Abstract
Novel psychoactive substances are continuously developed to circumvent legislative and regulatory efforts. A new synthetic cathinone, 4-methoxy-α-PVP, was identified for the first time in illegal products; however, the metabolism of this compound is not known. Complete metabolic profiles are needed for these novel psychoactive substances to enable identification of their intake and to link adverse effects to the causative agent. This study assessed 4-methoxy-α-PVP metabolic stability with human liver microsomes (HLMs) and identified its metabolites after HLM and hepatocyte incubations followed by high-resolution mass spectrometry (HRMS). A Thermo QExactive high-resolution mass spectrometer (HRMS) was used with full scan data-dependent mass spectrometry, with (1) and without (2) an inclusion list of predicted metabolite, and with full scan and all-ion fragmentation (3) to identify potential unexpected metabolites. In silico predictions were performed and compared to in vitro results. Scans were thoroughly mined with different data processing algorithms using WebMetabase (Molecular Discovery). 4-Methoxy-α-PVP exhibited a long half-life of 79.7 min in HLM, with an intrinsic clearance of 8.7 µL min−1 mg−1. In addition, this compound is predicted to be a low-clearance drug with an estimated human hepatic clearance of 8.2 mL min−1 kg−1. Eleven 4-methoxy-α-PVP metabolites were identified, generated by O-demethylation, hydroxylation, oxidation, ketone reduction, N-dealkylation, and glucuronidation. The most dominant metabolite in HLM and human hepatocyte samples was 4-hydroxy-α-PVP, also predicted as the #1 in silico metabolite, and is suggested to be a suitable analytical target in addition to the parent compound.
Keywords: 4-Methoxy-α-PVP, Novel psychoactive substances, Synthetic cathinones, Human hepatocytes, Human liver microsomes, In silico prediction
Introduction
In recent years, novel psychoactive substances (NPSs) appeared rapidly on the drug market in an effort to bypass controlled substance legislation. The European Monitoring Centre for Drugs and Drug Addiction (EMCDDA) reported 41 NPSs identified for the first time across Europe in 2010, 81 in 2013, and 101 in 2014 [1]. These NPSs are continuously developed to circumvent legislative and regulatory efforts, with limited available pharmacological and toxicological data. NPSs encompass a wide range of compounds including synthetic cannabinoids, phenethylamines, tryptamines, piperazines, ketamine, cathinones, and other plant-based psychoactive substances [2].
Synthetic cathinones emerged on the designer drug market as popular “legal” alternatives to illicit drugs in the late 2000s, and are marketed as “legal highs” and “not for human consumption”. They are stimulant-like drugs derived from cathinone, the active ingredient of the khat plant Catha edulis, with adverse effects including hyperthermia, agitation, confusion, psychosis, seizures, and tachycardia [3–7]. A variety of synthetic cathinones, alone and in combination with other illicit drugs, were detected in acute intoxications [4, 8, 9], impaired driving cases [10–13], and fatalities [4, 12, 14–23].
4-Methoxy-α-PVP (4-methoxy-α-pyrrolidinovalerophenone) was identified for the first time in illegal products purchased in Japan in 2013 [24] and also recently identified by Customs in Berlin [25]. 4-Methoxy-α-PVP is a substituted cathinone containing a pyrrolidinyl moiety and a methoxy group on the 4′ position of the aromatic ring (Fig. 1). Its structure is similar to other α-pyrrolidinophenone derivatives such as 3,4-methylenedioxypyrovalerone (MDPV) and α-pyrrolidinovalerophenone (α-PVP), both schedule I compounds [26, 27]. The mechanism of action of 4-methoxy-α-PVP is unknown; however, based on structural similarities to MDPV and α-PVP, it is hypothesized that 4-methoxy-α-PVP would act similarly as a monoamine transporter blocker increasing the amount of extracellular dopamine [28–31]. Extracellular dopamine increases pose a higher risk for addiction [28, 31]. In addition, the high lipophilicity of the pyrrolidine ring increases permeability through the blood–brain barrier, and hence increased potency and abuse potential [29, 32, 33].
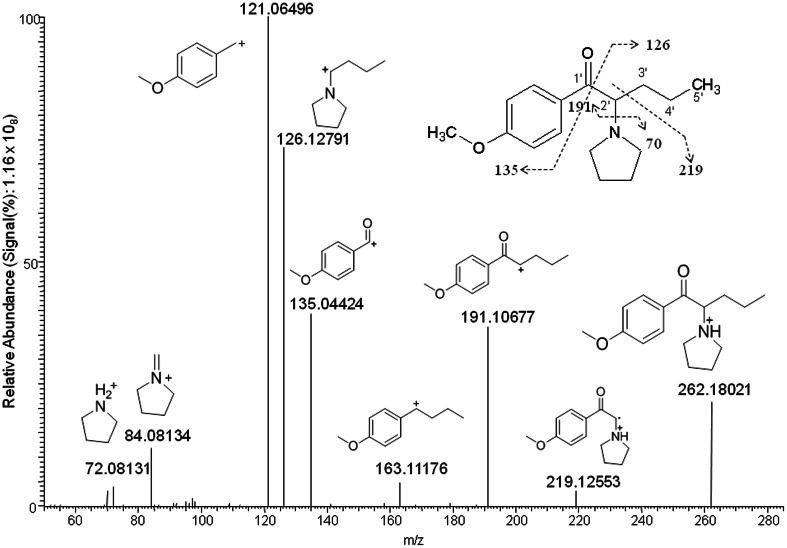
Fig. 1.
Product ion mass spectrum of 4-methoxy-α-PVP and its fragmentation pattern
The metabolic pathways of other α-pyrrolidinophenones, including MDPV, α-PVP, α-pyrrolidinobutiophenone (α-PBP), α-pyrrolidinopropiophenone (α-PPP), 4-methyl-α-pyrrolidinopropiophenone (MPPP), and 4-methoxy-α-pyrrolidinopropiophenone (MOPPP), were previously investigated [34–42]. There are no metabolism studies for 4-methoxy-α-PVP. A recent review of the pharmacology of α-pyrrolidinophenones outlined the major metabolic pathways of other structurally similar compounds including reduction of the keto moiety to the corresponding alcohol, hydroxylation followed by oxidation of the pyrrolidine ring to the lactam (2″-oxo), hydroxylation and carboxylation of the 4′-methyl group, O-demethylation of the 4′-methoxy group, and demethylenation followed by O-methylation of the 3′, 4′-methylenedioxy moiety [43].
A promising approach to elucidate metabolites of NPSs includes in silico metabolite predictions, and human liver microsome (HLM) and human hepatocyte incubations, followed by analysis with high-resolution mass spectrometry (HRMS) and software-assisted data mining [44, 45]. Complete metabolic profiles are needed for 4-methoxy-α-PVP to enable identification of intake and link adverse effects to the causative agent. We evaluated 4-methoxy-α-PVP in silico metabolism predictions, assessed metabolic stability with HLMs, and identified metabolites after HLM and human hepatocyte incubations followed by HRMS.
Materials and methods
Chemicals and reagents
4-Methoxy-α-PVP HCl was purchased from Cayman Chemicals (Ann Arbor, MI, USA) and diclofenac was obtained from Toronto Research Chemicals (Toronto, Canada). Fifty-donor purpose-pooled HLMs, 10-donor purpose-pooled cryopreserved human hepatocytes, InVitro Gro HT medium for thawing, and InVitro Gro KHB (Krebs-Henseleit buffer) for washing and incubation were acquired from BioreclamationIVT (Baltimore, MD, USA), and NADPH-regenerating system (Solution A) and glucose-6-phosphate dehydrogenase (Solution B) were purchased from Corning Gentest (Woburn, MA, USA). LC–MS grade water and formic acid were from Fisher Scientific (Fair Lawn, NJ, USA), and LC–MS grade acetonitrile from Sigma Aldrich (St. Louis, MO, USA).
In silico metabolite prediction
MetaSite software (v.5.0.3; Molecular Discovery, Pinner, UK) was used to predict in silico 4-methoxy-α-PVP metabolites. The structure of 4-methoxy-α-PVP was imported into the software and predictions generated using the CYP450 liver model, reactivity correction, 39 common biotransformations, and a minimum mass threshold of 50 Da for predicted metabolites. The predicted sites of metabolism are a consensus from liver CYP isoforms and FMO3. The computational procedure considers both thermodynamic and kinetic factors. Potential metabolites were assigned a probability score representing the likelihood of being generated, with 100 % being the maximum score. In addition, the top predicted in silico metabolite (100 % probability score) was imported into the software to predict second-generation metabolites.
