosome would not be able to keep up with the fast moving Pol II (Figure 1). This would result in several alternative splice sites being presented to the spliceosome to choose from, and by default, it would choose the stronger 3′ splice site more often relative to the weaker site(s), leading to some exons being spliced out (Figure 1). In contrast, if the rate of Pol II elongation was hindered in some way, either due to chromatin factors such as nucleosomes or due to DNA methylation in the intragenic regions, the spliceosome machinery is then able to keep up with Pol II elongation, and splices all possible exons. In support of this model, experiments that used Pol II mutants that slowed down the rate of Pol II elongation (de la Mata et al. 2003), or that inserted DNA elements that ‘paused’ Pol II in reporter constructs (Robson-Dixon and Garcia-Blanco 2004) were able to favor “weak” exon inclusion in the fibronectin (FN1) and fibroblast growth factor receptor 2 (FGFR2) genes.
Figure 1. Interplay between chromatin remodelers, RNA Pol II and the splicing machinery during EMT.
The kinetic model of alternative splicing posits that the spliceosome cannot keep up with the fast-moving RNA Pol II, therefore it has to choose between the splice sites presented to it and exon 2 (in this example) gets excluded. On the other hand, DNA methylation (represented by black lollipops), nucleosome positioning or histone modification changes, or microRNAs can all influence the rate of RNA Pol II elongation, slowing it down so that the spliceosome can now keep up with transcription. This results in all the exons being included. During EMT, many different types of slicing events can occur.
The involvement of chromatin in alternative splicing was first suggested in a study that followed adenovirus E1B mRNA splicing in HeLa cells at early versus late time after infection (Adami and Babiss 1991). The E1B precursor mRNA was known to produce two alternatively spliced mRNAs- a 22S, which was found during early infection (pre-replicative phase), and a 13S, which appeared later (replicative phase) during viral infection. Suspecting a trans-activating factor that switched the splicing during the course of infection, they tested this hypothesis by infecting HeLa cells first with one mutant virus, allowing replication for 15 hours, and then adding a second mutant virus. Irrespective of the presence of the first virus, which had switched splicing to the 13S form, the newly infected virus nevertheless still produced the 22S isoform (Adami and Babiss 1991), indicating that a trans-activating factor that appeared after viral infection was probably not involved, as the first infection should have already resulted in expression of large amounts of the suspected factor. As there was very little variation in the genomic sequence of the viral templates, the question of how the adenovirus could produce two such different transcripts was raised. The authors proposed that if Pol II elongation was hindered due to more compact chromatin, it would allow the incorporation of the upstream 5′ splice site, forming a shorter mRNA, relative to when the downstream splice site was used.
A subsequent study supported this idea following their demonstration that the upstream E1a adenovirus splice site was utilized when the cells expressed a slow Pol II point mutant (de la Mata et al. 2003). Further support for the kinetic model came from the Groudine laboratory, who showed that the introduction of an in vitro methylated DNA sequence downstream of a gene promoter caused local decreases in histone acetylation and chromatin accessibility, thereby slowing down Pol II elongation (Lorincz et al. 2004). Although a slow rate of Pol II elongation often allows for alternative exon inclusion, there are exceptions. Depending on the recruitment and function of RNA binding proteins involved in splicing, a slow rate of Pol II elongation may favor alternative exon exclusion. The alternative exon 9 of the cystic fibrosis transmembrane conductance regulator (CFTR) gene is excluded when the Pol II elongation rate is slow (Dujardin et al. 2014). Under these conditions a negative factor CELF2 (CUGBP, Elav-Like Family Member 2; ETR-3) is recruited to the UG-repeat at the exon 9 3′ splice site, resulting in the displacement of the constitutive splicing factor U2AF2 (U2 small nuclear RNA auxiliary factor 2; also called U2AF65) from the polypyrimidine tract (Dujardin et al. 2014). Together these events result in exon 9 exclusion.
Since these discoveries, a plethora of studies have been published, detailing the involvement of epigenetic factors- including nucleosome positioning, histone modifications and histone variants, DNA methylation and non-coding RNAs- in alternative splicing (reviewed in (Kornblihtt 2006; Allemand et al. 2008; Allo et al. 2010; Luco et al. 2011; Khan et al. 2012; Braunschweig et al. 2013), some of which will be discussed here.
Alternative splicing in cancer and EMT
Alternative splicing regulates many genes involved in cancer progression and metastasis. The importance of alternative splicing in cancer is evident from the fact that DNA mutations and normal genetic variation can contribute to alternative splicing defects: over 100 different point mutations have been reported near mRNA splice junctions, which affect the pathology of diseases by altering splicing efficiency (Krawczak et al. 1992; Wang and Cooper 2007). Loss of splicing fidelity can result in isoforms in proteins controlling various facets of cancer (reviewed in (Brinkman 2004; Venables 2006). For example, the BCL-x gene produces two splice variants, one proapoptotic Bcl-x(s) form, and the other an antiapoptotic Bcl-x(L) form, via alternative 5′ splice site selection involving binding of SF3B1 (Splicing Factor 3B subunit 1, also called SAP155), a splicing factor, to a cis-element on the Bcl2l1 (B-Cell CLL/Lymphoma 2 – like 1; or Bcl-x) pre-mRNA (Massiello et al., 2006). Activation of the Bcl-x(s) 5′ splice site helped drive the splicing to the pro-apoptotic RNA isoform, thus increasing the effectiveness of chemotherapy. Another splicing protein, KHDRBS1 (KH Domain containing, RNA Binding, Signal transduction associated 1, also known as Sam68), was shown to bind the pre-mRNA for Bcl-x and affect its alternative splicing, and phosphorylation of KHDRBS1 was shown to play a role in how it affected the alternative splicing of Bcl-x (Paronetto et al., 2007). There are many additional examples of how alternative pre-mRNA splicing is involved in tumorigenesis and tumor progression, including several that are as yet unknown.
The role of alternative splicing during EMT has been increasingly well documented recently. Cancer in its advanced stages is characterized by the development of metastases, as a result of cells moving from the original mass to different locations and forming secondary tumors. Cellular migration during metastasis is a result of alteration in cell shape from an epithelial to a mesenchymal cell phenotype (EMT), which results in the acquisition of an invasive, mesenchymal phenotype by tumor cells and the reverse process, MET, which enables their differentiation into secondary tumors. EMT is regulated by a long list of transcription factors, including SNAI1 (SNAIL), SNAI2 (SLUG), ZEB1, ZEB2, E47, TWIST1, etc. which function as ‘master regulators’ of gene expression, repressing epithelial-specific genes such as E-cadherin, and allowing an upregulation of mesenchymal specific genes such as Vimentin (reviewed in (Barrallo-Gimeno and Nieto 2005; Kalluri and Weinberg 2009; Lim and Thiery 2012; Nieto and Cano 2012; Wang and Zhou 2013; Lamouille et al. 2014; Zheng and Kang 2014).
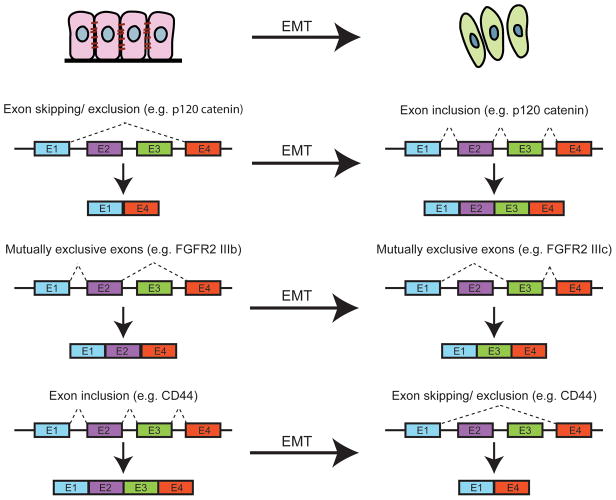
Several types of splicing events are known to occur during EMT (Figure 2 and Warzecha and Carstens 2012). One of the first EMT-related genes shown to be alternatively spliced is the fibroblast growth factor receptor 2 (FGFR2), during EMT in rat bladder carcinoma (Savagner et al. 1994). During EMT, the CD44 gene switches from several constitutive splicing variants (CD44v), found in the epithelial state, into a single, short isoform, CD44s (Figure 2), which is essential for EMT (Brown et al. 2011). This CD44 isoform switch is regulated by the splicing factor Epithelial Splicing Regulatory Protein 1 (ESRP1) (Warzecha et al. 2009; Brown et al. 2011). Both ESRP1 and its related protein ESRP2 are essential for maintaining the epithelial state, as loss of these proteins caused cells to transition from the epithelial to the mesenchymal state. Together, ESRP1 and ESRP2 regulate the splicing of several genes, including Fibroblast Growth Factor Receptor 2(FGFR2), CD44, Catenin (cadherin-associated protein), delta 1 (CTNND1, or p120-Catenin), and the Enabled homolog (ENAH) (Warzecha et al. 2009) during EMT, and loss of ESRP1 and 2 induced alternative splicing during EMT (Warzecha et al. 2010). During the process of EMT, the transcription repressor Snail was shown to bind to E-boxes in the ESRP1 promoter, causing repression of the ESRP1 gene transcription (Reinke et al. 2012). ESRP1, when bound to the intronic region flanking a CD44 variable exon, caused increased variable exon inclusion, and expression of ESRP1 caused downregulation of Snail-driven EMT (Reinke et al. 2012). More recently, a microarray-based analysis demonstrated that TGF-β induced alternative splicing events by downregulating ESRP1 and 2 via upregulation of two other EMT transcription factors, δEF1 and SIP1, which associated with the promoter of ESRP2 and repressed its expression (Horiguchi et al. 2012). Interestingly, ESRP1 and ESRP2 appear to effect alternative splicing by different mechanisms (Ishii et al. 2014). Knockdown of ESRP1 in head and neck cancer cell lines induced the expression of Rac1b, which is also known to increase Snail-induced EMT, (Radisky et al. 2005), thus affecting actin cytoskeleton dynamics. On the other hand, knockdown of ESRP2 caused a decrease in cell-cell adhesion by increasing the expression of EMT-related transcription factors δEF1 and SIP1, but not SNAIL, SLUG or TWIST (Ishii et al. 2014), suggesting that ESRP1 and ESRP2 might be redundant, or prevent EMT by different mechanisms.
