Abstract
Lactobacilli have been associated with a variety of immunomodulatory effects and some of these effects have been related to changes in gastrointestinal microbiota. However, the relationship between probiotic dose, time since probiotic consumption, changes in the microbiota, and immune system requires further investigation. The objective of this study was to determine if the effect of Lactobacillus casei 32G on the murine gastrointestinal microbiota and immune function are dose and time dependent. Mice were fed L. casei 32G at doses of 106, 107, or 108 CFU/day/mouse for seven days and were sacrificed 0.5h, 3.5h, 12h, or 24h after the last administration. The ileum tissue and the cecal content were collected for immune profiling by qPCR and microbiota analysis, respectively. The time required for L. casei 32G to reach the cecum was monitored by qPCR and the 32G bolus reaches the cecum 3.5h after the last administration. L. casei 32G altered the cecal microbiota with the predominance of Lachnospiraceae IS, and Oscillospira decreasing significantly (p < 0.05) in the mice receiving 108 CFU/mouse 32G relative to the control mice, while a significant (p < 0.05) increase was observed in the prevalence of lactobacilli. The lactobacilli that increased were determined to be a commensal lactobacilli. Interestingly, no significant difference in the overall microbiota composition, regardless of 32G doses, was observed at the 12h time point. A likely explanation for this observation is the level of feed derived-nutrients resulting from the 12h light/dark cycle. 32G results in consistent increases in Clec2h expression and reductions in TLR-2, alpha-defensins, and lysozyme. Changes in expression of these components of the innate immune system are one possible explanation for the observed changes in the cecal microbiota. Additionally, 32G administration was observed to alter the expression of cytokines (IL-10rb and TNF-α) in a manner consistent with an anti-inflammatory response.
Introduction
The human gastrointestinal tract hosts over 1014 cells made up of 500 to 1000 bacterial species collectively known as the human microbiom. The human microbiome is believed to play an important role in health and disease [1,2]. The gut microbiota is believed to serve as an organ which takes a part in many physiological and homeostatic functions [3–6]. It has been shown that a well balanced bidirectional interaction between the microbiota and the immune system exist. This complex relationship begins at birth, and has been shown to have a fundamental role in development and maturation of the immune system, but at the same time the immune system plays a role in shaping the microbiota composition and functions that will last a lifetime [3,7,8]. Disfunction in the balance has been linked to many diseases including inflammatory diseases such as inflammatory bowel disease (IBD) [9,10]. There are several interventions likely to modify the indigenous intestinal microbiota, including diet and antibiotics. Probiotics are one of the diet related interventions that have been shown to alter the microbial composition of the gut. Probiotics are live microorganisms, which when administered in adequate amounts, confer a health benefit on the host [11]. Probiotics have potential to influence gut dysbiosis and they have also been shown to help maintain immune health[12,13]. For example, C. difficile infection is believed to be caused by a collapse of the microbial community in the gut after an antibiotic assault that imbalances the microbiota [14,15]. Many patients return to the hospital with a recurrence of C. difficile infections. Probiotics could be a useful approach in restoring and supporting the “good” community in the gut, helping to restore balance the intestinal microbiota. A diverse and rapidly expanding set of health benefits have been ascribed to probiotics including: improved ability to tolerate lactose; reduction in gastrointestinal pathogens; reduction in colorectal cancer; decrease in incidence of cold and flu; and a reduction in the symptoms associated with the inflammation-related disorders, such as ulcerative colitis [16–18]. Probiotics come from a variety of genera, including Lactobacillus, Bifidobacterium, Propionibacterium, Escherichia, and Saccharomyces; however, the most commonly used strains are from the genus’ Lactobacillus and Bifidobacterium [19].
Lactobacillus are a component of the gut microbiota, are one of the major genera commonly used as probiotics and numerous health benefits have been associated with their use as probiotics [17,19–22] While a detailed mechanistic understanding of the probiotic effects of lactobacilli is lacking, it has commonly been thought that changes in the host gut microbiota are one reason for these beneficial health outcomes [23,24]. For example, Bruzzese et al. linked the consumption of Lactobacillus GG with improved cystic fibrosis and changes in the gut microbiota [24]. However, conflicting results concerning the linkage of changes in the gut microbiota and probiotic-related health impacts have also recently been reported [25]. A 2014 study by Aoki et al. demonstrated that L. casei Shirota helps to improve aberrant bowel movements in patients with gastrectomies. Interestingly, no change in the subjects’ fecal microbiota was detected [22]. Similarly Eloe-Fadrosh et al. demonstrated that consumption of L. rhamnosus GG administration to 12 health individuals, 65 to 80 years old, had an anti-inflammatory effect and linked this outcome not with changes in the composition of the gut microbiota, but rather with changes in activity of the commensal microbiota [23]. Understanding the seemingly contradictory results from these studies will require an understanding of the mechanism(s) by which probiotics alter the composition of the gut microbiota
Lactobacillus casei is a commonly utilized probiotic species, with more than 32 thousands servings per day consumed worldwide through dairy products like Yakult and DanActive [26]. L. casei are Gram (+), facultatively anaerobic, industrially important lactic acid bacteria that have been isolated from a variety of diverse habitats, including fermented dairy products (i.e., cheese) and plant materials (i.e., wine, pickle, and silage,), as well as the gastrointestinal (GI) tract, and oral cavity of humans and animals [27]. The L. casei species has been analyzed by Multilocus Sequence Typing (MLST) and determined to diverge into three major lineages approximately 1.5 million years ago [28]. Previously, our research group screened four different L. casei strains for their ability to survive the GI tract passage, adhere to ileum epithelium, and influence the intestinal microbiota in a piglet and a murine model [29]. L. casei 32G was found to survive passage through the GI tract and modify the ileum microbiota in both animal models. In this study, we monitored the abundance of the fed strain at different time points in the cecum and examined the impact of L. casei 32G dose and time since probiotic consumption on the mouse commensal cecal microbiota using 16S rRNA sequencing. Due to the well-described relationship between the gut microbiota and host immunity, we also examined the relationship between the probiotic strain, gut microbiota, and immune function.
Material and Methods
Bacterial strain
L. casei 32G was maintained at -80°C in MRS broth (BD Difco, Sparks, MD) with 25% (v/v) glycerol (Sigma-Aldrich, St. Louis, MO). Working cultures were prepared from frozen stocks by two sequential transfers in MRS broth and incubations were conducted statically at 37°C for 24 h and 18 h, respectively. The culture at early stationary phase was harvested by centrifugation at 5,000 rpm for 10 min at room temperature. The pellet was re-suspended in 0.85% NaCl (w/v) and the optical density at 600 nm (OD600) determined. A volume of washed cells (based upon the OD600) sufficient to yield a 5 ml cell suspension with an OD600 of 6.0 was harvested by centrifugation at 5,000 rpm and washed with 5 ml of 0.85% NaCl. The resulting pellet was suspended in 5 ml of 0.85% NaCl to obtain a final concentration of 109 CFU/ml. The culture was serially diluted in 0.85% NaCl to reach concentrations of 108 and 107 CFU/ml. The final culture solution that was kept at 4°C until the administration was enumerated daily on MRS agar to confirm the dose administered to the mice.
Animals
All procedures involving mice were conducted under the protocol #V01548 approved by the Animal Care and Use Committee of the University of Wisconsin-Madison. Healthy, male C57BL/6 mice aged 8 weeks were obtained from Jackson Laboratories (Bar Harbor, ME) and group housed at University of Wisconsin-Madison Animal Health and Biomedical Science facility. Housing conditions were controlled at 25°C, 20–44% relative humidity with a 12 h light/dark cycle. Mice were fed ad libitum water and mouse chow (Harlan Teklad 7964 rodent diet, Madison, WI) throughout the study. The animals (n:96) included in this study were divided into 5 groups; each group (n:30) was administered daily 100 μl of either 0.85% NaCl (control), 107, 108, or 109 CFU/ml of L. casei 32G by oral gavage for seven days. Therefore, the delivered doses were 106 (low dose), 107 (medium dose), and 108 (high dose) CFU/day/mouse. L. casei 32G was fed for 7 days to simulate long-term daily consumption of a probiotic, as is typically suggested for probiotic consumption.