Metabolic stability of 4-methoxy-α-PVP in HLMs
To investigate the metabolic stability of 4-methoxy-α-PVP, 1 µmol/L drug was incubated with 50-donor purpose-pooled HLMs. Water bath incubations were performed on a Precision reciprocal shaking bath (Winchester, VA, USA). The reaction mixture contained 780 µL purified water, 100 µL 0.5 mol/L potassium phosphate buffer pH 7.4, 10 µL Solution B, and 10 µL 4-methoxy-α-PVP (100 µmol/L). Samples were vortexed to homogenize the solutions, and 50 µL HLMs (20 mg protein/mL suspension) were added to the mixture and pre-incubated at 37 °C for approximately 3 min. Organic solvent percentage (methanol) was 1 %. The reaction was initiated with the addition of 50 µL Solution A, and 100 µL samples were collected at 0, 3, 8, 13, 20, 30, 45, and 60 min. The reaction was stopped with an equal volume (100 µL) of cold acetonitrile. Samples were then centrifuged at 15,000×g for 5 min at 4 °C and subsequently stored at −80 °C prior to analysis.
Incubation of 4-methoxy-α-PVP with cryopreserved primary human hepatocytes
For the human hepatocyte experiments, 10 µmol/L 4-methoxy-α-PVP was incubated at 37 °C with pooled cryopreserved human hepatocytes. Hepatocytes were incubated in a Forma™ Steri-Cycle™ CO2 incubator from Thermo Scientific (Fremont, CA, USA). The cells were thawed in and washed with InVitro Gro HT medium and centrifuged at 50×g for 5 min at room temperature. The supernatant was aspirated, and InVitro Gro KHB was added to wash the hepatocytes again. After centrifugation and removal of the supernatant, the cell pellet was re-suspended in 2 mL buffer. Cell viability was assessed with the Trypan blue exclusion method assuring >80 % viability. The reaction mixture contained 250 µL 4-methoxy-α-PVP in buffer (20 µmol/L) and 250 µL cell suspension, yielding a final drug concentration of 10 µmol/L and final cell concentration of 1 × 106 cells/mL. Samples (500 µL) were collected at 0, 1, and 3 h based on HLM half-life calculations, and the reaction stopped with 500 µL cold acetonitrile. Specimens were stored at −80 °C prior to analysis. Diclofenac also was incubated as a positive control for human hepatocyte functional viability.
Instrumentation
Liquid chromatography–high resolution mass spectrometry (LC-HRMS) was performed on a Thermo Scientific NCS-3500RS Ultimate 3000 Binary Rapid system coupled to a Thermo Scientific QExactive mass spectrometer (Thermo Scientific). The Ultimate 3000 RSLCnano system consisted of a degasser, a tertiary loading pump, a binary eluting pump, a column oven, and an RS Autosampler. The QExactive contained a heated electrospray ionization source (HESI-II) and was operated in positive ionization mode. The spray voltage was 3 kV, capillary temperature 350 °C, heater temperature 425 °C, S-lens RF level 50, sheath gas flow rate 50, auxiliary gas flow rate 13 and sweep gas 3 (manufacturer’s units). Nitrogen was used for spray stabilization, for collision-induced dissociation experiments in the HCD cell, and as the damping gas in the C-trap. The instrument was calibrated in the positive and negative modes every 25 h.
LC-HRMS for HLM samples
Chromatographic separation of HLM samples, diluted 100-fold with mobile phase A (0.1 % formic acid in water), was achieved with an Accucore C18 column (2.6 µm, 100 × 2.1 mm) and C18 guard cartridge (4 × 2.0 mm) with a 0.4 mL/min flow rate at 35 °C. Gradient elution was performed with 2 % B (0.1 % formic acid in acetonitrile) for 2 min, increased to 10 % B at 10 min, then ramped to 30 % B at 14 min, from 30 to 95 % in 2 min, held at 95 % for 1 min, and returned to initial conditions over 1 min. A 2-min equilibration followed, yielding a total run time of 20 min.
Full scan data-dependent MSMS (ddMS2) data were collected from m/z 100 to 600 at 70,000 resolution with automatic gain control (AGC) target 1.0 × 106, and maximum injection time 250 ms. Dd MS2 scans were triggered at the apex of the peak (3–8 s) when an underfill ratio of 5 % was reached (intensity 8.3 × 104) for the 5 most intense peaks per cycle (TopN of 5). MS scans were acquired at a resolution of 17,500 with AGC of 2 × 105, maximum injection time of 120 ms, and stepped normalized collision energy (NCE) 50 ± 30 %.
LC-HRMS for human hepatocytes
Hepatocyte samples were diluted fivefold with mobile phase A and chromatographically separated using a Synergi 4 Hydro-RP column (80Å, 150 × 2 mm) and C18 guard cartridge (4 × 2.0 mm) within 30 min at 0.4 mL/min flow rate. Initial conditions (2 % B) were held for 2 min, increased to 95 % B for 18 min, held at 95 % B for 5 min, and returned to initial conditions over 1 min. A 4-min equilibration followed, yielding a total run time of 30 min.
Hepatocytes incubations were analyzed with three different MS methods: full scan and ddMS2, with (1) and without (2) an inclusion list of predicted metabolites, and with a full scan and all-ion-fragmentation (AIF) method (3). Full scan and ddMS2 data (without an inclusion list) were acquired as previously described for HLM samples. In addition, an inclusion list was generated by MetaSite software based on its in silico metabolism predictions and imported into the full scan ddMS2 acquisition method. DdMS2 scans were triggered if the precursor ions from the inclusion list were detected above 8.3 × 104 intensity threshold. To identify potential unexpected metabolites, full scan and AIF data were acquired. AIF experiments were performed at 35,000 resolution over a scan range of m/z 100–600, with AGC target 5 × 105, maximum injection time of 120 ms, and a stepped NCE 50 ± 30 %.
Data analysis
In vitro microsomal half-life (T1/2) and microsomal intrinsic clearance (CLint, micr) of 4-methoxy-α-PVP were calculated on the model described by Baranczewski et al. [46]. This microsomal intrinsic clearance was then scaled to whole liver dimensions to calculate the intrinsic clearance (CLint) by multiplying two factors: the content of microsomal protein/g liver tissue (~45 mg/g) and liver weight/kg body weight (~20 g/kg), according to McNaney et al. [47]. Human hepatic clearance (CLH) and extraction ratio were calculated [46] without consideration of plasma protein binding.
Mass spectra acquired by the QExactive were analyzed with WebMetaBase software (v.2.0.2, Molecular Discovery) for metabolite candidates in the HLM and hepatocyte incubations. Raw data were submitted as a batch, which included a substrate (parent compound, 4-methoxy-α-PVP) structure mol file, a blank file (mobile phase), a substrate file (neat standard) to analyze the substrate fragmentation pattern, and raw data files for each incubation time point for hepatocyte incubations (t = 0, 1, 3 h) and for HLMs (t = 0, 3, 8, 13, 20, 30, 45, 60 min). Processing parameters for hepatocyte incubations were as follows: “hepatocyte metabolic system”; retention time window 2–25 min; chromatogram, MS, and MS/MS autofilter thresholds of 0.98; same peak tolerance of 0.010; three metabolite generations, minimum metabolite generation mass of 50 Da, and expected metabolites were rescue enabled (selects peaks with an equivalent m/z to a known metabolite regardless of peak area, provided there is MS2 data associated with the peaks). For the HLM incubations, similar processing parameters were chosen, except the metabolic system selected was HLMs and the retention time window was 2–18 min due to the different chromatography. The software auto-detects chromatographic peaks related to the parent compound and its metabolites, proposes potential structures based on fragmentation patterns for each detected peak and Markush structures (chemical structures with functional groups highlighted based on site reactivity), and ranks potential structures compatible with extracted fragment information. Structures of potential metabolite candidates were assigned based on retention times, mass shift between theoretical mass and observed mass (<5 ppm), peak abundance and fragmentation pattern. Metabolites were thoroughly examined against literature reports of structurally similar compounds and were eventually compared to the in silico predictions.