Figure 2. Examples of some splicing events during EMT.
During EMT, many different types of slicing events occur. For example, p120 catenin switches from a short form to a longer form that includes additional exons. FGFR2 switches between isoforms containing two mutually exclusive exons, Exon IIIb or IIIc. CD44 produces multiple splice variants from 11 different exons, while after EMT, only the short variant (CD44s) is produced due to exon skipping.
Besides ESRP1 and 2, another splicing factor, the Serine/arginine-rich Splicing Factor 1 protein (SRSF1, or SF2/ASF), is also involved in the cell’s decision to proceed with EMT (Valacca et al. 2010). Valacca and colleagues showed that SRSF1 is regulated during EMT and its reverse process mesenchymal to epithelial transition (MET) by alternative splicing, specifically through the splicing regulator KHDRBS1 (Sam68) (Valacca et al. 2010). Further, KHDRBS1 modulation of SRSF1 splicing appears to be controlled by epithelial cell-derived soluble factors that act through the ERK1/2 signaling pathway to regulate KHDRBS1 phosphorylation (Valacca et al. 2010). When overexpressed, some splice variants can act as hyper-oncogenic proteins, which often correlate with poor prognosis. For example, the matrix metalloproteinase 3 (MMP3) induces alternative splicing of the small GTPase Rac1, switching expression to Rac1b, its longer, more active form containing 19 additional amino acids. Rac1b in turn increases levels of reactive oxygen species, leading to increased expression of the transcription factor Snail, resulting in EMT (Radisky et al. 2005).
A global view of the EMT splicing program was provided in a study (Shapiro et al., 2011), where the authors overexpressed the EMT- inducing transcription factor, Twist1, in the human mammary epithelial cell line (HMLE) to induce EMT (Shapiro et al. 2011). They showed that several types of splicing factors, including ESRPs, RNA binding protein, fox (RBFOX) family members, muscleblind-like splicing regulator 1 (MBNL), Elav-like family (CELF), heterogeneous nuclear ribonucleoprotein (hnRNP), and phosphotyrosine-binding domain (PTB) family members regulated alternative splicing events in Twist1-induced EMT (Shapiro et al. 2011). Perhaps the most clinically relevant finding of their paper was that EMT-associated alternative transcripts correlated with the aggressive and metastatic phenotype of breast cancer cell lines, and were also found to be expressed in primary human breast cancer samples. Given the vast array of splicing factors involved in alternative splicing, it will be of great importance to delineate their respective roles during the process of EMT.
Chromatin and alternative splicing during EMT
Despite substantial independent evidence for the involvement of chromatin in EMT, and alternative splicing in EMT, there are no studies to date that link chromatin and alternative splicing together in the context of EMT. Yet, they are probably very closely connected, and determining the role of chromatin and epigenetic proteins in alternative splicing during EMT will undoubtedly shed light on the mechanisms driving EMT and cancer metastasis. Here, we will discuss what is known regarding the involvement of chromatin and alternative splicing during EMT, and attempt to connect the two mechanisms together.
1. Nucleosome positioning
Studies based on genome-wide analyses demonstrated that nucleosomes are preferentially positioned in exons (Andersson et al. 2009; Hon et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009; Wilhelm et al. 2011; De Conti et al. 2013), and that this positioning was conserved across species, which supported the idea that nucleosome positioning may play a role in alternative splicing. Two groups (Schwartz et al. 2009; Tilgner et al. 2009) used computational approaches to analyze experimental micrococcal nuclease sequencing (MNase-Seq) datasets to determine the distribution of nucleosomes in the human genome. Both found that the exons had much sharper peaks of nucleosome occupancy, while the introns were relatively less occupied, irrespective of gene transcription (Schwartz et al. 2009; Tilgner et al. 2009). Using computational prediction models for nucleosome occupancy, exonic regions were found to be enriched for sequences that favored nucleosome positioning, while the sequences flanking the exons were depleted of these high-affinity nucleosome sequences (Schwartz et al., 2009). As most exons are short (~150bp or so), and also have high GC content (Zhu et al. 2009), it was suggested that nucleosomes, which occupy ~147bp of DNA and also have higher occupancy over GC-rich regions (Kiyama and Trifonov 2002; Segal et al. 2006; Peckham et al. 2007), were preferentially placed over exonic regions.
The question then arises as to whether nucleosomes are simply present over exons due to sequence preferences, or do they actually play a role in splicing? If the idea that nucleosomes actually play a role in splicing is right, then a single nucleosome should be sufficient to demarcate an exon boundary, and suggest a direct mechanism connecting splicing with nucleosome positioning. This would also mean that the strength of the nucleosome positioning would dictate splicing- and exons that are conditionally spliced would have well-positioned nucleosomes. The analyses suggested that nucleosome positioning changed relative to the size of exonic sequence: while short exons (less than 90bp) had poor nucleosome occupancy, as the size of exon increased to ~250 nt, the nucleosome occupancy peak increased, and was centered in the middle of the exon (Tilgner et al., 2009). Secondly, the exons with stronger splice acceptor sites were found to have less clearly demarcated peaks of nucleosome occupancy, relative to the weaker sites, when comparing exons of similar size (Tilgner et al., 2009). These observations suggested that nucleosomes are not simply placed over exons by chance, and potentially play a regulatory role. Additional support for the hypothesis that nucleosome positioning over exons plays a role in splicing was provided by a recent study that showed more stable positioning of nucleosomes over exons that were preferentially spliced when breast cancer cells were stimulated with progesterone. The exons that were excluded or skipped following hormone treatment were (a) depleted of positioned nucleosomes, and (b) contained binding sites for the splicing regulator heterogeneous nuclear ribonucleoprotein A/B (hnRNPAB), depletion of which by siRNA prevented hormone-induced alternative splicing (Iannone et al. 2015).
One hypothesis is that nucleosomes could act as roadblocks to help slow down the rate of Pol II elongation, thus providing more time for the spliceosome machinery to cotranscriptionally recognize splice signals. Support for the nucleosome roadblock hypothesis was provided by a study that used optical tweezers to follow individual Pol II complexes transcribing through nucleosome-dense DNA. The nucleosomes were found to serve as effective barriers to Pol II, as they slowed down elongating Pol II complexes (Hodges et al. 2009). While the above results point to the notion that chromatin structure primes alternative splicing events, additional studies are needed to clearly distinguish between the nucleosome occupancy over exons as simply correlative versus playing an active role in alternative splicing. To date, there is no published literature on nucleosome positioning changes during EMT, and how this changes during alternative splicing. It would undoubtedly be of interest to determine whether the genes that undergo alternative splicing during EMT also undergo changes in nucleosome positioning.
2. Histone modifications
An alternative explanation for the question of why nucleosomes are positioned over exons could be that these nucleosomes are associated with certain types of histone modifications that help to regulate splicing. In support of this idea, different groups working with various types of cells have identified several histone modifications over exons. The histone mark H3K36Me3 was shown to correlate with nucleosome occupancy over exonic regions (Andersson et al. 2009; Nahkuri et al. 2009; Schwartz et al. 2009; Spies et al. 2009; Tilgner et al. 2009; Zhou et al. 2012; Simon et al. 2014). Additional marks that were shown to be enriched over exons include H2BK5Me1, H3K27me1, H3K27me2, H3K27me1, and H3K79me1 (Andersson et al. 2009), H3K4Me1 (Spies et al. 2009) and H4K20me1 (Hon et al. 2009). However, another group claimed that H4K20me1 was not correlated with nucleosome positioning (Tilgner et al. 2009). The histone mark H3S10p, which was shown to interact with the serine/arginine-rich proteins SRSF1 and SRSF3 (also known as SRp20) is also thought to be involved in regulating alternative splicing, possibly via this interaction (Loomis et al. 2009). H3K9A and H3K36Me3 were shown to be associated with exon skipping events (Schor et al. 2009), while H3K9Me2 and H3K27Me3 were associated with alternative splicing and reduced Pol II (Allo et al. 2009).
Given the vast array of histone marks that are correlated with alternative splicing events, the question then arises as to whether all these histone marks influence alternative splicing, or are they simply ‘along for the ride’, and are more a consequence of splicing changes? If histone marks truly play a role in modulating splicing, then it stands to reason that manipulating the writers of the marks should cause a change in alternative splicing. Indeed, Luco et al found exactly that- by modulating levels of SET2 (H3K36me3 methyltransferase) and ASH2 (H3K4me3 methyltransferase) they found that loss of these histone marks over the exons also changed the splicing patterns (Luco et al. 2010).
The observations that intron-containing genes contain higher levels of H3K4Me3 and H3K36Me3 than do intronless genes are consistent with splicing affecting the location and intensity of these two histone marks (de Almeida et al. 2011; Bieberstein et al. 2012). Pre-mRNA splicing regulates SetD2 recruitment and H3K36me3 levels along the body of transcribed genes (de Almeida et al. 2011; Kim et al. 2011). The splicing inhibitors, meayamycin or spliceostatin A, reduced H3K36me3 levels and position along the gene body without altering elongation rates or chromatin-associated RNA (de Almeida et al. 2011; Kim et al. 2011). H3K4Me3 and H3K9ac are located primarily at the first exon-intron boundary (Bieberstein et al. 2012). Similar to H3K36Me3, the intensity and location of H3K4Me3 along the gene body is dependent on pre-mRNA splicing (Bieberstein et al. 2012). Inhibition of pre-mRNA splicing with spliceostatin A resulted in the loss of the H3K4me3 located at the exon 1 5′ splice site (Bieberstein et al. 2012).