Sample collection
Six mice from each group were euthanized by CO2 asphyxiation at 0.5h, 3.5h, 12h, 24h and 72h after administration of the last probiotic dose to evaluate the effect of time and immediately after euthanization the intestinal tract was removed for analysis. The cecum content was collected and the samples were immediately put on ice, and then frozen at -20°C until processed for microbial DNA extraction. Approximately 2 cm-tissue from the distal ileum was collected for RNA isolation and preserved in RNAlater (Ambion, Carlsbad, CA) overnight at 4°C. After the overnight treatment, the samples were stored at -80°C until processing.
Determination of lactic acid content in the cecum content
The amount of D- and L-lactic acid in the cecum digesta was determined using D-Lactic acid/L-lactic acid kit from R-Biopharm (Darmstadt, Switzerland). A 0.9 ml sample of digesta was centrifuged at 20,000 ×g for 1 h and the supernatant was recovered. Proteins present in the supernatant were precipitated using the Carrez clarification reagent (85 mM K4[Fe(CN)6] and 250 mM ZnSO4) as described for the D-Lactic acid/L-lactic acid kit and the pH adjusted to 8.0 using 1N NaOH. Subsequently, D- and L-lactate were quantified as directed by the supplier, except that the total volume of the assay was decreased from 3 ml to 600 μl, while maintaining the proportions described in the manufacturer’s instructions for each reagent.
DNA extraction
The cecum digesta was homogenized in 1.5ml of PBS and total DNA from 200 μl of the homogenate was isolated using the QIAamp DNA Stool Mini Kit (Qiagen Sciences, MD) with modifications to the manufacturer’s instructions. These modifications included an initial mechanical cell disruption step by inclusion of 0.1 mm glass beads (Sigma-Aldrich) followed by exposure to six 1 min beating at maximum speed in a Mini-beadbeater-96 (Biospec Products, Inc., Bartlesville, OK) with intervals of 2 min on ice. Subsequently, a heat treatment step was performed for 5 min at 95°C. The DNA was further purified by phenol:chloroform:isoamyl alcohol (25:24:1, pH 8) extraction, phase separation using Phase Lock Gels (5 PRIME) and ethanol precipitation using pellet paint co-precipitant (EMD Millipore). DNA was quantified by Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA). Extracted DNA was used to perform 16S rRNA sequencing and L. casei 32G detection by qPCR.
Ion Torrent PGM Sequencing and Microbiota Analysis
Partial 16S rRNA sequences were determined on a 318 v2 chip using the Ion Torrent Personal Genome Machine System at University of Wisconsin-Madison, Biotechnology Center. Briefly, the V1-V2 region was amplified using forward primers that contained a sample-specific bar-code with an Ion A adapter and a key sequence, while the associated reverse primer contained a truncated P1 (trP1) adapter. The sequence of these primers were: forward (8FM—5'–CCA TCT CAT CCC TGC GTG TCT CCG AC T CAG BBB BBB BBB BBB BAG AGT TTG ATC MTG GCT CAG—3') with the Ion A adapter in italics, the key sequence in italics and underlined, the 13 bp bar code designated as Bs, and the 16S primer sequence in capital letters; reverse (357R - 5'–CCT CTC TAT GGG CAG TCG GTG ATC TGC TGC CTY CCG TA- 3') with the trP1 adapter in italics and the 16S primer sequence in capital letters. All PCR reactions were quality-controlled for amplicon saturation by gel electrophoresis. Equal quantities of each of the amplicons were pooled and purified using AxyPrep Mag PCR beads (Corning, Inc.). The resulting products were quantified using PicoGreen (Invitrogen) and Qubit fluorometer (Invitrogen) before sequencing. The sequences were deposited in the NCBI BioProject database with the study identification number SRP062166. The data processing pipeline removed low-quality reads that: 1) did not completely match the PCR primer and barcode; 2) were shorter than 300 bp or longer than 400 bp in length; or 3) had an average quality score <22. Data analysis was performed in QIIME 1.8 framework [30]. Operational Taxonomic Units (OTUs) were chosen with QIIME picking OTU workflow based upon sequence similarity with a 97% similarity threshold. Taxonomic identities were assigned using greengenes version 13_5 [31].
Lactobacillus sequences extracted from the microbiota dataset were blasted against NCBI database to identify the species with highest identity.
RNA isolation and Gene Expression Analysis
Tissue samples from the distal small intestine were homogenized in UltraPure guanidine isothiocyanate solution (Invitrogen) using a tissue grinder with a smooth pestle (Thomas Scientific, Swedesboro, NJ). RNA was isolated using PureLink RNA mini kit (Invitrogen) as recommended by the supplier. Concentrations and purity of RNA samples were determined with a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA) and the integrity was checked by the control reaction included in the Bio-Rad prime PCR assay. Total RNA was treated with DNase I (Invitrogen) to remove DNA contamination and subsequently converted into cDNA using iScriptTM cDNA synthesis kit (Bio-Rad, Hercules, CA) according to manufacturer’s protocol. qPCR was performed using the primers shown at Table 1 and the customized 96-well prime PCR assays (Bio-rad) were used to screen 29 different genes of interest. SsoFast™ EvaGreen® Supermix (Bio-Rad) was used under the following conditions: initial denaturation at 95°C for 2 min, followed by 40 cycles of 5 sec at 95°C and 30 sec at 60°C. Data were acquired in the final step at 95°C for 5 sec and melting curves (65 to 95°C) were generated at the end for each set of primers. Gene expression was normalized to β-actin and relative gene expression was calculated by 2-ΔΔCt method [36].
Table 1. Primers used in qPCR analysis.
| Gene * | Forward | Reverse | Reference |
|---|---|---|---|
| Defa-rs1c | 5'-CACCACCCAAGCTCCAAATACACAG-3' | 5'-ATCGTGAGGACCAAAAGCAAATGG-3' | [32] |
| Defcr1 | 5’-TCAAGAGGCTGCAAAGGAAGAGAAC-3’ | 5’-TGGTCTCCATGTTCAGCGACAGC-3’ | [32] |
| Defcr4 | 5’-CCAGGGGAAGATGACCAGGCTG-3’ | 5’-TGCAGCGACGATTTCTACAAAGGC-3’ | [32] |
| Lyz1-2 | 5’-GCCAAGGTCTACAATCGTTGTGAGTTG-3’ | 5’-CAGTCAGCCAGCTTGACACCACG-3’ | [32] |
| Pla2g2 | 5’-AGGATTCCCCCAAGGATGCCAC-3’ | 5’-CAGCCGTTTCTGACAGGAGTTCTGG-3’ | [32] |
| sPLA2-IIA | 5’-ACAGGTCCAAGGGAACATTG-3’ | 5’-TCTGGTTTGCAGAACAGGTG-3’ | [33] |
| Occludin | 5’-CCCTGACCACTATGAAACAG-3’ | 5’-TTGATCTGAAGTGATAGGTG-3’ | [34] |
| ZO-1 | 5’-CCTAAGACCTGTAACCATCT-3’ | 5’-CTGATAGA- TATCTGGCTCCT-3’ | [34] |
| ZO-2 | 5’-CTAGACCCCCAGAGCCCCAGAAA-3’ | 5’-TCGCAGGAGTCCACGCATACAAG-3’ | [35] |
* The common names: cryptdin-related sequence (Defa-rs1c); cryptdin-1 (Defcr1); cryptdin-4 (Defcr4); Lysozyme (Lyz1); zonula occludens (ZO).
Lactobacillus casei 32G detection by qPCR
Total DNA extracted from cecum digesta was used to detect L. casei 32G, the fed microorganism. A deletion in the 32G dltX gene was identified and a strain specific primer to the junction on the deletion was designed. The primer set was checked for sequence similarity by the BLAST (NCBI). The sequences of the primers are as follows; 5′- AAG TGA ACA GAC ACG CAT CG -3′ and 5′- AAC GCC TGT CAG CTT CAT CT-3′. The primers were tested by qPCR with an annealing temperature gradient. The following conditions were applied: initial denaturation at 95°C for 30 sec, followed by 40 cycles of 5 sec at 95°C and 5 sec at 57°C. A melting curve was generated at the end for the primer set. Samples considered positive for the presence of 32G yielded a single sharp melting curve. A matrix based standard curve was created. The cecum content suspended with PBS was spiked with 108, 107, 106, 105, 104 and 102 CFU/ml of L. casei 32G culture, which was confirmed by enumerating on MRS, and DNA was extracted from each sample. DNA was amplified by qPCR in triplicate.