Results
In silico metabolite prediction
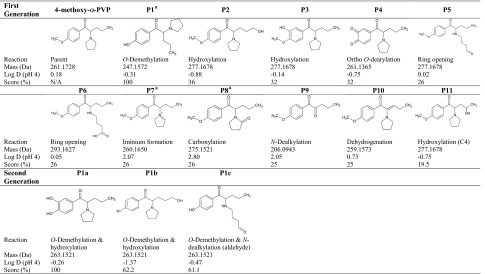
Eleven predicted in silico metabolites were generated with MetaSite software and are summarized in Table 1 (scores > 20 %), with three second-generation metabolites predicted from the highest ranked in silico metabolite. The top three metabolites predicted in silico were the O-demethylated metabolite, the metabolite hydroxylated at C5′ position, and the metabolite hydroxylated at the phenyl ring. Using the top predicted first-generation metabolite (4-hydroxy-α-PVP), three second-generation metabolites were predicted including the aromatic hydroxylated metabolite, the metabolite hydroxylated at C5′ position, and the metabolite resulting from ring opening and oxidation (Table 1).
Table 1.
In silico predicted metabolites for 4-methoxy-α-PVP in decreasing order based on score
Second generation metabolites were generated by importing first generation metabolites with scores of 100 % into the in silico software (only P1 had a score of 100 %)
aIdentified in 3 h human hepatocyte incubation
Metabolic stability assessment with HLMs
4-Methoxy-α-PVP exhibited an in vitro T1/2 of 79.7 ± 1.3 min in HLMs, with a microsomal intrinsic clearance CLint, micr of 8.7 µL min−1 mg−1 and intrinsic clearance CLint of 8.2 mL min−1 kg−1. Hepatic clearance was calculated to 5.8 mL min−1 kg−1 and the extraction ratio was 0.29.
Identification of HLM metabolites
In the HLM samples, the O-demethylated metabolite (M3 in the hepatocyte samples) was the only metabolite detected. Peak areas increased from the 20 to 60 min incubation times.
Metabolite profiling with human hepatocytes
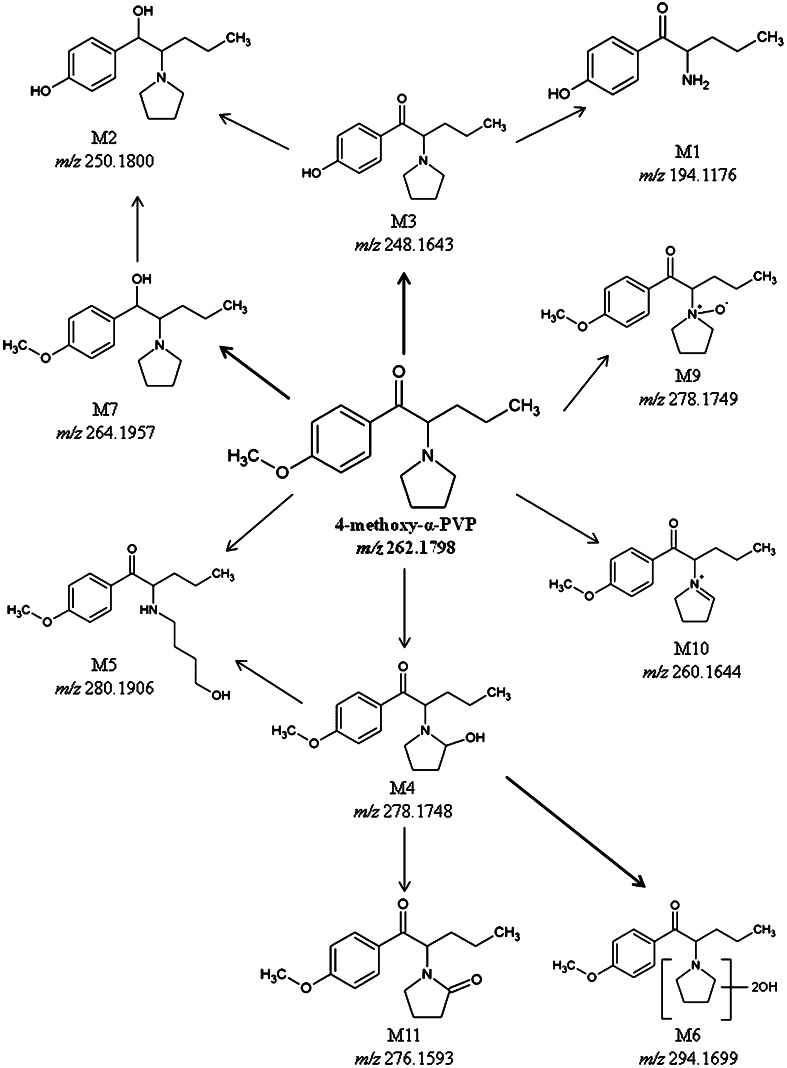
Diclofenac (positive control) and its metabolites, 4-hydroxydiclofenac and diclofenac β-d-acyl glucuronide, were observed in the hepatocyte samples confirming hepatocyte metabolic activity. Accurate full scan and product ion spectra were obtained and thoroughly data-mined, identifying eleven 4-methoxy-α-PVP metabolites (Table 2) with mass measurement errors <1.5 ppm. Metabolites eluted between 4.9 and 13.8 min with 4-methoxy-α-PVP parent at 9.1 min (Table 2). Ten phase I metabolites (M1–M7, M9–M11) and one phase II metabolite (M8) were identified in the hepatocyte samples. In the 1-h sample, M1, M3–M7, and M9 were the only metabolites detected with 91.0 % of 4-methoxy-α-PVP unchanged. All metabolites were found in the 3-h sample with 78.7 % of the parent compound remaining. Consistent with ongoing metabolism, 4-methoxy-α-PVP peak areas decreased during incubation while in general metabolite peak areas increased from 1 to 3 h incubation. The metabolic reactions observed included: O-demethylation (M3), O-demethylation + N-dealkylation (M1), O-demethylation + ketone reduction (M2), hydroxylation (M4, M6), pyrrolidine ring opening + hydroxylation (M5), ketone reduction (M7), carbonylation (M11), aliphatic N-oxidation (M9), and iminium ion formation (M10). One glucuronide also was noted (M8); however, a product ion mass spectrum for M8 was not obtained, as the intensity threshold criterion was not met. Based on observed peak areas, M3 was the most intense metabolite in hepatocyte incubations, followed by M6 and M7.
Table 2.