Similar to the histone code, is there a ‘splicing code’ that exists? In other words, are there sets of histone marks that change specifically during alternative splicing? To address this question, Podlaha and colleagues used computational methods, to analyze the correlation between histone modifications, transcription start-site switching and splicing on a genome-wide level using published RNA-seq data from 9 normal and cancer cell lines (Podlaha et al. 2014). Histone variants and marks including histone H2A.Z, histone marks H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K9me3, H3K27ac, H3K27me3, H3K36me3, H3K79me2, and H4K20me were analyzed across all exons in the same cell line, and across the same exon in all cell lines. Similar to other groups, the authors found strong positive correlation of the histone H3K36Me3 mark with splicing in protein-coding genes; while H3K4me2 and H3K4me3 were most negatively correlated with alternative splicing.
Using the patterns of histone marks found associated with splicing, the authors attempted to predict splicing patterns in a different cell line based on just ChIP-seq data using the histone marks that were correlated with splicing. Interestingly, these computer-based methods enabled the prediction of alternative splicing patterns with great accuracy based simply on epigenetic patterns (Podlaha et al. 2014). The idea that we can use histone marks to accurately predict alternative-splicing patterns is a powerful one, and could be used to predict early changes in tumor progression, for instance. Interestingly, some of the histone marks that are predicted to be changing during alternative splicing also change during EMT. The first genome-wide study to look at epigenetic changes during the course of EMT was by MacDonald et al, who showed that global changes in histone marks during EMT that were mainly localized to large organized heterochromatin K9 modifications (LOCKs) (McDonald et al. 2011). The authors used the non-cancerous mouse AML12 cell line induced to undergo EMT by addition of TGF-β, and determined genome-wide epigenetic changes. They found a global reduction in the heterochromatin mark H3K9Me2, and an increase in the euchromatin mark H3K4Me3 and in the transcriptional mark H3K36Me3 (McDonald et al. 2011). Given the large number of alternative splicing events that occur during EMT (Shapiro et al. 2011), it would be interesting to determine whether the global increase seen in H3K36me3 in the AML12 cell line occurs over exonic regions that are alternatively spliced during EMT. Would these marks be retained and transmitted to daughter cells, as the tumors divide? Further, it would be extremely interesting to determine whether there is a temporal order to these histone marks during the process of EMT, and specifically in the context of splicing during EMT.
3. Chromatin remodeling factors
Given the correlation between nucleosome positioning over exonic regions, it is conceivable that proteins and mechanisms that regulate nucleosome positioning should also affect alternative splicing. For instance, chromatin remodeling factors such as the Brahma (Brm) subunit of the chromatin remodeling factor SWI/SNF (switch/sucrose non- fermentable) can interact with Pol II, spliceosomal snRNPs U1 and U5, and the RNA-binding protein KHDRBS1, and as a result, promote exon inclusion into the mRNA of the CD44 gene (Batsche et al. 2006). KHDRBS1, when it is phosphorylated by the extracellular-regulated kinase (ERK) mitogen-activated protein (MAP) kinase, can bind to and regulate the splicing of the CD44 gene, although it is unknown whether this can occur in the context of EMT (Batsche et al. 2006), although it has been suggested. However, KHDRBS1 is also known to play a role in EMT, by regulating levels of the alternative splicing factor SRSF1 through alternative splicing via the nonsense-mediated mRNA decay pathway (Valacca et al. 2010).
To date, no one has drawn a direct correlation between a chromatin remodeler that is responsible for alternative splicing during EMT. However, there are likely to be several chromatin remodelers that are directly involved in altering splicing isoforms during EMT. For instance, BAF250/ARID1, a large subunit of the mammalian SWI/SNF complex, is known to exist in two isoforms BAF250a/ARID1a and BAF250b/ARID1b, of which BAF250b was shown to interact with Smad2/3 in response to the cytokine transforming growth factor β (TGF-β); however whether this occurs in the context of EMT is unknown. CHD1 (chromatin remodeling ATPase) can bind to spliceosomal components and knockdown of CHD1 along with reduction of H3K4Me3, which CHD1 binds to, was shown to alter splicing efficiency (Sims et al. 2007; Teoh and Sharrocks 2014). Little is known about the functional consequence of CHD1 and spliceosome interactions during EMT.
Perhaps the most well-studied chromatin remodeling complex in the context of EMT is the Histone deacetylase (HDAC) family. Many studies implicated Snail and Slug in the repression of E-cadherin during EMT by recruitment of HDAC- containing complexes to the promoter (Bolos et al. 2003; Peinado et al. 2004; Peinado et al. 2007). Khan et al showed that HDAC1 and HDAC2 were recruited to the Myeloid Cell Leukemia 1 (MCL1) gene and catalyzed dynamic histone acetylation of the exon 2 nucleosome (Khan et al. 2014). The use of HDAC inhibitors caused a loss of HDACs and hence increased histone acetylation at exon 2, resulting in exclusion of the MCL1 exon2. The shorter form of MCL1 that was produced as a result of this alternative splicing was pro-apoptotic. Additionally, they showed by mass spectrometry and subsequent validation by immunoprecipitation, that HDAC1 and the non-phosphorylated form of HDAC2 co-precipitated with SRSF1 (Khan et al. 2014).
In addition to their role in splicing, several chromatin remodeling proteins have isoforms that are themselves generated by alternative splicing (some of these are listed in Table 1). For example, the histone methyltransferase capabilities of two human enzymes, G9A and Suppressor of Variegation 3–9 Homolog 2 (SUV39H2) were affected by alternative splicing in a variety of cell lines (Mauger et al. 2015). Given its involvement in repression of genes during EMT in cancer, it would be interesting to see whether G9A is alternatively spliced in the context of EMT (Dong et al. 2012; Liu et al. 2015). The coactivator-associated arginine methyltransferase 1 (CARM1; also called PRMT4) was shown to have several isoforms that are regulated by alternative splicing, and it also serves to regulate alternative splicing of CD44; however, whether this happens in the context of EMT is not yet known (Matsuda et al. 2007; Wang et al. 2013). HDAC6 has two splice variants, hHDAC6p131 and hHDAC6p114 that are involved in TGF-β signaling during EMT. Similar studies implicated other chromatin complexes in the repression of E-cadherin during EMT, including Enhancer of Zeste Homolog 2 (EZH2) -containing Polycomb repressive complex 2 (PRC2) complex (Herranz et al. 2008), the Ajuba/Lim-domain proteins (Langer et al. 2008; Hou et al. 2010), Lysine specific demethylase (LSD1) (Lin et al. 2010a; McDonald et al. 2011; Ferrari-Amorotti et al. 2013) and more recently Protein arginine Methyltransferase 7 (PRMT7) (Yao et al. 2014). However, none of these so far have been implicated in alternative splicing during the process of EMT. Further studies will undoubtedly uncover novel roles for these alternatively spliced chromatin modifiers in EMT and other processes.
Table 1.
Chromatin remodeling and epigenetic proteins involved in alternative splicing and EMT.