Statistical analysis
For microbiota data, the statistical difference between treatments was examined using the Monte-Carlo test in package ade4 [37] of R 2.14.0 [38] as described by de Carcer et al [39]. The values of zero were replaced with the detection limit, which is determined by the ratio of one to the lowest read number in the data set. The dominant genera that increased or decreased in abundance were identified by running the correspondence analysis provided in the package ade4 of R 2.14.0 as described by de Carcer et al [39]. The Benjamini-Hochberg procedure was applied to control the false discovery rate. Statistical difference for relative gene expression was assessed with the Wilcoxon rank sum test (Mann–Whitney test) using JMP version 10 (SAS Institute Inc., Cary, NC) and was presented as mean ± SEM. Statistical difference was determined at a P value of 0.05 or less.
Results and Discussion
Detection of Lactobacillus casei 32G in mice after oral administration
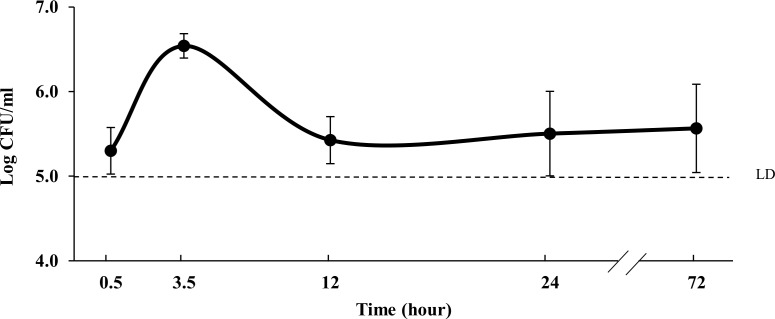
To initiate research on the mechanism(s) by which L. casei 32G influences cecal microbiota composition and modulates the immune system, transit through the murine GI tract was followed using qPCR. A qPCR primer set specific to 32G was designed based on the publically available genome on NCBI. To determine the limit of detection and establish a standard curve, cecum content obtained from control mice were spiked with known quantities of L. casei 32G. 32G was detected in samples containing 105 CFU/ml or greater quantities of 32G, indicating that this was the limit of detection of the method (S1 Fig). To evaluate GI transit of 32G, cecum samples were collected from mice fed with the high dose (108 CFU/day) of L. casei 32G at different time points after the last administration. L. casei 32G was detected in all time points at levels ranging from 5.3 to 6.5 log CFU/ml, with the highest concentration being observed at the at the 3.5h time point (Fig 1). It is important to note that this analysis does not provide information concerning the viability of the organisms detected. These results are similar to those described by Daniel et al., who examined transit of L. plantarum using bioluminescence imaging. Their results indicate that L. plantarum took about 90 min to reach the cecum and localized there until 4h after the last administration [40]. L. planatarum was also monitored previously by Marco et al. by enumerating lactobacilli in mouse feces and by measuring the level of 16S rRNA in gut compartments after a single oral administration; however, L. plantarum was not selectively monitored [41]. To determine if 32G could be detected from all three dose levels, qPCR analysis was conducted with the 3.5h cecum samples from all three doses. L. casei 32G was detected in 0, 2, and 4 of the 4 samples examined from the 106, 107, and 108 CFU/day samples, respectively (S1 Table). These results demonstrate that only mice fed a high dose of 32G contained greater than 105 CFU/ml of 32G in their cecal contents and that 32G was present at the highest level 3.5 h after administration.
Fig 1. L. casei 32G detection in cecum digesta of mouse fed with 108 CFU/mouse at 0.5h, 3.5h, 12h, 24h, and 72h after last administration.
(n:6/time point). LD, limit of detection.
Lactobacillus casei 32G alters the cecal microbiota composition depending on dose and time
The interaction between the commensal GI microbiota and immune system is an area of intense research interest [4,12]. This interest has resulted in a rapid increase in the number of investigations on the impact of probiotics on the GI microbiota; however, only rarely has effect of dose and time since probiotic consumption been examined. In this study, we examined the impact of L. casei 32G dose and time since probiotic consumption on the mouse commensal cecal microbiota using 16S rRNA sequencing. Ion Torrent PGM sequencing of cecal content was conducted to assess the influence of L. casei 32G dose and time on the mice GI microbiota. The sequencing resulted in a total of 2,239,643 filtered reads from 96 mice cecum digesta samples; the number of reads varied from 7,535 to 42,559 with an average of 18,664 reads per sample. After the taxonomic status of each read was assigned, 9 phyla, 16 classes, 29 orders, 48 families, 72 genera were identified. To assess whether sufficient sequence reads had been collected to accurately determine the diversity of organisms present, shannon and chao1 index were examined; the results of this analysis are presented in S2 Fig. These results indicate that sufficient sequence reads were obtained to accurately describe the diversity present in these samples. Additionally a principal coordinate analysis (PCA) plots using weighted Unifrac distances were generated to provide a visual overview of gut microbial dynamics in response to administration of L. casei 32G (S3 Fig).
The cecum microbiota of the mice fed with 32G at 106 (low), 107 (medium) and 108 CFU/day (high) doses at different time points after the last administration were compared with the control mice at genus level. As determined by Monte-Carlo analysis, the overall microbiota of the 32G-fed mice did not differ significantly (p < 0.05) from the controls for the low and the medium dose. However, the overall microbiota differed significantly (p < 0.05) at the 3.5h time points for high dose, (Fig 2). The mouse-to-mouse variation in the cecum microbiota is presented in S4 Fig. The dominant genera that increased or decreased in abundance were identified by correspondence analysis. When the samples were compared at the genus level by correspondence analysis, significant (p < 0.05) changes were observed between all samples and their controls, with a 12, 23, and 25 changes at the genus level for the 106, 107, and 108 CFU/day doses, respectively (Table 2). These results demonstrate that 32G dose has a significant (p < 0.05) influence on the composition of the cecum microbiota.
Fig 2. Comparison of predominant genera in the microbiota of mice cecums sorted based on time.
(A) The major microbial communities of mouse cecum content at genus level in the control group and L. casei 32G groups; 106 CFU/ mouse (low), 107 CFU/ mouse (medium) and 108 CFU/ mouse (high), at 0.5, 3.5, 12, and 24 h after the last administration. Only genera with over 5% of the total bacteria are presented (n: 6 for each bar). (B) Pair wise comparison of each group. Statistical p-values were assessed using a Monte-Carlo test with 10000 replicates. The values with p≤0.05 are highlighted.
Table 2. Bacterial genera detected a in cecum content of mice administered saline (control) or Lactobacillus casei 32G once per day by oral gavage sorted based on time.