4-Methoxy-α-PVP phase I metabolites identified after incubation with human hepatocytes, sorted by retention time (RT). Rank was based on mass spectrometric peak areas. Fragments are expressed in nominal mass
| ID | Metabolic reaction | RT (min) | Elemental composition | m/z | Mass error (ppm) | Characteristic fragments | MS % peak areas | Ranking | ||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 h | 3 h | 1 h | 3 h | |||||||
| M1 | O-Demethylation + N-dealkylation | 4.87 | C11H15NO2 | 194.1177 | 0.52 | 135, 107, 58 | 2.2e7 | 2.9e7 | 4 | 6 |
| M2 | Ketone reduction + O-demethylation | 7.26 | C15H23NO2 | 250.1800 | 0.80 | 189, 232, 107, 70 | – | 1.3e7 | – | 9 |
| M3 | O-Demethylation | 7.48 | C15H21NO2 | 248.1643 | 0.81 | 107, 126, 121, 72, 177 | 4.7e8 | 7.5e8 | 1 | 1 |
| M4 | Hydroxylation | 8.51 | C16H23NO3 | 278.1748 | 1.08 | 121, 142, 135, 191 | 1.2e7 | 2.7e7 | 6 | 7 |
| M5 | Pyrrolidine ring opening + hydroxylation | 8.65 | C16H25NO3 | 280.1906 | 0.36 | 121, 190, 262, 148, 135, 144 | 1.4e7 | 1.2e7 | 5 | 10 |
| M6 | Di-hydroxylation | 8.78 | C16H23NO4 | 294.1699 | 0.31 | 121, 87, 140, 276, 135, 191 | 8.0e7 | 1.9e8 | 2 | 2 |
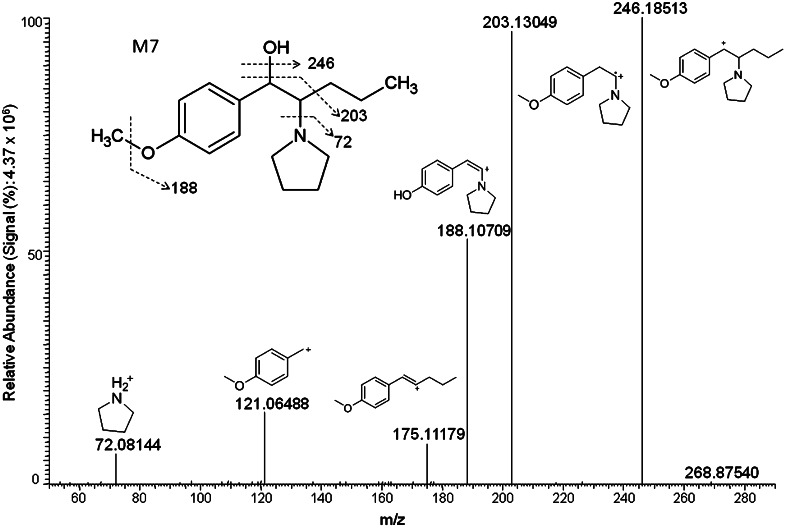
| M7 | Ketone reduction | 8.96 | C16H25NO2 | 264.1957 | 0.38 | 246, 203, 188, 121, 175, 72 | 4.8e7 | 1.1e8 | 3 | 3 |
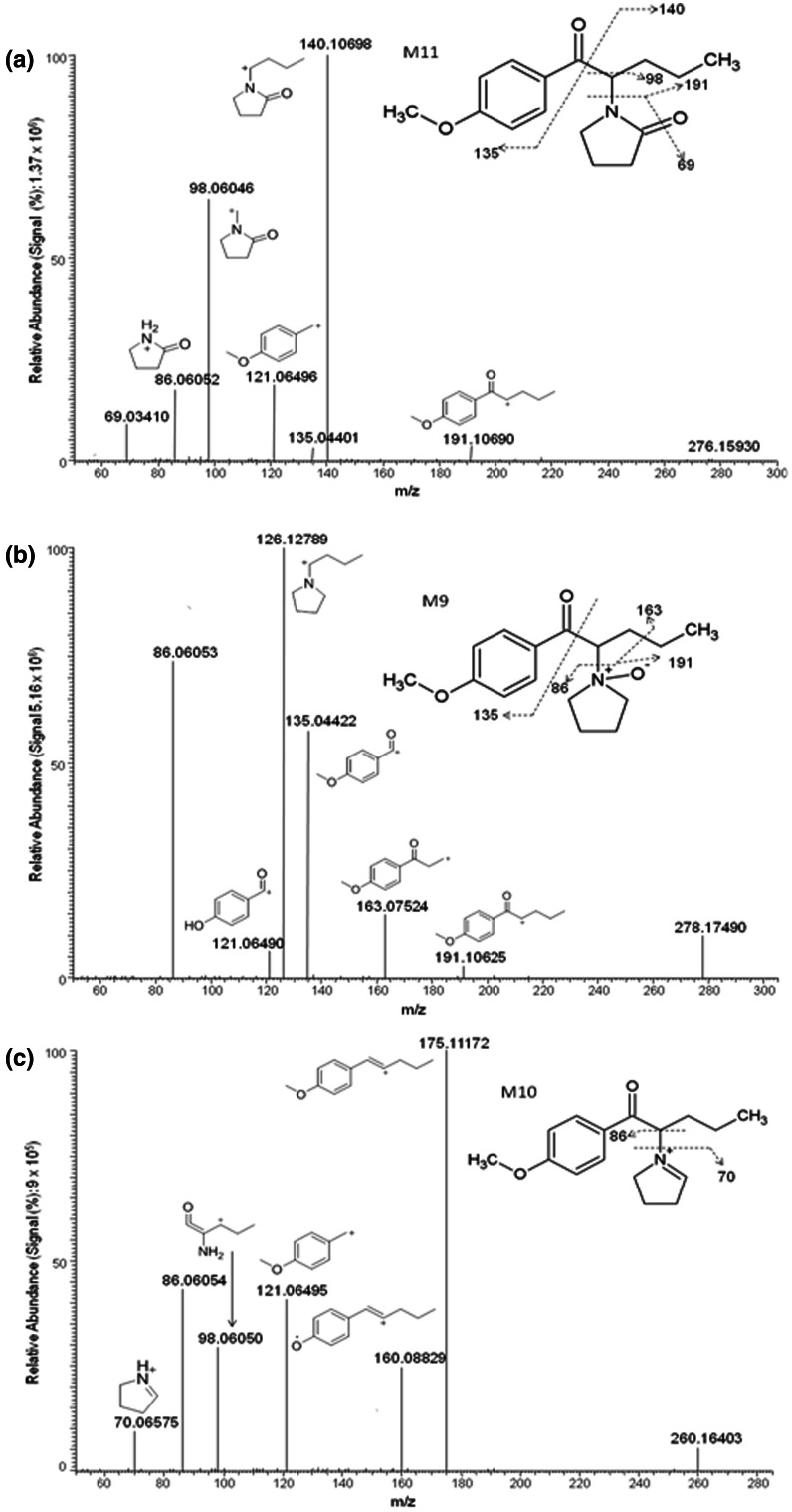
| M9 | Aliphatic N-oxidation | 9.81 | C16H23NO3 | 278.1749 | 0.72 | 126, 86, 135, 163, 121 | 9.3e6 | 1.8e7 | 7 | 8 |
| M10 | Iminium ion formation | 11.48 | C16H21NO2 | 260.1644 | 1.54 | 175, 86, 121, 70 | – | 3.6e7 | – | 5 |
| M11 | Carbonylation | 13.83 | C16H21NO3 | 276.1593 | 0.36 | 140, 98, 121, 135, 69, 191 | – | 3.9e7 | – | 4 |
| Parent | 9.10 | C16H23NO2 | 262.1798 | 1.53 | 121, 126, 135. 191, 84, 163, 219, 72, 70 | 6.6e9 | 4.4e9 | – | – | |
All metabolites were identified with full scan and ddMS2, with the exception of M8. This glucuronidated metabolite was identified with our full scan and dd-MS2 with the in silico generated inclusion list. Other than M8, this acquisition method did not detect any additional metabolites and only found 5/10 metabolites (M3, M4, M6, M7, and M11) identified with full scan and ddMS2 . The full scan and AIF method did not detect any additional unexpected metabolites. Compared to full scan and ddMS2, the full scan and AIF did not identify M5 (pyrrolidine ring opening + hydroxylation), the lowest prevalence metabolite other than M8.
Discussion
A variety of different product ions were observed for 4-methoxy-α-PVP (Fig. 1), with m/z 121 the most intense fragment ion at the given collision energy conditions. The base peak is generated by cleavage between the β-keto (C1′) and aromatic ring. Cleavage of the bond between the β-keto (C1′) and alpha (C2′) carbon generated the next highest intensity ions at m/z 126 and 135, similar to cleavage that was previously reported for α-PVP [37]. Fragmentation of the bond between the pyrrolidine ring and alpha carbon (C2′) produced m/z 191 and 72 ions. Other distinct product ions included m/z 219 (loss of C3H7 radical) and 84. The unique fragmentation pattern of 4-methoxy-α-PVP was used for metabolite structure elucidation.