| Chromatin regulator | Functions of main protein product | Isoforms produced by alternative splicing and function, if known. | Function in EMT, if known | References |
|---|---|---|---|---|
| BAF250/ARID1 | Large subunit of mammalian SWI/SNF complex | 2 isoforms: BAF250a/ARID1a and BAF250b/ARID1b | Isoform BAF250b interacts with Smad2/3 in response to transforming growth factor β (TGF-β). | (Luo et al. 2008) |
| BMI1 | Component of the Polycomb repressor complex (PRC1), RING finger protein | Three isoforms | Needed for Twist1-induced EMT Involved in a negative feedback loop with Zeb1 and miR200 to induce EMT |
(Kim et al. 2006) (Yang et al. 2010) (Liu et al. 2014) |
| BRG1 and hBRM | Subunits of mammalian SWI/SNF complex | Several predicted by RefSeq | BRG1 is crucial for expression of Wnt target genes, such as WNT1-inducible signaling protein-1 (WISP1), which induces EMT | (Griffin et al. 2011) (Konigshoff et al. 2009) |
| CARM1 (PRMT4) | Arginine methylase (H3R8) | 4 isoforms in human, mouse & rat. | CARM1-exon v3 regulates alternative splicing of CD44; however, whether this happens in the context of EMT is not yet known. | (Matsuda et al. 2007) |
| CHD1 | Chromatin remodeling ATPase | 2 isoforms; can bind to spliceosomal components knockdown of CHD1 was shown to alter splicing | Unknown | (Sims et al. 2007). |
| DNMT1 | DNA methylation | 4 isoforms DNMT1o/s/p specific to oocyte, somatic or spermatocye DNMT1b (exon4); Localization, translational regulation, sex-specific expression exon 4: similar kinetic parameters on DNA methylation | DNMT1 interacts with Snail to repress E-cadherin (Espada et al); and fructose-1,6-biphosphatase (FBP1) in basal-like breast cancer (BLBC) (Dong et al, 2013). | (Mertineit et al. 1998) (Bonfils et al. 2000) |
| DNMT3B | DNA methylation | 40 isoforms DNMT3B4 B5 lacking exons21+22 DNMT3B3-like splice variant lacking exon 5. Exons21+22 encoding catalytic domain Exon5 increases DNA binding or influences the 3D structure of the adjacent PWWP domain |
Involved in transforming growth factor beta (TGF-β) EMT induction. TGF-β induces DNMT3B, through down-regulation of miR-29a. |
(Robertson et al. 1999) (Van Emburgh and Robertson 2011) (Kogure et al. 2014) |
| EZH2 | HMTase of H3K27 | 2 isoforms EZH2α EZH2β brings Exon4 skipping and Exon8 3′SS. EZH2β represses a predominantly unique subset of gene targets from EZH2α |
EZH2 forms a repressive complex with SNAIL and ubiquitin E3 ligase Ring1B, leading gene repression or silencing. Overexpression of SNAIL and SLUG activates EZH2 by repressing of miR-101. |
(Grzenda et al. 2013) (Chen et al. 2014) (Zheng et al. 2015) |
| G9A | HMTase of H3K9me1/2 | 2 isoforms produced by inclusion or skipping of exon 10. Exclusion of exon 10 does not affect HMTase activity, and may control a different function. | Regulates many genes with functions related to tumor growth, tumor suppression, or metastasis. Interacts with Snail to mediate E-cadherin repression in human breast cancer and Head and neck squamous cell carcinoma. |
(Mauger et al. 2015) (Dong et al. 2012) (Liu et al. 2015) |
| HDAC1, 2 and 6 | Histone deacetylases | HDAC 1 leads to gene repression during TGF-β induction of EMT. HDAC 1 &2 form a gene repressive complex with SNAIL to inhibit E-cadherin during EMT. HDAC6: functions in TGF-β induced EMT through deacetylation of alpha-tubulin and inactivating SMAD3. HDAC6 has two splice variants, hHDAC6p131 and hHDAC6p114 that are involved in TGF-β EMT signaling. |
Regulation of several EMT marker genes, such as E-cadherin, ZO-1, alpha-SMA, alpha-tubulin, etc. | (Glenisson et al. 2007) (Shan et al. 2008) (Lei et al. 2010) (Wu et al. 2011) (Peinado et al. 2004) |
| JMJD1A/2B/2C | Histone demethylases | JMJD1A has a prospective splice variant of 135kDa that is induced by hypoxia in LNCaP and HeLa cell lines (Beyer). Function unknown. A 8 kb isoform of JMJD1A was observed in thymus and heart of mice (Knebel). Function not investigated. |
Regulation of Adrenomedullin, EDN1, HMOX1, GDF15 affecting tumor growth. JMJD2B: Induces EMT by interacting and increasing nuclear localization of β-catenin, resulting in binding to the promoter and activation of vimentin through H3K9 demethylation (Zhao). |
(Pollard et al. 2008) (Krieg et al. 2010) (Knebel et al. 2006) |
| LSD1 | H3K4 Demethylase | 4 isoforms. 4 amino acid exon E8a brings the phosphorylated threonine important for protein conformation and activity |
Regulates genes in LOCKS during EMT Interacts with SNAIL and HMG20A, leading to gene repression during EMT (Rivero). |
(Zibetti et al. 2010) (Toffolo et al. 2014) (McDonald et al. 2011) (Rivero et al. 2015) (Lin et al. 2010) |
| MBD2 | Methyl-CpG binding protein | 2 isoforms generated by SRSF2-mediated alternative splicing. MBD2a preferentially interacts with repressive NuRD chromatin remodeling factors and promotes hPSC differentiation. MBD2c enhances reprogramming of fibroblasts to pluripotency. |
MBD2 may interact with δEF1 to regulate DNA methylation at E-cadherin promoter during EMT (Fukagawa). | (Lu et al. 2014) (Fukagawa et al. 2015) |
| MTA1 | Mi-2/NuRD complex subunit | Mouse has several alternative splicing variants Splice variant MTA1s lacks a nuclear localization sequence and is found mostly in cytoplasm. |
In colorectal cancer, eukaryotic initiation factor 5A2 (EIF5A2) induces MTA1, which induces EMT (Zhu). Induced by TGF-β1 signaling. MTA1 leads to induction of FosB, which represses E-cadherin (Pakala) MTA1 also leads to β-catenin activation to promote EMT in nasopharyngeal carcinoma (Lin). |
(Yaguchi et al. 2005) (Kumar et al. 2002) (Zhu et al. 2012) (Pakala et al. 2011) (Lin et al. 2014) |
| MTA3 | Mi-2/NuRD complex subunit | Long (MTA3L) and short (MTA3) isoforms. MTA3 more abundant than MTA3L in cancer cell lines. | MTA3 represses SNAI1, a master regulator of EMT. | (Fujita et al. 2003) |
| PRMT7 | Protein arginine methyltransferase | PRMT7 has two isoforms (PRMT7α) and (PRMT7β). | PRMT7 is found to have higher expression in breast cancer tumors that are more invasive. Overexpression of PRMT7 induced EMT by interacting with YY1 and HDAC3 to repress E cadherin. |
(Yao et al. 2014) |
| SET8/KMT5A | SET-domain containing histone methyltransferase | 2 isoforms: 42kDa isoform interacts with a tumor suppressor, Lethal 3 malignant brain tumor 1 (L3MBTL1). 39kDa form interacts with SIRT2, a tumor suppressor. |
Induces H4K20me to activate the expression of AXIN2 (an axin homolog that stabilizes b-catenin) and c-Myc, and contributes to tumorigenesis. SET8 physically associates with Twist1 to induce H4K20me on the promoters of CDH1 and CDH2, leading to EMT and metastasis. | (Li et al. 2011) (Yang et al. 2012) (Kalakonda et al. 2008) (Serrano et al. 2013) |
4. DNA methylation
DNA methylation, the addition of a methyl group to the cytosine base of DNA, correlates with closed chromatin structures and, as a consequence, with reduced transcription (Lorincz et al. 2004). Genome-wide analyses in plants and human cells revealed an enrichment of DNA methylation (5-methyl cytosine, 5-mC) in nucleosome-associated DNA and thus also in exonic compared to intronic regions (Hodges et al. 2009; Chodavarapu et al. 2010). Although the function of DNA methylation in gene promoter regions was well established to play a role in transcriptional repression, the function of the evolutionarily conserved widespread distribution of DNA methylation in gene body regions was incompletely understood until recently.
A mechanism by which DNA methylation regulates alternative splicing by preventing CCCTC-binding factor (CTCF) binding was proposed by Shukla et al (2011), who investigated DNA methylation-induced skipping of CD45 exon 5 during lymphocyte development (Shukla et al. 2011). Binding of the chromatin insulator protein CTCF, which recognizes specific DNA sequences within CD45 exon 5, was shown to slow down RNA polymerase II elongation, thus promoting exon 5 inclusion in CD45 pre-mRNA. They demonstrated that DNA methylation prevented CTCF binding, thus allowing faster RNA polymerase II elongation, which caused exon 5 skipping (Shukla et al. 2011). The prevention of CTCF binding by DNA methylation was not merely specific to CD45, however, as genome-wide analyses of CTCF binding and splicing changes following CTCF depletion suggest that the interplay between CTCF binding and DNA methylation at the 5′ of exons can regulate a substantial number of alternative splicing events (Shukla et al. 2011).
Keiji Zhao and colleagues demonstrated that DNA methylation was enriched in exons that were included as a result of alternative splicing, and that inhibition of DNA methylation resulted in aberrant splicing of these exons (Maunakea et al. 2013). Further, they showed that the methyl-CpG-binding protein (MeCP2), was enriched in the alternatively spliced exons, particularly those that are also highly methylated, and inhibition of DNA methylation disrupts specific targeting of MeCP2 to exons. Loss of MeCP2 was accompanied by increased histone acetylation over the exonic regions, and aberrant alternative splicing. A similar effect was seen when HDAC inhibitors were used, thus also increasing histone acetylation over exons, indicating that intragenic DNA methylation and/or histone deacetylation enhance exon recognition by the splicing machinery. Alternatively spliced exons were found to display lower enrichment of 5-mC compared to constitutive ones (Gelfman et al. 2013), reinforcing the idea that DNA methylation can contribute to mark exon recognition by the spliceosome. The global impact of DNA methylation was investigated recently in embryonic stem cells, and it was determined that DNA methylation could either enhance or silence exon recognition. Interestingly, a subset of these exons with DNA methylation were bound by Heterochromatin protein 1 (HP1), which regulated alternative splicing of the exon in a DNA methylation dependent manner, by recruiting splicing factors to the exon (Yearim et al. 2015). Overall, these studies highlight the role of DNA methylation in alternative splicing, and suggest that it can drive alternative splicing in two ways: (a) by preventing binding of proteins such as CTCF, thus inhibiting Pol II elongation; and (b) by increasing binding of proteins like HP1 and MeCP2, which can potentially target the spliceosome to those exons.
The first genome-wide study to look at epigenetic changes during the course of EMT did not detect changes in DNA methylation(McDonald et al. 2011). However the study only considered a short time (24h) during the EMT process. Changes in DNA methylation during EMT were however shown in a recent study using ovarian cancer cells that were triggered to undergo EMT by addition of TGF-β (Cardenas et al. 2014). The authors used the Infinium HumanMethylation450 BeadChip to identify several genes that were hypermethylated at 48 and 120 h after TGF-β stimulation, and also showed changes in gene expression corresponding to the change in methylation (Cardenas et al. 2014). Another study using Twist-induced EMT as a model system demonstrated hypermethylation in some genomic regions, and widespread global DNA hypomethylation (Malouf et al. 2013).
Besides cytosine methylation, 5-hydroxycytosine methylation (5hmC) has also been shown to be involved in alternative splicing. For instance, the DNA coding for constitutive exons in the frontal cortex of the human brain were shown to contain higher levels of 5hmC relative to exons that were alternatively spliced (Khare et al. 2012). More recently, loss of Tet methylcytosine dioxygenase 1 (TET1) following cocaine administration was shown to result both in loss of 5hmC and, surprisingly, gain of 5hmC at several genomic locations, and the change in 5hmC correlated with differences in alternative splicing (Feng et al. 2015). Specifically, an increase in 5hmC was correlated with upregulation of alternative splice isoforms, while loss of 5hmC at splice sites was more likely to be associated with isoforms downregulated after cocaine administration (Feng et al. 2015).