| Taxon | Percentage (mean ± SE) b c | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5h | 3.5h | 12h | 24h | |||||||||||||
| Control | Low | Medium | High | Control | Low | Medium | High | Control | Low | Medium | High | Control | Low | Medium | High | |
| S24-7 IS | 66.2±15.2 | 61.4±27.7 | 70.4±13.0 | 64.3±25.3 | 75.0±15.8 | 81.3±17.5 | 65.9±5.9 | 69.7±15.6 | 84.6±10.3 | 86.2±5.9 | 78.8±20.3 | 74.1±14.0 | 53.6±22.0 | 70.8±20.7 | 52.9±31.2 | 60.9±18.1 |
| Lachnospiraceae IS | 15.9±9.9 | 26.1±25.0 | 13.8±9.6 | 13.2±8.5 | 8.9±7.2 | 8.0±10.2 | 13.9±8.3 | 2.7±1.6 | 7.1±6.0 | 3.3±2.5 | 12.2±14.6 | 8.8±8.4 | 28.8±19.7 | 12.3±11.3 | 10.7±12.2 | 10.2±6.7 |
| Oscillospira | 10.0±4.7 | 3.8±1.6 | 5.9±3.9 | 4.3±4.1 | 3.3±3.0 | 2.7±3.6 | 2.0±1.3 | 0.8±0.6 | 2.4±2.8 | 1.0±0.6 | 2.1±2.7 | 1.9±1.6 | 7.2±3.3 | 7.1±6.2 | 4.9±4.9 | 4.2±3.1 |
| Ruminococcaceae Other | 2.1±1.2 | 3.2±2.3 | 1.2±0.8 | 2.4±1.9 | 0.6±0.3 | 1.4±0.6 | 0.5±0.2 | 1.5±1.0 | 2.0±1.4 | 3.3±1.9 | 2.0±2.1 | 8.3±4.1 | 2.3±1.4 | 3.2±1.9 | 2.6±2.5 | 2.2±0.9 |
| Ruminococcus | 1.3±0.6 | 1.1±0.7 | 0.4±0.2 | 1.2±1.4 | 0.7±0.4 | 0.5±0.4 | 0.2±0.1 | 0.7±0.8 | 1.0±0.5 | 1.9±1.4 | 0.7±0.8 | 1.2±0.7 | 0.8±0.3 | 1.0±0.3 | 0.6±0.5 | 1.4±1.2 |
| Clostridia Other | 1.2±0.8 | 1.0±0.6 | 0.4±0.4 | 0.7±0.7 | 0.3±0.1 | 0.6±0.6 | 0.2±0.1 | 0.2±0.1 | 0.5±0.5 | 0.6±0.3 | 0.3±0.2 | 0.6±0.5 | 0.9±0.3 | 0.8±1.2 | 0.7±0.7 | 0.8±0.6 |
| Lactobacillus | 0.9±1.6 | 0.5±0.9 | 5.0±4.6 | 11.0±16.4 | 6.8±7.7 | 3.6±6.9 | 15.0±9.6 | 22.2±14.6 | 0.4±0.5 | 0.6±0.9 | 0.6±0.4 | 1.9±2.7 | 3.7±4.3 | 1.1±1.0 | 24.8±17.3 | 16.9±19.5 |
| Lachnospiraceae Other | 0.9±0.4 | 0.9±0.5 | 0.4±0.2 | 0.7±0.6 | 0.3±0.2 | 0.3±0.4 | 0.2±0.1 | 0.2±0.1 | 0.4±0.5 | 0.7±0.8 | 0.3±0.2 | 0.6±0.5 | 0.8±0.4 | 1.1±1.5 | 0.5±0.5 | 0.8±0.4 |
| Clostridiales Other | 0.6±0.3 | 0.6±0.4 | 0.5±0.4 | 0.4±0.4 | 0.1±0.1 | 0.3±0.3 | 0.2±0.1 | 0.1±0.1 | 0.3±0.3 | 0.2±0.1 | 0.3±0.2 | 0.6±0.5 | 0.6±0.3 | 0.5±0.3 | 0.6±0.8 | 0.4±0.2 |
| Ruminococcaceae IS | 0.2±0.1 | 0.3±0.4 | 0.9±0.6 | 0.7±0.9 | 0.3±0.2 | 0.2±0.3 | 0.6±0.3 | 0.8±0.4 | 0.2±0.1 | 0.6±0.7 | 1.0±0.6 | 0.8±0.5 | 0.2±0.1 | 0.4±0.2 | 0.4±0.3 | 0.5±0.2 |
| Firmicutes Other | 0.2±0.1 | 0.1±0.2 | 0.2±0.1 | 0.2±0.2 | 0.2±0.1 | 0.2±0.2 | 0.2±0.1 | 0.2±0.1 | 0.2±0.2 | 0.2±0.1 | 0.2±0.1 | 0.1±0.1 | 0.2±0.1 | 0.4±0.3 | 0.4±0.2 | 0.4±0.2 |
| Bacteria Other | 0.2±0.0 | 0.1±0.1 | 0.3±0.1 | 0.2±0.1 | 0.2±0.1 | 0.2±0.1 | 0.4±0.2 | 0.3±0.1 | 0.2±0.1 | 0.1±0.0 | 0.2±0.1 | 0.4±0.2 | 0.2±0.1 | 0.2±0.1 | 0.3±0.1 | 0.5±0.1 |
| Erysipelotrichaceae IS | 0.1±0.1 | 0.2±0.2 | 0.1±0.1 | 0.0±0.0 | 0.0±0.0 | 0.1±0.0 | 0.1±0.1 | 0.0±0.0 | 0.1±0.1 | 0.1±0.1 | 0.2±0.2 | 0.0±0.0 | 0.0±0.0 | 0.1±0.1 | 0.1±0.0 | 0.1±0.0 |
| Turicibacter | 0.0±0.1 | 0.1±0.2 | 0.0±0.0 | 0.3±0.4 | 2.7±2.6 | 0.2±0.2 | 0.1±0.2 | 0.4±0.7 | 0.4±0.5 | 0.8±0.9 | 0.8±1.1 | 0.4±0.5 | 0.1±0.1 | 0.3±0.3 | 0.0±0.0 | 0.0±0.1 |
| Coprobacillaceae IS | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.1±0.1 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 0.2±0.2 | 0.0±0.0 | 0.0±0.0 |
| Adlercreutzia | BQL | 0.1±0.1 | 0.2±0.1 | 0.0±0.0 | 0.2±0.5 | 0.1±0.2 | 0.3±0.2 | 0.1±0.0 | 0.1±0.2 | 0.1±0.2 | 0.2±0.1 | 0.0±0.0 | 0.1±0.0 | 0.2±0.3 | 0.2±0.2 | 0.1±0.1 |
| Coriobacteriales IS | BQL | BQL | BQL | BQL | 0.0±0.0 | BQL | BQL | BQL | BQL | BQL | BQL | BQL | 0.0±0.0 | 0.0±0.0 | BQL | 0.2±0.2 |
| Number of alterations d | - | 2 | 8 | 4 | - | 3 | 6 | 6 | - | 2 | 2 | 6 | - | 5 | 7 | 9 |
aOnly genera that were present at ≥1% in a sample are included in this table.
bThe detection limit was 0.00003 and this value was used to calculate the p-value.
cGenera that differ from control within each group are shown in bold (p≤0.05).
dThe number of genera that differed from the control for that treatment.
IS: Incertae Sedis.
BQL: Below quantifiable limit.
The most abundant genus found in the cecum content of all of the samples was Bacteroidales S24-7 Incertae Sedis (IS), a poorly studied genus, which accounted for 53%-86% of the total microbiota. The dominant genera (> 3% of the total microbiota) in rank order of the cecum microbiota in the control mice were S24-7 IS, Lachnospiraceae IS, Oscillospira and Lactobacillus, together these genera comprise 93% of the total microbiota (Table 2). Our qPCR results demonstrated that 32G reaches the cecum 3.5h after administration; therefore, the influence of the 32G administration on the mouse cecum microbiota was evaluated with the 3.5h samples. The high dose samples were selected as this dose resulted in the largest number of alterations to the cecum microbiota. The predominance of Lachnospiraceae IS, and Oscillospira decreased significantly (p < 0.05) in the mice receiving 108 CFU/mouse 32G relative to the control mice; in contrast, a significant (p < 0.05) increase was observed in the prevalence of Lactobacillus (Table 2, Fig 2). Lachnospiraceae IS, a butyrate producing superfamily, decreased from 8.9% to 2.7% [42,43]. A decrease in Lachnospiraceae IS was also observed in our previous study done on 32G fed piglets [29]. Lachnospiraceae are highly abundant in human gut [43] and are known to decrease the severity of colitis and the degree of Clostridium difficile colonization [44,45]. Ravussin et al. reported that the greatest difference in cecal microbiota between diet induced obese mice and the control mice was the level of Lachnospiraceae, which were found at a much higher level in obese mice [46]. Oscillospira is the other genus that decreased (from 3.3% to 0.8%) at the 3.5h time point as a result of 32G administration. Oscillospira, an uncultured group, are normally found in rumen of cattle and sheep, have been detected in human large intestine [47], and have been shown to be depleted in obese patients [48]. Lactobacillus increased (from 6.8% to 22.2%) significantly (p < 0.05) in high dose mice at 3.5h, in comparison to their control mice [49]. In IBS patients, commensal Lactobacillus are depleted; similarly stress has been observed to result in decreases in lactobacilli in animals [5]. Previous studies have observed that Lachnospiraceae and Oscillospira prevalence typically increase or decrease together and that Lactobacillus typically changes in opposite direction [50,51], as was observed in this study. There are a number of possible explanations for these observations including: microbe-microbe interactions; microbe-host interactions; and the impact of diet on the composition of the microbiota.