Identification of O-demethylated metabolites
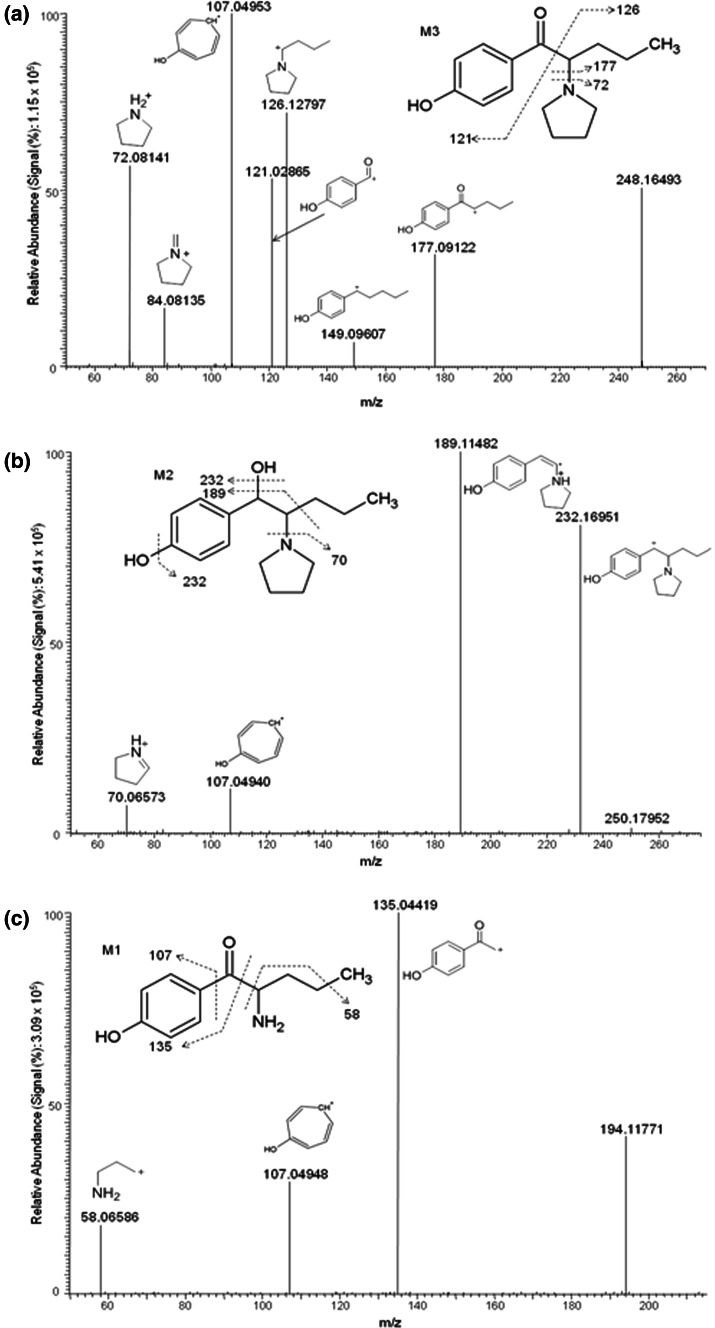
4-Hydroxy-α-PVP (M3), the product of O-demethylation of the 4-methoxy moiety, was the most intense metabolite in the HLMs and 1 and 3 h hepatocyte samples (Table 2). Figure 2 illustrates the product ion spectra for all O-demethylated metabolites. M3 product ion mass spectrum showed three distinct fragments of the precursor 4-methoxy-α-PVP molecule (m/z 126, 72, and 84) suggesting no modifications to the pyrrolidine ring and aliphatic chain of the molecule. The characteristic fragments of M3 included m/z 107 and 121 indicating O-demethylation at the methoxy group and subsequent formation of the tropylium ion and bond cleavage between the β-keto (C1′) and alpha (C2′) carbons. Further reduction of M3 at the keto moiety yielded M2 at m/z 250. M2 product ion spectrum showed a characteristic m/z 189 fragment produced by cleavage of the carbonyl and terminal C3H7 radical, and a m/z 232 fragment from loss of water. In addition, N-dealkylation of the pyrrolidine ring of M3 yielded M1. This metabolite product ion spectrum showed characteristic fragments m/z 135, 107, and 58 suggesting modifications to the pyrrolidine ring and 4-methoxy moiety.
Fig. 2.
Product ion mass spectra and assigned fragmentation patterns for 4-methoxy-α-PVP metabolites generated by O-demethylation. a 4-Hydroxy-α-PVP M3, b O-demethylated and ketone reduced metabolite M2, and c the O-demethylated and N-dealkylated metabolite M1
Identification of hydroxylated metabolites
Three hydroxylated metabolites, one monohydroxylated (M4), one di-hydroxylated (M6) and one pyrrolidine ring opening and hydroxylated metabolite (M5) were identified. Figure 3 illustrates the product ion spectra for all hydroxylated metabolites. Hydroxylations occurred at the pyrrolidine ring, as three characteristic fragments from 4-methoxy-α-PVP were identified in each metabolite (m/z 121, 135, and 191). M6 was the second most intense peak after 1 and 3 h incubations showing fragments at m/z 158, 140, 276, 98, and 87, suggesting di-hydroxylation at the pyrrolidine ring. M4 showed distinct fragments including m/z 142, 100, and 70 supporting a monohydroxylation on the pyrrolidine ring. The exact position of the hydroxyl group is unclear, although, previous metabolism studies identified 2″-OH-α-PVP in the structurally similar compound α-PVP [37, 40, 41]. It is expected that similar metabolism could be observed with M4. The last identified hydroxyl metabolite (M5) was the product of the pyrrolidine ring opening and hydroxylation on the terminal end, similar to what was reported for α-PVP [48]. Characteristic fragments were m/z 262, 144, and 219.
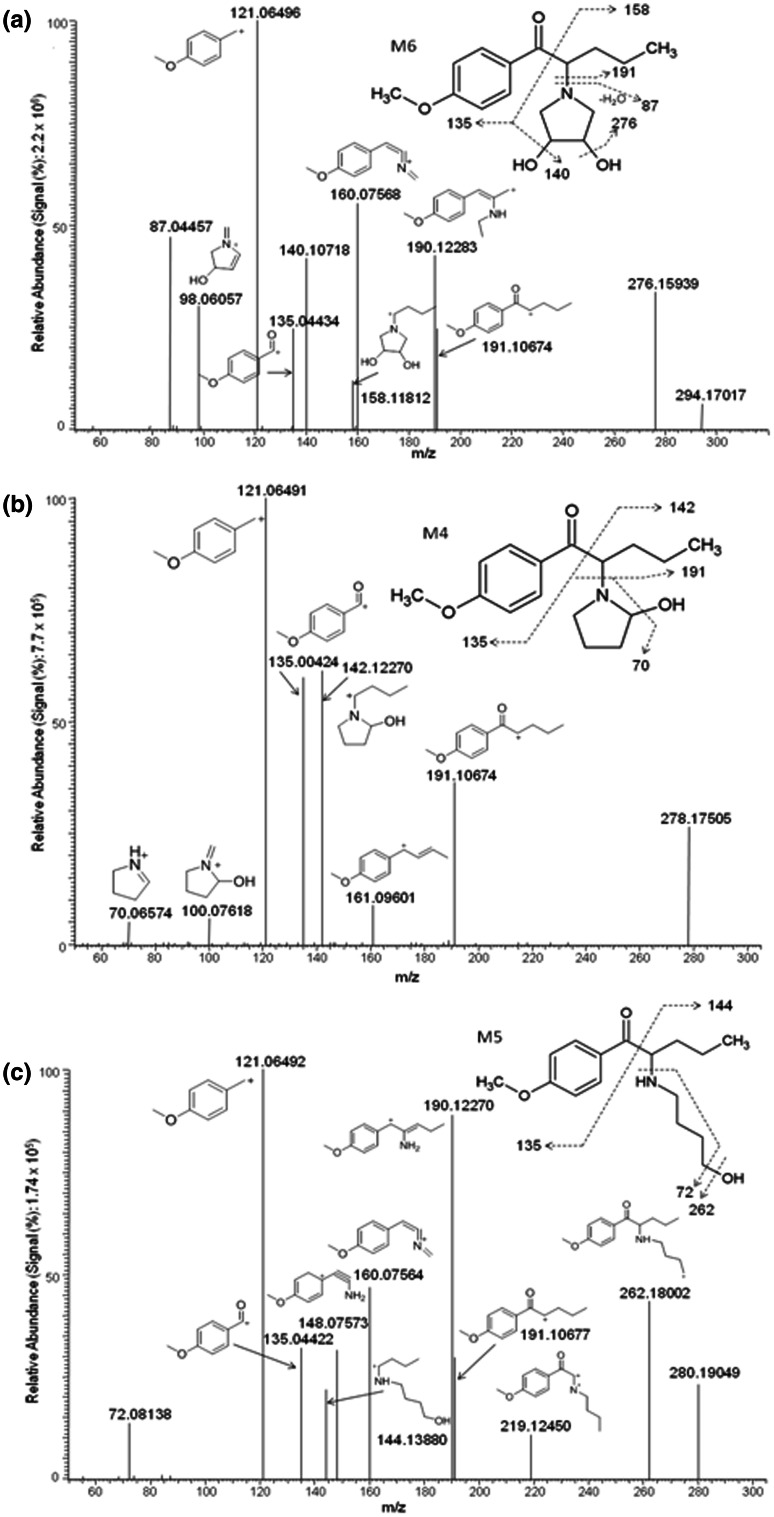
Fig. 3.