TET1, which regulates 5-hmC (see section on DNA methylation) was also shown to regulate hypoxia-induced EMT by acting as a co-activator for the hypoxia-inducible factors (HIF-1α and HIF-2α), to enhance their transactivation activity (Tsai et al. 2014). The TET family of methylcytosine dioxygenases can also interact with the microRNA miR-22, which targets it to the promoter of the anti-metastatic miR-200, thereby silencing it, leading to metastasis and poor patient outcome (Song et al. 2013). To date, none of these studies directly examined the interplay between DNA methylation or hydroxymethylation and alternative splicing events during EMT. Given the evidence for the involvement of DNA methylation in alternative splicing, and the fact that alternative splicing plays significant roles in driving the EMT phenotype, it stands to reason that these mechanisms might be linked.
5. MicroRNAs and other Noncoding RNAs
MicroRNAs (miRNAs) are small, 21–24nt regulatory RNAs that influence the stability and translational efficiency of target mRNAs, thus altering gene expression profiles (Eulalio et al. 2008; Bartel 2009; Adams et al. 2014). As many miRNA genes are located in intronic regions, the miRNA processing machinery and the spliceosome machinery often interact, and there is possibly a lot of interplay between the two. In support of this idea, the microRNA processing proteins Drosha and DiGeorge Syndrome Critical Region Gene 8 (DGCR8) were found to co-sediment with the supraspliceosome, (Agranat-Tamir et al. 2014) a ~21 MDa complex of four native spliceosomes connected by the pre-mRNA (Azubel et al. 2006; Shefer et al. 2014). The authors focused their study on two alternative splicing events, which extend exon 14 of the gene coding for the mini chromosome maintenance 7 protein (MCM7), where the miR-106b-25 cluster is located. If the miRNAs were included in the splicing event as the extended exon 14, then they were not made into miRNAs. On the other hand, inhibiting splicing resulted in increased miRNAs expression, as this region was not excluded, and was processed into miRNA. Similarly, knockdown of Drosha increased splicing. Interestingly, the members of the miR-106b-25 cluster are key modulators of the TGF-β signaling pathway in tumors, and also induce EMT (Petrocca et al. 2008; Smith et al. 2012). Thus, it would be interesting to determine whether this alternative splicing event also occurs during EMT.
Additionally, miRNAs and noncoding RNAs themselves can regulate alternative splicing events by modulating expression of splicing factors. For instance, during heart development, miR-23a/b was shown to downregulate vital regulators of splicing events in the heart, CELF1 (CUGBP) and embryonically lethal abnormal vision-type RNA binding protein 3 (ETR-3) -like factor (Kalsotra et al. 2010). The microRNA miR-132 targets polypyrimidine tract-binding protein 2 (PTBP2), a splicing factor that is involved in neurodevelopment (Smith et al. 2011). A study by Ameyar-Zazoua et al., 2012 found that the Argonaute protein, which is involved in miRNA gene silencing, regulates alternative splicing by interacting with splicing factors and chromatin remodelers to help with spliceosome placement at splice sites and modifying RNA polymerase transcription rates (Ameyar-Zazoua et al. 2012; Batsche and Ameyar-Zazoua 2015).
Several microRNAs have been identified as playing important pathophysiological roles in cancer, and more specifically, in EMT (Bullock et al. 2012; Diaz-Lopez et al. 2014). For instance, the miR-200 family of microRNAs, which is downregulated in breast tumors, was shown to repress expression of the Snail and Zeb family members (Burk et al. 2008; Gregory et al. 2008a; Liu et al. 2014; Perdigao-Henriques et al. 2015), which induce EMT. The microRNA miR-10b is highly expressed in metastatic tumors and is upregulated by Twist, an EMT protein (Ma et al. 2007). Interestingly, miR-10b was shown to also regulate alternative splicing events that are critical for retinoic acid-induced SH-SY5Y neuroblastoma cell differentiation by targeting the SRSF1 splicing factor (Meseguer et al. 2011). Further, miR-10b was required for the associated changes in migration, invasion, and in vivo metastasis. Therefore, it might very well be important for regulating similar alternative splicing events in the EMT process. Importantly, an antagonist of miR-10b (antagomir) was used successfully to inhibit tumor growth and metastasis formation in a mouse model of breast cancer (Ma et al. 2007). Other antagomirs have been used successfully to inhibit tumor growth and metastasis formation (reviewed in Gregory et al. 2008b; Wright et al. 2010; Zhang and Ma 2012; Diaz-Lopez et al. 2014). Given the success of antagomir-based therapy, combined therapies targeting both miRNAs and/or splicing factors could have improved effects on cancer metastasis.
Lastly, long non-coding RNAs (lncRNAs), a class of RNAs longer than 200 nt have been reported to be involved in the regulation of transcription, chromatin remodeling, post-transcriptional RNA processing and cancer metastasis (Tano and Akimitsu 2012; Adams et al. 2014; Yang et al. 2014; Holoch and Moazed 2015). Several of these long non-coding RNAs have been associated with EMT and metastasis (De Craene and Berx 2013; Hu et al. 2014; Richards et al. 2015). For instance, metastasis associated lung adenocarcinoma transcript 1 (MALAT1, an 8000 nt lncRNA) was shown to increase bladder cancer metastasis by induction of EMT (Ying et al. 2012; Fan et al. 2014). Interestingly, MALAT1 is also a modulator of alternative splicing- by regulating the levels of phosphorylated SR proteins, it can control the concentration gradient of how much SR proteins are available to the splicing machinery (Tripathi et al. 2010). This is possibly true during the process of EMT as well. Another lncRNA, HOX transcript antisense RNA (HOTAIR, 2200 nt), is required for EMT, and to maintain cancer cells in a stem-cell like state (Padua Alves et al. 2013). It is also associated with poor prognosis in colon (Wu et al. 2014) and cervical (Kim et al. 2015) cancer progression. HOTAIR is thought to act by downregulating the expression of miR-7, a tumor suppressor protein (Zhang et al. 2014), thereby leading to cancer progression. Recently, six transcript variants of HOTAIR were identified, and it was suggested that these could be functionally very different based on the presence or absence of the PRC2-interacting domain (Mercer et al. 2012; Loewen et al. 2014). Genome-wide profiling of lncRNAs during TGF-β induced (Richards et al. 2015) or Twist-induced (Hu et al. 2014) EMT has resulted in the identification of several more lncRNAs that are regulated in EMT. Whether these lncRNAs are involved in alternative splicing, however, remains to be determined.
CONCLUSIONS
EMT is an extremely complex process, involving multiple layers of regulation that are tightly controlled by various factors. Given that so many different aspects are involved in EMT, from transcription to cellular movement to loss of apico-basal polarity, proper coordination of these events is essential for the transition from epithelial to mesenchymal cell state. At the level of transcription alone, the master regulators have to actively coordinate recruitment of chromatin remodelers to the promoters of genes such as E-cadherin at the right time, to start the process of EMT. Many different chromatin remodelers have been shown to be involved during the EMT process, and all of these were shown to be critical for repression at the E-cadherin locus alone (Peinado et al. 2004; Herranz et al. 2008; Langer et al. 2008; Lei et al. 2010; Lin et al. 2010a; Yang et al. 2010; Dong et al. 2012; Yang et al. 2012; Yao et al. 2014; Fukagawa et al. 2015; Liu et al. 2015). Can so many factors all be present at one locus at the same time? We believe that this is probably a highly dynamic process, as suggested for the dynamics of TFs at other loci (Sung et al. 2014; Voss and Hager 2014). Further, the ability of cancer cells to not rely on one factor alone, but count on alternative factors to substitute for the absence of any one factor, might be beneficial to their survival. Additionally, alternative splicing might be a really easy way to change the function of a protein, e.g. from pro-apoptotic to anti-apoptotic, as we saw in the case of Bcl-x (Massiello et al., 2006). Therefore, from the clinical viewpoint, it is important to think about attacking these cells on multiple fronts to be able to fully confront the problem of cancer metastasis.
Like transcription and chromatin remodeling, alternative splicing is tightly controlled during the process of EMT. For instance, alternative splicing of a subset of genes has to be regulated such that they exclude exons and switch from longer to shorter isoforms [e.g. SLC37A2, KIF13A, FLNB, and MBNL1 genes, (Shapiro et al. 2011)] during EMT, while other groups of genes do the reverse and show a greater degree of exon inclusion [e.g. PLEKHA1, MLPH, ARHGEF11, CLSTN1 and PLOD2 genes; (Shapiro et al. 2011)]. Yet other genes undergo a variety of other types of splicing changes, including alternative splice site selection, retaining introns or using alternative exons (Shapiro et al. 2011) during EMT. How do these events occur simultaneously within a cell? One possibility is that different types of splicing events might recruit different types of chromatin remodelers, e.g. exon inclusion events might require remodeler A, vs. exon exclusion events, which might require remodeler B. Alternatively, there might be certain transcription factors that can specifically target different sets of chromatin remodelers to different genes. Intriguingly, despite being closely related, there are no studies to date that directly link alternative splicing and chromatin together in the context of EMT. We believe that determining the role of chromatin and epigenetic proteins in alternative splicing during the EMT process will help define the mechanisms driving EMT and cancer metastasis, thus setting the stage for novel clinical interventions of this devastating disease.
Acknowledgments
This work was supported by grants from the National Institutes of Health [5P20GM104360 to AD]; University of North Dakota School of Medicine Faculty Seed Grant [AD], the Canadian Breast Cancer Foundation (JRD) and a Canada Research Chair (JRD). We thank Min Wu and Sergei Nechaev for their critical evaluations, as well as members of Dhasarathy and Davie labs for helpful discussions. We apologize to those researchers whose work could not be cited due to space considerations, but nevertheless have made valuable contributions to this field.