Although this study was not designed to evaluate the influence of the 12h dark/light photoperiodic cycle on the composition of the cecum microbiota, the lack of significant differences in the overall microbiota composition, regardless of 32G dose, of all samples at 12h was compelling (Figs 2 and S5, S2 Table). Previous researchers have demonstrated that C57BL/6 mice, the same strain used in this study, had their highest food consumption during early dark cycle when fed ad libitum [52,53]. Based on the 24-hour Zeitgeber time units (ZT), where the lights are turned on as ZT0 and off at ZT12, our 12h time point corresponds to ZT13, which is four hours after increased food intake starts. At this time point the microbiota composition likely reflects the presence of a low level of feed-derived nutrients, as time is required for both transit of the food and changes in microbial composition. This suggests that the light/dark cycle has a significant impact on the composition of the cecum microbiota, likely due to the level of feed derived-nutrients [54], and hence must be taken into consideration when designing experiments that follow microbiota composition.
The increase in prevalence of Lactobacillus in the intestinal microbiota was not directly due to the fed microorganism, Lactobacillus casei 32G
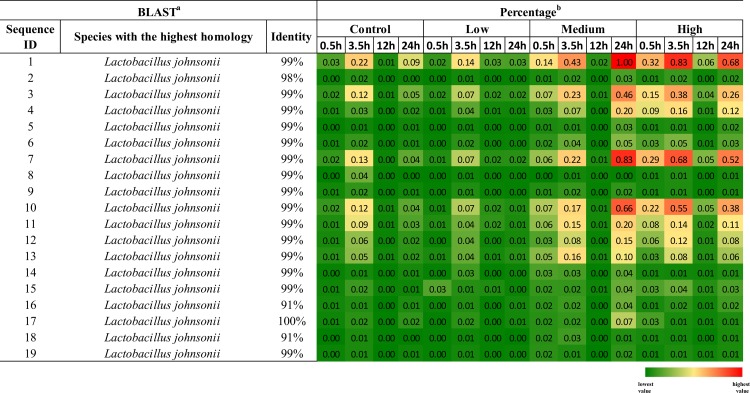
Lactobacillus was observed to increase at all time points other than 12h in mice fed 107 or 108 CFU L. casei 32G. This increase could either be due directly to the presence of 32G or indirectly by 32G altering the conditions present in the cecum, thereby allowing the commensal lactobacilli to increase in prevalence. To investigate whether the lactobacilli that increased in prevalence in the intestinal microbiota were L. casei, we extracted the representative genera sequences from the QIIME pipeline. Nineteen distinct Lactobacillus sequences were present more than 500 times in the overall dataset, these sequences were blasted against the NCBI database and the species with the highest homology are presented in Fig 3. In all cases, Lactobacillis johnsonii was the species with the highest homology to the sequences present in these samples, although the sequence identity was relatively low (91%) for two of the Lactobacillus sequences. L. casei sequence identity with these reads varied from 80 to 84% and there were always at least 10 species with higher identity than L. casei. These findings suggest that the Lactobacillus which increased in prevalence were L. johnsonii, not L. casei 32G. L. johnsonii are commensal lactobacilli present in the murine gut [55,56]. One explanation of these results is that feeding of L. casei 32G at 107 and 108 CFU/mouse daily altered the environment present in the cecum such that the growth of commensal lactobacilli was favored.
Fig 3. The species of Lactobacillus detected in the cecums of L. casei fed mice.
The species of Lactobacillus with the highest 16S rRNA sequence identity to those present in the cecums of L. casei fed mice Lactobacillus sequences detected were blasted against NCBI database to identify the species with the highest identity (a). The heat map depicts the relative abundance of Lactobacillus sequences detected in the cecums of L. casei fed mice. The percentage was calculated from the average values for each sample (n:6/time point) and a graded color scale is utilized to visualize the relative abundance (b).
Lactic acid concentration and pH of the cecum content
Intestinal pH is one of the factors involved in modification of gut microbiota [1,39,57,58]. The pH of the cecum digesta obtained from each group of mice is presented in S3 Table. Interestingly, there was no significant difference in the mice fed with L. casei 32G, when compared to the control mice, at any dose at 3.5h time point, when the 32G bolus passes the cecum. Considering that lactic acid is the primary metabolic end product of L. casei carbohydrate fermentation [59], we measured the lactate concentration in the cecum samples collected from the mice subjected to the high dose 32G at 3.5h. There were also no significant differences in the D-lactate, L-lactate or total lactate concentrations (data not shown). Moreover, there was a slight increase in the pH of the samples from the medium (0.5 and 24h) and high dose (0.5h) which could be result of products by commensals in the gut. These results suggest that neither pH nor lactic acid had a major role in the observed 32G associated changes in cecal microbiota.
Intestinal innate immune profile associated with L. casei 32G administration
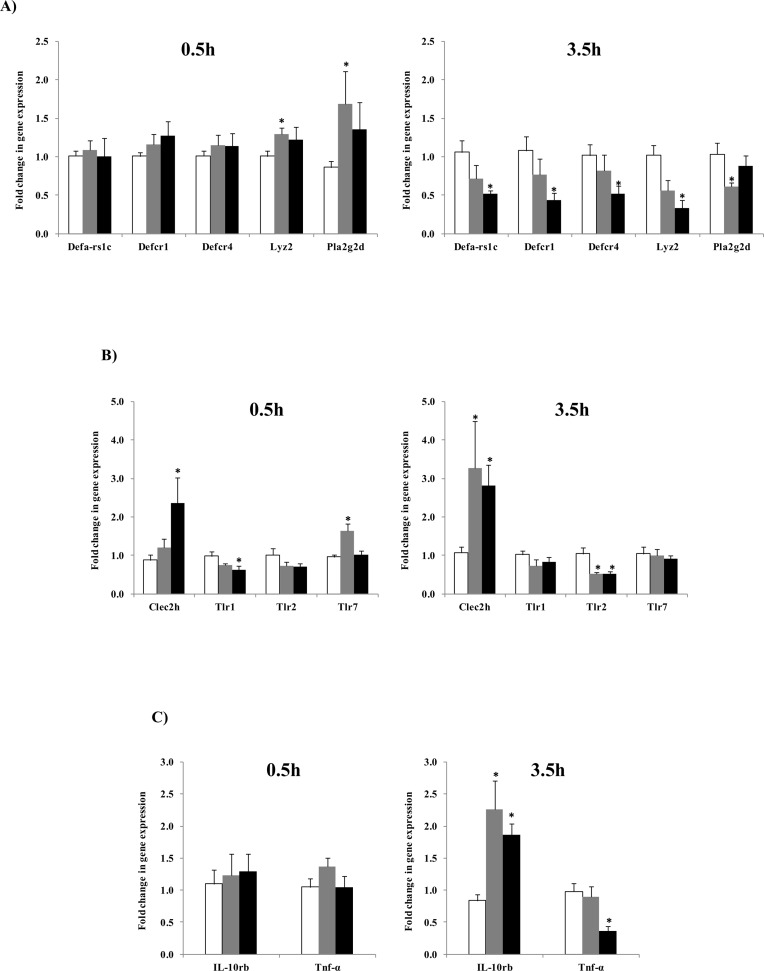
The relationship between intestinal microbiota and host immune system functions in two directions. Typically, this interaction is well balanced and a break down in balance can lead to gastrointestinal inflammation and metabolic disorders [60]. Probiotics have been shown to alter the expression of genes involved in host innate immunity [61,62], hence we investigated influence of L. casei 32G on the innate immune response. Total RNA was isolated from ileal tissue to screen for changes in genes encoding components of the innate immune system resulting from 32G administration. To examine the influence of dose we included mice receiving the low (106 CFU/mouse) and high (108 CFU/mouse) dose of 32G. To examine the influence of time we chose to focus on the 0.5 and 3.5 h time points, as these were the times the 32G bolus reached the cecum (3.5h) and the longest time since the innate immune system had encountered a high level of 32G (0.5h). We targeted 38 genes associated with the innate immune system. Eight of the genes (IL-12a, IL-12b, IL-12rb1, IL-12rb2, IL-22, Pla2g1b, Ifng, and TLR6) were below the limit of detection in all samples. Eighteen of the targeted genes (Lyz1-2, Pla2g2, sPLA2-IIA, Occludin, ZO-1, IL-10ra, Lyz1, Nf-kB1, Pla2g10, Pla2g12a, Reg3β, Reg3γ, Tgfβ1, TLR3, TLR4, TLR5, TLR8 and TLR9) showed no statistical differences from the control in all samples (data not shown). Twelve of the targeted genes (Defa-rs1c, Defcr1, Defcr4, Lyz2, Pla2g2d, Clec2h, Tlr1, Tlr2, Tlr7, IL-10rb, Tnf-α, and ZO-2) were expressed statistically (p<0.05) different from the control mice in some samples and are presented in Fig 4.