Product ion mass spectra and assigned fragmentation pattern for hydroxylated 4-methoxy-α-PVP for metabolites: a the di-hydroxylated metabolite on the pyrrolidine ring M6, b the monohydroxylated metabolite on the pyrrolidine ring M4, and c the ring opening and hydroxylated metabolite M5. The exact location of the di-hydroxylated metabolite is unknown; however, the position of the hydroxyl group at the 2″ in M4 is suggested based on previously identified α-pyrrolidine metabolites
Metabolite generated by ketone reduction
The third most prevalent metabolite in the 1 and 3 h hepatocyte samples was the ketone reduced metabolite, M7 (Fig. 4). The base peak m/z 246 resulted from the loss of water. Other characteristic fragments included m/z 203, 188, 175, and 121 and 72 as observed with the precursor molecule.
Fig. 4.
Product ion mass spectrum and fragmentation pattern for the metabolite generated by ketone reduction M7
Other identified metabolites
Two metabolites were products of oxidation reactions (M11 and M9) (Fig. 5). M11 was the result of carbonylation of the pyrrolidine ring, a common biotransformation for α-pyrrolidinophenones [43]. Characteristic fragments of this late eluting compound included m/z 140, 98, 86, 121, and 191. The amide is less basic than the tertiary amine in the parent compound and, therefore, is less likely to be protonated and as water soluble as the parent. The N-oxide metabolite (M9) also eluted after the parent compound, with the simultaneous appearance of m/z 126 and 135 and an m/z 86 fragment that was not observed in the monohydroxylated metabolite (M4), thus suggesting this oxidation reaction. Other distinct fragments included m/z 163, 121, and 191. In addition to these oxidative metabolites, the formation of the iminium ion was proposed (M10). This compound also was predicted in silico (P7). Fragments included m/z 175, 121, 86, 98,160, and 70. The m/z 70 fragment suggests the formation of an iminium ion at the pyrrolidine ring; otherwise fragment m/z 72 should be observed as in the parent compound (Fig. 1) after cleavage of the pyrrolidine ring.
Fig. 5.
4-Methoxy-α-PVP metabolites and assigned fragmentation patterns for metabolites generated by a carbonylation of the pyrrolidine ring M11, b aliphatic N-oxidation M9, and c iminium ion formation M10. The location of the carbonyl moiety at the 2″-position is based on previously identified α-pyrrolidine lactams
Prevalence of 4-methoxy-α-PVP metabolites
Most likely, the most intense MS peak areas observed in vitro will be the major 4-methoxy-α-PVP metabolites in vivo, or the precursors of second-generation in vivo metabolites. The most abundant metabolite in the hepatocyte samples at both 1 and 3 h was 4-hydroxy-α-PVP [M3, retention time (RT) 7.48 min], resulting from O-demethylation. This also was the only metabolite observed in the HLM samples and the highest predicted in silico metabolite. The next most abundant hepatocyte metabolites were di-hydroxylations of the pyrrolidine ring (M6, RT 8.78 min) and ketone reduction to the corresponding alcohol (M7, RT 8.96 min). Other major metabolites of 4-methoxy-α-PVP may exist and may have been missed in human hepatocyte and HLM incubations due to short incubation times, and the long half-life of parent 4-methoxy-α-PVP. Based on our results, 4-hydroxy-α-PVP is the major metabolite of 4-methoxy-α-PVP and could be used in combination with the parent compound as potential markers. A metabolic pathway for 4-methoxy-α-PVP in hepatocytes is illustrated in Fig. 6.
Fig. 6.
Proposed metabolic pathways for 4-methoxy-α-PVP in human hepatocytes. Position of di-hydroxylation on the pyrrolidine ring in M6 is unknown and represented with Markush structures
Metabolic stability assessment in HLMs
4-Methoxy-α-PVP exhibited a long half-life (79.7 ± 1.3 min) in vitro, identified by the slow 9 and 21.3 % decrease of parent compound at 1 and 3 h in the hepatocyte samples, respectively. Based on the intrinsic clearance, extraction ratio and half-life observed in this study, 4-methoxy-α-PVP is predicted to be slowly metabolized. Low clearance compounds typically have CLint < 15 mL min−1 kg−1 [47] and extraction ratios <0.3 [49]. It is important to note that predicted hepatic clearance can vary among individuals based on enzymatic polymorphisms.
Comparison with other synthetic cathinones
In general, 4-methoxy-α-PVP followed the metabolic patterns identified in other structurally similar synthetic cathinones, i.e. multiple hydroxylation products, carbonylation, ring opening and oxidation reactions, ketone reduction, and O-demethylation [43]. The major metabolic pathway identified in our study was O-demethylation of the 4-methoxy group, similar to MOPPP (a 4-methoxy-α-pyrrolidinophenone with a three-carbon alkyl chain) in HLMs by Springer et al. [36].
Several studies investigated the metabolism of α-PVP, the closest analog to 4-methoxy-α-PVP. An in vivo study examining rat urine following a 20 mg/kg α-PVP dose reported multiple hydroxylation products of α-PVP, carbonylation of the 2′-position of the pyrrolidine ring, ring opening and oxidation reactions, as well as degradation of the pyrrolidine ring to the corresponding primary amine [37]. For α-PVP and α-PBP in authentic human urine specimens, Uralets et al. [42] reported N-dealkylation to the primary amine metabolite that subsequently underwent ketone reduction. The authors suggested that the pyrrolidine ring hinders direct ketone reduction. No information was provided about the time of urine collection and how long after dosing the specimens were collected. However, ketone reduction was identified by Tyrkko et al. [39] while investigating the phase I metabolites of α-PVP in vitro using HLMs and in authentic human urine. Similarly, Shima et al. [40] found that the two major metabolic pathways of α-PVP identified in human urine were the reduction of the ketone to the corresponding alcohol and the carbonylation of the 2′-position of the pyrrolidinophenone ring to produce the 2″-oxo metabolite, which was also confirmed by Namera et al. [41]. Alpha-PVP and α-PBP do not contain the 4-methoxy moiety, and therefore, cannot produce O-demethylated metabolites.
We identified all α-PVP metabolic biotransformations, as discussed above, in our study of the novel 4-methoxy-α-PVP, with the exception of N-dealkylation of the pyrrolidine ring to the corresponding primary amine. Since formation of the primary amine metabolite was one of the major pathways for α-PVP, we carefully checked all our raw data, and although we did not identify this metabolite, we did find the metabolite resulting from degradation of the pyrrolidine ring in combination with O-demethylation (M1). In agreement with Tyrkko et al. [39] and Shima et al. [40], we also identified a metabolite resulting from direct reduction of the ketone moiety for 4-methoxy-α-PVP (M7) as also observed for α-PVP. Ketone reduction and carbonylation of the pyrrolidine ring were ranked 3rd and 4th among the most prevalent metabolites, respectively.
It is important to note that although incubation with hepatocytes reflects liver metabolism, it cannot reflect additional processes involved in drug elimination including extrahepatic metabolism, enterohepatic circulation, and renal filtration, re-absorption, and secretion. In addition, distribution processes can alter parent and metabolite patterns and concentrations that hepatocytes cannot simulate. Metabolites identified in these experiments may be present in human urine and blood after 4-methoxy-α-PVP intake; however, it is important to confirm these metabolites in authentic specimens as hepatocytes only reflect one aspect of drug elimination.
In silico prediction of 4-methoxy-α-PVP metabolites
In silico metabolite prediction software was used to predict 4-methoxy-α-PVP metabolic candidates and evaluate them against metabolites identified in vitro. Eleven phase I metabolites with a probability score >20 % were predicted in silico, while ten phase I metabolites were identified in the 3 h hepatocyte samples. Three in silico metabolites (>20 %) matched metabolites identified in the 3 h hepatocyte samples, namely M3, which corresponds to P1 scoring 100 % in silico and also the predominant hepatocyte metabolites, M10 (→P7) and M11 (→P8) (Table 1). Metabolites resulting from hydroxylation of the pyrrolidine ring (M4) and ketone reduction (M7) also were predicted in silico, although scores were less than 20 % (19.1 and 8.2 %, respectively). Five hepatocyte biotransformations were not predicted in silico; however, these metabolites involved combinations of phase I biotransformations, which the software does not readily predict. Manufacturers suggest analyzing the top predicted metabolites (100 % scores) in the software in order to identify potential second-generation metabolites. In our case, none of the predicted second-generation metabolites were identified in vitro in this study; a possible reason might be the low clearance of the parent compound. Although in silico prediction software has limitations, it is a valuable investigative tool for metabolite identification, especially for novel psychoactive substances. Limited information for these compounds exists; thus in silico prediction software can aid in developing a chromatographic method as it suggests metabolites that may be less polar than the parent. In addition, the software also can be used to import an inclusion list for MS acquisition methods.