BIBLIOGRAPHY
- Adami G, Babiss LE. DNA template effect on RNA splicing: two copies of the same gene in the same nucleus are processed differently. EMBO J. 1991;10(11):3457–3465. doi: 10.1002/j.1460-2075.1991.tb04910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24(16):R762–76. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agranat-Tamir L, Shomron N, Sperling J, Sperling R. Interplay between pre-mRNA splicing and microRNA biogenesis within the supraspliceosome. Nucleic Acids Res. 2014;42(7):4640–4651. doi: 10.1093/nar/gkt1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: a menage a trois. Curr Opin Genet Dev. 2008;18(2):145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16(7):717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- Allo M, Schor IE, Munoz MJ, de la Mata M, Agirre E, Valcarcel J, Eyras E, Kornblihtt AR. Chromatin and alternative splicing. Cold Spring Harb Symp Quant Biol. 2010:75103–111. doi: 10.1101/sqb.2010.75.023. [DOI] [PubMed] [Google Scholar]
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol. 2011;18(12):1435–1440. doi: 10.1038/nsmb.2143. [DOI] [PubMed] [Google Scholar]
- Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC, Mathieu J, Hamiche A, Ait-Si-Ali S, Muchardt C, Batsche E, Harel-Bellan A. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19(10):998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19(10):1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azubel M, Habib N, Sperling R, Sperling J. Native spliceosomes assemble with pre-mRNA to form supraspliceosomes. J Mol Biol. 2006;356(4):955–966. doi: 10.1016/j.jmb.2005.11.078. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132(14):3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsche E, Ameyar-Zazoua M. The influence of Argonaute proteins on alternative RNA splicing. Wiley Interdiscip Rev RNA. 2015;6(1):141–156. doi: 10.1002/wrna.1264. [DOI] [PubMed] [Google Scholar]
- Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bhat-Nakshatri P, Song EK, Collins NR, Uversky VN, Dunker AK, O’Malley BW, Geistlinger TR, Carroll JS, Brown M, Nakshatri H. Interplay between estrogen receptor and AKT in estradiol-induced alternative splicing. BMC Med Genomics. 2013;621 doi: 10.1186/1755-8794-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberstein NI, Carrillo Oesterreich F, Straube K, Neugebauer KM. First exon length controls active chromatin signatures and transcription. Cell Rep. 2012;2(1):62–68. doi: 10.1016/j.celrep.2012.05.019. [DOI] [PubMed] [Google Scholar]
- Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116(Pt 3):499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- Bonfils C, Beaulieu N, Chan E, Cotton-Montpetit J, MacLeod AR. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J Biol Chem. 2000;275(15):10754–10760. doi: 10.1074/jbc.275.15.10754. [DOI] [PubMed] [Google Scholar]
- Bradley T, Cook ME, Blanchette M. SR proteins control a complex network of RNA-processing events. RNA. 2015;21(1):75–92. doi: 10.1261/rna.043893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152(6):1252–1269. doi: 10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman BM. Splice variants as cancer biomarkers. Clin Biochem. 2004;37(7):584–594. doi: 10.1016/j.clinbiochem.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J Clin Invest. 2011;121(3):1064–1074. doi: 10.1172/JCI44540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock MD, Sayan AE, Packham GK, Mirnezami AH. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell. 2012;104(1):3–12. doi: 10.1111/boc.201100115. [DOI] [PubMed] [Google Scholar]
- Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busa R, Sette C. An emerging role for nuclear RNA-mediated responses to genotoxic stress. RNA Biol. 2010;7(4):390–396. doi: 10.4161/rna.7.4.12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas H, Vieth E, Lee J, Segar M, Liu Y, Nephew KP, Matei D. TGF-beta induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics. 2014;9(11):1461–1472. doi: 10.4161/15592294.2014.971608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu H, Zou X, Wang J, Zhu Y, Chen H, Shen B, Deng X, Zhou A, Chin YE, Rauscher F, Jr, Peng C, Hou Z. Snail recruits Ring1B to mediate transcriptional repression and cell migration in pancreatic cancer cells. Cancer Res. 2014;74(16):4353–4363. doi: 10.1158/0008-5472.CAN-14-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell. 2007;26(6):867–881. doi: 10.1016/j.molcel.2007.05.036. [DOI] [PubMed] [Google Scholar]
- de Almeida SF, Grosso AR, Koch F, Fenouil R, Carvalho S, Andrade J, Levezinho H, Gut M, Eick D, Gut I, Andrau JC, Ferrier P, Carmo-Fonseca M. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat Struct Mol Biol. 2011;18(9):977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- De Conti L, Baralle M, Buratti E. Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA. 2013;4(1):49–60. doi: 10.1002/wrna.1140. [DOI] [PubMed] [Google Scholar]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13(2):97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Diaz-Lopez A, Moreno-Bueno G, Cano A. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag Res. 2014:6205–216. doi: 10.2147/CMAR.S38156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122(4):1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G, Lafaille C, de la Mata M, Marasco LE, Munoz MJ, Le Jossic-Corcos C, Corcos L, Kornblihtt AR. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell. 2014;54(4):683–690. doi: 10.1016/j.molcel.2014.03.044. [DOI] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol Cell. 2006;21(6):849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell. 2008;132(1):9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, Liu Y. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20(6):1531–1541. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, Koo JW, Ribeiro E, Labonte B, Laitman BM, Estey D, Stockman V, Kennedy P, Courousse T, Mensah I, Turecki G, Faull KF, Ming GL, Song H, Fan G, Casaccia P, Shen L, Jin P, Nestler EJ. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18(4):536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari-Amorotti G, Fragliasso V, Esteki R, Prudente Z, Soliera AR, Cattelani S, Manzotti G, Grisendi G, Dominici M, Pieraccioli M, Raschella G, Chiodoni C, Colombo MP, Calabretta B. Inhibiting interactions of lysine demethylase LSD1 with snail/slug blocks cancer cell invasion. Cancer Res. 2013;73(1):235–245. doi: 10.1158/0008-5472.CAN-12-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113(2):207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Fukagawa A, Ishii H, Miyazawa K, Saitoh M. deltaEF1 associates with DNMT1 and maintains DNA methylation of the E-cadherin promoter in breast cancer cells. Cancer Med. 2015;4(1):125–135. doi: 10.1002/cam4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfman S, Cohen N, Yearim A, Ast G. DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure. Genome Res. 2013;23(5):789–799. doi: 10.1101/gr.143503.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta. 2007;1773(10):1572–1582. doi: 10.1016/j.bbamcr.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Sorting out the complexity of SR protein functions. RNA. 2000;6(9):1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008a;10(5):593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008b;7(20):3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc Natl Acad Sci U S A. 2011;108(6):2282–2287. doi: 10.1073/pnas.1013751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Svingen P, Mathison A, Calvo E, Iovanna J, Xiong Y, Faubion W, Urrutia R. Functional characterization of EZH2beta reveals the increased complexity of EZH2 isoforms involved in the regulation of mammalian gene expression. Epigenetics Chromatin. 2013;6(1):3. doi: 10.1186/1756-8935-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol. 1994;239(3):357–365. doi: 10.1006/jmbi.1994.1377. [DOI] [PubMed] [Google Scholar]
- Henriques R, Mas P. Chromatin remodeling and alternative splicing: pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin Cell Dev Biol. 2013;24(5):399–406. doi: 10.1016/j.semcdb.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, Garcia de Herreros A, Peiro S. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28(15):4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325(5940):626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holoch D, Moazed D. RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet. 2015;16(2):71–84. doi: 10.1038/nrg3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5(11):e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi K, Sakamoto K, Koinuma D, Semba K, Inoue A, Inoue S, Fujii H, Yamaguchi A, Miyazawa K, Miyazono K, Saitoh M. TGF-beta drives epithelial-mesenchymal transition through deltaEF1-mediated downregulation of ESRP. Oncogene. 2012;31(26):3190–3201. doi: 10.1038/onc.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Peng H, White DE, Wang P, Lieberman PM, Halazonetis T, Rauscher F., Jr 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 2010;70(11):4385–4393. doi: 10.1158/0008-5472.CAN-10-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Yang J, Hou Y, Zhang H, Zeng Z, Zhao L, Yu T, Tang X, Tu G, Cui X, Liu M. LncRNA expression signatures of twist-induced epithelial-to-mesenchymal transition in MCF10A cells. Cell Signal. 2014;26(1):83–93. doi: 10.1016/j.cellsig.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Iannone C, Pohl A, Papasaikas P, Soronellas D, Vicent GP, Beato M, Valcarcel J. Relationship between nucleosome positioning and progesterone-induced alternative splicing in breast cancer cells. RNA. 2015 doi: 10.1261/rna.048843.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H, Saitoh M, Sakamoto K, Kondo T, Katoh R, Tanaka S, Motizuki M, Masuyama K, Miyazawa K. Epithelial splicing regulatory proteins 1 (ESRP1) and 2 (ESRP2) suppress cancer cell motility via different mechanisms. J Biol Chem. 2014;289(40):27386–27399. doi: 10.1074/jbc.M114.589432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson IJ. A reappraisal of non-consensus mRNA splice sites. Nucleic Acids Res. 1991;19(14):3795–3798. doi: 10.1093/nar/19.14.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12(1):5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, Rice JC, Nimer SD. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27(31):4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24(7):653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan DH, Gonzalez C, Cooper C, Sun JM, Chen HY, Healy S, Xu W, Smith KT, Workman JL, Leygue E, Davie JR. RNA-dependent dynamic histone acetylation regulates MCL1 alternative splicing. Nucleic Acids Res. 2014;42(3):1656–1670. doi: 10.1093/nar/gkt1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan DH, Jahan S, Davie JR. Pre-mRNA splicing: role of epigenetics and implications in disease. Adv Biol Regul. 2012;52(3):377–388. doi: 10.1016/j.jbior.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Khare T, Pai S, Koncevicius K, Pal M, Kriukiene E, Liutkeviciute Z, Irimia M, Jia P, Ptak C, Xia M, Tice R, Tochigi M, Morera S, Nazarians A, Belsham D, Wong AH, Blencowe BJ, Wang SC, Kapranov P, Kustra R, Labrie V, Klimasauskas S, Petronis A. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nat Struct Mol Biol. 2012;19(10):1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Menet JS, Tolan M, Rosbash M. Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA. 2012;18(12):2174–2186. doi: 10.1261/rna.034090.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MTn, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25(23):2502–2512. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, Kim YT. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–530. doi: 10.3892/ijo.2014.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108(33):13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Paylor SW, Magnuson T, Schumacher A. Juxtaposed Polycomb complexes co-regulate vertebral identity. Development. 2006;133(24):4957–4968. doi: 10.1242/dev.02677. [DOI] [PubMed] [Google Scholar]