Fig 4. Fold change in gene expression of target genes of the mouse ileum in the control group (white bar) and L. casei 32G groups; Low dose, 106 CFU/ mouse (grey bar) and High dose, 108 CFU/ mouse (black bar) at 0.5h and 3.5h after the last L. casei 32 administration.
(A) Change in AMPs, (B) Change in PRRs, (C) Change in cytokines; * p<0.05: significant differences from the control, (n: 6/group).
Intestinal cells sense microbial ligands through pattern recognition receptors (PRRs) that contribute to cross-talk between the gut microbiota and the innate immune system [63]. We screened intestinal PRRs including Toll-like receptors (TLRs) and C-type lectin-like receptor 2h (Clec2h) to evaluate the effect of 32G on PRRs. Levels of TLR3, 4, 5, 8 and 9 expression were not significantly different than that observed in the control mice at any of the doses or time points examined. TLR6 expression was lower than the limit of detection at all doses and times examined. Alterations were observed in the expression of Clec2h, TLR1, TLR2, and TLR7 (Fig 4). The discussion of these results will focus on Clec2h and TLR2, as consistent significant (p<0.05) results were observed. TLR2 is a receptor that recognizes Gram (+) bacteria [58,64]. TLR2 was lower at the 3.5h time point in mice fed the 106 and 108 CFU/mouse doses, while no significant differences were observed at the 0.5h time point. A similar result was observed in an in vitro study where a vaginal epithelial cell line subjected to Lactobacillus strains (L. rhamnosus GR-1W and L. reuteri RC-14). The epithelial cells had a reduction in TLR2 expression compared to control [65]. The authors hypothesized that exposure to elevated of Gram (+) bacteria might create a state of hyporesponsiveness and results in a decrease in TLR2 expression [66]. Another PRR we targeted, Clec2h (C-type lectin receptor), was significantly (p < 0.05) up-regulated in the high dose at 0.5h and in both doses at the 3.5h time point. The function of Clec2h is poorly defined; however, it is thought to have a role in regulating innate immune responses [67]. These results suggest that administration of 32G alters the expression of genes known to be involved in the regulation of the innate immune system.
Antimicrobial peptides (AMPs) are components of innate immune system that are active in intestinal mucosal defense and have an important role in shaping the composition of the intestinal microbiota [68,69]. We examined the expression of different AMPs in mouse small intestine to evaluate the influence of L. casei 32G administration on expression of genes encoding AMPs. Significant (p<0.05) changes were observed in the expression of the alpha defensins (Defa-rs1c, Defcr1, Defcr4), lysozyme and Pla2g2d (Fig 4A). The discussion of these results will focus upon the alpha defensins and lysozyme as consistent significant (p<0.05) results were observed. Administration of 32G resulted in a dose dependent reduction in expression of alpha defensins and lysozyme-2 at the 3.5h time point, while their expression were not different than control at 0.5h. Salzman et al. showed that paneth cells alpha defensins are essential in homeostatic control regulating and shaping the composition of intestinal microbiota [32]. In addition, Menendez et al. found that alpha defensin gene expression was sensitive to oral antibiotic administration, suggesting that expression of alpha defensins is dependent upon the commensal microbiota [70]. The results suggest that 32G administration resulted in altering the expression of intestinal AMPs, thereby altering the composition the cecal microbiota. Alternatively, AMPs have been shown to be regulated by short-chain fatty acids, such as butyrate, which are produced by microbial fermentation in the gut [71–74]. This explanation is supported by observed significant (p < 0.05) decrease in the Lachnospiraceae, a butyrate producing family, in mice microbiota fed with high dose 32G at the 3.5h time point [75]. While our results suggest a role for AMPs in the 32G mediated changes in the composition of the cecal microbiota, other explanations are also possible and further research is required to determine the mechanism(s) by which 32G alters the cecal microbiota.
Cytokines are critical in regulation and development of the innate immune response [76]. Effect of 32G administration on both anti-inflammatory and pro-inflammatory cytokines was analyzed in this study. Of the cytokines examined, significant (p<0.05) changes in expression were only observed with IL10rb and TNF-α. Expression of IL-10rb, an anti-inflammatory marker, was significantly (p<0.05) increased at the 3.5h time point with both doses, relative to the control. In contrast, we observed a significant (p<0.05) reduction in expression of TNF-α, which serves as an indicator of a pro-inflammatory response, with the high dose at the 3.5h [10]. IL-10rb is a receptor shared by IL-10 subfamily; it is required for the activation of the IL-10 subfamily and a deficiency in IL-10rb has been shown to result in inflammatory bowel disease [77]. IL-10 regulates the inflammatory response by suppressing the production of pro-inflammatory cytokines such as TNF-α [78]. This is a possible explanation for the reduction of TNF-α expression observed in this study. Another possible explanation for the observed reduction in TNF-α is the reduction in TLR2 expression, as TLR2 expression is positively correlated with TNF-α expression [79,80]. These results suggest that administration of 32G has an anti-inflammatory effect on the murine immune system.
Conclusion
After administering L. casei 32G to mice at one of three different doses (106, 107, or 108 CFU/day/mouse) we were able to detect L. casei 32G by qPCR and monitor its abundance at different time points. We demonstrated that only mice fed a high dose of 32G contained greater than 105 CFU/ml of 32G in their cecal contents and that 32G was present at the highest level 3.5 h after administration. Our data revealed that L. casei 32G administration was capable of altering murine cecal microbiota and the alteration was dose and time dependent. Additionally, there was a lack of significant differences in the overall microbiota composition, regardless of 32G dose, in all treatments at 12h. These results suggest that the light/dark cycle has a significant (p < 0.05) impact on the compositions of the microbiota, likely due to the level of feed derived-nutrients, and therefore the light/dark cycle must be taken into consideration when designing experiments that follow microbiota composition. Lactobacillus, one of the dominant genera, increased in cecum content of mice fed with medium and high dose of L. casei 32G. We demonstrated that the Lactobacillus, which increased in prevalence in the cecal microbiota was not L. casei 32G. Our results indicate that feeding of L. casei 32G at 107 and 108 CFU/day/mouse altered the environment, released small peptides or metabolites, present in the cecum such that the growth of commensal lactobacilli was favored. Due to the interaction between gut microbiota and host immunity we examined the effect of 32G on the murine immune system. Interactions between PRRs, AMPs and the gut microbiota result in immune homeostasis [81,82]. The administration of L. casei 32G alters the gut microbiota composition in a dose and time dependent manner, potentially via changes in the expression of PRRs and AMPs. The 32G induced changes in the gut microbiota result in changes in the expression of cytokines, thereby resulting in an anti-inflammatory modulation of the murine immune system. Future research will evaluate the L. casei strain specificity for the ability to alter the composition of the murine gut microbiota and modulate immune system.
Supporting Information
The standard curves were generated by amplification of DNA isolated from cecum content spiked with 108, 107, 106, 105, or 104 CFU/ml of L. casei 32G culture, (n: 3 for each concentration). A) Standard curve. B) Melting curves.
(PDF)
(PDF)
Symbols represent data from individual mice, color-coded by the indicated metadata.
(PDF)
(PDF)
The major microbial communities of mouse cecum content at genus level in the control group and L. casei 32G groups; 106 CFU/ mouse (low), 107 CFU/ mouse (medium) and 108 CFU/ mouse (high), at 0.5, 3.5, 12, and 24 h after the last administration. Only genera with over 5% of the total bacteria are presented (n: 6 for each bar).