Conclusions
4-Methoxy-α-PVP followed the metabolic patterns comparable to other pyrrolidinophenones, including O-demethylation, ketone reduction, multiple hydroxylation products, ring opening and oxidation reactions, N-dealkylation, and carbonylation to the corresponding lactam. The combination of different HRMS acquisition methods proved useful for identifying potential metabolites, as these are the first data identifying 4-methoxy-α-PVP metabolites that are good targets for documenting drug intake in forensic and clinical investigations. Based on our results, 4-hydroxy-α-PVP, the predominant metabolite, and the parent compound are good markers to identify consumption. This study demonstrated the applicability of using in vitro techniques coupled with HRMS methods to elucidate the metabolic profiles of emerging NPSs and comparison with in silico metabolite predictions. It is necessary to elucidate the metabolic pathways of these NPSs to enable linkage to adverse effects and recent intake.
Acknowledgments
The authors would like to acknowledge Timothy Moeller from BioreclamationIVT for his assistance with the human hepatocyte incubations, and Ismael Zamora from Molecular Discovery for his help with MetaSite and WebMetabase software. This research was supported by the Intramural Research Program of the National Institute on Drug Abuse, National Institutes of Health.
Compliance with ethical standards
Conflict of interest
There are no financial or other relations that could lead to a conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.EMCDDA (2015) New psychoactive substances in Europe—An update from the EU Early Warning System. Lisbon
- 2.United Nations Office on Drugs and Crime . World Drug Report 2013. Vienna: United Nations; 2013. [Google Scholar]
- 3.Pearson JM, Hargraves TL, Hair LS, Massucci CJ, Frazee CC, 3rd, Garg U, Pietak BR. Three fatal intoxications due to methylone. J Anal Toxicol. 2012;36:444–445. doi: 10.1093/jat/bks043. [DOI] [PubMed] [Google Scholar]
- 4.Spiller HA, Ryan ML, Weston RG, Jansen J. Clinical experience with and analytical confirmation of “bath salts” and “legal highs” (synthetic cathinones) in the United States. Clin Toxicol. 2011;49:499–505. doi: 10.3109/15563650.2011.590812. [DOI] [PubMed] [Google Scholar]
- 5.Tekulve K, Alexander A, Tormoehlen L. Seizures associated with synthetic cathinone exposures in the pediatric population. Pediatr Neurol. 2014;51:67–70. doi: 10.1016/j.pediatrneurol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Vardakou I, Pistos C, Spiliopoulou C. Drugs for youth via Internet and the example of mephedrone. Toxicol Lett. 2011;201:191–195. doi: 10.1016/j.toxlet.2010.12.014. [DOI] [PubMed] [Google Scholar]
- 7.Winstock A, Mitcheson L, Ramsey J, Davies S, Puchnarewicz M, Marsden J. Mephedrone: use, subjective effects and health risks. Addiction. 2011;106:1991–1996. doi: 10.1111/j.1360-0443.2011.03502.x. [DOI] [PubMed] [Google Scholar]
- 8.Wood DM, Davies S, Puchnarewicz M, Button J, Archer R, Ovaska H, Ramsey J, Lee T, Holt DW, Dargan PI. Recreational use of mephedrone (4-methylmethcathinone, 4-MMC) with associated sympathomimetic toxicity. J Med Toxicol. 2010;6:327–330. doi: 10.1007/s13181-010-0018-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thornton SL, Gerona RR, Tomaszewski CA. Psychosis from a bath salt product containing flephedrone and MDPV with serum, urine, and product quantification. J Med Toxicol. 2012;8:310–313. doi: 10.1007/s13181-012-0232-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pedersen AJ, Reitzel LA, Johansen SS, Linnet K. In vitro metabolism studies on mephedrone and analysis of forensic cases. Drug Test Anal. 2013;5:430–438. doi: 10.1002/dta.1369. [DOI] [PubMed] [Google Scholar]
- 11.Kriikku P, Wilhelm L, Schwarz O, Rintatalo J. New designer drug of abuse: 3,4-methylenedioxypyrovalerone (MDPV). Findings from apprehended drivers in Finland. Forensic Sci Int. 2011;210:195–200. doi: 10.1016/j.forsciint.2011.03.015. [DOI] [PubMed] [Google Scholar]
- 12.Marinetti LJ, Antonides HM. Analysis of synthetic cathinones commonly found in bath salts in human performance and postmortem toxicology: method development, drug distribution and interpretation of results. J Anal Toxicol. 2013;37:135–146. doi: 10.1093/jat/bks136. [DOI] [PubMed] [Google Scholar]
- 13.Knoy JL, Peterson BL, Couper FJ. Suspected impaired driving case involving alpha-pyrrolidinovalerophenone, methylone and ethylone. J Anal Toxicol. 2014;38:615–617. doi: 10.1093/jat/bku073. [DOI] [PubMed] [Google Scholar]
- 14.Hasegawa K, Suzuki O, Wurita A, Minakata K, Yamagishi I, Nozawa H, Gonmori K, Watanabe K. Postmortem distribution of α-pyrrolidinovalerophenone and its metabolite in body fluids and solid tissues in a fatal poisoning case measured by LC–MS–MS with the standard addition method. Forensic Toxicol. 2014;32:225–234. doi: 10.1007/s11419-014-0227-8. [DOI] [Google Scholar]
- 15.Gil D, Adamowicz P, Skulska A, Tokarczyk B, Stanaszek R. Analysis of 4-MEC in biological and non-biological material–three case reports. Forensic Sci Int. 2013;228:e11–e15. doi: 10.1016/j.forsciint.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 16.McIntyre IM, Hamm CE, Aldridge L, Nelson CL. Acute methylone intoxication in an accidental drowning–a case report. Forensic Sci Int. 2013;231:e1–e3. doi: 10.1016/j.forsciint.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Carbone PN, Carbone DL, Carstairs SD, Luzi SA. Sudden cardiac death associated with methylone use. Am J Forensic Med Pathol. 2013;34:26–28. doi: 10.1097/PAF.0b013e31827ab5da. [DOI] [PubMed] [Google Scholar]
- 18.Saito T, Namera A, Osawa M, Aoki H, Inokuchi S. SPME–GC–MS analysis of α-pyrrolidinovaleorophenone in blood in a fatal poisoning case. Forensic Toxicol. 2013;31:328–332. doi: 10.1007/s11419-013-0183-8. [DOI] [Google Scholar]
- 19.Murray BL, Murphy CM, Beuhler MC. Death following recreational use of designer drug “bath salts” containing 3,4-Methylenedioxypyrovalerone (MDPV) J Med Toxicol. 2012;8:69–75. doi: 10.1007/s13181-011-0196-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maskell PD, De Paoli G, Seneviratne C, Pounder DJ. Mephedrone (4-methylmethcathinone)-related deaths. J Anal Toxicol. 2011;35:188–191. doi: 10.1093/anatox/35.3.188. [DOI] [PubMed] [Google Scholar]
- 21.Dickson AJ, Vorce SP, Levine B, Past MR. Multiple-drug toxicity caused by the coadministration of 4-methylmethcathinone (mephedrone) and heroin. J Anal Toxicol. 2010;34:162–168. doi: 10.1093/jat/34.3.162. [DOI] [PubMed] [Google Scholar]
- 22.Wikstrom M, Thelander G, Nystrom I, Kronstrand R. Two fatal intoxications with the new designer drug methedrone (4-methoxymethcathinone) J Anal Toxicol. 2010;34:594–598. doi: 10.1093/jat/34.9.594. [DOI] [PubMed] [Google Scholar]
- 23.Torrance H, Cooper G. The detection of mephedrone (4-methylmethcathinone) in 4 fatalities in Scotland. Forensic Sci Int. 2010;202:e62–e63. doi: 10.1016/j.forsciint.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Uchiyama N, Matsuda S, Kawamura M, Kikura-Hanajiri R, Goda Y. Identification of two new-type designer drugs, piperazine derivative MT-45 (I-C6) and synthetic peptide Noopept (GVS-111), with synthetic cannabinoid A-834735, cathinone derivative 4-methoxy-α-PVP, and phenethylamine derivative 4-methylbuphedrine from illegal products. Forensic Toxicol. 2014;32:9–18. doi: 10.1007/s11419-013-0194-5. [DOI] [Google Scholar]