- Kiyama R, Trifonov EN. What positions nucleosomes?--A model. FEBS Lett. 2002;523(1–3):7–11. doi: 10.1016/s0014-5793(02)02937-x. [DOI] [PubMed] [Google Scholar]
- Knebel J, De Haro L, Janknecht R. Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J Cell Biochem. 2006;99(1):319–329. doi: 10.1002/jcb.20945. [DOI] [PubMed] [Google Scholar]
- Kogure T, Kondo Y, Kakazu E, Ninomiya M, Kimura O, Shimosegawa T. Involvement of miRNA-29a in epigenetic regulation of transforming growth factor-beta-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Hepatol Res. 2014;44(8):907–919. doi: 10.1111/hepr.12188. [DOI] [PubMed] [Google Scholar]
- Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Gunther A, Eickelberg O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119(4):772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol. 2006;13(1):5–7. doi: 10.1038/nsmb0106-5. [DOI] [PubMed] [Google Scholar]
- Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum Genet. 1992;90(1–2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- Krecic AM, Swanson MS. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11(3):363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30(1):344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Wang RA, Mazumdar A, Talukder AH, Mandal M, Yang Z, Bagheri-Yarmand R, Sahin A, Hortobagyi G, Adam L, Barnes CJ, Vadlamudi RK. A naturally occurring MTA1 variant sequesters oestrogen receptor-alpha in the cytoplasm. Nature. 2002;418(6898):654–657. doi: 10.1038/nature00889. [DOI] [PubMed] [Google Scholar]
- Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer EM, Feng Y, Zhaoyuan H, Rauscher F, Jr, Kroll KL, Longmore GD. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev Cell. 2008;14(3):424–436. doi: 10.1016/j.devcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem. 2015 doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen S, Marsellach FX, Codlin S, Schmidt A, Clement-Ziza M, Beyer A, Bahler J, Orengo C, Pancaldi V. Stress induces remodelling of yeast interaction and co-expression networks. Mol Biosyst. 2013;9(7):1697–1707. doi: 10.1039/c3mb25548d. [DOI] [PubMed] [Google Scholar]
- Lei W, Zhang K, Pan X, Hu Y, Wang D, Yuan X, Shu G, Song J. Histone deacetylase 1 is required for transforming growth factor-beta1-induced epithelial-mesenchymal transition. Int J Biochem Cell Biol. 2010;42(9):1489–1497. doi: 10.1016/j.biocel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Li Z, Nie F, Wang S, Li L. Histone H4 Lys 20 monomethylation by histone methylase SET8 mediates Wnt target gene activation. Proc Natl Acad Sci U S A. 2011;108(8):3116–3123. doi: 10.1073/pnas.1009353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Thiery JP. Epithelial-mesenchymal transitions: insights from development. Development. 2012;139(19):3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010a;29(35):4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010b;29(11):1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Wan X, Jiang R, Deng L, Gao Y, Tang J, Yang Y, Zhao W, Yan X, Yao K, Sun B, Chen Y. Epstein-Barr virus-encoded latent membrane protein 2A promotes the epithelial-mesenchymal transition in nasopharyngeal carcinoma via metastatic tumor antigen 1 and mechanistic target of rapamycin signaling induction. J Virol. 2014;88(20):11872–11885. doi: 10.1128/JVI.01867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13(9):815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- Liu S, Ye D, Guo W, Yu W, He Y, Hu J, Wang Y, Zhang L, Liao Y, Song H, Zhong S, Xu D, Yin H, Sun B, Wang X, Liu J, Wu Y, Zhou BP, Zhang Z, Deng J. G9a is essential for EMT-mediated metastasis and maintenance of cancer stem cell-like characters in head and neck squamous cell carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sanchez-Tillo E, Lu X, Huang L, Clem B, Telang S, Jenson AB, Cuatrecasas M, Chesney J, Postigo A, Dean DC. The ZEB1 transcription factor acts in a negative feedback loop with miR200 downstream of Ras and Rb1 to regulate Bmi1 expression. J Biol Chem. 2014;289(7):4116–4125. doi: 10.1074/jbc.M113.533505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7(1):90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33(4):450–461. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Dickerson DR, Schmitt M, Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nat Struct Mol Biol. 2004;11(11):1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- Lu Y, Loh YH, Li H, Cesana M, Ficarro SB, Parikh JR, Salomonis N, Toh CX, Andreadis ST, Luckey CJ, Collins JJ, Daley GQ, Marto JA. Alternative splicing of MBD2 supports self-renewal in human pluripotent stem cells. Cell Stem Cell. 2014;15(1):92–101. doi: 10.1016/j.stem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144(1):16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, Mermel C, Barbie DA, Awad T, Zhou X, Nguyen T, Piqani B, Li C, Golub TR, Meyerson M, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105(51):20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Malouf GG, Taube JH, Lu Y, Roysarkar T, Panjarian S, Estecio MR, Jelinek J, Yamazaki J, Raynal NJ, Long H, Tahara T, Tinnirello A, Ramachandran P, Zhang XY, Liang S, Mani SA, Issa JP. Architecture of epigenetic reprogramming following Twist1-mediated epithelial-mesenchymal transition. Genome Biol. 2013;14(12):R144. doi: 10.1186/gb-2013-14-12-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda H, Paul BD, Choi CY, Shi YB. Contrasting effects of two alternative splicing forms of coactivator-associated arginine methyltransferase 1 on thyroid hormone receptor-mediated transcription in Xenopus laevis. Mol Endocrinol. 2007;21(5):1082–1094. doi: 10.1210/me.2006-0448. [DOI] [PubMed] [Google Scholar]
- Mauger O, Klinck R, Chabot B, Muchardt C, Allemand E, Batsche E. Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Chepelev I, Cui K, Zhao K. Intragenic DNA methylation modulates alternative splicing by recruiting MeCP2 to promote exon recognition. Cell Res. 2013;23(11):1256–1269. doi: 10.1038/cr.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald OG, Wu H, Timp W, Doi A, Feinberg AP. Genome-scale epigenetic reprogramming during epithelial-to-mesenchymal transition. Nat Struct Mol Biol. 2011;18(8):867–874. doi: 10.1038/nsmb.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Gerhardt DJ, Dinger ME, Crawford J, Trapnell C, Jeddeloh JA, Mattick JS, Rinn JL. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol. 2012;30(1):99–104. doi: 10.1038/nbt.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertineit C, Yoder JA, Taketo T, Laird DW, Trasler JM, Bestor TH. Sex-specific exons control DNA methyltransferase in mammalian germ cells. Development. 1998;125(5):889–897. doi: 10.1242/dev.125.5.889. [DOI] [PubMed] [Google Scholar]
- Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF) J Biol Chem. 2011;286(6):4150–4164. doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8(20):3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- Nieto MA, Cano A. The epithelial-mesenchymal transition under control: global programs to regulate epithelial plasticity. Semin Cancer Biol. 2012;22(5–6):361–368. doi: 10.1016/j.semcancer.2012.05.003. [DOI] [PubMed] [Google Scholar]
- Padua Alves C, Fonseca AS, Muys BR, de Barros ELBR, Burger MC, deSouza JE, Valente V, Zago MA, Silva WAJ. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31(12):2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- Pakala SB, Singh K, Reddy SD, Ohshiro K, Li DQ, Mishra L, Kumar R. TGF-beta1 signaling targets metastasis-associated protein 1, a new effector in epithelial cells. Oncogene. 2011;30(19):2230–2241. doi: 10.1038/onc.2010.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15(10):1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17(8):1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24(1):306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Perdigao-Henriques R, Petrocca F, Altschuler G, Thomas MP, Le MT, Tan SM, Hide W, Lieberman J. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene. 2015 doi: 10.1038/onc.2015.69. [DOI] [PubMed] [Google Scholar]
- Petrocca F, Vecchione A, Croce CM. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008;68(20):8191–8194. doi: 10.1158/0008-5472.CAN-08-1768. [DOI] [PubMed] [Google Scholar]
- Podlaha O, De S, Gonen M, Michor F. Histone modifications are associated with transcript isoform diversity in normal and cancer cells. PLoS Comput Biol. 2014;10(6):e1003611. doi: 10.1371/journal.pcbi.1003611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Loenarz C, Mole DR, McDonough MA, Gleadle JM, Schofield CJ, Ratcliffe PJ. Regulation of Jumonji-domain-containing histone demethylases by hypoxia-inducible factor (HIF)-1alpha. Biochem J. 2008;416(3):387–394. doi: 10.1042/BJ20081238. [DOI] [PubMed] [Google Scholar]
- Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, Werb Z, Bissell MJ. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razanau A, Xie J. Emerging mechanisms and consequences of calcium regulation of alternative splicing in neurons and endocrine cells. Cell Mol Life Sci. 2013;70(23):4527–4536. doi: 10.1007/s00018-013-1390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial-mesenchymal transition. J Biol Chem. 2012;287(43):36435–36442. doi: 10.1074/jbc.M112.397125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Zhang G, Li ZP, Permuth-Wey J, Challa S, Li Y, Kong W, Dan S, Bui MM, Coppola D, Mao WM, Sellers TA, Cheng JQ. Long Non-coding RNAs (LncRNA) Regulated by Transforming Growth Factor (TGF) beta: LncRNA-HIT-MEDIATED TGFbeta-INDUCED EPITHELIAL TO MESENCHYMAL TRANSITION IN MAMMARY EPITHELIA. J Biol Chem. 2015;290(11):6857–6867. doi: 10.1074/jbc.M114.610915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S, Ceballos-Chavez M, Bhattacharya SS, Reyes JC. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene. 2015 doi: 10.1038/onc.2014.446. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, Jones PA. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27(11):2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson-Dixon ND, Garcia-Blanco MA. MAZ elements alter transcription elongation and silencing of the fibroblast growth factor receptor 2 exon IIIb. J Biol Chem. 2004;279(28):29075–29084. doi: 10.1074/jbc.M312747200. [DOI] [PubMed] [Google Scholar]
- Roscigno RF, Garcia-Blanco MA. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA. 1995;1(7):692–706. [PMC free article] [PubMed] [Google Scholar]
- Savagner P, Valles AM, Jouanneau J, Yamada KM, Thiery JP. Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol Biol Cell. 1994;5(8):851–862. doi: 10.1091/mbc.5.8.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106(11):4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442(7104):772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, Fondevila D, Munoz P, Kruger M, Tischfield JA, Vaquero A. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27(6):639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan B, Yao TP, Nguyen HT, Zhuo Y, Levy DR, Klingsberg RC, Tao H, Palmer ML, Holder KN, Lasky JA. Requirement of HDAC6 for transforming growth factor-beta1-induced epithelial-mesenchymal transition. J Biol Chem. 2008;283(30):21065–21073. doi: 10.1074/jbc.M802786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7(8):e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15(17):7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Nguyen H, Geng C, Hinman MN, Luo G, Lou H. Calcium-mediated histone modifications regulate alternative splicing in cardiomyocytes. Proc Natl Acad Sci U S A. 2014;111(46):E4920–8. doi: 10.1073/pnas.1408964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shefer K, Sperling J, Sperling R. The Supraspliceosome - A Multi-Task Machine for Regulated Pre-mRNA Processing in the Cell Nucleus. Comput Struct Biotechnol J. 2014;11(19):113–122. doi: 10.1016/j.csbj.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20(13):1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479(7371):74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS, Prins JF, Lieb JD, Rathmell WK, Davis IJ. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24(2):241–250. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJr, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28(4):665–676. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31(50):5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Smith PY, Delay C, Girard J, Papon MA, Planel E, Sergeant N, Buee L, Hebert SS. MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum Mol Genet. 2011;20(20):4016–4024. doi: 10.1093/hmg/ddr330. [DOI] [PubMed] [Google Scholar]
- Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, Richardson AL, Pandolfi PP. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154(2):311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36(2):245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung MH, Guertin MJ, Baek S, Hager GL. DNase footprint signatures are dictated by factor dynamics and DNA sequence. Mol Cell. 2014;56(2):275–285. doi: 10.1016/j.molcel.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3219 doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell. 1996;84(5):801–811. doi: 10.1016/s0092-8674(00)81057-0. [DOI] [PubMed] [Google Scholar]
- Teoh PL, Sharrocks AD. WDR5, ASH2L, and RBBP5 control the efficiency of FOS transcript processing. Cell Mol Biol Lett. 2014;19(2):215–232. doi: 10.2478/s11658-014-0190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigo R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res. 2012;22(9):1616–1625. doi: 10.1101/gr.134445.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16(9):996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Toffolo E, Rusconi F, Paganini L, Tortorici M, Pilotto S, Heise C, Verpelli C, Tedeschi G, Maffioli E, Sala C, Mattevi A, Battaglioli E. Phosphorylation of neuronal Lysine-Specific Demethylase 1LSD1/KDM1A impairs transcriptional repression by regulating interaction with CoREST and histone deacetylases HDAC1/2. J Neurochem. 2014;128(5):603–616. doi: 10.1111/jnc.12457. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YP, Chen HF, Chen SY, Cheng WC, Wang HW, Shen ZJ, Song C, Teng SC, He C, Wu KJ. TET1 regulates hypoxia-induced epithelial-mesenchymal transition by acting as a co-activator. Genome Biol. 2014;15(12):513. doi: 10.1186/s13059-014-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J Cell Biol. 2010;191(1):87–99. doi: 10.1083/jcb.201001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Emburgh BO, Robertson KD. Modulation of Dnmt3b function in vitro by interactions with Dnmt3L, Dnmt3a and Dnmt3b splice variants. Nucleic Acids Res. 2011;39(12):4984–5002. doi: 10.1093/nar/gkr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147(5):1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP. Unbalanced alternative splicing and its significance in cancer. Bioessays. 2006;28(4):378–386. doi: 10.1002/bies.20390. [DOI] [PubMed] [Google Scholar]
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014;15(2):69–81. doi: 10.1038/nrg3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8(10):749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- Wang L, Charoensuksai P, Watson NJ, Wang X, Zhao Z, Coriano CG, Kerr LR, Xu W. CARM1 automethylation is controlled at the level of alternative splicing. Nucleic Acids Res. 2013;41(14):6870–6880. doi: 10.1093/nar/gkt415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhou BP. Epithelial-mesenchymal Transition---A Hallmark of Breast Cancer Metastasis. Cancer Hallm. 2013;1(1):38–49. doi: 10.1166/ch.2013.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT) Semin Cancer Biol. 2012;22(5–6):417–427. doi: 10.1016/j.semcancer.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29(19):3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell. 2009;33(5):591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Aligianni S, Codlin S, Watt S, Bahler J. Differential patterns of intronic and exonic DNA regions with respect to RNA polymerase II occupancy, nucleosome density and H3K36me3 marking in fission yeast. Genome Biol. 2011;12(8):R82. doi: 10.1186/gb-2011-12-8-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7) doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JA, Richer JK, Goodall GJ. microRNAs and EMT in mammary cells and breast cancer. J Mammary Gland Biol Neoplasia. 2010;15(2):213–223. doi: 10.1007/s10911-010-9183-z. [DOI] [PubMed] [Google Scholar]
- Wu MZ, Tsai YP, Yang MH, Huang CH, Chang SY, Chang CC, Teng SC, Wu KJ. Interplay between HDAC3 and WDR5 is essential for hypoxia-induced epithelial-mesenchymal transition. Mol Cell. 2011;43(5):811–822. doi: 10.1016/j.molcel.2011.07.012. [DOI] [PubMed] [Google Scholar]
- Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, Qiu GQ, Peng ZH, Yan DW. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32(1):395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- Yaguchi M, Wada Y, Toh Y, Iguchi H, Kono A, Matsusue K, Takiguchi S. Identification and characterization of the variants of metastasis-associated protein 1 generated following alternative splicing. Biochim Biophys Acta. 2005;1732(1–3):8–14. doi: 10.1016/j.bbaexp.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Yang F, Sun L, Li Q, Han X, Lei L, Zhang H, Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31(1):110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci. 2014;39(1):35–43. doi: 10.1016/j.tibs.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MH, Hsu DS, Wang HW, Wang HJ, Lan HY, Yang WH, Huang CH, Kao SY, Tzeng CH, Tai SK, Chang SY, Lee OK, Wu KJ. Bmi1 is essential in Twist1-induced epithelial-mesenchymal transition. Nat Cell Biol. 2010;12(10):982–992. doi: 10.1038/ncb2099. [DOI] [PubMed] [Google Scholar]
- Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, Hou P, Gao Y, Zhao L, Wang G, Zhang Y, Liu DX, Huang B, Lu J. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74(19):5656–5667. doi: 10.1158/0008-5472.CAN-14-0800. [DOI] [PubMed] [Google Scholar]
- Yearim A, Gelfman S, Shayevitch R, Melcer S, Glaich O, Mallm JP, Nissim-Rafinia M, Cohen AH, Rippe K, Meshorer E, Ast G. HP1 Is Involved in Regulating the Global Impact of DNA Methylation on Alternative Splicing. Cell Rep. 2015;10(7):1122–1134. doi: 10.1016/j.celrep.2015.01.038. [DOI] [PubMed] [Google Scholar]
- Ying L, Chen Q, Wang Y, Zhou Z, Huang Y, Qiu F. Upregulated MALAT-1 contributes to bladder cancer cell migration by inducing epithelial-to-mesenchymal transition. Mol Biosyst. 2012;8(9):2289–2294. doi: 10.1039/c2mb25070e. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cai K, Wang J, Wang X, Cheng K, Shi F, Jiang L, Zhang Y, Dou J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells. 2014;32(11):2858–2868. doi: 10.1002/stem.1795. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ma L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012;31(3–4):653–662. doi: 10.1007/s10555-012-9368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Kang Y. Multilayer control of the EMT master regulators. Oncogene. 2014;33(14):1755–1763. doi: 10.1038/onc.2013.128. [DOI] [PubMed] [Google Scholar]
- Zheng M, Jiang YP, Chen W, Li K, Liu X, Gao SY, Feng H, Wang SS, Jiang J, Ma XR, Cen X, Tang YJ, Chen Y, Lin YF, Tang YL, Liang XH. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget. 2015 doi: 10.18632/oncotarget.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lu Y, Tian W. Epigenetic features are significantly associated with alternative splicing. BMC Genomics. 2012;13123 doi: 10.1186/1471-2164-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Zhang W, Yang S, Chen JQ, Tian D. Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics. 2009;1047 doi: 10.1186/1471-2164-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ, Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, Guan XY, Xie D. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymaltransition. Gut. 2012;61(4):562–575. doi: 10.1136/gutjnl-2011-300207. [DOI] [PubMed] [Google Scholar]
- Zibetti C, Adamo A, Binda C, Forneris F, Toffolo E, Verpelli C, Ginelli E, Mattevi A, Sala C, Battaglioli E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J Neurosci. 2010;30(7):2521–2532. doi: 10.1523/JNEUROSCI.5500-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]