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We appreciate the technical contributions from Dr. Ekkarat Phrommao, Elena Vinay-Lara, and Jessie Heidenreich.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the DuPont Inc (Grant no. PRJ19MQ) and by the WARF (Grant no. PRJ66JZ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013;5: 1–19. 10.1002/emmm.201201773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312: 1355–9. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Proctor LM. The human microbiome project in 2011 and beyond. Cell Host Microbe. Elsevier; 2011;10: 287–291. 10.1016/j.chom.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 4. Kamada N, Núñez G. Regulation of the immune system by the resident intestinal bacteria. Gastroenterology. 2014;146: 1477–88. 10.1053/j.gastro.2014.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayer E a, Savidge T, Shulman RJ. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology. 2014;146: 1500–12. 10.1053/j.gastro.2014.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nieuwdorp M, Gilijamse PW, Pai N, Kaplan LM. Role of the microbiome in energy regulation and metabolism. Gastroenterology. Elsevier, Inc; 2014;146: 1525–33. 10.1053/j.gastro.2014.02.008 [DOI] [PubMed] [Google Scholar]
- 7. Aagaard K, Ma J, M.Antony K, RadhikaGanu, Petrosino J, Versalovic J. The Placenta Harbors a Unique Microbiome. Sci Transl Med. 2014;6: 237ra65 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. DiGiulio DB, Romero R, Amogan HP, Juan Pedro Kusanovic, Bik EM, Gotsch F, et al. Microbial Prevalence, Diversity and Abundance in Amniotic Fluid During Preterm Labor: A Molecular and Culture-Based Investigation. PLoS One. 2008;3: e3056 10.1371/journal.pone.0003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ohno H. Impact of commensal microbiota on the host pathophysiology: focusing on immunity and inflammation. Semin Immunopathol. 2015;37: 1–3. 10.1007/s00281-014-0472-2 [DOI] [PubMed] [Google Scholar]
- 10. Sommer F, Bäckhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. 2013;11: 227–238. 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 11.FAO/WHO. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Córdoba, Argentina; 2001.
- 12. Owyang C, Wu GD. The gut microbiome in health and disease. Gastroenterology. 2014;146: 1433–6. 10.1053/j.gastro.2014.03.032 [DOI] [PubMed] [Google Scholar]
- 13. Ceapa C, Wopereis H, Rezaïki L, Kleerebezem M, Knol J, Oozeer R. Influence of fermented milk products, prebiotics and probiotics on microbiota composition and health. Best Pract Res Clin Gastroenterol. 2013;27: 139–155. 10.1016/j.bpg.2013.04.004 [DOI] [PubMed] [Google Scholar]
- 14. Chen X, Dong M, Sun X. Mechanisms of action and applications of probiotics for the treatment of Clostridium difficile infection. Formatex. 2013;2: 1154–1163. [Google Scholar]
- 15. Kondepudi KK, Ambalam P, Karagin PH, Nilsson I, Wadström T, Ljungh Å, et al. A novel multi-strain probiotic and synbiotic supplement for prevention of Clostridium difficile infection in a murine model. Microbiol Immunol. 2014;58: 1–24. 10.1111/1348-0421.12184 This [DOI] [PubMed] [Google Scholar]
- 16. Salminen SJ, Gueimonde M, Isolauri E. Probiotics That Modify Disease Risk. J Nutr. 2005; 1294–1298. [DOI] [PubMed] [Google Scholar]
- 17. Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics. 2009;124: e172–e179. 10.1542/peds.2008-2666 [DOI] [PubMed] [Google Scholar]
- 18. Hörmannsperger G, Clavel T, Hoffmann M, Reiff C, Kelly D, Loh G, et al. Post-translational inhibition of IP-10 secretion in IEC by probiotic bacteria: impact on chronic inflammation. PLoS One. 2009;4: e4365 10.1371/journal.pone.0004365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kearney N, Stanton C, Desmond C, Coakley M, Collins KJ, Fitzgerald G, et al. Handbook of fermented functional foods 2nd ed. Farnworth ER, editor. Boca Raton: CRC Press; 2008. [Google Scholar]
- 20. Anukam KC, Reid G. Probiotics: 100 years (1907–2007) after Elie Metchnikoff’s observation. Commun Curr Res Educ Top Trends Appl Microbiol. 2007; 466–474. [Google Scholar]
- 21. Dietrich CG, Kottmann T, Alavi M. Commercially available probiotic drinks containing Lactobacillus casei DN-114001 reduce antibiotic-associated diarrhea. World J Gastroenterol. 2014;20: 15837–44. 10.3748/wjg.v20.i42.15837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aoki T, Asahara T, Matsumoto K, Takada T, Chonan O, Nakamori K, et al. Effects of the continuous intake of a milk drink containing Lactobacillus casei strain Shirota on abdominal symptoms, fecal microbiota, and metabolites in gastrectomized subjects. Scand J Gastroenterol. 2014;49: 552–63. 10.3109/00365521.2013.848469 [DOI] [PubMed] [Google Scholar]
- 23. Eloe-Fadrosh EA, Brady A, Crabtree J, Drabek EF, Ma B, Mahurkar A, et al. Functional Dynamics of the Gut Microbiome in Elderly People during Probiotic Consumption. Am Soc Microbiol. 2015;6: 1–12. 10.1128/mBio.00231-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruzzese E, Callegari ML, Raia V, Viscovo S, Scotto R, Ferrari S, et al. Disrupted intestinal microbiota and intestinal inflammation in children with cystic fibrosis and its restoration with Lactobacillus GG: a randomised clinical trial. PLoS One. 2014;9: e87796 10.1371/journal.pone.0087796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Derrien M, Vlieg JETVH. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23: 354–366. 10.1016/j.tim.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 26.Yakult 80th Annual Report. Global Yakult [Internet]. Tokyo, Japan; 2015. Available: http://www.yakult.co.jp/english/ir/management/pdf/ar2015.pdf
- 27. Kandler O, Weiss N. Genus Lactobacillus In: Sneath PHA, Mair NS, Sharpe ME, Holt JG, editors. Bergey’s Manual of Systematic Bacteriology. 9th ed. Baltimore: Williams & Wilkins; 1986. pp. 1063–1065. [Google Scholar]
- 28. Cai H, Rodríguez BT, Zhang W, Broadbent JR, Steele JL. Genotypic and phenotypic characterization of Lactobacillus casei strains isolated from different ecological niches suggests frequent recombination and niche specificity. Microbiology. 2007;153: 2655–65. 10.1099/mic.0.2007/006452-0 [DOI] [PubMed] [Google Scholar]
- 29.Tandee K. Evaluation of potential probiotic Lactobacillus casei strains. PhD. Thesis. University of Wisconsin-Madison. 2013. Available: http://depot.library.wisc.edu/repository/fedora/1711.dl:LS3GUH4GRCPJD8Y/datastreams/REF/content
- 30. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high- throughput community sequencing data. Nat Methods. 2010;7: 335–336. 10.1038/nmeth0510-335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72: 5069–72. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11: 76–83. 10.1038/ni.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Busch R a, Heneghan AF, Pierre JF, Wang X, Kudsk K a. The enteric nervous system neuropeptide, bombesin, reverses innate immune impairments during parenteral nutrition. Ann Surg. 2014;260: 432–44. 10.1097/SLA.0000000000000871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Corridoni D, Pastorelli L, Mattioli B, Locovei S, Ishikawa D, Arseneau KO, et al. Probiotic bacteria regulate intestinal epithelial permeability in experimental ileitis by a TNF-dependent mechanism. PLoS One. 2012;7: e42067 10.1371/journal.pone.0042067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2: e1308 10.1371/journal.pone.0001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods. 2001;25: 402–8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 37. Dray S, Dufour A-B. Journal of Statistical Software. J Stat Softw Sept. 2007;22: 1–20. [Google Scholar]
- 38.TheRCoreTeam. R : A Language and Environment for Statistical Computing. 2013.