- 25.Roesner P (2014) Designer drugs online news: 4-methoxy-2-PVP. http://designer-drugs.de. Accessed 29 Dec 2014
- 26.US DEA. Establishment of drug codes for 26 substances. Final rule. Fed Regist. 2013;78:664–666. [PubMed] [Google Scholar]
- 27.US DEA. Schedules of controlled substances: temporary placement of 10 synthetic cathinones into schedule I. Final rule. Fed Regist. 2014;79:12938–12943. [PubMed] [Google Scholar]
- 28.Baumann MH, Partilla JS, Lehner KR, Thorndike EB, Hoffman AF, Holy M, Rothman RB, Goldberg SR, Lupica CR, Sitte HH, Brandt SD, Tella SR, Cozzi NV, Schindler CW. Powerful cocaine-like actions of 3,4-methylenedioxypyrovalerone (MDPV), a principal constituent of psychoactive ‘bath salts’ products. Neuropsychopharmacology. 2013;38:552–562. doi: 10.1038/npp.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simmler LD, Buser TA, Donzelli M, Schramm Y, Dieu LH, Huwyler J, Chaboz S, Hoener MC, Liechti ME. Pharmacological characterization of designer cathinones in vitro. Br J Pharmacol. 2013;168:458–470. doi: 10.1111/j.1476-5381.2012.02145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaizaki A, Tanaka S, Numazawa S. New recreational drug 1-phenyl-2-(1-pyrrolidinyl)-1-pentanone (alpha-PVP) activates central nervous system via dopaminergic neuron. J Toxicol Sci. 2014;39:1–6. doi: 10.2131/jts.39.1. [DOI] [PubMed] [Google Scholar]
- 31.Marusich JA, Antonazzo KR, Wiley JL, Blough BE, Partilla JS, Baumann MH. Pharmacology of novel synthetic stimulants structurally related to the “bath salts” constituent 3,4-methylenedioxypyrovalerone (MDPV) Neuropharmacology. 2014;87:206–213. doi: 10.1016/j.neuropharm.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppola M, Mondola R. 3,4-Methylenedioxypyrovalerone (MDPV): chemistry, pharmacology and toxicology of a new designer drug of abuse marketed online. Toxicol Lett. 2012;208:12–15. doi: 10.1016/j.toxlet.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Dargan PI, Sedefov R, Gallegos A, Wood DM. The pharmacology and toxicology of the synthetic cathinone mephedrone (4-methylmethcathinone) Drug Test Anal. 2011;3:454–463. doi: 10.1002/dta.312. [DOI] [PubMed] [Google Scholar]
- 34.Springer D, Peters FT, Fritschi G, Maurer HH. Studies on the metabolism and toxicological detection of the new designer drug 4′-methyl-alpha-pyrrolidinopropiophenone in urine using gas chromatography-mass spectrometry. J Chromatogr B. 2002;773:25–33. doi: 10.1016/S1570-0232(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 35.Springer D, Fritschi G, Maurer HH. Metabolism of the new designer drug alpha-pyrrolidinopropiophenone (PPP) and the toxicological detection of PPP and 4′-methyl-alpha-pyrrolidinopropiophenone (MPPP) studied in rat urine using gas chromatography-mass spectrometry. J Chromatogr B. 2003;796:253–266. doi: 10.1016/j.jchromb.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Springer D, Staack RF, Paul LD, Kraemer T, Maurer HH. Identification of cytochrome P450 enzymes involved in the metabolism of 4′-methoxy-alpha-pyrrolidinopropiophenone (MOPPP), a designer drug, in human liver microsomes. Xenobiotica. 2003;33:989–998. doi: 10.1080/00498250310001602775. [DOI] [PubMed] [Google Scholar]
- 37.Sauer C, Peters FT, Haas C, Meyer MR, Fritschi G, Maurer HH. New designer drug α-pyrrolidinovalerophenone (PVP): studies on its metabolism and toxicological detection in rat urine using gas chromatographic/mass spectrometric techniques. J Mass Spectrom. 2009;44:952–964. doi: 10.1002/jms.1571. [DOI] [PubMed] [Google Scholar]
- 38.Meyer MR, Wilhelm J, Peters FT, Maurer HH. Beta-keto amphetamines: studies on the metabolism of the designer drug mephedrone and toxicological detection of mephedrone, butylone, and methylone in urine using gas chromatography-mass spectrometry. Anal Bioanal Chem. 2010;397:1225–1233. doi: 10.1007/s00216-010-3636-5. [DOI] [PubMed] [Google Scholar]
- 39.Tyrkko E, Pelander A, Ketola RA, Ojanpera I. In silico and in vitro metabolism studies support identification of designer drugs in human urine by liquid chromatography/quadrupole-time-of-flight mass spectrometry. Anal Bioanal Chem. 2013;405:6697–6709. doi: 10.1007/s00216-013-7137-1. [DOI] [PubMed] [Google Scholar]
- 40.Shima N, Katagi M, Kamata H, Matsuta S, Sasaki K, Kamata T, Nishioka H, Miki A, Tatsuno M, Zaitsu K, Ishii A, Sato T, Tsuchihashi H, Suzuki K. Metabolism of the newly encountered designer drug α-pyrrolidinovalerophenone in humans: identification and quantitation of urinary metabolites. Forensic Toxicol. 2014;32:59–67. doi: 10.1007/s11419-013-0202-9. [DOI] [Google Scholar]
- 41.Namera A, Konuma K, Kawamura M, Saito T, Nakamoto A, Yahata M, Ohta S, Miyazaki S, Shiraishi H, Nagao M. Time-course profile of urinary excretion of intravenously administered α-pyrrolidinovalerophenone and α-pyrrolidinobutiophenone in a human. Forensic Toxicol. 2014;32:68–74. doi: 10.1007/s11419-013-0203-8. [DOI] [Google Scholar]
- 42.Uralets V, Rana S, Morgan S, Ross W. Testing for designer stimulants: metabolic profiles of 16 synthetic cathinones excreted free in human urine. J Anal Toxicol. 2014;38:233–241. doi: 10.1093/jat/bku021. [DOI] [PubMed] [Google Scholar]
- 43.Zaitsu K, Katagi M, Tsuchihashi H, Ishii A. Recently abused synthetic cathinones, α-pyrrolidinophenone derivatives: a review of their pharmacology, acute toxicity, and metabolism. Forensic Toxicol. 2014;32:1–8. doi: 10.1007/s11419-013-0218-1. [DOI] [Google Scholar]
- 44.Castaneto M, Wohlfarth A, Pang S, Zhu M, Scheidweiler K, Kronstrand R, Huestis M. Identification of AB-FUBINACA metabolites in human hepatocytes and urine using high-resolution mass spectrometry. Forensic Toxicol. 2015;33:295–310. doi: 10.1007/s11419-015-0275-8. [DOI] [Google Scholar]
- 45.Wohlfarth A, Castaneto MS, Zhu M, Pang S, Scheidweiler KB, Kronstrand R, Huestis MA. Pentylindole/pentylindazole synthetic cannabinoids and their 5-fluoro analogs produce different primary metabolites: metabolite profiling for AB-PINACA and 5F-AB-PINACA. AAPS J. 2015 doi: 10.1208/s12248-015-9721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baranczewski P, Stanczak A, Sundberg K, Svensson R, Wallin A, Jansson J, Garberg P, Postlind H. Introduction to in vitro estimation of metabolic stability and drug interactions of new chemical entities in drug discovery and development. Pharmacol Rep. 2006;58:453–472. [PubMed] [Google Scholar]
- 47.McNaney CA, Drexler DM, Hnatyshyn SY, Zvyaga TA, Knipe JO, Belcastro JV, Sanders M. An automated liquid chromatography-mass spectrometry process to determine metabolic stability half-life and intrinsic clearance of drug candidates by substrate depletion. Assay Drug Dev Technol. 2008;6:121–129. doi: 10.1089/adt.2007.103. [DOI] [PubMed] [Google Scholar]
- 48.Friscia M, Wolf S, Mohr ALA, Diamond FX, Yeakel JK,Logan BK (2015) Identification of major metabolites in human blood and urine associated with the ingestion of alpha PVP. Paper presented at the American Academy of Forensic Sciences, Orlando, Florida, February 19, 2015
- 49.Lave T, Dupin S, Schmitt C, Valles B, Ubeaud G, Chou RC, Jaeck D, Coassolo P. The use of human hepatocytes to select compounds based on their expected hepatic extraction ratios in humans. Pharm Res. 1997;14:152–155. doi: 10.1023/A:1012036324237. [DOI] [PubMed] [Google Scholar]