- 39. Aguirre de Cárcer D, Cuív PO, Wang T, Kang S, Worthley D, Whitehall V, et al. Numerical ecology validates a biogeographical distribution and gender-based effect on mucosa-associated bacteria along the human colon. ISME J. 2011;5: 801–9. 10.1038/ismej.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Daniel C, Poiret S, Dennin V, Boutillier D, Pot B. Bioluminescence imaging study of spatial and temporal persistence of Lactobacillus plantarum and Lactococcus lactis in living mice. Appl Environ Microbiol. 2013;79: 1086–94. 10.1128/AEM.03221-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marco ML, Bongers RS, De Vos WM, Kleerebezem M. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl Environ Microbiol. 2007;73: 124–132. 10.1128/AEM.01475-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang H, Dibaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106: 2365–2370. 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biddle A, Stewart L, Blanchard J, Leschine S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity. 2013;5: 627–640. 10.3390/d5030627 [DOI] [Google Scholar]
- 44. Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2: 145–158. 10.4161/gmic.2.3.16333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Britton R a, Young VB. Role of the Intestinal Microbiota in Resistance to Colonization by Clostridium difficile. Gastroenterology. 2014;146: 1547–53. 10.1053/j.gastro.2014.01.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity. 2012;20: 738–47. 10.1038/oby.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mackie RI, Aminov RI, Hu W, Klieve a. V., Ouwerkerk D, Sundset M a., et al. Ecology of Uncultivated Oscillospira Species in the Rumen of Cattle, Sheep, and Reindeer as Assessed by Microscopy and Molecular Approaches. Appl Environ Microbiol. 2003;69: 6808–6815. 10.1128/AEM.69.11.6808-6815.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57: 601–9. 10.1002/hep.26093 [DOI] [PubMed] [Google Scholar]
- 49. Zakostelska Z, Kverka M, Klimesova K, Rossmann P, Mrazek J, Kopecny J, et al. Lysate of probiotic Lactobacillus casei DN-114 001 ameliorates colitis by strengthening the gut barrier function and changing the gut microenvironment. PLoS One. 2011;6: e27961 10.1371/journal.pone.0027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Walters W a, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588: 4223–4233. 10.1016/j.febslet.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kohl KD, Amaya J, Passement C a, Dearing MD, McCue MD. Unique and shared responses of the gut microbiota to prolonged fasting: a comparative study across five classes of vertebrate hosts. FEMS Microbiol Ecol. 2014;2: 1–12. 10.1111/1574-6941.12442 [DOI] [PubMed] [Google Scholar]
- 52. Yoon J-A, Han D-H, Noh J-Y, Kim M-H, Son GH, Kim K, et al. Meal time shift disturbs circadian rhythmicity along with metabolic and behavioral alterations in mice. PLoS One. 2012;7: e44053 10.1371/journal.pone.0044053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sanchez-Alavez M, Klein I, Brownell SE, Tabarean I V, Davis CN, Conti B, et al. Night eating and obesity in the EP3R-deficient mouse. Proc Natl Acad Sci USA. 2007;104: 3009–14. 10.1073/pnas.0611209104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell. 2014;159: 514–29. 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 55. Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, et al. Comparative genomics of lactic acid bacteria reveals a niche-specific gene set. Proc Natl Acad Sci USA. 2006;103: 15611–15616. 10.1186/1471-2180-9-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pridmore RD, Berger B, Desiere F, Vilanova D, Barretto C, Pittet A-C, et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc Natl Acad Sci USA. 2004;101: 2512–2517. 10.1073/pnas.0307327101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sohn M, Harding MJ, Park J, Kim D, Maher SE, Chae W, et al. Correction for Choi et al., Cell-permeable Foxp3 protein alleviates autoimmune disease associated with inflammatory bowel disease and allergic airway inflammation. Proc Natl Acad Sci USA. 2010;107: 21943–21943. 10.1073/pnas.1016618107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schuijt TJ, van der Poll T, de Vos WM, Wiersinga WJ. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol. 2013;21: 221–229. 10.1016/j.tim.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 59. Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics. 2012;13: 533 10.1186/1471-2164-13-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Iliev ID, Funari V a, Taylor KD, Nguyen Q, Reyes CN, Strom SP, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012;336: 1314–7. 10.1126/science.1221789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kekkonen R a., Lummela N, Karjalainen H, Latvala S, Tynkkynen S, Järvenpää S, et al. Probiotic intervention has strain-specific anti-inflammatory effects in healthy adults. World J Gastroenterol. 2008;14: 2029–2036. 10.3748/wjg.14.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thomas CM, Versalovic J. Probiotics-host communication: Modulation of signaling pathways in the intestine. Gut Microbes. 2010;1: 148–163. 10.4161/gmic.1.3.11712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chu H, Mazmanian SK. Innate immune recognition of the microbiota promotes host-microbial symbiosis. Nat Immunol. 2013;14: 668–675. 10.1038/ni.2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11: 443–51. [DOI] [PubMed] [Google Scholar]
- 65. Wagner RD, Johnson SJ. Probiotic lactobacillus and estrogen effects on vaginal epithelial gene expression responses to Candida albicans. J Biomed Sci. 2012;19: 58 10.1186/1423-0127-19-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Otte J-M, Cario E, Podolsky DK. Mechanisms of cross hyporesponsiveness to toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology. 2004;126: 1054–1070. 10.1053/j.gastro.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 67. Vogler I, Steinle A. Vis-à-vis in the NKC: genetically linked natural killer cell receptor/ligand pairs in the natural killer gene complex (NKC). J Innate Immun. 2011;3: 227–35. 10.1159/000324112 [DOI] [PubMed] [Google Scholar]
- 68. Cunliffe R. α-Defensins in the gastrointestinal tract. Mol Immunol. 2003;40: 463–467. 10.1016/S0161-5890(03)00157-3 [DOI] [PubMed] [Google Scholar]
- 69. Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3: 710–20. 10.1038/nri1180 [DOI] [PubMed] [Google Scholar]
- 70. Menendez A, Willing BP, Montero M, Wlodarska M, So CC, Bhinder G, et al. Bacterial stimulation of the TLR-MyD88 pathway modulates the homeostatic expression of ileal Paneth cell α-defensins. J Innate Immun. 2013;5: 39–49. 10.1159/000341630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schauber J, Svanholm C, Termen S, Iffland K, Menzel T, Scheppach W, et al. Expression of the cathelicidin LL—37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut. 2003;52: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schwab M, Reynders V, Loitsch S, Steinhilber D, Schröder O, Stein J. The dietary histone deacetylase inhibitor sulforaphane induces human beta-defensin-2 in intestinal epithelial cells. Immunology. 2008;125: 241–51. 10.1111/j.1365-2567.2008.02834.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sunkara LT, Zeng X, Curtis AR, Zhang G. Cyclic AMP synergizes with butyrate in promoting β-defensin 9 expression in chickens. Mol Immunol. 2014;57: 171–80. 10.1016/j.molimm.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 74. Zeng X, Sunkara LT, Jiang W, Bible M, Carter S, Ma X, et al. Induction of porcine host defense peptide gene expression by short-chain fatty acids and their analogs. PLoS One. 2013;8: e72922 10.1371/journal.pone.0072922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014; 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- 76. Lacy P, Stow JL. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood. 2011;118: 9–18. 10.1182/blood-2010-08-265892 [DOI] [PubMed] [Google Scholar]
- 77. Gertz EM, Ph D, Schäffer AA, Noyan F, Perro M, Sc M, et al. Inflammatory Bowel Disease and Mutations Affecting the Interleukin-10 Receptor. N Engl J Med. 2009;361: 2033–2045. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Batista ML, Rosa JC, Lopes RD, Lira FS, Martins E, Yamashita a S, et al. Exercise training changes IL-10/TNF-alpha ratio in the skeletal muscle of post-MI rats. Cytokine. 2010;49: 102–8. 10.1016/j.cyto.2009.10.007 [DOI] [PubMed] [Google Scholar]
- 79. Kirschning CJ, Schuman RR. Toll-Like Receptor Family Members and Their Ligands. Beutler B, Wagner H, editors. Germany: Springer-Verlag Berlin Heidelberg; 2002. [Google Scholar]
- 80. Paul WE, editor. Fundamental Immunology. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2003. [Google Scholar]
- 81. Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. MyD88-mediated signals induce the bactericidal lectin RegIII gamma and protect mice against intestinal Listeria monocytogenes infection. J Exp Med. 2007;204: 1891–900. 10.1084/jem.20070563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper L V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci USA. 2008;105: 20858–63. 10.1073/pnas.0808723105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The standard curves were generated by amplification of DNA isolated from cecum content spiked with 108, 107, 106, 105, or 104 CFU/ml of L. casei 32G culture, (n: 3 for each concentration). A) Standard curve. B) Melting curves.
(PDF)
(PDF)
Symbols represent data from individual mice, color-coded by the indicated metadata.
(PDF)
(PDF)
The major microbial communities of mouse cecum content at genus level in the control group and L. casei 32G groups; 106 CFU/ mouse (low), 107 CFU/ mouse (medium) and 108 CFU/ mouse (high), at 0.5, 3.5, 12, and 24 h after the last administration. Only genera with over 5% of the total bacteria are presented (n: 6 for each bar).
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.